Abstract
The E6 protein of the high-risk human papillomaviruses (HPVs) and the cellular ubiquitin-protein ligase E6AP form a complex which causes the ubiquitination and degradation of p53. We show here that HPV16 E6 promotes the ubiquitination and degradation of E6AP itself. The half-life of E6AP is shorter in HPV-positive cervical cancer cells than in HPV-negative cervical cancer cells, and E6AP is stabilized in HPV-positive cancer cells when expression of the viral oncoproteins is repressed. Expression of HPV16 E6 in cells results in a threefold decrease in the half-life of transfected E6AP. E6-mediated degradation of E6AP requires (i) the binding of E6 to E6AP, (ii) the catalytic activity of E6AP, and (iii) activity of the 26S proteasome, suggesting that E6-E6AP interaction results in E6AP self-ubiquitination and degradation. In addition, both in vitro and in vivo experiments indicate that E6AP self-ubiquitination results primarily from an intramolecular transfer of ubiquitin from the active-site cysteine to one or more lysine residues; however, intermolecular transfer can also occur in the context of an E6-mediated E6AP multimer. Finally, we demonstrate that an E6 mutant that is able to immortalize human mammary epithelial cells but is unable to degrade p53 retains its ability to bind and degrade E6AP, raising the possibility that E6-mediated degradation of E6AP contributes to its ability to transform mammalian cells.
Strong epidemiologic, virologic, and biochemical evidence indicates that the high-risk subgroup of genital human papillomaviruses (HPVs; including HPV types 16, 18, 31, 33, and 39) is the causative agent in the development of >90% of cervical cancers (reviewed in reference 64). The HPV16 E6 and E7 proteins are necessary and sufficient for immortalization of primary human keratinocytes in culture (17, 20, 36). HPV16 E6 also cooperates with activated ras to transform baby mouse kidney cells (51) and can transform NIH 3T3 cells (45) and immortalize human mammary epithelial cells (2) independent of E7.
HPV16 E6 and E7 bind and promote the degradation of the tumor suppressor proteins p53 (44, 59) and pRB (5, 8, 26), respectively. HPV16 E6 induces p53 degradation by forming a complex with the cellular ubiquitin-protein ligase E6AP (22), which is then able to bind and ubiquitinate p53 (42). E6AP is a component of the ubiquitin-proteasome pathway which targets proteins for degradation by covalently linking them to multimeric chains of the 8-kDa protein ubiquitin. In this pathway, ubiquitin is first activated in an ATP-dependent reaction and then passed from the ubiquitin-activating enzyme (E1) to a ubiquitin-conjugating enzyme (E2). Ubiquitin is then transferred to lysine residues of the target protein with the aid of a ubiquitin-protein ligase (E3). Multiubiquitinated proteins are subsequently recognized and degraded by the 26S proteasome (18). While some classes of E3 proteins, such as the SCF complexes, function as adaptors that bring E2 enzymes into complex with their substrates (9, 47), E6AP belongs to a class of ubiquitin-protein ligases called HECT E3 proteins which directly transfer ubiquitin to their substrates (21, 43). The catalytic domain of HECT proteins is a conserved 350-amino-acid region defined by its homology to the E6AP carboxy terminus (HECT). The HECT domain binds to specific E2 enzymes (31) and contains an active site cysteine residue that forms a thiolester bond with ubiquitin (21, 37). X-ray crystallography studies have determined that the HECT domain is a bilobed structure, with a larger N-terminal lobe which interacts with the ubiquitin-conjugating enzyme and a smaller C-terminal lobe containing the catalytic cysteine residue (19).
The HPV16 E6 protein binds to E6AP within its putative substrate recognition domain, directing E6AP to ubiquitinate p53 (23). Current evidence indicates that p53 is targeted by E6AP only in cells that express E6 (3, 53). Recently, a number of potential E6-independent substrates of E6AP have been identified, including the human homolog of the yeast RAD23 protein involved in nucleotide excision repair (HHR23A) (32), the src-family kinase Blk (39), and the MCM7 subunit of replication licensing factor (30). In addition, E6AP auto-ubiquitination has been recently documented in vitro (38).
It is conceivable that the redirection of E6AP activity toward p53 by E6 might inhibit the targeting of the normal substrates of E6AP and that such an alteration of E6AP activity could account for some of the transforming activity of E6. Indeed, recent studies suggest that E6 contributes to tumorigenesis via mechanisms distinct from its ability to induce p53 degradation. For example, transgenic mice expressing E6 from the epithelium-specific K14 promoter developed epidermal hyperplasia which was not observed in p53-null mice, and K14E6/p53-null mice also developed epithelial hyperplasia (48). Other experiments have shown that an E6 mutant which is unable to bind or degrade p53 can still promote colony formation and growth on soft agar (27), as well as induce cell cycle reentry in cells even when p53 transactivation activity is inhibited (50). Another p53-independent activity of HPV16 E6 is its ability to activate telomerase (29), which has been shown to contribute to transformation of primary human cells in conjunction with other oncogenes (15).
Here we show that E6 promotes the ubiquitination and degradation of E6AP. Both in vitro and in vivo experiments demonstrate that this degradation requires the binding of E6 to E6AP and the catalytic activity of E6AP. Our experiments suggest that E6 can induce both the intramolecular and intermolecular self-ubiquitination of E6AP and the subsequent degradation of E6AP by the 26S proteasome. Finally, we show that an E6 mutant which is still able to immortalize human mammary epithelial cells but is unable to degrade p53 retains its ability to degrade E6AP. Our results suggest that degradation of E6AP may contribute to the transforming activity of the high-risk HPV E6 proteins.
MATERIALS AND METHODS
Plasmid constructs.
The pCMV4-HA-E6AP constructs (p4053-4055 for isoforms I to III, respectively; the “p-numbers” represent the plasmid numbers in our laboratory DNA plasmid bank) were created by subcloning the E6AP isoforms (62) into the pCMV4 expression vector (1) at the BglII and HindIII sites. Individual cDNAs encoding each of the three isoforms of E6AP were originally cloned into the pGEM-1 expression plasmid. The PCR was used to add sequence encoding the influenza virus hemagglutinin (HA) epitope to the N terminus of each E6AP isoform. The C-A mutation was introduced into the pCMV4-HA-E6AP isoforms I (C820A) and II (C843A) by replacing the wild-type fragments of these isoforms with the EcoRV/HindIII fragment from a plasmid carrying the catalytically inactive C833A point mutant of the 95-kDa form of E6AP in the pCMV4 vector which has been previously described (53). The ΔE6 mutation was introduced into E6AP isoforms I and II by replacing the wild-type BstBI/EcoRV fragment with the corresponding fragment from pCMV4 E6AP 95-kDa ΔE6 (53). Expression plasmids for BPV1 E2-TA (p2450) and E2-TR (p1153) proteins (6, 49, 60), as well as the AU1 epitope-tagged HPV16E6 expression plasmid (46), have also been described previously. The puromycin resistance plasmid pBabe-puro was obtained from Stratagene.
Protein expression.
35S-labeled E6AP protein was generated by in vitro transcription-translation in the presence of l-[35S]methionine in rabbit reticulocyte lysate according to the manufacturer's instructions (Promega). Wheat E1 and Arabidopsis thaliana Ubc8 were expressed in Escherichia coli BL21 using the pET vector system as described previously (22, 42, 53). Expression and harvesting of E6, E6AP, and p53 protein via baculovirus vectors in High Five insect cells (Invitrogen) were performed as detailed previously (21, 41, 42).
In vitro ubiquitination reactions.
