Abstract
All baculovirus genomes sequenced to date encode a homolog of an alkaline nuclease that has been characterized in the Herpesviridae. In this report we describe the characterization of the alkaline nuclease (AN) homolog of the Autographa californica multinucleocapsid nucleopolyhedrovirus (AcMNPV) (open reading frame 133). His-tagged AN constructs were expressed in recombinant baculoviruses and affinity purified, and then their enzymatic activity was characterized. AN was found to degrade linear DNA at alkaline pH, preferred Mg2+ over Mn2+, had optimal activity at 35°C, and did not appear to have a salt requirement. To rule out contamination by the endogenous baculovirus gene product or a cellular enzyme, point mutations were introduced into a highly conserved domain of the gene. These mutations were found to markedly reduce or eliminate most of the activity of the affinity-purified enzyme. An antibody generated against the protein was used to analyze its expression by Western blot analysis. AN was found to be expressed at low levels by 12 h postinfection, with maximal expression at 24 h postinfection. Attempts to generate a virus with this gene inactivated were unsuccessful, suggesting that AN may be encoded by an essential gene.
Baculoviruses are a large family of viruses that infect invertebrates, particularly insects of the order Lepidoptera. They contain circular, supercoiled, double-stranded DNA genomes of 100 to 180 kb. These genomes are punctuated by repeated sequences called homologous regions that function as origins of DNA replication in transient assays (18, 26). Evidence suggests that genome replication occurs through a rolling-circle intermediate, resulting in large concatemers that are resolved into unit-length molecules during virion maturation (21, 25, 34). Although a number of genes have been identified that are involved in DNA replication (17, 23), neither cis-acting genome sequences nor the genes required for proper genome processing have been identified.
The complete sequences of a number of baculovirus genomes have recently been reported (1, 2, 12, 15, 19). All encode a homolog of an alkaline nuclease present in members of the Herpesviridae (reviewed in reference 9). In herpes simplex virus type 1 (HSV-1), the alkaline nuclease (AN) has the properties of an exonuclease and functions optimally at pH 9 on linear DNA (4, 9, 10, 16). Recombinant cell lines expressing AN have been used to produce HSV-1 AN deletion mutants. These virus synthesize wild-type (wt) levels of DNA and produce encapsidated genomes. However, the virions are of low infectivity in certain cell lines and are not detected in the infected cell cytoplasm, suggesting that AN is required for the production of viable nucleocapsids that are capable of exit from the nucleus into the cytoplasm (30). This may involve the processing of branched replication intermediates into genomic DNA that can be encapsidated (10, 24). It has also been suggested that it may play a role in the generation of 3′OH-terminal single-stranded DNA tails that are thought to be involved in repair of breaks in homologous DNA regions as part of a DNA recombination system (16).
Because AN homologs appear to be universally present in baculovirus genomes, are likely to play an essential role in baculovirus replication, and may be involved in the final processing steps leading to the production of mature genomes, we initiated investigations of this enzyme. In this report, we describe the purification and characterization of the Autographa californica multinucleocapsid nucleopolyhedrovirus (AcMNPV) AN and compare it properties to those of the herpesvirus enzyme.
MATERIALS AND METHODS
Virus and cell lines.
Spodoptera frugiperda (Sf-9) cells (32) were cultured in TNM-FH medium (14) supplemented with 10% fetal bovine serum, penicillin G (50 U/ml), streptomycin (50 μg/ml; Whittaker Bioproducts), and amphotericin B (Fungizone; 375 ng/ml; Flow Laboratories). Cell culture maintenance was carried out according to published procedures (31). Sf-9 cells were also cultured in Sf-900 II medium (Gibco-BRL) as previously described (11). AcMNPV (strain E-2) was used for wt infections.
Enzymes, radioisotopes, DNA purification, PCR, and DNA sequencing.
Restriction and DNA-modifying enzymes were purchased from Life Technologies and New England Biolabs and were used according to the manufacturer's instructions. Isotopes were purchased from New England Nuclear, Inc. DNA sequence analysis and PCR were carried out as described previously (22). DNA was purified using Qiagen columns (Qiagen, Inc.).
Recombinant baculovirus and construction of mutants.
Recombinant baculoviruses were produced using pBlueBacHis2B vector and BacNBlue linear DNA (Invitrogen) as instructed by the manufacturer. To remove the BamHI site in pBlueBacHis2B, the plasmid was digested with BamHI, blunted with T4 DNA polymerase, and religated. The vector was then digested with XhoI and PstI and ligated to a SalI (nucleotide [nt] 112551)-to-NsiI (nt 114840) (2) fragment containing the AcMNPV AN homolog. The SalI site is 9 nt upstream of the predicted translational initiation codon of AcMNPV AN, whereas the NsiI site is about 1,000 nt downstream of the stop codon. This resulted in a His6-tagged fusion protein with a predicted mass of 52.6 kDa and the sequence MPRGSHHHHHHGMASMTGGQQMGRDLYDDDDKDASELDIM upstream of the wt ATG.
