Abstract
Hepatocellular Carcinoma (HCC), the predominant primary hepatic malignancy, is the prime contributor to mortality. Despite the availability of multiple surgical interventions, patient outcomes remain suboptimal. Immunotherapies have emerged as effective strategies for HCC treatment with multiple clinical advantages. However, their curative efficacy is not always satisfactory, limited by the dysfunctional T cell status. Thus, there is a pressing need to discover novel potential biomarkers indicative of T cell exhaustion (Tex) for personalized immunotherapies. One promising target is Cyclin-dependent kinase inhibitor 2 (CDKN2) gene, a key cell cycle regulator with aberrant expression in HCC. However, its specific involvement remains unclear. Herein, we assessed the potential of CDKN2 expression as a promising biomarker for HCC progression, particularly for exhausted T cells. Our transcriptome analysis of CDKN2 in HCC revealed its significant role involving in HCC development. Remarkably, single-cell transcriptomic analysis revealed a notable correlation between CDKN2 expression, particularly CDKN2A, and Tex markers, which was further validated by a human cohort study using human HCC tissue microarray, highlighting CDKN2 expression as a potential biomarker for Tex within the intricate landscape of HCC progression. These findings provide novel perspectives that hold promise for addressing the unmet therapeutic need within HCC treatment.
Keywords: Clinical biomarker, Cyclin-dependent kinase inhibitor 2, Hepatocellular carcinoma, T cell exhaustion, Tissue microarray
INTRODUCTION
HCC, which accounts for most primary cancers of the liver, stands as the fourth leading cause of cancer-induced mortality worldwide (1). Owing to the restricted discernment achieved through the prevailing early diagnostic methodologies and the aggressive nature characterizing HCC, a substantial proportion of patients are commonly diagnosed at advanced stages, wherein surgical or localized therapeutic interventions are ineffective (2). In this regard, researchers are striving to advance immune checkpoint inhibitor (ICIs) therapies to reactivate antitumor immune response for long-lasting tumor remissions (3, 4). However, unexpectedly, the efficacy of ICIs is occasionally suboptimal.
Several studies have pointed toward a considerable proportion of CD8+ T cells demonstrate functional hyporesponsiveness, rendering them ineffective for cancer elimination. This signifies the existence of a dysfunctional T cell status defined as Tex (5). Tex encompasses overall dysfunctional conditions that emerge within a repressive tumor microenvironment with extended exposure to antigens, and is characterized by reduced capacity of T cells to secrete cytokines and increased expression of inhibitory receptors (6). The upregulation of diverse inhibitory receptors, typified by programmed cell death protein 1 (PD-1), has been recognized as a pivotal hallmark of Tex, with its co-expression on CD8+ T cells significantly impacting the severity of functional deficits. Blocking these suppressive receptors is supposed to be a viable strategy to enhance the curative efficacy of ICIs (7). Therefore, the foremost imperative is to discern the distinct T cell states in individual patients to provide personalized therapeutic evidence for immunotherapies. Nevertheless, there is a conspicuous scarcity of effective biomarkers for identifying Tex status, highlighting the pressing need for further endeavors in this domain.
Notably, recent investigations have implied the existence of another tolerogenic pathway that disrupts T cell function and responsiveness, known as T cell senescence (Tes). Tes has been reported to involve in multiple types of cancers, such as HCC, gastric cancer, and lung cancer (8), with common attributes in senescent somatic cells. Current research has demonstrated the significance of Tes in regulating HCC tumorigenesis, development, and immune infiltration, wherein a gene family, CDKN2, holds significant involvements (9). CDKN2 gene is cyclin-dependent kinase inhibitor with main functions of cell cycle regulation, which manifests marked dysregulation in diverse cancer types, with clarified correlation with unfavorable clinical prognosis (10, 11). However, despite its emphasized impact across multiple cancers, studies on the mechanistic pathways and significance of CDKN2 in HCC are still lacking clarity. Intriguingly, a previous study confirmed that CDKN2B is an essential target through which PD-1 promotes Tex progression (12), which extremely contrasts with its conventional function. Nevertheless, hitherto, studies elucidating the putative relevance of CDKN2 gene in Tex during HCC progression, beyond its impact on Tes, remains unclear.