For in vitro ubiquitination reactions utilizing in vitro-translated proteins, 4 μl of 35S-labeled translation reactions were incubated for 30 minutes at 25°C in the presence of 50 ng of E1, 50 ng of E2 (A. thaliana Ubc8), 6 μg of ubiquitin (Sigma), 4 mM ATP, and partially purified baculovirus-expressed E6 proteins or an equivalent control fraction from cells infected with nonrecombinant virus. Preparation of E1 and E2 proteins has been described previously (42). The total reaction volume was 40 μl in a buffer containing 25 mM Tris (pH 8.0) and 125 mM NaCl. The HPV16 E6 and HPV16 E6 SAT8–10 were expressed in insect cells (High Five cells; Invitrogen) using recombinant baculoviruses prepared using the BaculoGold system (Pharmingen). E6 proteins were partially purified from insect cell lysates by using single-step high-salt elution (25 mM Tris, pH 6.8; 500 mM NaCl; 1 mM dithiothreitol) from a Bio-Rad S column. The reactions were terminated by the addition of standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and boiled for 5 min. Proteins were resolved by SDS-PAGE and visualized by autoradiography.
For in vitro ubiquitination assays using baculovirus-expressed E6AP, E6AP proteins were partially purified by anion-exchange chromatography, as described previously (42). A total of 20 ng of partially purified E6AP proteins was incubated under the conditions described above, with partially purified baculovirus-expressed HPV16 E6 protein or an equivalent control fraction from cells infected with nonrecombinant virus. The reactions were terminated by addition of SDS-PAGE loading buffer, boiled for 5 min, and resolved on an SDS–10% PAGE gel. Proteins were transferred to Immobilon P (Millipore) and probed with a rabbit polyclonal E6AP antibody and a horseradish peroxidase-linked goat anti-rabbit secondary antibody. Chemiluminescent signal was detected with Renaissance reagents (DuPont-NEN).
Cell cultures and transfection.
Human cervical carcinoma C33A cells and U2OS osteosarcoma cells were cultured in DMEM (Dulbecco modified Eagle medium; GIBCO) supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2. The Saos-2 osteosarcoma cell line was cultured in DMEM plus 15% FBS. C33A and U2OS cells were transfected using standard calcium phosphate procedures as described previously (53); Saos-2 cells were transfected using the FuGene6 transfection reagent according to manufacturer's instructions (Roche Molecular Biochemicals).
Pulse-chase analysis.
For analysis of transfected E6AP, 106 C33A cells were seeded onto a 60-mm plate for transfection for each time point. Transfected cells were washed twice with phosphate-buffered saline (PBS) and then placed in DMEM without methionine or cysteine at 37°C for 30 min. The cells were labeled with 100 μCi of [35S]methionine per plate for 30 min, washed three times with PBS, and chased with DMEM containing 10% fetal calf serum. At selected time points, the cells were scraped into PBS, pelleted, resuspended in Nonidet P-40 lysis buffer (100 mM Tris-HCl, pH 7.4; 120 mM NaCl; 1% Nonidet P-40; 1 mM phenylmethylsulfonyl fluoride; 1 μg of aprotinin per ml; 1 μg of leupeptin per ml). The lysates were incubated for 15 min on ice with occasional vortexing and then centrifuged at 16,000 × g for 10 min. For each experiment, equal amounts of total protein as measured by Bradford assay were immunoprecipitated overnight at 4°C using the 12CA5 antibody directed against the HA epitope. Extracts were then incubated for 1 h with 40 μl of a mixture of protein A and protein G agarose beads, which were then washed five times with ice-cold radioimmunoprecipitation assay buffer (20 mM Tris-HCl, pH 7.5; 2 mM EDTA; 150 mM NaCl; 0.25% SDS; 1% Nonidet P-40; 1% deoxycholate; 1 mM phenylmethylsulfonyl fluoride; 1 μg of aprotinin per ml; 1 μg of leupeptin per ml). Immunoprecipitated E6AP was resolved on an SDS–7.5% PAGE gel, which was dried and exposed to film.
Pulse-chase analysis of endogenous E6AP in HeLa and C33A cells was performed similarly, except that 2 × 106 cells were seeded onto 60-mm plates for each time point the day before 35S labeling.
Puromycin selection of transfected cells.
Human cervical carcinoma HeLa and C33A cells were seeded at 1 × 105 to 2 × 105 per 60-mm plates 1 day before transfection. The cells were cotransfected using the FuGene 6 system with 5 μg of E6, E7, or E2 plasmid; 1 μg of pBABE-puro selection plasmid; and 4 μg of salmon sperm DNA as a carrier DNA. At 24 h after transfection, the cells were split into plates (2 by 60 mm) and placed under selection in medium containing 0.4 μg of puromycin per ml for HeLa cells or 1.0 μg of puromycin per ml for C33A cells. The cells were washed once with PBS and fed with fresh selection medium every 3 days.
Proteasome inhibition experiments.
At 36 h after transfection, cells were treated with DMEM plus 10% FBS containing the proteasome inhibitor Z-L3VS at a concentration of 50 μM. Control cells were treated with an equal amount of dimethyl sulfoxide (DMSO). The Z-L3VS was a gift from Hidde Ploegh, Harvard Medical School.
Western blot analysis.
Whole-cell extracts were prepared from transfected cells in Nonidet P-40 lysis buffer, as described above. A 50-μg portion of each sample was boiled in SDS sample buffer and resolved on a 7.5% (for E6AP and p53) or 13% (for E6 and p16) SDS-PAGE gel. Proteins were transferred to polyvinylidene difluoride membranes (NEN) according to standard procedures, and membranes were probed with the appropriate antibodies followed by detection using chemiluminescence reagents (NEN). The 12CA5 antibody was used to detect HA-tagged E6AP, while the AU1 antibody (Babco) was used for detection of AU-tagged E6. The monoclonal anti-p16INK4A was a gift from Philip W. Hinds, Harvard Medical School. For p53 detection, the monoclonal antibody Ab-6 (Oncogene) was used. Antitubulin was purchased from Santa Cruz Biotechnology.
RESULTS
HPV16 E6 decreases steady-state levels of E6AP in vivo.
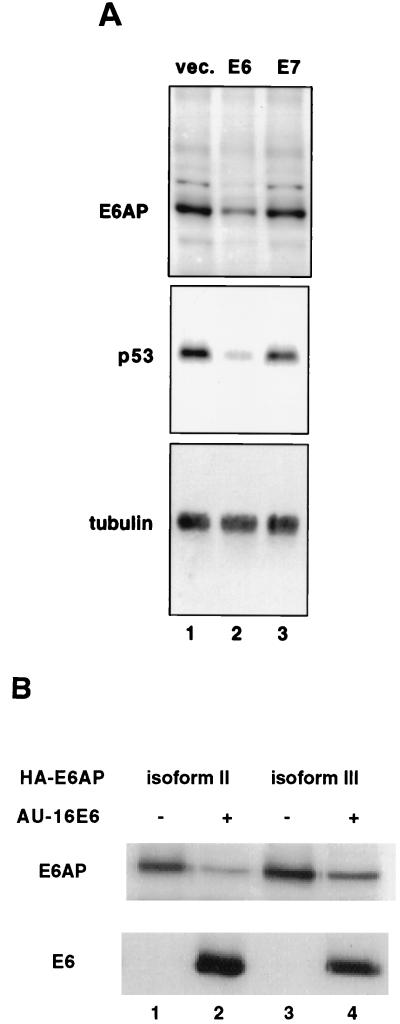
During the course of our analysis of E6-mediated p53 degradation, we noticed that the steady-state levels of E6AP were lower in cells expressing the HPV16 E6 protein. Expression of HPV16 E6 in C33A cells resulted in a substantial decrease in the levels of endogenous E6AP (Fig. 1A, upper panel), in addition to the expected decrease in p53 levels (Fig. 1A, middle panel). By contrast, no change in E6AP levels was observed in cells transfected with HPV16 E7 or with vector alone. As a control, the blot was also probed with antibodies to tubulin, the levels of which are not affected by E6 (Fig. 1A, lower panel). To assess the effect of E6 on each of the three full-length E6AP isoforms, we examined the levels of the individual HA-tagged E6AP isoforms when cotransfected with AU1-tagged HPV16 E6 (46). The levels of all three E6AP isoforms were substantially decreased in vivo when coexpressed with AU1-E6 (Fig. 1B, compare lanes 1 and 2 for isoform II and lanes 3 and 4 for isoform III; see below for isoform I [Fig. 6A, compare lanes 1 and 3]).