Mutant AN construction took advantage of unique StyI (nt 112975) and BamHI (nt 113032) (2) sites that flanked a motif (motif II) which is predicted to encode a metal binding domain (30) and is conserved between baculoviruses and herpesviruses. Two oligomers were synthesized for each mutant so that they could be annealed and inserted into DNA cut with these two enzymes. The double-stranded oligomers (translation products are indicated above the nucleotide sequences; mutant nucleotide and amino acids are underlined) were AN-G141A L A L H A A S P D A . . .+5′CTTGGCTTTGCACGCCGCTTCGCCCGATGCGTATTTTTCTCTCGCCGACGGAACGTG3′−3′ CGAAACGTGCGGCGAAGCGGGCTACGCATAAAAAGAGAGCGGCTGCCTTGCACCTAG5′
andAN-S146A L G L H A A A P D A . . .+5′CTTGGGTTTGCACGCCGCTGCGCCCGATGCGTATTTTTCTCTCGCCGACGGAACGTG3′−3′ CCAAACGTGCGGCGACGCGGGCTACGCATAAAAAGAGAGCGGCTGCCTTGCACCTAG5′
After cloning of each mutant sequence, the alteration was confirmed by DNA sequence analysis (1).
Deletion of AN.
We attempted to delete the AN gene from AcMNPV. To accomplish this, the β-galactosidase gene under the Drosophila heat shock promoter (hsp) (pAcDZ1 [33]) was cut with XbaI and SmaI to isolate the hsp-lacZ-containing fragment, the XbaI end was blunted with T4 DNA polymerase, and the fragment was inserted into pAN1 (see below) between the StyI (nt 112975) and HpaI (nt 113148) sites such that about 175 nt, including the highly conserved motif II, were deleted. The resultant plasmid, pANlacZ, was linearized and transfected with wt AcMNPV DNA into Sf-9 cells to construct recombinant virus.
Preparation of extracts of Sf-9 cells.
The His-tagged enzyme expressed in Sf-9 cells was purified using a modification of a published procedure (30). Log-phase Sf-9 cells (about 2 × 106/ml) in about 100 ml of Sf-900 II medium (Life Technologies) in a 2-liter tissue culture flask were infected at a multiplicity of infection of about 10 and incubated on a shaker at 27°C. After 1 h of incubation, the volume was increased to 200 ml with Sf-900 II and incubated at 27°C for 48 h. Cells were then harvested by centrifugation (3,000 rpm for 15 min), resuspended in 10 ml of phosphate-buffered saline, pH 7.4 (Sigma Chemical Co.), and centrifuged again; the pellet was stored at −80°C. For enzyme purification, the frozen pellets were resuspended in 8 ml of buffer A (20 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 5 mM β-mercaptoethanol, 80 mM KCl, 0.2% NP-40), freeze-thawed twice, and then incubated on ice for 20 min in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF) and the following proteinase inhibitors from Life Technologies: aprotinin (10 μg/ml), leupeptin (7 μg/ml), and pepstatin (7 μg/ml). Cells were homogenized 20 to 25 times in a Dounce homogenizer with a type B pestle and centrifuged at 10,000 × g in a Sorvall GSA rotor, and the pellet and supernatant were saved. The pellet was treated again as described above, and the supernatants were combined and then centrifuged at 100,000 × g in an SW28 rotor. The supernatant (about 16 ml) was precipitated with 3.2 g of ammonium sulfate (20%) for 45 min at 10°C. The precipitate was pelleted by centrifugation at 17,000 × g in a Sorvall SS-34 rotor for 30 min. An additional 5.6 g of ammonium sulfate was added to the supernatant (to about 55%) and incubated for 1 h at 10°C. This preparation was centrifuged as described above, and the two pellets were suspended in 5 ml of buffer B (20 mM Tris-HCl, 150 mM NaCl, 5 mM β-mercaptoethanol, 20% glycerol [pH 8.0]) and dialyzed overnight against buffer B. The dialysate was then centrifuged at 17,000 × g to remove insoluble material, and then the preparation was affinity purified on TALON resin (Clontech, Inc.) essentially as recommended by the manufacturer. The resin (150 μl) that had been washed with 10 ml of wash buffer (buffer B containing 0.1% Triton X-100) was mixed with the dialysate and rotated for 30 min at 4°C. The resin was centrifuged at 1,000 rpm in an International centrifuge for 5 min. The supernatant was removed and designated the flowthrough. The resin was then treated with four 1-ml aliquots of wash buffer (wash fractions 1 to 4 [W1 to W4] and 1-ml solutions of wash buffer containing the following imidazole concentrations: four times, 10 mM (W5 to W8); four times, 30 mM (elution fractions 1 to 4 [E1 to E4]), four times, 50 mM (E5 to E8); and two applications, 100 mM (E9 and E10). Protein concentration was determined by Coomassie blue staining, Western analysis, and spectrophotometric quantification using a Coomassie Plus protein assay kit (Pierce, Inc.). Fractions E6 and E7 were used for the assays described below.
Cloning, expression, and antibody production against bacterially expressed His-tagged AN.
