Table 3.
Chemical structure descriptors that were significantly correlated with bioactivity.
| Descriptora | Structureb | Descriptor Meaning | ρc | P adj d | Chemicals with Descriptor | Cell Type (Phenotype) |
|---|---|---|---|---|---|---|
| SGR10703 |
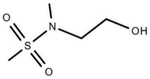
|
N-attached double bonded heteroatoms | −0.70 | 1.08E-05 | MeFOSE PFHxSA | PHHs (Mitochondrial Intensity) |
| SGR10587 | Sulfonamide | |||||
| SGR10099 | Heteroatom-nitrogen bond | |||||
| SGR10668 | Heteroatom-bonded methyl group | −0.70 | 1.08E-05 | MeFOSE MePF2EtOA | ||
| SGR10199 | Two oxygens, 5 bonds apart | −0.57 | 3.11E-02 | MeFOSE PFBOH MePF2EtOA | ||
| SGR10032 | Methyl group | −0.57 | 3.11E-02 | MeFOSE, PFPE-1, PFMBA | ||
| SGR10343 |
|
Any primary amine | −0.56 | 4.17E-02 | AmFPrOH, PFHxSA, PFOAMD | iCell Heps (Cytoplasmic Integrity) |
| SGR10704 |
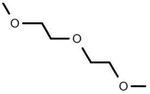
|
Polyethers | −0.69 | 2.84E-05 | PFPE-6, C7F3ETOH | HUVECs (All Nuclei Mean Area) |
| SGR10013 | Any carbon | −0.57 | 2.46E-02 | All tested PFAS (C # varies) | iCell Cardio (Min POD) | |
| SGR10029 | Any heteroatom | −0.57 | 2.79E-02 | All tested PFAS (heteroatom # varies) | ||
| SGR10308 | H-bond acceptors | −0.57 | 3.18E-02 | All tested PFAS (# of H-bond acceptors varies) |
Saagar descriptors can be found in Table S4.
Combined (top) or individual Saagar descriptor structures are shown, where applicable. Individual Saagar descriptors listed herein can be visualized using the hyperlinks in Table S8.
Spearman (ρ) correlation coefficients are shown.
Multiple comparison adjustment was done using Holm–Bonferroni method (see Methods section).