FIG. 1.
HPV16 E6 lowers E6AP levels in vivo. (A) E6 but not E7 lowers levels of endogenous E6AP in C33A cells. C33A cells were cotransfected with puromycin resistance plasmid along with empty vector (lane 1), vector expressing E6 (lane 2), or vector expressing E7 (lane 3). After selection of puromycin-resistant cells, protein extracts were resolved by SDS-PAGE and analyzed by immunoblotting with rabbit polyclonal E6AP antibody (upper panel), anti-p53 antibody (middle panel), or antitubulin antibody (lower panel). (B) HPV16 E6 decreases the steady-state levels of transfected E6AP full-length isoforms. C33A cells were transfected with plasmids encoding AU1-tagged HPV16 E6 (lanes 2 and 4) and HA-tagged E6AP isoforms II (lanes 1 and 2) or III (lanes 3 and 4). Cells not receiving the E6 plasmid were transfected with an equivalent amount of empty vector. After cell lysis, protein extracts were resolved by SDS-PAGE and immunoblotted with the 12CA5 antibody to detect HA-tagged E6AP or with the AU1 antibody to detect AU1-tagged E6.
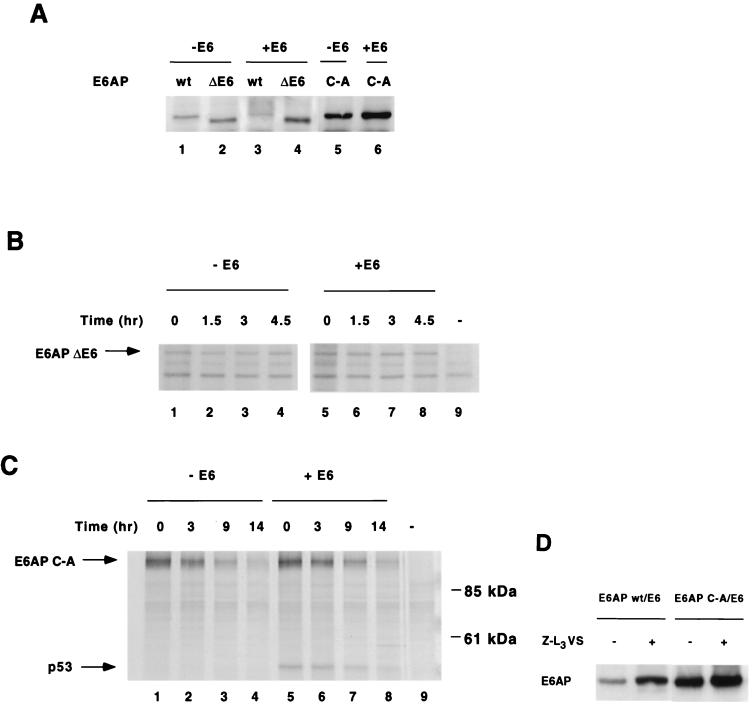
FIG. 6.
E6-mediated E6AP degradation in human cells requires E6-E6AP interaction, E6AP catalytic activity, and proteasome function. (A) Effect of E6 on steady-state levels of E6AP wild-type, ΔE6, and C-A. HA-E6AP isoform I (wild-type, ΔE6, or C-A mutant) was transfected with or without AU1-E6 into C33A cells. Cells were harvested 48 h posttransfection and immunoblotted for HA-E6AP with the 12CA5 antibody. (B and C) E6 expression does not affect the degradation rate of transfected E6AP ΔE6 (B) or C-A mutants (C) in C33A cells. Pulse-chase analysis of E6AP ΔE6 and C-A in the absence or presence of E6 was performed as in Fig. 2. Endogenous p53 immunoprecipitates with E6AP C-A in the presence of E6 as shown. (D) Steady-state levels of E6AP wild-type and C-A mutant in the presence of the proteasome inhibitor Z-L3VS. C33A cells were cotransfected with AU1-E6 and either HA-E6AP wild type or C-A. At 36 h posttransfection, transfected cells were treated for 14 h with either 50 μM Z-L3VS in DMSO or DMSO alone as a negative control. Cells were harvested, separated by SDS-PAGE and immunoblotted with the 12CA5 antibody.
HPV16 E6 increases the rate of degradation of E6AP.
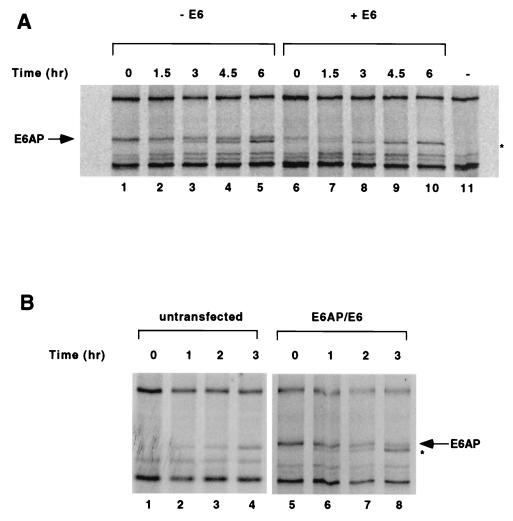
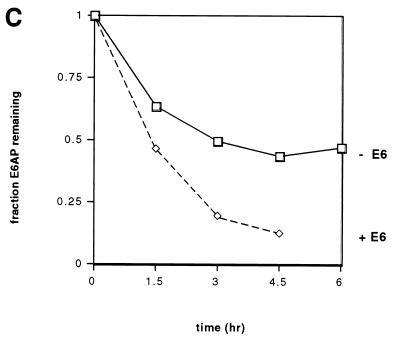
We next examined whether the decrease in E6AP levels in the presence of E6 was due to an increase in the rate of E6AP degradation. C33A cells transfected with HA-tagged E6AP and AU1-tagged E6 were metabolically labeled with [35S]methionine, and the kinetics of E6AP degradation were measured by pulse-chase analysis. As shown in Fig. 2A and C, E6 expression resulted in an approximately threefold reduction in the half-life of transfected E6AP, from 4.5 h in the absence of E6 (Fig. 2A, lanes 1 to 5) to 1.5 h in the presence of E6 (Fig. 2A, lanes 6 to 10).
FIG. 2.