The AN open reading frame (ORF) was cloned as a SalI (nt 112551)-NsiI (114840) (2) fragment inserted into the XhoI and PstI sites of pKS(−) to construct pAN1. pAN1 was cut with KpnI and XbaI; the insert containing the AN gene was gel purified and inserted into pHT4 cut with the same enzymes. pHT4 was constructed by cloning AcMNPV DNA polymerase gene, with a NcoI site at the initial ATG and extended to the downstream SacI site, into NcoI and SacI sites of pTrc-7Hpro (7). The resultant plasmid was digested with NcoI and KpnI, blunted with T4 DNA polymerase, and then religated to produce pHT-AN. This His7-tagged fusion contained the sequence MMHHHHHHHAMGPPLDIM upstream of the AN ATG and has a predicted molecular size of about 50.4 kDa. For protein production, Escherichia coli BL21 cells transformed with pHT-AN were inoculated into 2 ml of Luria-Bertani (LB) broth (29) containing 50 μl of ampicillin per ml. After 3 h at 37°C, this culture was used to seed a 100-ml LB culture and incubated for 3 to 4 h until the cells reached an optical density at 600 nm of 0.6 to 1.0. Isopropyl-β-d-thiogalactopyranoside (IPTG) (dissolved in 2% ethanol) was added to a final concentration of 1 mM, and the culture was incubated overnight at 22°C. Cells were harvested and washed with 10 ml of phosphate-buffered saline and the pellet was stored at −80°C. Protein purification was adapted from reference 4. The pellet was resuspended in 20 ml of lysis buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM β-mercaptoethanol, 1 mM PMSF, 20% sucrose) containing 1 mg of lysozyme per ml, incubated on ice for 20 min, subjected to two freeze-thaw cycles, and then sonicated for 30 s. This sample was centrifuged at 20,000 × g for 15 min, and the supernatant was transferred to a fresh tube. The pellet was resuspended in 20 ml of lysis buffer and centrifuged as above. The two supernatants were combined and mixed with 150 μl of TALON (Clontech) resin previously treated with wash buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM β-mercaptoethanol, 1 mM PMSF, 0.1% Triton X-100, 10 mM imidazole) and rotated for 1 h at 4°C. The suspension was centrifuged at 1,000 rpm and washed sequentially with 2-ml aliquots of wash buffer containing the following concentrations of imidazole: 25 mM, four times; 100 mM, twice; and 200 mM, once. The 100 and 200 mM eluates were pooled, and the protein content and purity were assayed by Coomassie blue staining of polyacrylamide gels after polyacrylamide gel electrophoresis (PAGE) and spectrophotometrically using a Coomassie Plus protein assay kit (Pierce). A rabbit was injected with 100 μg of this preparation in complete Freund's adjuvant. Beginning 3 weeks after the first inoculation, the animal was subjected to three boosts of 30 μg at 14-day intervals in incomplete Freund's adjuvant. One week after the final boost, the animal was bled and the serum was prepared for use in this study.
Assay for AN activity using [3H]DNA.
E. coli DNA was labeled with [3H]thymidine as described elsewhere (13), and assays for AN activity were similar to the procedures of Goldstein and Weller (9). Two micrograms (about 240,000 cpm) of labeled single-stranded DNA plus 4 μg of single-stranded salmon sperm DNA were mixed with 10 ng of affinity-purified AN. Standard conditions used were 200 μl of 50 mM Tris (pH 9.0) per ml–5 mM Mg2+–0.1 mg of bovine serum albumin (BSA) per ml at 37°C for 20 min. The digested DNA was mixed with 0.25 mg of BSA, precipitated with 5% trichloroacetic acid and microcentrifuged for 5 min. The supernatant was mixed with 3.5 ml of scintillation cocktail (formula A-989; Packard, Inc.) and counted.
RESULTS AND DISCUSSION
Baculovirus AN homologs.
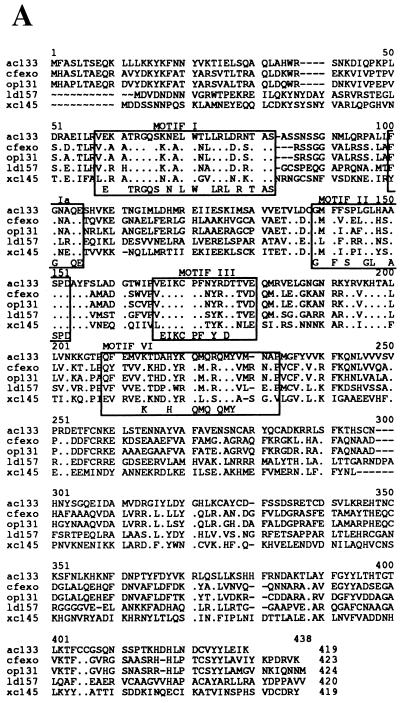
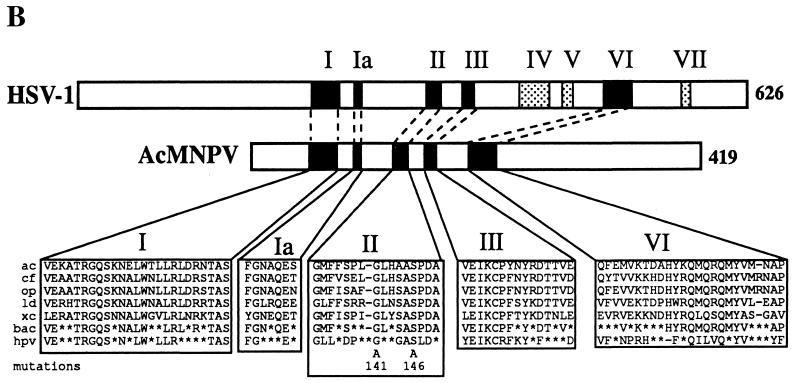
Five baculovirus homologs of the herpesvirus AN are aligned in Fig. 1A. These ORFs are from AcMNPV (orf133) (2), Orgyia pseudotsugata MNPV (OpMNPV) (orf131) (1), Lymantria dispar MNPV (LdMNPV) (orf157) (19), Choristoneura fumiferana MNPV (CfMNPV) (A. Poloumienko and P. Krell, unpublished data [GenBank accession no. AAB53344]), and a granulovirus of Xestia c-nigrum (XcGV) (orf145) (12). The following amino acid sequence identities of the predicted baculovirus ORFs were observed: OpMNPV and CfMNPV, 80%; OpMNPV and CfMNPV to AcMNPV, 53%; OpMNPV, CfMNPV, and AcMNPV to LdMNPV, 37 to 40%; and XcGV to the NPVs, about 30%. The sequence identity is concentrated in the N-terminal 240 amino acids with six regions of limited sequence variation. Several of these regions are related to conserved motifs found in herpesvirus AN (9, 16). The baculovirus motif I, Ia, II, and III sequences are 15/16, 3/3, 5/11, and 6/11 identical to the corresponding alphaherpesvirus consensus sequence (Fig. 1B). There