In our study, we aimed to estimate the potential implications of senescence-associated factor CDKN2 in HCC, particularly in Tex. Our observations indicated that CDKN2 gene assume pivotal roles in the pathogenesis and advancement of HCC. Remarkably, CDKN2 expression, notably that of CDKN2A, held significant promise as a potential biomarker of Tex in HCC. These findings not only provide novel insights for in-depth mechanical explorations, but also yield promising avenues for potential immunotherapeutic interventions.
RESULTS
Expression patterns of CDKN2 gene family in human HCC
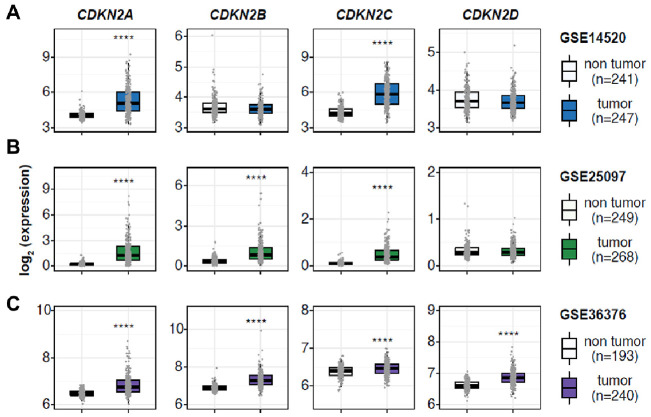
To assess the potential implications of CDKN2 gene in HCC, we analyzed the RNA sequencing data and corresponding clinicopathological information sourced from three independent Gene Expression Omnibus (GEO) cohorts (Fig. 1). Across these datasets, CDKN2A and CDKN2C consistently exhibited elevated expression levels in HCC compared to normal tissues. Conversely, CDKN2B expression was remarkably upregulated in GSE25097 and GSE36376, while displaying a mild but discernible increasing trend in GSE14520. In contrast, CDKN2D showed varying ambiguity and even slightly conflicting expression profiles, which necessitated further assessment within specific clinical settings. Collectively, although conclusive remarks regarding the expression profiles of certain CDKN2 remained elusive, elevated expression of CDKN2A and CDKN2C provided evidence of the implication of CDKN2 in HCC pathogenesis.
Fig. 1.
CDKN2 gene expression profiles in normal liver and hepatocellular carcinoma (HCC) indicating the implication of CDKN2 in HCC development. (A-C) Boxplots showing relative mRNA levels of CDKN2 gene in patients with HCC belonging to three independent cohorts. The horizontal line crossing each box designates the median, while the top and bottom edges represent the first (25%) and third quartiles (75%), respectively. The dots indicate the expression value of each subject. ****P < 0.0001.
CDKN2 gene expression was associated with survival outcomes in patients with HCC
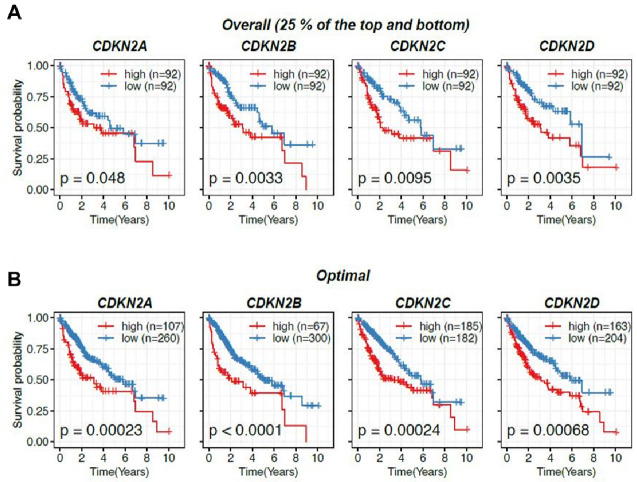
To further investigate the implication of CDKN2 gene in HCC advancement, Kaplan-Meier assay of CDKN2 expression in patients with HCC sourced from The Cancer Genome Atlas (TCGA) liver hepatocellular carcinoma (LIHC) cohorts was performed. Notably, our investigation of overall survival illuminated a positive association between CDKN2 expression and adverse prognosis in HCC. Individuals with HCC were initially grouped into high- and low-expression cohorts based on a conventional and commonly recognized classification, as previous described (13). Individuals with higher expression levels of CDKN2 gene exhibited markedly diminished overall survival rates compared to those with lower expression levels (Fig. 2A), suggesting that CDKN2 was a promising indicator of decreased survival rates in HCC.