HPV16 E6 accelerates the degradation of E6AP. (A) Pulse-chase analysis of E6AP isoform I in the absence (lanes 1 to 5) or presence (lanes 6 to 10) of E6. C33A cells were transfected with HA-E6AP isoform I either with or without AU1-16E6. As a negative control, one plate of cells was transfected with an equivalent amount of empty vector (lane 11). At 48 h posttransfection, cells were labeled with [35S]methionine and chased with nonradioactive media for the indicated times. Cell extracts were harvested, and equal amounts of each sample were immunoprecipitated overnight with the 12CA5 antibody and a mixture of protein A- and protein G-agarose beads. Immunoprecipitated proteins were resolved by SDS-PAGE and visualized by autoradiography. (B) Pulse-chase analysis of untransfected C33A cells (lanes 1 to 4) or cells transfected with HA-E6AP isoform I and AU1-E6 (lanes 5 to 8). The band immediately below E6AP in panels A and B (marked by asterisk) increases with time even in the absence of transfected HA-E6AP (Fig. 2B, lanes 1 to 4) and is likely an endogenous protein not related to E6AP that cross-reacts with the 12CA5 antibody. (C) Quantitation of data shown in panel A. Intensities of the gel bands were quantified using the NIH Image 1.60/68k program.
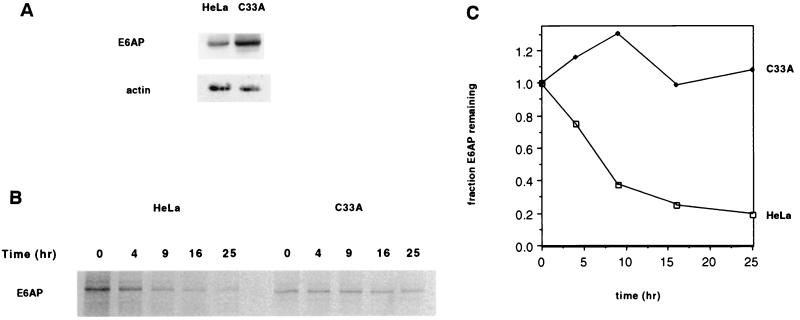
Endogenous E6AP is degraded more rapidly in HPV-positive HeLa cells than in HPV-negative C33A cells.
To test the physiologic relevance of the enhanced proteolysis of E6AP by E6, we examined whether endogenous E6AP was degraded more quickly in HPV-positive cells than in HPV-negative cells. We found that this was indeed the case by comparing E6AP levels in HPV18-positive HeLa cervical carcinoma cells and in HPV-negative C33A cervical carcinoma cells. Western blot analysis revealed that the steady-state levels of endogenous E6AP were 3.2 times greater in HPV-negative C33A cells than in HeLa cells (Fig. 3A). Furthermore, pulse-chase analysis demonstrated that in HeLa cells E6AP was degraded with a half-life of 7 h, whereas in C33A cells E6AP was stable over the entire 25 h of the experiment (Fig. 3B and C). Thus, while endogenous E6AP was more stable than transfected E6AP (Fig. 2), as has been documented previously (38), this result is nevertheless consistent with E6-mediated degradation of endogenous E6AP.
FIG. 3.
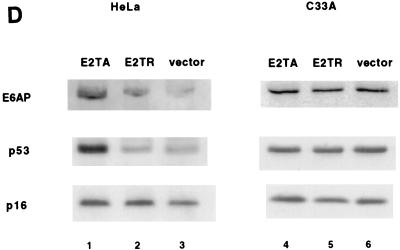
Endogenous E6AP is more rapidly degraded in HPV-positive HeLa cells than in HPV-negative C33A cells. (A) Comparison of endogenous E6AP levels in HeLa and C33A cells. A total of 50 μg of protein extract from HeLa and C33A cells were protein extracts were resolved by SDS-PAGE and analyzed by immunoblotting with anti-E6AP antibody (upper panel) or anti-actin (lower panel). (B) Pulse-chase analysis of E6AP in HeLa and C33A cells. Cells were labeled with [35S]methionine and chased with nonradioactive media for the indicated times. Cell extracts were harvested, and equal amounts of each sample were immunoprecipitated with polyclonal anti-E6AP, separated by SDS-PAGE, and visualized by autoradiography. (C) Quantitation of data in panel B. (D) Stabilization of E6AP protein levels in HeLa cells, but not in C33A cells, transfected with BPV1 E2. Cells were transfected with 1 μg of pBabe-puro plus 5 μg of either BPV1 E2TA or E2TR expression plasmids or else vector alone. Cells were maintained under puromycin-containing medium for 5 days. After harvesting of the cell extracts, 50 μg of protein per sample was resolved by SDS-PAGE and immunoblotted with anti-E6AP (upper panel), anti-p53 (middle panel), or anti-p16 (lower panel).
Repression of the E6 expression in HeLa cells leads to increased levels of E6AP.
To further test the hypothesis that endogenous HPV18 E6 stimulates degradation of E6AP in HeLa cells, we reasoned that this degradation should be attenuated by expression of the BPV1 E2 protein, which has been shown to repress expression of endogenous HPV E6 and E7 from the P105 promoter in HeLa cells (25, 40, 54, 55). HeLa cells were cotransfected with E2-expressing plasmid and a puromycin resistance plasmid, and after 5 days under puromycin selection the cells were harvested and assayed for E6AP protein levels by Western blot. Expression of full-length BPV1 E2 protein (E2-TA) resulted in a 2.8-fold elevation of E6AP levels compared to cells transfected with E2-TR (Fig. 3D, lanes 1 and 2, upper panel), a truncated form of BPV E2 which does not downregulate E6 expression (11, 14). Consistent with previous reports (7, 24), the decrease in HPV18 E6 expression caused by E2-TA also resulted in a sixfold increase in p53 levels compared to cells transfected with E2-TR (Fig. 3D, compare lanes 1 and 2, middle panel). As a control, we examined the levels of the tumor suppressor p16INK4A in HeLa cells which are unaffected by E2-TA expression (S. I. Wells, D. A. Francis, J. J. Dowhanick, J. D. Benson, P. M. Howley, submitted for publication) (Fig. 3D, lanes 1 to 3, lower panel).
By contrast, expression of E2-TA did not result in elevated E6AP levels (Fig. 3D, lanes 4 to 6, upper panel) or elevated p53 levels (Fig. 3D, lanes 4 to 6, middle panel) in HPV-negative C33A cells, indicating that the effect of E2-TA on E6AP most likely involves E6 through repression of the integrated HPV18P105 promoter. Since E2-TA also represses expression of HPV18 E7, this experiment by itself does not rule out the possibility that E7 also promotes E6AP degradation. However, this is an unlikely possibility given that expression of E6, and not E7, resulted in E6AP degradation when these proteins were expressed individually in C33A cells (Fig. 1A). These experiments therefore provide strong support for our hypothesis that E6 expression results in a more rapid turnover of E6AP.
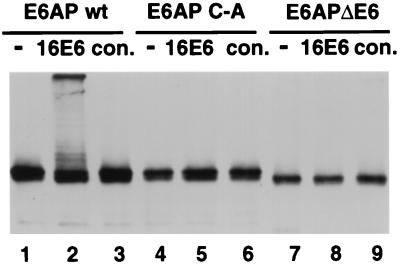
E6-mediated E6AP ubiquitination in vitro requires the catalytic activity of E6AP.