is a highly conserved domain at amino acids 190 to 200 that does not appear to correspond to a herpesvirus motif. In addition, there is a conserved region that shows limited homology to motif VI. The baculovirus predicted proteins are about 200 amino acids shorter than those from HSV-1 (Fig. 1B). This is evident in a significantly truncated baculovirus amino-terminal region upstream of motif I (218 amino acids in HSV-1, versus 56 in AcMNPV). A form of the HSV-1 AN has been described that results from initiation at an internal ATG codon such that it too lacks a significant portion (126 amino acids) of this N-terminal region (5). It has been found to retain its enzymatic activity and is capsid associated. However, the predicted baculovirus AN sequences all also lack motifs IV, V, and VII. In addition, the region between motifs III and VI contains 151 amino acids in HSV-1, versus about 40 in AcMNPV (Fig. 1B), but the distance from motif VI to the C terminus is longer in AcMNPV (180 amino acids, versus 112 in HSV-1).
FIG. 1.
Comparison of baculovirus AN sequences. (A) Alignment of baculovirus sequences. Indicated are: AcMNPV (ac133) (2), CfMNPV (cfexo) (Poloumienko and Krell, unpublished), OpMNPV (op131) (1), LdMNPV (ld157) (19), and XcGV (xc145) (12). The domains conserved with the Herpesviridae AN are boxed. Dots indicate positions where all amino acids are identical, and dashes indicate gaps in the alignment. (B) Comparison of conserved domains between HSV-1 (9) and baculovirus AN. Below are shown the sequences with the consensus Baculoviridae (bac) (four out of five identical) and Herpesviridae (hpv) sequences indicated. Asterisks indicate nonconsensus amino acids. The mutations within motif II that were constructed and tested are also shown.
Expression and purification of AcMNPV orf133.
Since amino-terminal regions of the homologs of AcMNPV orf133 are highly variable (Fig. 1), we reasoned that alteration of amino-terminal amino acids would be unlikely to affect the activity of the enzyme. We used a SalI site at nt 112551 on the AcMNPV genome (2) which is 9 nt upstream of the orf133 ATG and an NsiI site at nt 114845 that is about 1,000 nt downstream of the stop codon in our cloning protocols. A His7-tagged construct was expressed in bacteria, and the protein was processed using a renaturing protocol that was successfully used to generate active HSV-1 AN (4). However, we were unsuccessful in generating an active protein using this technique. Therefore, we constructed a recombinant baculovirus expressing a His6-tagged version of orf133.
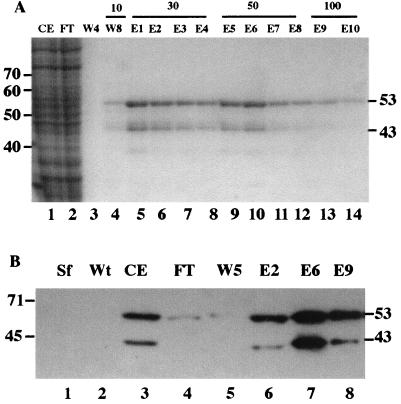
Sf-9 cells infected with the recombinant AcMNPV expressing orf133 were processed using a combination of ammonium sulfate precipitation and affinity chromatography (Fig. 2A). The resuspended ammonium sulfate precipitate is shown in Fig. 2A, lane 1. After binding of this extract to the affinity resin, the unbound material looked similar to the starting material (lane 2). The resin was washed initially without imidazole (lane 3) and then with buffer containing 10 mM imidazole (lane 4). A slight amount of protein was evident in the latter wash (lane 4). When treated with 30 mM imidazole, two major Coomassie blue-stained bands of about 43 and 53 kDa were eluted (lane 5). Subsequent washes with 30 and 50 mM imidazole showed similar elution profiles (lanes 5 to 12). Elution with 100 mM imidazole yielded only a small amount of these two bands.
FIG. 2.
Purification and characterization of His-tagged alkaline exonuclease. Sf-9 cells were infected with baculovirus expressing the His-tagged AN gene and affinity purified as described in Materials and Methods. (A) PAGE analysis of affinity purification. Lane 1, infected cell extract (CE) after dialysis; lane 2, flowthrough (FT) from the TALON resin; lane 3, W4 without imidazole; lanes 4 to 14, W8 with 10 mM imidazole and elution with imidazole at 30 mM (lanes 5 to 8; E1 to E4), 50 mM (lanes 9 to 12; E5 to E8), and 100 mM (lanes 13 and 14; E9 and E10). Lanes 1 and 2 represent 5 μl of the 5.0-ml dialysate before and after binding the affinity resin; lanes 3 to 14 represent 15 μl from 1-ml fractions. Samples were analyzed by PAGE through a 10% gel and stained with Coomassie brilliant blue. The positions of selected size standards (Life Technologies 10-kDa ladder) are shown on the left, and the estimated masses (in kilodaltons) of the polypeptides binding to the affinity resin are shown at the right. (B) Identification of His-tagged polypeptides. The enzyme was characterized using the INDIA HisProbe-HRP reagent (Pierce) according to the manufacturer's instructions. Lane 1, uninfected Sf-9 cells (Sf); lane 2, wt AcMNPV-infected Sf-9 cells (Wt). Lanes 3 to 8 are from the samples described for panel A.