Fig. 2.
CDKN2 gene expression was associated with survival outcomes in patients with HCC. (A) Kaplan-Meier curves showing the correlation between CDKN2 gene expression and overall survival by comparing patients with HCC demonstrating high and low CDKN2 expression. (B) Kaplan-Meier curves showing the differential survival probability between CDKN2 expression subgrouping divided using optimized values obtained from the R package, multipleROC.
To validate our findings and ensure the avoidance of any bias, patients with HCC were further categorized based on the optimal cutoff of CDKN2 expression values determined by multiple receiver operating characteristic curve (multipleROC) analysis (14). Consistent with our previous results, patients with HCC exhibiting elevated CDKN2 expression consistently demonstrated significantly worse prognosis than those with lower expression levels of CDKN2 (Fig. 2B). These findings emphasize the considerable involvement of CDKN2 in HCC progression.
Single-cell transcriptomic analysis of CD8+ T cell indicated a correlation between CDKN2A expression and Tex
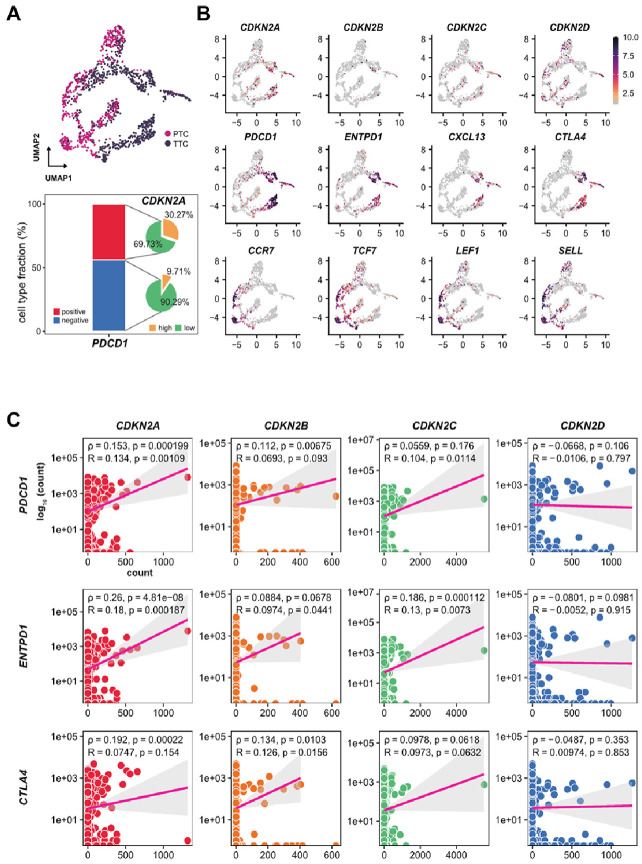
To explore the mechanisms that underlie the correlation between CDKN2 gene expression and HCC clinical outcomes, we carried out a systematic single-cell transcriptomic analysis. Specifically, we focused on CD8+ T cells originating from individuals with HCC, with a particular focus on assessing their correlation with immune checkpoints. UMAP-based trajectory assay confirmed the existence of CD8+ T cells in PTC and TTC (Fig. 3A). Intriguingly, the expression patterns of CDKN2B and CDKN2C did not exhibit substantial associations with distinct cellular subsets, registering comparatively modest levels. On the other hand, CDKN2D exhibited a relatively similar expression pattern to that of PTC rather than TCC. Conversely, CDKN2A demonstrated distinct similarities in specific subpopulations within the TCC. Notably, CDKN2A was expressed in a subpopulation of CD8+ T cells originating from HCC cells. These observations prompted us to delve deeper into the intricate association between CDKN2 expression and immune checkpoints, which are pivotal regulators of Tex progression. The visualization results indicated a consistent association between CDKN2A expression and Tex markers, encompassing PDCD1, ENTPD1, CXCL13, and CTLA4. Contrastingly, CDKN2D displayed concurrent expression with Naïve T cell markers, encompassing CCR7, TCF7, LEF1, and SELL; whereas CDKN2B and CDKN2C did not display relatively similar expression correlations. Additionally, specific analysis yielded supportive evidence that CDKN2A was substantially present among PD-1 positive T cells (30.27%), juxtaposed with a meager presence within PD-1 negative T cells (9.71%). To further validate the correlation between CDKN2 and Tex, we assessed the relationship between each gene and exhausted T cell markers including PDCD1, ENTPD1, and CTLA4 (Fig. 3B). CDKN2A, CDKN2B, and CDKN2C exhibited positive correlations with these immune checkpoints, with CDKN2A demonstrating the most pronounced association. In line with our prior findings, CDKN2D did not show any correlation with these markers indicative of Tex, suggesting its potential involvement in naïve T cells rather than Tex (Fig. 3C). Collectively, drawing upon the existing analysis, we speculate that CDKN2A may be linked to Tex progression within the context of HCC. These observations provide a plausible rationale for adverse clinical outcomes associated with elevated CDKN2 expression, particularly CDKN2A, within the scope of our study and in the context of prior investigations.