To determine if the in vivo effect of E6 on E6AP stability might be due to enhanced ubiquitination of E6AP, baculovirus-expressed and partially purified HPV16 E6 and E6AP proteins were incubated together in the presence of purified E1 enzyme, E2 enzyme, ATP, and ubiquitin. As shown in Fig. 4, E6 induced the multiubiquitination of E6AP (lane 2). This ubiquitination was dependent upon the active-site cysteine (C820) of E6AP (lane 5), suggesting that isopeptide-linked ubiquitination was the result of transfer of thiolester-linked ubiquitin from the active-site cysteine to one or more lysine residues of E6AP. Deletion of the 18-amino-acid E6 binding domain also abrogated E6-induced ubiquitination (lane 8). Therefore, the effect of E6 on E6AP turnover appears to be the result of self-catalyzed multiubiquitination of E6AP.
FIG. 4.
E6-dependent E6AP ubiquitination requires the active-site cysteine and the E6-binding domain of E6AP. Partially purified baculovirus-expressed epitope tagged wild-type E6AP (wt; lanes 1 to 3), E6AP carrying the active-site cysteine to alanine mutation (C-A; lanes 4 to 6), and E6AP with an internal deletion of 18 amino acids corresponding to the E6 binding domain (ΔE6; lanes 7 to 9) were incubated with E1 protein, E2 protein (A. thaliana Ubc8), ubiquitin, and ATP either without (−) or with baculovirus-expressed HPV16 E6 protein (16E6) or an equivalent control fraction from cells infected with wild-type baculovirus alone (con.). E6AP was detected by SDS-PAGE and immunoblotting with antibody against the HA epitope.
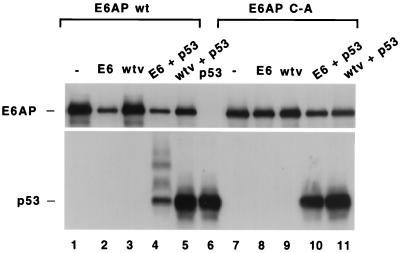
Figure 5 shows that the same effects could be observed by coinfection of E6- and E6AP-expressing baculoviruses in insect cells, which lack endogenous E6AP activity (36). As anticipated, baculovirus-expressed p53 was ubiquitinated and degraded when coexpressed with wild-type E6AP and HPV16 E6 (Fig. 5, compare lanes 4 and 5, middle panel), whereas E6 did not induce p53 ubiquitination in the presence of catalytically inactive E6AP C-A (compare lanes 10 and 11). Wild-type E6AP protein levels (lane 2 versus lane 3) but not E6AP C-A (lane 8 versus lane 9) were correspondingly reduced in the presence of HPV16 E6.
FIG. 5.
HPV16 E6 stimulates the degradation of both E6AP and p53 in an insect cell system. High Five insect cells (derived from Tricoplusia ni; Invitrogen) were infected with combinations of baculoviruses expressing wild-type E6AP (wt; lanes 1 to 5) or the active-site cysteine-to-alanine mutation of E6AP (C-A; lanes 7 to 11), HPV16 E6, p53, and nonrecombinant virus (wtv) as indicated. Lane 6 represents cells infected only with p53-expressing virus. Cell extracts were made 36 h postinfection and analyzed by SDS-PAGE and immunoblotting with anti-p53 antibody and rabbit polyclonal E6AP antibody.
Degradation of E6AP in mammalian cells requires interaction with E6 and catalytic activity of E6AP and is proteasome dependent.
To verify that E6-mediated degradation of E6AP occurs via E6AP catalytic activity in mammalian cells, we cotransfected AU1-tagged HPV16 E6 and HA-tagged E6AP into human cell lines. As observed previously, wild-type E6AP levels were greatly reduced in the presence of E6 in C33A cells (Fig. 6A, compare lanes 1 and 3). In accord with in vitro results (Fig. 4), E6 expression did not decrease the steady-state levels of E6AP ΔE6, in which the E6-binding region has been deleted (Fig. 6A, compare lanes 2 and 4), and E6 did not affect the kinetics of E6AP ΔE6 degradation, as determined by pulse-chase analysis (Fig. 6B, compare lanes 1 to 4 and 5 to 8) in C33A cells. E6 expression also did not significantly affect the steady-state levels (Fig. 6A, compare lanes 5 and 6) or stability (Fig. 6C, compare lanes 1 to 4 and 5 to 8) of E6AP C-A. While it must be noted that in other cell lines we observed a slight decrease in E6AP C-A levels in the presence of E6 (see Fig. 8 below and Discussion), these results nonetheless suggest that E6-E6AP interaction also results in E6AP self-ubiquitination in mammalian cells.
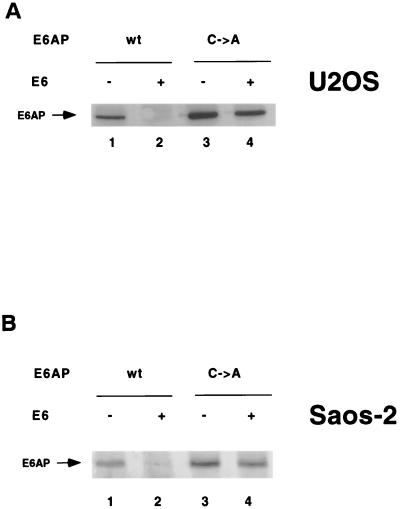
FIG. 8.
Expression of HPV16 E6 induces degradation of wild-type and mutant E6AP in Saos-2 (A) and U2OS (B) cells. HA-E6AP (wild-type or C-A mutant) was transfected with or without AU1-tagged E6 into tissue culture cells. Cells were harvested at 48 h posttransfection and immunoblotted for HA-E6AP with the 12CA5 antibody. U2OS cells were transfected with E6AP isoform I; Saos-2 cells were transfected with E6AP isoform II.
To determine whether E6 was causing degradation of E6AP by the proteasome, we studied the effect of the proteasome inhibitor Z-L3VS (4) on E6AP levels. C33A cells were cotransfected with E6AP and E6 as before and treated for 14 h with 50 μM Z-L3VS or with DMSO as a mock control. In the presence of E6 expression, Z-L3VS treatment resulted in a 360% increase in wild-type E6AP (Fig. 6D, lanes 1 and 2), while E6AP C-A levels only increased 25% during Z-L3VS treatment (Fig. 6D, lanes 3 and 4), suggesting that E6AP C-A, which is incapable of self-ubiquitination, is not efficiently degraded by the proteasome in the presence of E6. These results are therefore also consistent with E6-stimulated E6AP self-ubiquitination and subsequent degradation by the 26S proteasome.
HPV16 E6-induced E6AP ubiquitination is primarily, but not exclusively, an intramolecular reaction.
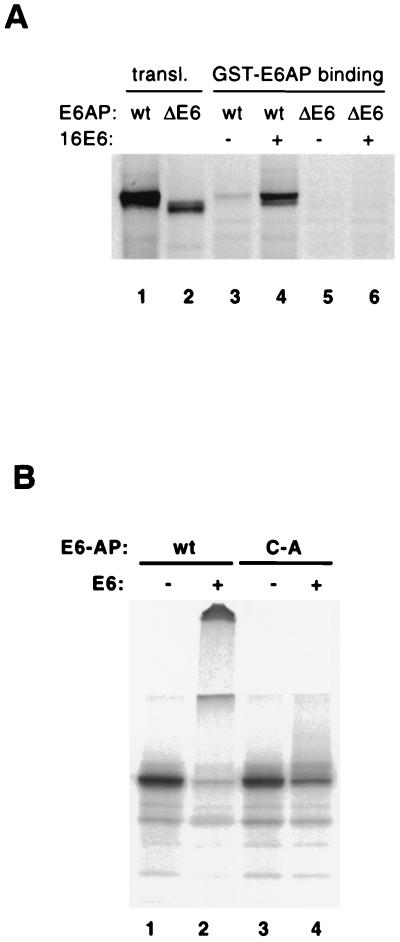
The simplest model for E6-induced self-ubiquitination of E6AP is that ubiquitination is the result of an intramolecular transfer of ubiquitin from the active-site cysteine of E6AP to lysine residues on the same E6AP molecule. However, the possibility of an intermolecular ubiquitin transfer within an E6-mediated E6AP multimer was suggested by binding experiments showing that glutathione S-transferase (GST)-E6AP bound to in vitro translated E6AP in the presence of HPV16 E6 (Fig. 7A, lane 4). Binding was dependent on the 18-amino-acid E6 binding domain of E6AP (Fig. 7A, compare lanes 4 and 6). Therefore, it was conceivable that self-catalyzed ubiquitination of E6AP could be the result of ubiquitin transfer from one E6AP molecule to another within an E6-mediated dimer or higher multimer.