To determine whether these bands were composed of His-tagged recombinant AN, we used a reagent that specifically stains His-tagged proteins (Fig. 2B). We found that extracts from uninfected Sf-9 cells (lane 1) or wt AcMNPV-infected cells (lane 2) showed no evidence of a reactive His-tagged protein. However, when fractions described above were examined, we found that the starting material before affinity purification contained a His-tagged protein (lane 3), as did fractions E2, E6, and E9. This demonstrated that the two major bands of 43 and 53 kDa were His-tagged molecules and indicated that the His-tagged An was present as two species; one full length (53 kDa) and a shorter (43-kDa) form that apparently retained the N-terminal His tag and therefore likely lacked the carboxyl terminus.
Nuclease activity of affinity-purified AN.
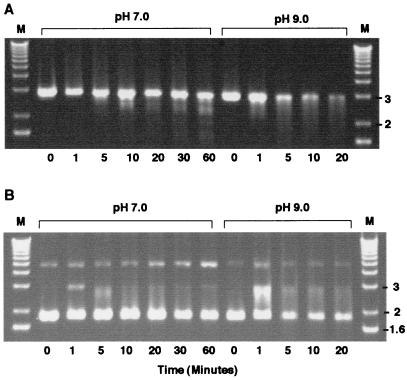
To determine if the purified His-tagged protein had nuclease activity, we tested it with both linear and supercoiled DNA templates at neutral and alkaline pH (Fig. 3). We found that under the conditions that we examined, most of the linear template was degraded in 20 min at pH 9.0, whereas at pH 7.0 most of the starting material remained at 60 min (Fig. 3A). In contrast, the effect on supercoiled templates was not nearly so dramatic. At pH 9, there appeared to be an initial conversion of some DNA to linear-sized fragments at 1 min (Fig. 3B). Although this linear DNA was subsequently degraded, substantial amounts of the supercoils remained after 20 min. The effect at pH 7 was even less than at pH 9, with some conversion to linear-sized DNA at 1 min; however, by 60 min almost all of the supercoiled DNA remained. Therefore, the purified AN has a strong preference for linear over supercoiled DNA and showed the highest levels of activity at an alkaline pH, suggesting that the baculovirus-encoded protein is an alkaline exonuclease.
FIG. 3.
Characterization of AN activity on DNA at pH 7.0 and 9.0. (A) Time course of digestion of linear pKS(−) DNA at the times and pH indicated. Qiagen column-purified pKS(−) was linearized with EcoRI. DNA (0.2 μg) was mixed with 10 ng of affinity-purified enzyme and incubated for various times in 200 μl of the standard buffer (50 mM Tris-HCl, 5 mM MgCl2) at 37°C and the indicated pH. The digests were then electrophoresed on a 1.0% agarose gel. (B) Time course of digestion of supercoiled pKS DNA at the times and pH indicated. Supercoiled DNA (0.2 μg) for each sample was processed as described above. Positions of markers (M) are indicated in kilobases.
Characterization of the AcMNPV alkaline exonuclease.
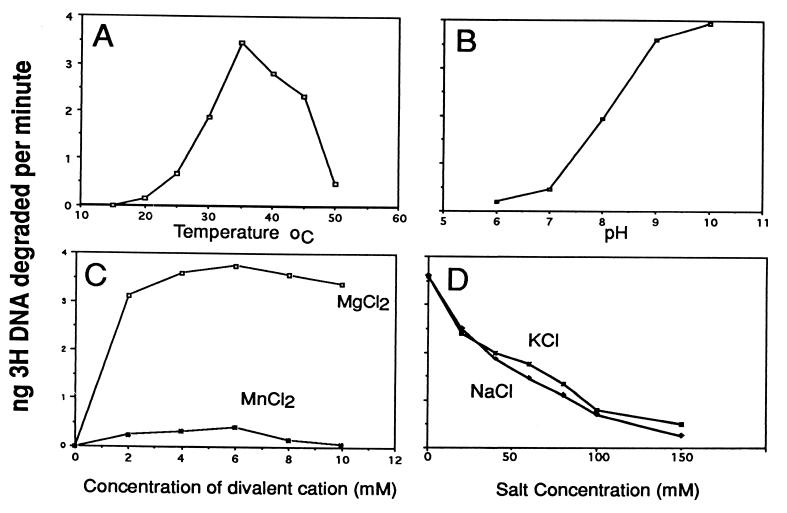
We characterized the properties of the baculovirus AN in more detail by quantifying its ability to hydrolyze 3H-labeled linear DNA (Fig. 4). We found that the optimal temperature was 35°C (Fig. 4A), and as expected from Fig. 3, the highest activity was observed in an alkaline pH range of 9 to 10 (Fig. 4B). Divalent cations were required for activity, and the enzyme showed optimal activity over a broad range (2 to 10 mM); Mg2+ gave about a 10-fold-higher level of activity than Mn2+ (Fig. 4C). The enzyme did not appear to require salt, and increasing concentrations were inhibitory (Fig. 4D). These optimal conditions are similar to those reported for HSV-1 AN with the exception of salt concentration, for which the HSV-1 enzyme showed a broad optimum of up to 40 mM (4).