Fig. 3.
CDKN2A positively associated with immune checkpoints in exhausted CD8+ T cells in patients with HCC. (A) UMAP-based Trajectory Analysis demonstrating the developmental trajectory of CD8+ T cells in peripheral blood (PTC) and HCC tissues (TTC). (B) UMAPs visualization representing the diverse expression patterns of CDKN2 gene in diverse cell types and cell type-specific transcriptional profiles of typical genes in exhausted and naïve status. (C) The correlation analysis of each CDKN2 gene expression with exhausted T cell markers in the context of HCC.
Histologic analysis of human HCC suggested CDKN2A expression as a potential biomarker and positive association with the functional state of Tex
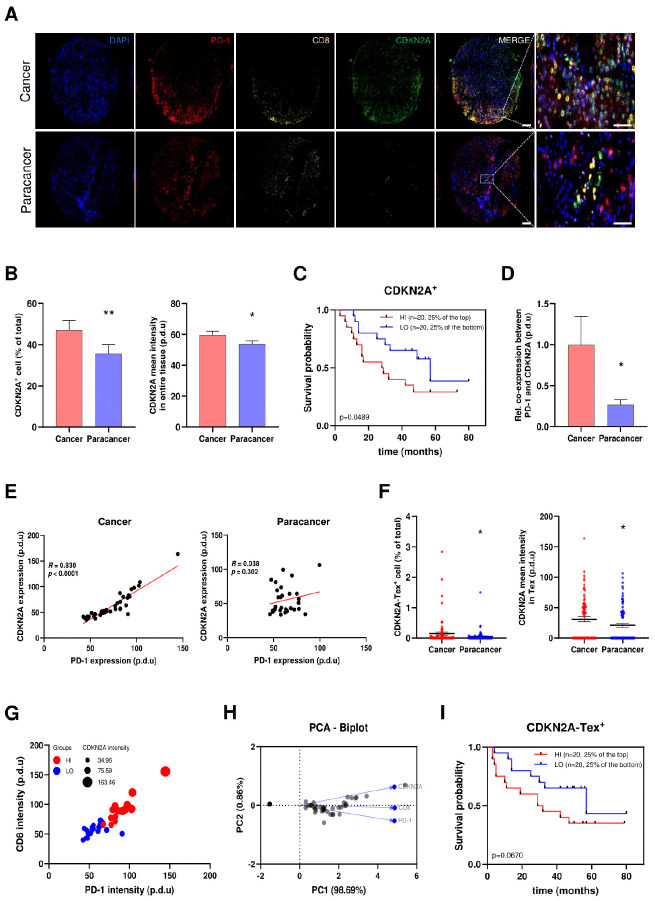
For further validation, we recruited a cohort of 79 patients with HCC and performed further validation. Considering the difficulty in obtaining normal liver tissues from healthy individuals, we collected para-cancerous tissues from patients with HCC that represented normal liver tissues along with HCC specimens. All collected human samples were processed into microarrays, followed by immunofluorescence staining and analysis of clinicopathological characteristics. Elevated CDKN2A expression in HCC was validated through representative co-staining images (Fig. 4A), which was substantiated by higher percentage of CDKN2A positive (CDKN2A+) cells and robust fluorescence intensity, in comparison to para-cancer cells (Fig. 4B). Collectively, these findings suggested that CDKN2A held certain potential involving in HCC development. To further verify the implication of CDKN2A with regard to the clinical outcomes of patients with HCC, we subsequently divided the patients into high (top 25%, marked as HI) and low (bottom 25%, marked as LO) expression cohorts based on CDKN2A expression levels. Kaplan-Meier analysis highlighted a negative association between CDKN2A expression and the overall poor prognosis of HCC individuals, thus corroborating our findings from the bioinformatic analysis (Fig. 4C).