FIG. 7.
HPV16 E6 induces degradation of a mixture of wild-type and mutant E6AP and mediates E6AP dimerization in vitro. (A) E6AP dimerization in the presence of E6. In vitro-translated, 35S-labeled wild-type (lanes 3 and 4) and ΔE6 (lanes 5 and 6) forms of E6AP were incubated with GST-E6AP protein in the presence or absence of E6 and then immobilized on glutathione-Sepharose beads. The beads were washed and analyzed by SDS-PAGE, followed by autoradiography. (B) Degradation of wild-type and mutant E6AP in vitro. E6AP wild type (wt) and the active-site Cys-to-Ala mutant (C-A) were translated in rabbit reticulocyte lysate in the presence of [35S]methionine. The translation reactions were incubated for 15 min at room temperature with E1 protein, E2 protein (A. thaliana Ubc8), ubiquitin, ATP, and baculovirus-expressed HPV16 E6 protein (+) or an equivalent control fraction (−). Reaction products were analyzed by SDS-PAGE and autoradiography.
To distinguish between the intra- and intermolecular transfer models, we generated 35S-labeled wild-type and catalytically inactive E6AP in rabbit reticulocyte lysate. If E6-mediated self-ubiquitination was exclusively intramolecular, wild-type E6AP would be predicted to be multiubiquitinated and degraded, whereas E6AP C-A should be unaffected by E6. In contrast, if the transfer were exclusively intermolecular, both wild-type E6AP and the inactive E6AP C-A should be ubiquitinated to a similar degree, since rabbit reticulocyte lysate contains endogenous E6AP that is functionally equivalent to human E6AP (22). Figure 7B shows that wild-type E6AP was efficiently ubiquitinated and degraded in rabbit reticulocyte lysate (lane 2), whereas E6AP C-A was affected to a much lesser, but still detectable, degree (lane 4).
A potential trivial explanation for the data in Fig. 7B was that the E6AP C-A mutant might be poorly ubiquitinated in vitro in the presence of E6 simply by losing its ability to associate with wild-type E6AP. However, we have found that E6AP C-A associates with GST-E6AP in the presence of E6 to a similar degree as observed for the wild-type–wild-type interaction seen in Fig. 7A (S. L. Beaudenon and J. M. Huibregtse, unpublished data). Therefore it appears that E6-induced E6AP ubiquitination is indeed primarily, but not exclusively, the result of intramolecular transfer of ubiquitin from the active-site cysteine to one or more lysine residues on the same E6AP molecule.
To assess whether intramolecular ubiquitination of E6AP occurs in vivo, we compared the levels of transfected E6AP in U2OS and Saos-2 cells in the absence or presence of cotransfected E6. E6AP C-A levels in U2OS (Fig. 8A) and Saos-2 cells (Fig. 8B) were slightly decreased in the presence of E6 (compare lanes 3 and 4 in Fig. 8A and B), probably due to ubiquitination by endogenous wild-type E6AP, whereas the levels of transfected wild-type E6AP were substantially reduced by coexpression of E6 (lanes 1 and 2 in Fig. 8A and B). Thus, E6-mediated E6AP self-ubiquitination in vivo may also be primarily intramolecular, with intermolecular ubiquitination also occurring to some degree.
In addition, the fact that E6-mediated degradation of E6AP occurred in both U2OS cells, which express low levels of wild-type p53, and in p53-null Saos-2 cells shows that E6 stimulates E6AP ubiquitination either in the absence or presence of p53. Therefore, although it has been previously reported that high concentrations of p53 can inhibit E6AP self-ubiquitination in vitro in the presence of E6 (38), we conclude that normal endogenous levels of wild-type p53 do not interfere with E6-mediated degradation of E6AP in vivo.
An HPV16 E6 mutant deficient for p53 degradation but retaining immortalization ability is able to stimulate the ubiquitination and degradation of E6AP.
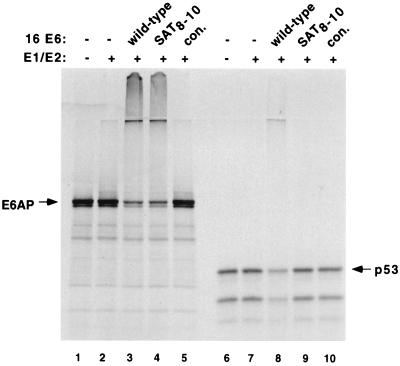
The HPV16 E6 SAT8–10 mutant has been previously shown to retain its ability to immortalize human mammary epithelial cells (MECs) (27), to induce colony formation and growth on soft agar (50), and to induce telomerase activity (27). We therefore tested the ability of E6AP SAT8–10 to degrade E6AP in vitro. Figure 9 demonstrates that E6 SAT8–10 promotes the ubiquitination of E6AP to a similar extent as wild-type E6 (lane 3 versus lane 4) but, unlike wild-type E6, the SAT8–10 mutant was not able to degrade p53 (lane 8 versus lane 9). While further analyses of E6 mutants are required, these results raise the possibility that the degradation of E6AP by E6 may contribute to the ability of E6 to immortalize primary human cells.
FIG. 9.
HPV16E6 SAT8–10 mutant ubiquitinates E6AP and not p53 in vitro. Wild-type E6AP (95-kDa protein) and p53 were translated in rabbit reticulocyte lysate in the presence of [35S]methionine. The translation reactions were incubated for 30 min at room temperature without (−) or with E1 protein, E2 protein (A. thaliana Ubc8), ubiquitin, or ATP in the absence (−) or presence of partially purified baculovirus-expressed HPV16 E6, HPV16E6 SAT8–10 mutant, or an equivalent control fraction from cells infected with wild-type virus alone (con.). Reaction products were analyzed by SDS-PAGE and autoradiography.
DISCUSSION
Recent evidence suggests that certain components of the ubiquitin-proteasome machinery itself, such as the F-box proteins Cdc4p, Grr1p, and Met30p, are short-lived proteins whose activity is regulated by ubiquitination (12, 63). The E6AP ubiquitin-protein ligase has also been shown to auto-ubiquitinate in vitro, and it has been proposed that E6AP auto-ubiquitination may play a role in regulation of E6AP activity (38). We demonstrate here that the HPV16 E6 enhances the self-ubiquitination and degradation of E6AP.