FIG. 4.
Effects of temperature (A), pH (B), divalent cations Mg2+ and Mn2+ (C), and salt concentration (D) on purified AcMNPV alkaline exonuclease activity. For these experiments, single-stranded DNA (2 μg of labeled DNA plus 4 μg of salmon sperm DNA) was mixed with 10 ng of purified AN. Samples were prepared for each condition tested and then incubated for 20 min. Standard conditions were 37°C, 50 mM Tris (pH 9.0), 5 mM MgCl, and 0.1 mg of BSA per ml in 200 μl unless otherwise indicated. The digested DNA was mixed with 0.25 mg of BSA, then precipitated with 5% trichloroacetic acid, and centrifuged for 5 min in a microcentrifuge; the supernatants were mixed with 3.5 ml of scintillation cocktail (formula A-989; Packard) and counted. All samples were done in triplicate, and each point represents an average of the values. Very little deviation from the average was observed.
The fact that the baculovirus-encoded enzyme has properties similar to those of the enzyme characterized from HSV-1 suggests that the shared motifs (I, Ia, II, III, and VI) may contribute to this activity, but the other motifs common to the Herpesviridae but lacking in the Baculoviridae (IV, V, and VII) (Fig. 1) may be involved in some other function.
Mutations in a highly conserved domain.
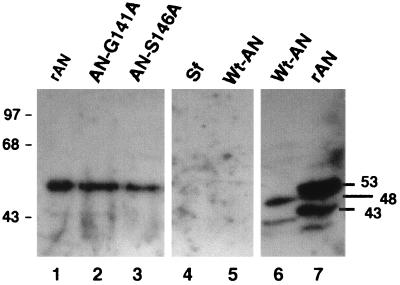
To ensure that the enzyme activity that we observed was due to the affinity-purified recombinant AN (rAN) and not contamination from the endogenous AN encoded by the virus or from a cellular enzyme, we constructed a number of mutants with single amino acids altered at positions in motif II that are conserved in all baculovirus and herpesvirus sequences (Fig. 1B). Selected mutations in this region of the HSV-1 AN led to inactivation of the enzyme (9). We produced three mutations in this region: amino acid mutations G141A and S146A and deletion of amino acids 142 to 148. The His-tagged rAN and two of the mutants were expressed and could be detected by Western blot analysis using a rabbit antiserum that we generated against a bacterially expressed form of the protein (Fig. 5, lanes 1 to 3). However, the mutant with amino acids 142 to 148 deleted was apparently unstable and was not evident in extracts of cells infected with recombinant viruses expressing this construct. An immunoreactive band was not observed in affinity-purified extracts from uninfected Sf-9 cells or cells infected with wt AcMNPV (lane 4 or 5, respectively). In contrast, the antiserum reacted with an appropriate-sized band in extracts that had not been affinity purified from cells infected with either wt virus or the recombinant virus expressing His-tagged rAN (lane 6 or 7, respectively).
FIG. 5.
Western blot analysis of wt and mutant AN. AN was purified as described in Materials and Methods. Samples include His-tagged rAN (lane 1) and the mutants indicated (lanes 2 and 3). Controls show that there is no material binding to the affinity resin from extracts of uninfected Sf-9 cells (lane 4) or wt AcMNPV-infected cells (lane 5). For these control assays (lanes 4 and 5), the cells were carried through the purification protocol, fractions E6 and E7 (Fig. 2) were pooled, and about three times the volume used for the His-tagged enzyme-containing extracts (about 24 μl) was loaded onto the gel. Lanes 6 and 7 contain 10 μl of dialysate from cells infected with wt AcMNPV and recombinant AcMNPV expressing AN, respectively. Samples were electrophoresed through sodium dodecyl sulfate–10% polyacrylamide gels (20) and electroblotted onto polyvinylidene difluoride membranes (Micron Separations, Inc.) for 2 h at 185 mA; then, Western blot analyses were carried out as previously described (27). Samples were treated with 1:1,000 dilution of the antiserum, and the second antibody (goat anti-rabbit conjugated to horseradish peroxidase; Promega) was used at 1:2,500. The positions of selected size standards are shown in kilodaltons on the left. The estimated values for the major immunoreactive bands are shown on the right.
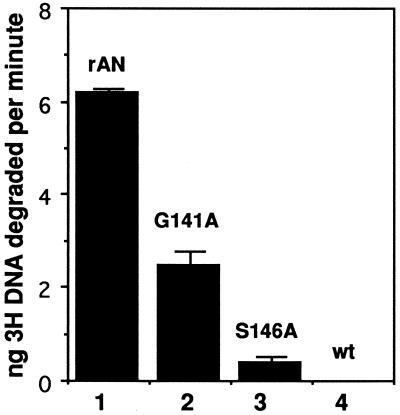
We then examined the ability of the affinity-purified mutant protein to hydrolyze linear [3H]DNA (Fig. 6). We found that G141A showed about 35% the activity of the rAN, whereas S146A showed less than 10% the activity of the nonmutant enzyme. No activity was evident from affinity-purified extracts of cells that had been infected with wt virus indicating that, as expected, the native enzyme which would lack the His tag was not affinity purified by our protocol (Fig. 6, lane 4). These data indicated that the wt enzyme was not a major contaminant of our affinity-purified preparations, and as with HSV-1 AN (9), motif II is critical for the activity of the baculovirus enzyme.