Fig. 4.
Microarray-based analysis verified the correlation between CDKN2A and T cell exhaustion (Tex) status in the context of HCC. (A) Representative immunofluorescence staining indicating the differential expression patterns of CDKN2A in HCC compared to normal tissues, as well as the co-expression among CDKN2A and Tex markers PD-1 and CD8. Scale bar: 200 μm (Merged figure), 50 μm (Enlarged figure). (B) The expression patterns of CDKN2A in HCC and normal tissues. (C) Kaplan-Meier curves verifying the implication of CDKN2A in HCC advancement. (D) The normalized co-expression percentage between PD-1 and CDKN2A in cancer and adjacent paracancer tissues. (E) The co-expression feature plot of CDKN2A and PD-1 in cancer and paracancer tissues, respectively. (F) The expression patterns of CDKN2A in Tex. (G) Bubble chart from multivariate analysis displaying the correlation among CDKN2A, CD8+, and PD-1 expression. (H) Principal Component Analysis (PCA) Biplot revealing the correlation among CDKN2A, CD8+, and PD-1. PC: Principal Component. (I) Kaplan-Meier curves elucidating the implication of CDKN2A-Tex in HCC progression. *P < 0.05, **P < 0.01.
To substantiate the underlying implications of CDKN2A in Tex, we assessed the correlation between the expression profiles of CDKN2A and typical Tex markers PD-1. An evidently elevated co-expression percentage between PD-1 and CDKN2A was observed in cancer tissues compared to that in adjacent paracancer (Fig. 4D), consistent with the findings in the co-expression feature plot of CDKN2A-PD-1 (Fig. 4E). Meanwhile, the proportion of CDKN2A-PD-1-CD8+ positive (CDKN2A-Tex+) cells, along with the intensity of CDKN2A expression within these CDKN2A-Tex+ cells, was increased in HCC compared to normal tissues (Fig. 4F). Notably, bubble chart derived from multivariate analysis revealed concurrent high expression of CDKN2A in conjunction with elevated expression of PD-1 and CD8+ in CDKN2A-Tex+ cells, concomitant with an analogous pattern observed in the low expression cohort (Fig. 4G). The results from the Principal Component Analysis (PCA) Biplot revealed an impressive PC1 (98.069%) of total variance with similar expression trends among these factors (Fig. 4H), in conjunction with the bubble chart, suggesting the existence of mutual relationships between CDKN2A and Tex. Moreover, analysis of survival rates displayed a distinct pattern, indicating that elevated levels of CDKN2A-Tex+ was inversely correlated with the prognosis of HCC individuals (Fig. 4I).
DISCUSSION
Tumor suppressive mechanism inherent to CDKN2 gene is contemplated to restrict the growth of cancers in the early stage (15). Loss of CDKN2 gene has been verified as a significant effector for diverse cancers like HCC (16). Conversely, overexpression of this tumor suppressor was observed to accelerate the onset of multiple cancers (17). Additionally, elevated CDKN2 expression has been identified in HCC (18) and demonstrated a positive correlation with enhanced HCC risk (19, 20), being consistent in our present study and previous reports while contradictory compared to its inherent suppressive features. However, the origin of this contradiction is not entirely understood. Meanwhile, the extent of their involvement in the development of HCC, whether through senescence or other mechanisms, also remains unclear.
Recent research revealed that CDKN2 has been implicated in triggering T cell dysfunction through Tes induction (21), with its overexpression being a reasonable mechanism for promoting HCC progression. Apart from Tes, emerging investigations have revealed another mechanism termed Tex, demonstrating certain impacts in HCC progression (22, 23); however, the specific molecular mechanisms remain largely undefined. Both dysfunctional states exhibit overlapping features but possess distinct regulatory mechanisms. The failure of ICIs in treating patients with glioblastoma indicates that T cell subsets in this specific tumor is predominantly senescent rather than exhausted (24). In contrast, the mutual representation of both states is also evident within tumors. For instance, T cells expressing Tes biomarkers exhibit an adverse response to PD-1 inhibitors in patients with advanced lung cancer, implying that Tes confers resistance to exhaustion inhibition (25). This emphasizes the intricate mechanistic crosstalk between Tes and Tex. Remarkably, PD-1 has been identified as a contributor to T cell dysfunction through the upregulation of Tes marker, CDKN2B (12). Despite debates regarding the expression of exhaustion markers such as PD-1 and LAG-3 on Tes (26, 27), we questioned whether CDKN2 might also participate in the regulation of Tex in a manner relatively independent of Tes.