We suspect that E6 might trigger the self-ubiquitination reaction by altering the conformation of E6AP such that the catalytic domain of E6AP has access to suitable lysine residues elsewhere on the protein. The self-ubiquitination reaction appears to be primarily the result of an intramolecular transfer of ubiquitin from the active-site cysteine of E6AP to one or more lysine residues of the same E6AP molecule, where it is linked via isopeptide bonds. Multiple ubiquitins are linked to E6AP, although whether one or more lysine residues of E6AP are targeted or whether a multiubiquitin chain is assembled at a single E6AP lysine residue is unknown. The fact that E6 can stimulate the relatively inefficient ubiquitination of the E6AP C-A mutant in the presence of wild-type E6AP suggests that the self-ubiquitination reaction can also proceed via an intermolecular transfer of ubiquitin, from one molecule of E6AP to another, in the context of an E6-mediated dimer or higher multimer. This model is supported by in vitro binding experiments that show that GST-E6AP binds E6AP in the presence of E6.
An intermolecular mechanism of self-ubiquitination is supported by our in vivo observation that E6 expression results in slightly decreased levels of the E6AP C-A mutant protein in both U2OS and Saos-2 cells, presumably because E6 induces ubiquitination of the transfected E6AP C-A mutant protein by the endogenous wild-type protein. In contrast, E6 expression did not result in a decrease in E6AP C-A levels in C33A cells, even though wild-type E6AP was efficiently degraded. This observation suggests that there may be some cell line variability to the extent of an intramolecular versus intermolecular transfer of ubiquitin in E6-stimulated E6AP auto-ubiquitination. It will be of interest to determine the molecular basis of this cell line variability.
We have not yet ruled out the possibility that E6 also contributes to E6AP destabilization through more indirect methods, perhaps by inducing E6AP to degrade an inhibitor of E6AP turnover, by altering E6AP localization, or by recruiting modifying enzymes that could activate E6AP self-ubiquitination. However, the requirement of E6AP catalytic activity for E6-mediated degradation of E6AP in vivo and for degradation of partially purified E6AP in vitro provides compelling evidence that E6 destabilizes E6AP by promoting a self-ubiquitination reaction.
The finding that E6 promotes the self-ubiquitination of E6AP raises the possibility that other E6AP-binding proteins may also affect E6AP stability. It will be interesting to determine whether E6AP substrates affect E6AP turnover and what factors might account for any differences in the ability of various E6AP-binding proteins to affect E6AP stability. Just as the turnover of the src-family kinases is increased upon activation (16), it is conceivable that an increase in E6AP activity might also stimulate its ability to ubiquitinate itself. In effect, the auto-ubiquitination of the catalytically active form of E6AP could provide a negative feedback loop to limit E6AP activity once a substrate has been ubiquitinated.
While it is clear that the ability of E6 to cause the self-ubiquitination of E6AP does not prevent E6AP-mediated degradation of p53, it remains possible that E6-induced self-ubiquitination of E6AP may impair its ability to ubiquitinate other substrates such as the src-family kinases (39) or HHR23A and -B, which are involved in nucleotide excision repair (35, 52). It has been reported that E6-expressing fibroblasts are deficient in nucleotide excision repair (10). Although this deficiency could be accounted for in part by E6-mediated p53 degradation, the possibility remains that disruption of HHR23 function by E6 may also compromise the capacity for DNA repair. Alterations in the function of one of the src-family kinases or the HHR23 proteins by HPV16 E6 via destabilization of E6AP could also contribute to the transforming potential of the E6 protein.
Alternatively, it is possible that, while E6 does promote E6AP self-ubiquitination, the E6-E6AP interaction could alter E6AP conformation in a way that enhances its ability to ubiquitinate some of its substrates. This is an intriguing possibility; however, to date there is no evidence that ubiquitination of any E6AP substrate (other than p53) is stimulated by E6. We are currently performing experiments to assess whether the stability of established substrates of E6AP such as HHR23A and blk is affected by E6. Several proteins including E6TP1 and hMCM7 have been reported to be degraded in the presence of E6 (13, 30), but it has not been determined whether E6AP is involved in their degradation.
While much of our analysis has focused on HPV16 E6, we have also demonstrated that E6AP is degraded in HPV18-positive HeLa cells and that expression of wild-type BPV1 E2, which represses expression of endogenous E6, significantly increases the levels of endogenous E6AP in HeLa cells (Fig. 3). Furthermore, in vitro and in vivo experiments have indicated that E6AP self-ubiquitination is stimulated by 18E6, as well as the E6 proteins of HPV33 and 39, which are also cancer-associated HPVs (S. L. Beaudenon, A. L. Talis, and W. H. Kao, unpublished data). Therefore, it appears that HPV18 E6 accounts for the destabilization of E6AP in HeLa cells.
We have also shown that the E6 SAT8–10 mutant, which retains its ability to transform human mammary epithelial cells, is unable to degrade p53 but is able to degrade E6AP. E6, however, is known to bind to a number of proteins, and some of these interactions have been proposed as potential mechanisms for E6-mediated oncogenesis (13, 28). Proteins that interact with E6 include hDLG (28), which has been identified as a tumor suppressor gene in Drosophila spp. (34, 61); paxillin (56, 58), which may be involved in the transduction of signals from plasma membrane to focal adhesions and actin cytoskeleton (57); and E6TP1, a protein with high homology to GTPase-activating proteins (13). While the significance of these interactions is still being assessed, it is possible that these interactions may contribute to p53-independent oncogenic properties of E6. Thus far, no interaction between HPV16 E6 and any single protein has been proven to be sufficient to account for all of the transformation properties of E6. These properties may therefore be the result of the ability of E6 to interact with multiple proteins.
It is not yet known whether the E6 SAT8–10 mutant is able to bind to any other E6-interacting proteins, or whether other E6 mutants retain their ability to degrade E6AP. Thus, no definitive answer can yet be given as to whether E6-mediated degradation of E6AP plays any role in transformation. It must be noted, however, that two additional HPV16 E6 mutants (F2V and Y54H) are deficient in their ability to degrade p53 in vitro at 37°C but retain E6AP binding as well as their ability to immortalize MECs (33); if these mutants also retain their ability to degrade E6AP, it would certainly strengthen the case for a potential role for E6AP degradation in the p53-independent transformation functions of E6.
We are beginning to appreciate the complexities of the interactions between E6AP, E6, p53, and the normal cellular substrates of E6AP. Here we have demonstrated that both E6AP and p53 are ubiquitinated and degraded through contact with E6, with E6AP degradation apparently occurring through self-ubiquitination. Further investigations into the nature of the E6AP, E6, and p53 interaction should enhance our understanding of E6-mediated transformation, E6AP-regulated pathways, and of the ubiquitin-proteasome pathway in general.
ACKNOWLEDGMENTS
W.H.K. and S.L.B. contributed equally to this work.
We are grateful to Amanda Chan, Lisa Decker, Kimya Harris, and Susanne Schmid for helpful suggestions and comments on the manuscript.
This research was supported by grants from the National Institutes of Health (R37CA64888-06 and P01CA50661 to P.M.H. and R01CA 72943 to J.M.H.).