FIG. 6.
Quantification of activity of mutant and wt AN. For details, see Materials and Methods. All samples were done in triplicate, and the error bars represent 1 standard deviation. The experiment is representative of at least two different infection and extract preparations for each mutant virus. Lane 4 is a sample from wt AcMNPV-infected cells processed as described in the legend to Fig. 5.
Construction of a mutant of AcMNPV with AN deleted.
In an attempt to construct a mutant of AcMNPV with the AN gene inactivated, we constructed a plasmid with the β-galactosidase gene under the control of the Drosophila heat shock promoter inserted within the AN gene such that the gene was disrupted and a portion was deleted. We used cotransfection with the wt virus and linearized plasmid DNA to produce the deletion mutants. We isolated and extensively plaque purified a number of isolates expressing β-galactosidase. However, upon PCR analysis, we found that although the β-galactosidase-expressing construct was present in the genome, the wt gene was also present. In addition, attempts to delete the AN homolog from the Bombyx mori NPV genome have been unsuccessful (S. J. Gomi, personal communication). The data from both of these viruses suggest that this enzyme plays a vital role in the baculovirus replication cycle.
Characterization of AN expression in AcMNPV-infected cells.
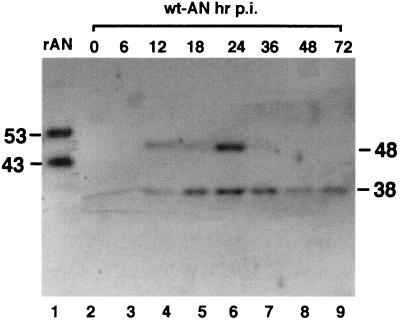
The anti-AN antiserum that we prepared against bacterially expressed, His-tagged AN was used to examine the time course of infection of AN in AcMNPV-infected cells (Fig. 7). As a control we used the affinity-purified recombinant AN (Fig. 7, lane 1). We found that a polypeptide of the predicted molecular mass (48 kDa) was first observed at about 12 h postinfection (p.i.) and the peak level of expression occurred at 24 h p.i., consistent with late gene expression. There is a possible RNA polymerase II promoter (TATTT) and mRNA start site consensus sequence (CAGT) (3) starting about 140 nt upstream of the predicted initiation codon. There is also a late promoter element (GTAAG) located 22 nt upstream of the ATG (2). This suggests that AcMNPV AN may be expressed as both an early and a late gene. There was also a smaller, 38-kDa band present at a lower concentration that may represent a breakdown product similar to that observed with rAN (Fig. 2); however, an immunoreactive band of this size was also observed in extracts of uninfected cells.
FIG. 7.
Western blot analysis of AN expression in extracts of infected insect cells. Monolayers of Sf-9 cells were infected with AcMNPV at a multiplicity of infection of 10 and prepared as previously described (28). Lane 1, rAN (300 ng); lanes 3 to 10, time course of wt AN expression in AcMNPV-infected Sf-9 cells. The hour postinfection is indicated above the lanes; sizes are indicated in kilodaltons.
We previously reported the presence of a nuclease active against linear DNA templates in nuclear extracts of AcMNPV-infected cells that were used for in vitro transcription assays (8). Although the extracts from uninfected or early infected cells lacked the nuclease activity, those at 24 h p.i. showed a high level of activity. The reaction conditions for these investigations were buffered at pH 8.4, which is well within the active range of the AN we have described in this report. A nuclease activity associated with baculovirus infection was also reported by others (6). It is likely that the AcMNPV AN that we have characterized is responsible for the nuclease activities described in these reports.
ACKNOWLEDGMENTS
We thank Doug Leisy for reviewing the manuscript and Doug Grossenbach and Dennis Hruby for assistance in characterization of His-tagged proteins.
This project was supported by a grant from the NSF (MCB-9630769).
Footnotes
Technical Report no. 11656 from the Oregon State University Agricultural Experiment Station.