To address this apprehension, the pivotal core was to ascertain whether there is an association between CDKN2 and Tex, and if so, to elucidate the specific nature of this relationship. Single-cell transcriptomic analysis intuitionally indicated the positive correlation between expression of CDKN2 gene, particularly CDKN2A, and exhausted T cell markers. This finding aligned with and further elucidated prior research, affirming the existing connection between CDKN2A and immune infiltrates within the HCC TME (19). However, regrettably, due to the lack of proteomics dataset from patients with HCC, herein, we were unable to analyze the protein expression of CDKN2 in HCC via the public available database. In this case, we conducted human cohort study using HCC microarray to verify the involvement of CDKN2 in HCC progression. Additionally, the analogous expression patterns between CDKN2A and Tex markers, as well as the significant co-expression observed in HCC tissues compared to those of normal, jointly substantiated the existence of mutual relationships between CDKN2A and Tex. HCC patients with higher CDKN2A expression in terms of Tex have adverse prognosis, implying that CDKN2A might also function as an indicator of Tex state, thereby synergistically impairing the clinical outcomes of HCC patients. These verifications provide compelling evidence to reinforce the notion that CDKN2A holds potential as a biomarker indicating Tex status within the HCC context. While, limited by the absence of publicly available data regarding the impacts of CDKN2A expression on the efficacy of immune checkpoint blockade therapy or T cell therapy, herein we are unable to corroborate our findings using existing datasets. Further experimental investigations are essential to uncover the underlying mechanisms, thus providing insightful targets for advanced HCC therapy.
Conclusively, we conducted an assessment on the expression profiles of CDKN2 gene in HCC. Our results emphasize the potential contribution of CDKN2A in Tex, implying its regulatory involvement in HCC progression. Further in-depth mechanical studies at molecular level are imperative to unravel the precise modulation of CDKN2 in HCC and explore promising therapeutic interventions targeting the CDKN2 gene to address this malignancy.
MATERIALS AND METHODS
Data acquisition and processing
Hepatic transcriptomic datasets including normal liver and HCC tissues were downloaded from the NCBI GEO databases (28-30). Additionally, RNA sequencing and clinical data of the LIHC cohort were obtained from TCGA data portal. To ensure the integrity and comparability of the HCC transcriptomic data sourced from TCGA, a normalization procedure was executed via the method of Fragments Per Kilobase of Exon per Million.
Gene Ontology (GO) enrichment assay
The complete transcriptomes of 20 individuals exhibiting the highest or lowest expression levels for each CDKN2 gene were obtained from LIHC. Subsequently, the absolute fold changes (F.C. = Abs [mean expressions of highest / lowest]) were calculated to pinpoint the top associated phenotype ontology terms. Gene sets were regarded as statistically enriched when P-value and false discovery rate were below 0.05.
Trajectory analysis and gene expression assessment
The data applied for the analysis was obtained from public GEO database (31), which encompassed single-cell RNA sequencing data originating from peripheral blood (PTC), adjacent normal tissues, and tumor tissues (TTC) of 6 individuals with HCC. The trajectory assay was performed by the phate R package.
Validation of bioinformatic analysis using human samples from HCC microarray
Human liver HCC microarray (HLivH180Su11) was obtained from Shanghai Outdo Biotech Company, consisted of para-cancer and cancer tissue from HCC patients. Immunofluorescence staining was conducted as previously described (5), targeting PD-1, CD8+, and CDKN2A. Staining images and quantifications were scanned and assessed by the TissueFAXS.