REFERENCES
- 1.Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 2.Band V, De Caprio J A, Delmolino L, Kulesa V, Sager R. Loss of p53 protein in human papillomavirus type 16 E6-immortalized human mammary epithelial cells. J Virol. 1991;65:6671–6676. doi: 10.1128/jvi.65.12.6671-6676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer-Romero P, Glass S, Rolfe M. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene. 1997;14:595–602. doi: 10.1038/sj.onc.1200872. [DOI] [PubMed] [Google Scholar]
- 4.Bogyo M, McMaster J S, Gaczynska M, Tortorella D, Goldberg A L, Ploegh H. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc Natl Acad Sci USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer S N, Wazer D E, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 6.Dostatni N, Lambert P F, Sousa R, Ham J, Howley P M, Yaniv M. The functional BPV-1 E2 trans-activating protein can act as a repressor by preventing formation of the initiation complex. Genes Dev. 1991;5:1657–1671. doi: 10.1101/gad.5.9.1657. [DOI] [PubMed] [Google Scholar]
- 7.Dowhanick J J, McBride A A, Howley P M. Suppression of cellular proliferation by the papillomavirus E2 protein. J Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyson N, Howley P M, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 9.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 10.Ford J M, Baron E L, Hanawalt P C. Human fibroblasts expressing the human papillomavirus E6 gene are deficient in global genomic nucleotide excision repair and sensitive to ultraviolet irradiation. Cancer Res. 1998;58:599–603. [PubMed] [Google Scholar]
- 11.Francis D A, Schmid S I, Howley P M. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J Virol. 2000;74:2679–2686. doi: 10.1128/jvi.74.6.2679-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galan J M, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Q, Srinivasan S, Boyer S N, Wazer D E, Band V. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol Cell Biol. 1999;19:733–744. doi: 10.1128/mcb.19.1.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodwin E C, Naeger L K, Breiding D E, Androphy E J, DiMaio D. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J Virol. 1998;72:3925–3934. doi: 10.1128/jvi.72.5.3925-3934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 16.Harris K F, Shoji I, Cooper E M, Kumar S, Oda H, Howley P M. Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc Natl Acad Sci USA. 1999;96:13738–13743. doi: 10.1073/pnas.96.24.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawley-Nelson P, Vousden K H, Hubbert N L, Lowy D R, Schiller J T. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 19.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley P M, Huibregtse J M, Pavletich N P. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 20.Hudson J B, Bedell M A, McCance D J, Laiminis L A. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. J Virol. 1990;64:519–526. doi: 10.1128/jvi.64.2.519-526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huibregtse J M, Scheffner M, Howley P M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huibregtse J M, Scheffner M, Howley P M. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang E S, Naeger L K, DiMaio D. Activation of the endogenous p53 growth inhibitory pathway in HeLa cervical carcinoma cells by expression of the bovine papillomavirus E2 gene. Oncogene. 1996;12:795–803. [PubMed] [Google Scholar]
- 25.Hwang E S, Riese D J D, Settleman J, Nilson L A, Honig J, Flynn S, DiMaio D. Inhibition of cervical carcinoma cell line proliferation by the introduction of a bovine papillomavirus regulatory gene. J Virol. 1993;67:3720–3729. doi: 10.1128/jvi.67.7.3720-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones D L, Thompson D A, Munger K. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 27.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 28.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klingelhutz A J, Foster S A, McDougall J K. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 30.Kuhne C, Banks L. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J Biol Chem. 1998;273:34302–34309. doi: 10.1074/jbc.273.51.34302. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S, Kao W H, Howley P M. Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J Biol Chem. 1997;272:13548–13554. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Talis A L, Howley P M. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Chen J J, Gao Q, Dalal S, Hong Y, Mansur C P, Band V, Androphy E J. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J Virol. 1999;73:7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lue R A, Marfatia S M, Branton D, Chishti A H. Cloning and characterization of hdlg: the human homologue of the Drosophila discs large tumor suppressor binds to protein 4.1. Proc Natl Acad Sci USA. 1994;91:9818–9822. doi: 10.1073/pnas.91.21.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek P J, Bootsma D, et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munger K, Phelps W C, Bubb V, Howley P M, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nuber U, Schwarz S, Kaiser P, Schneider R, Scheffner M. Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP and RSP5. J Biol Chem. 1996;271:2795–2800. doi: 10.1074/jbc.271.5.2795. [DOI] [PubMed] [Google Scholar]
- 38.Nuber U, Schwarz S E, Scheffner M. The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur J Biochem. 1998;254:643–649. doi: 10.1046/j.1432-1327.1998.2540643.x. [DOI] [PubMed] [Google Scholar]
- 39.Oda H, Kumar S, Howley P M. Regulation of the src family tyrosine kinase blk through E6AP-mediated ubiquitination. Proc Natl Acad Sci USA. 1999;96:9557–9562. doi: 10.1073/pnas.96.17.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romanczuk H, Thierry F, Howley P M. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol. 1990;64:2849–2859. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheffner M, Huibregtse J M, Howley P M. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc Natl Acad Sci USA. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 43.Scheffner M, Nuber U, Huibregtse J M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 44.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 45.Sedman S A, Barbosa M S, Vass W C, Hubbert N L, Haas J A, Lowy D R, Schiller J T. The full-length E6 protein of human papillomavirus type 16 has transforming and trans-activating activities and cooperates with E7 to immortalize keratinocytes in culture. J Virol. 1991;65:4860–4866. doi: 10.1128/jvi.65.9.4860-4866.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman L, Schlegel R. Serum- and calcium-induced differentiation of human keratinocytes is inhibited by the E6 oncoprotein of human papillomavirus type 16. J Virol. 1996;70:3269–3279. doi: 10.1128/jvi.70.5.3269-3279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 48.Song S, Pitot H C, Lambert P F. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J Virol. 1999;73:5887–5893. doi: 10.1128/jvi.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spalholz B A, Vande Pol S B, Howley P M. Characterization of the cis elements involved in basal and E2-transactivated expression of the bovine papillomavirus P2443 promoter. J Virol. 1991;65:743–753. doi: 10.1128/jvi.65.2.743-753.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spitkovsky D, Aengeneyndt F, Braspenning J, von Knebel Doeberitz M. p53-independent growth regulation of cervical cancer cells by the papillomavirus E6 oncogene. Oncogene. 1996;13:1027–1035. [PubMed] [Google Scholar]
- 51.Storey A, Banks L. Human papillomavirus type 16 E6 gene cooperates with EJ-ras to immortalize primary mouse cells. Oncogene. 1993;8:919–924. [PubMed] [Google Scholar]
- 52.Sugasawa K, Ng J M, Masutani C, Maekawa T, Uchida A, van der Spek P J, Eker A P, Rademakers S, Visser C, Aboussekhra A, Wood R D, Hanaoka F, Bootsma D, Hoeijmakers J H. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol Cell Biol. 1997;17:6924–6931. doi: 10.1128/mcb.17.12.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talis A L, Huibregtse J M, Howley P M. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998;273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 54.Thierry F, Howley P M. Functional analysis of E2-mediated repression of the HPV18 P105 promoter. New Biol. 1991;3:90–100. [PubMed] [Google Scholar]
- 55.Thierry F, Yaniv M. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 1987;6:3391–3397. doi: 10.1002/j.1460-2075.1987.tb02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong X, Howley P M. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc Natl Acad Sci USA. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turner C E. Paxillin: a cytoskeletal target for tyrosine kinases. Bioessays. 1994;16:47–52. doi: 10.1002/bies.950160107. [DOI] [PubMed] [Google Scholar]
- 58.Vande Pol S B, Brown M C, Turner C E. Association of bovine papillomavirus type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif. Oncogene. 1998;16:43–52. doi: 10.1038/sj.onc.1201504. [DOI] [PubMed] [Google Scholar]
- 59.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 60.Winokur P L, McBride A A. Separation of the transcriptional activation and replication functions of the bovine papillomavirus-1 E2 protein. EMBO J. 1992;11:4111–4118. doi: 10.1002/j.1460-2075.1992.tb05504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woods D F, Bryant P J. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto Y, Huibregtse J M, Howley P M. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics. 1997;41:263–266. doi: 10.1006/geno.1997.4617. [DOI] [PubMed] [Google Scholar]
- 63.Zhou P, Howley P M. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- 64.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]