REFERENCES
- 1.Ahrens C H, Russell R, Funk C J, Evans J T, Harwood S H, Rohrmann G F. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. doi: 10.1006/viro.1997.8448. [DOI] [PubMed] [Google Scholar]
- 2.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 3.Blissard G W, Rohrmann G F. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology. 1989;170:537–555. doi: 10.1016/0042-6822(89)90445-5. [DOI] [PubMed] [Google Scholar]
- 4.Bronstein J C, Weber P C. Purification and characterization of herpes simplex virus type 1 alkaline exonuclease expressed in Escherichia coli. J Virol. 1996;70:2008–2013. doi: 10.1128/jvi.70.3.2008-2013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronstein J C, Weller S K, Weber P C. The product of the UL12.5 gene of herpes simplex virus type 1 is a capsid-associated nuclease. J Virol. 1997;71:3039–3047. doi: 10.1128/jvi.71.4.3039-3047.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidoff A N, Mendelow B V. Identification of endonuclease activity in HIV-1 gp120 preparations produced using baculovirus expression systems. BioTechniques. 1997;23:296–299. doi: 10.2144/97232st07. [DOI] [PubMed] [Google Scholar]
- 7.Evans J T, Rohrmann G F. The baculovirus single-stranded DNA binding protein, LEF-3, forms a homotrimer in solution. J Virol. 1997;71:3574–3579. doi: 10.1128/jvi.71.5.3574-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glocker B, Hoopes R R, Rohrmann G F. In vitro transactivation of baculovirus early genes by nuclear extracts from Autographa californica nuclear polyhedrosis virus-infected Spodoptera frugiperda cells. J Virol. 1992;66:3476–3484. doi: 10.1128/jvi.66.6.3476-3484.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein J N, Weller S K. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology. 1998;244:442–457. doi: 10.1006/viro.1998.9129. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein J N, Weller S K. In vitro processing of herpes simplex virus type 1 DNA replication intermediates by the viral alkaline nuclease, UL12. J Virol. 1998;72:8772–8781. doi: 10.1128/jvi.72.11.8772-8781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harwood S H, Li L, Ho P S, Preston A K, Rohrmann G F. AcMNPV late expression factor-5 interacts with itself and contains a zinc ribbon domain that is required for maximal late transcription activity and is homologous to elongation factor TFIIS. Virology. 1998;250:118–134. doi: 10.1006/viro.1998.9334. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa T, Ko R, Okano K, Seong S, Goto C, Maeda S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology. 1999;262:277–297. doi: 10.1006/viro.1999.9894. [DOI] [PubMed] [Google Scholar]
- 13.Hays J B, Martin S J, Bhatia K. Repair of nonreplicating UV-irradiated DNA: cooperative dark repair by Escherichia coli uvr and phr functions. J Bacteriol. 1985;161:602–608. doi: 10.1128/jb.161.2.602-608.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hink W F. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature. 1970;226:466–467. doi: 10.1038/226466b0. [DOI] [PubMed] [Google Scholar]
- 15.Ijkel W F J, van Strien E A, Jeldens J G M, Broer R, Zuidema D, Goldbach R W, Vlak J M. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J Gen Virol. 1999;80:3289–3304. doi: 10.1099/0022-1317-80-12-3289. [DOI] [PubMed] [Google Scholar]
- 16.Kehm E, Goksu M, Bayer S, Knopf C W. Herpes simplex virus type 1 DNase: functional analysis of the enzyme expressed by recombinant baculovirus. Intervirology. 1998;41:110–119. doi: 10.1159/000024922. [DOI] [PubMed] [Google Scholar]
- 17.Kool M, Ahrens C, Goldbach R W, Rohrmann G F, Vlak J M. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci USA. 1994;91:11212–11216. doi: 10.1073/pnas.91.23.11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kool M, Van Den Berg P M M M, Tramper J, Goldbach R W, Vlak J M. Location of two putative origins of DNA replication of Autographa californica nuclear polyhedrosis virus. Virology. 1993;192:94–101. doi: 10.1006/viro.1993.1011. [DOI] [PubMed] [Google Scholar]
- 19.Kuzio J, Pearson M N, Harwood S H, Funk C J, Evans J T, Slavicek J, Rohrmann G F. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Leisy D J, Rohrmann G F. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology. 1993;196:722–730. doi: 10.1006/viro.1993.1529. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Harwood S H, Rohrmann G F. Identification of additional genes that influence baculovirus late gene expression. Virology. 1999;255:9–19. doi: 10.1006/viro.1998.9546. [DOI] [PubMed] [Google Scholar]
- 23.Lu A, Miller L K. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oppenheimer D I, Volkman L E. Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch Virol. 1997;142:2107–2113. doi: 10.1007/s007050050229. [DOI] [PubMed] [Google Scholar]
- 26.Pearson M N, Bjornson R M, Pearson G D, Rohrmann G F. The Autographa californica baculovirus genome: evidence for multiple replication origins. Science. 1992;257:1382–1384. doi: 10.1126/science.1529337. [DOI] [PubMed] [Google Scholar]
- 27.Quant-Russell R L, Pearson M N, Rohrmann G F, Beaudreau G S. Characterization of baculovirus p10 synthesis using monoclonal antibodies. Virology. 1987;160:9–19. doi: 10.1016/0042-6822(87)90038-9. [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen C, Rohrmann G F. Characterization of the Spodoptera frugiperda TATA-binding protein: nucleotide sequence and response to baculovirus infection. Insect Biochem Mol Biol. 1994;7:699–708. doi: 10.1016/0965-1748(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Shao L, Rapp L M, Weller S K. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology. 1993;196:146–162. doi: 10.1006/viro.1993.1463. [DOI] [PubMed] [Google Scholar]
- 31.Smith G E, Summers M D. Analysis of baculovirus genomes with restriction endonucleases. Virology. 1978;89:517–527. doi: 10.1016/0042-6822(78)90193-9. [DOI] [PubMed] [Google Scholar]
- 32.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Bulletin no. 1555. College Station, Tex: Texas Agricultural Experiment Station; 1987. [Google Scholar]
- 33.Vaughn J L, Goodwin R H, Tompkins G J, McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera: Noctuidae) In Vitro. 1977;13:213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- 34.Vlak J M, Schouten A, Usmany M, Belsham G J, Klinge-Roode E C, Maule A J, Van Lent J W, Zuidema D. Expression of cauliflower mosaic virus gene I using a baculovirus vector based upon the p10 gene and a novel selection method. Virology. 1990;179:312–320. doi: 10.1016/0042-6822(90)90299-7. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Liu G, Carstens E B. Replication, integration, and packaging of plasmid DNA following cotransfection with baculovirus viral DNA. J Virol. 1999;73:5473–5480. doi: 10.1128/jvi.73.7.5473-5480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]