Data assessment and statistical analysis
All data were evaluated as previously described, via Rstudio, R version 4.2.3, and R packages (Data processing: reshape2, naniar, skimr, stringr, and dplyr; Boxplot and survival curve: ggpubr, ggplot2, survival, egg, gridExtra, pROC, multipleROC, and survminer). The R package multipleROC were performed to identify the optimal values of gene expression for survival probability assessment. The optimal values were subsequently employed to stratify the sample population into two distinct groups. The survival outcomes were displayed with Kaplan-Meier curves, produced via R packages, survival and survminer. The P-value was considered statistically significant below 0.05.
ACKNOWLEDGEMENTS
This study was performed according to the Declaration of Helsinki, and approved by the Ethics Committee of Shanghai Outdo Biotech Company (Protocol code: SHYJS-CP-1701002). The informed consent was obtained from all subjects involved in the study.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2022R1I1A1A01063460 and 2021R1A5A8029876 to Y.J.; 2023R1A2C3006220, 2022K2A9A1A06091879, and RS-2023-00261370 to D.R. and K.T.H). Moreover, the study was also supported by an Inha University Research Grant to D.R. and a “GIST Research Institute (GRI) IIBR” grant funded by the GIST to D.R. in 2023.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park M, Moon B, Kim JH, et al. Downregulation of SETD5 suppresses the tumorigenicity of hepatocellular carcinoma cells. Mol Cells. 2022;45:550–563. doi: 10.14348/molcells.2022.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin EC. Cancer immunotherapy: special issue of BMB Reports in 2021. BMB Rep. 2021;54:1. doi: 10.5483/BMBRep.2021.54.1.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee HO, Park WY. Single-cell RNA-Seq unveils tumor microenvironment. BMB Rep. 2017;50:283–284. doi: 10.5483/BMBRep.2017.50.6.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang Y, Wei S, Koo JM, et al. Integrative evaluation of the clinical significance underlying protein arginine methyltransferases in hepatocellular carcinoma. Cancers (Basel) 2023;15:4183. doi: 10.3390/cancers15164183.63ccdc10a98145c49950187e1d12343a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin HT, Jeong YH, Park HJ, Ha SJ. Mechanism of T cell exhaustion in a chronic environment. BMB Rep. 2011;44:217–231. doi: 10.5483/BMBRep.2011.44.4.217. [DOI] [PubMed] [Google Scholar]
- 7.Lee YJ, Lee JB, Ha SJ, Kim HR. Clinical perspectives to overcome acquired resistance to anti-programmed death-1 and anti-programmed death ligand-1 therapy in non-small cell lung cancer. Mol Cells. 2021;44:363–373. doi: 10.14348/molcells.2021.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, He T, Xue L, Guo H. Senescent T cells: a potential biomarker and target for cancer therapy. EBioMedicine. 2021;68:103409. doi: 10.1016/j.ebiom.2021.103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasakovski D, Xu L, Li Y. T cell senescence and CAR-T cell exhaustion in hematological malignancies. J Hematol Oncol. 2018;11:91. doi: 10.1186/s13045-018-0629-x.387b082ef1bd41ccab6750c26e2980c4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Guo Y, Zhao D, et al. Comprehensive analysis revealed that CDKN2A is a biomarker for immune infiltrates in multiple cancers. Front Cell Dev Biol. 2021;9:808208. doi: 10.3389/fcell.2021.808208.7c75df29b7b74f9fa41999776c5c1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chae J, Park WS, Kim MJ, et al. Genomic characterization of clonal evolution during oropharyngeal carcinogenesis driven by human papillomavirus 16. BMB Rep. 2018;51:584–589. doi: 10.5483/BMBRep.2018.51.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patsoukis N, Sari D, Boussiotis VA. PD-1 inhibits T cell proliferation by upregulating p27 and p15 and suppressing Cdc25A. Cell Cycle. 2012;11:4305–4309. doi: 10.4161/cc.22135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen SC, Lo CM, Wang SH, Su EC. RNA editing-based classification of diffuse gliomas: predicting isocitrate dehydrogenase mutation and chromosome 1p/19q codeletion. BMC Bioinformatics. 2019;20:659. doi: 10.1186/s12859-019-3236-0.5b649244e4044b36bab9daaecf0ba583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mo Z, Yu L, Cao Z, Hu H, Luo S, Zhang S. Identification of a hypoxia-associated signature for lung adenocarcinoma. Front Genet. 2020;11:647. doi: 10.3389/fgene.2020.00647.2dd8ad2ffd394245b39131862ec03720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao R, Choi BY, Lee MH, Bode AM, Dong Z. Implications of genetic and epigenetic alterations of CDKN2A (p16(INK4a)) in cancer. EBioMedicine. 2016;8:30–39. doi: 10.1016/j.ebiom.2016.04.017.6f3472b869134523a4988954c1e46071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreuger IZM, Slieker RC, van Groningen T, van Doorn R. Therapeutic strategies for targeting CDKN2A loss in melanoma. J Invest Dermatol. 2023;143:18–25. doi: 10.1016/j.jid.2022.07.016. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Yu X, Shen E. Overexpression of CDKN2B is involved in poor gastric cancer prognosis. J Cell Biochem. 2019;120:19825–19831. doi: 10.1002/jcb.29287. [DOI] [PubMed] [Google Scholar]
- 18.Khemlina G, Ikeda S, Kurzrock R. The biology of hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer. 2017;16:149. doi: 10.1186/s12943-017-0712-x.21140ded61b4497eb63f7f2d82efef3c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo JP, Wang J, Huang JH. CDKN2A is a prognostic biomarker and correlated with immune infiltrates in hepatocellular carcinoma. Biosci Rep. 2021;41:BSR20211103. doi: 10.1042/BSR20211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Wang XB, Qiu XP, Shuai Z, Wang C, Zheng F. CDKN2A promoter methylation and hepatocellular carcinoma risk: a meta-analysis. Clin Res Hepatol Gastroenterol. 2018;42:529–541. doi: 10.1016/j.clinre.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Erickson S, Sangfelt O, Heyman M, Castro J, Einhorn S, Grander D. Involvement of the Ink4 proteins p16 and p15 in T-lymphocyte senescence. Oncogene. 1998;17:595–602. doi: 10.1038/sj.onc.1201965. [DOI] [PubMed] [Google Scholar]
- 22.Seimiya T, Otsuka M, Fujishiro M. Overcoming T-cell exhaustion: new therapeutic targets in HCC immunotherapy. Hepatology. 2023;78:1009–1011. doi: 10.1097/HEP.0000000000000039. [DOI] [PubMed] [Google Scholar]
- 23.Barsch M, Salie H, Schlaak AE, et al. T-cell exhaustion and residency dynamics inform clinical outcomes in hepatocellular carcinoma. J Hepatol. 2022;77:397–409. doi: 10.1016/j.jhep.2022.02.032. [DOI] [PubMed] [Google Scholar]
- 24.Huff WX, Bam M, Shireman JM, et al. Aging- and tumor-mediated increase in CD8(+)CD28(-) T cells might impose a strong barrier to success of immunotherapy in glioblastoma. Immunohorizons. 2021;5:395–409. doi: 10.4049/immunohorizons.2100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrara R, Naigeon M, Auclin E, et al. Circulating T-cell immunosenescence in patients with advanced non-small cell lung cancer treated with single-agent PD-1/PD-L1 inhibitors or platinum-based chemotherapy. Clin Cancer Res. 2021;27:492–503. doi: 10.1158/1078-0432.CCR-20-1420. [DOI] [PubMed] [Google Scholar]
- 26.Suen H, Brown R, Yang S, et al. Multiple myeloma causes clonal T-cell immunosenescence: identification of potential novel targets for promoting tumour immunity and implications for checkpoint blockade. Leukemia. 2016;30:1716–1724. doi: 10.1038/leu.2016.84. [DOI] [PubMed] [Google Scholar]
- 27.Song Y, Wang B, Song R, et al. T-cell Immunoglobulin and ITIM domain contributes to CD8(+) T-cell immunosenescence. Aging Cell. 2018;17:e12716. doi: 10.1111/acel.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roessler S, Jia HL, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamb JR, Zhang C, Xie T, et al. Predictive genes in adjacent normal tissue are preferentially altered by sCNV during tumorigenesis in liver cancer and may rate limiting. PLoS One. 2011;6:e20090. doi: 10.1371/journal.pone.0020090.b53d4a768873417aa36a0629f6eecc3f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim HY, Sohn I, Deng S, et al. Prediction of disease-free survival in hepatocellular carcinoma by gene expression profiling. Ann Surg Oncol. 2013;20:3747–3753. doi: 10.1245/s10434-013-3070-y. [DOI] [PubMed] [Google Scholar]
- 31.Zheng C, Zheng L, Yoo JK, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]