Abstract
Background
Colon adenocarcinoma (COAD) is a serious public health issue due to high incidence and mortality rate. This study aimed to identify possible tumor antigens and necroptosis subtypes of COAD for the development of mRNA vaccines and the selection of appropriate patients for precision therapy.
Methods
Gene expression profiles and clinical information for COAD were obtained from The Cancer Genome Atlas and Gene Expression Omnibus, respectively. We comprehensively studied the alterations in necroptosis-related genes (NRGs) using cBioPortal, and screened the hub NRGs associated with the prognosis of patients with COAD using Gene Expression Profiling Interactive Analysis 2. Consensuses clustering analysis was performed to identify necroptosis subtypes. Weighted gene co-expression network analysis (WGCNA) was used to identify the co-expression modules of the NRGs. The necroptosis landscape of COAD was assessed using graph learning-based dimensionality reduction. Finally, a drug sensitivity analysis of the two necroptosis subtypes was performed.
Findings
Two tumor antigens, BLC-2-associated X protein (BAX) and interleukin 1 beta (IL1B) were identified based on their associations with prognosis of patients and antigen presenting cell infiltration. Two necroptosis subtypes (N1 and N2) were distinguished in patients with COAD, and they were characterized by their differential survival status and molecular expression levels of immune checkpoint proteins and immunogenetic cell death modulators. Furthermore, the necroptosis landscape of COAD indicated that individual patients had obvious heterogeneity. Co-expression modules were identified using WGCNA, and the hub NRGs were found to be involved in various immune processes. Drug sensitivity analysis indicated that there were significant differences in drug sensitivity between the N1 and N2 subtypes. Cell experiments suggested that both overexpression of BAX and IL1B promoted necroptosis of COAD cells and enhanced the cytotoxicity of CD8+ T cells.
Interpretation
BAX and IL1B are potential antigens for the development of anti-COAD mRNA vaccines, specifically for patients with the N2 subtype. Consequently, this study will guide the development of more effective immunotherapeutic approaches and the identification of appropriate patients.
Keywords: Colon adenocarcinoma, Necroptosis, Necroptosis-related genes, mRNA vaccine, Tumor antigen
Highlights
-
•
Necroptosis-related immune landscape can help identify potential mRNA vaccine development.
-
•
Necroptosis-related antigens is crucial for designing vaccination strategies.
-
•
Necroptosis subtypescan help tailor mRNA vaccines and immunotherapies to specific patient populations.
-
•
Necroptosis landscape aids in facilitating the selection of suitable patients for vaccination.
1. Introduction
Colorectal cancer is one of the most common cancers worldwide. Globally, 1,931,590 new colorectal cancer cases and 935,173 colorectal cancer-related deaths were reported in 2020 [1]. In 2023, it was estimated that there were 153,020 new cases and 52,550 deaths attributed to colorectal cancer in the United States [2]. Colon adenocarcinoma (COAD) represents the predominant pathological form of colon cancer worldwide. COAD is a heterogeneous disease, characterized by numerous molecular alterations that result in the dysregulation of several signaling pathways, and is associated with the initiation and development of tumors [3,4]. The therapeutic outcomes have improved owing to advances in surgical methods, chemotherapy, bio-targeted therapy and immune therapy. However, approximately 35 % of patients with COAD are diagnosed at an advanced stage, and their prognosis is poor. Therefore, new biomarkers for the early diagnosis and the treatment of patients with COAD must be explored.
In the past decade, many breakthroughs have been made in immunotherapy, including some large-scale clinical trials, such as those by Reck et al., which demonstrated that the use of immune checkpoint inhibitors (ICIs) in the treatment of non-small-cell lung cancer is an important advancement [5]. For COAD, some patients with microsatellite instability–high (MSIH) are also suitable for ICI therapy, which presents good clinical outcomes. However, approximately 5 % of patients present metastatic COAD, which means that most COAD patients cannot benefit from ICI therapy. When patients are unsuitable for immunotherapy, other treatments may be available. Tumor vaccines engineer against private tumor-specific mutations represent a potentially effective treatment option. Recently, Lu et al. demonstrated that the α-fetoprotein (AFP) vaccine effectively recruits CD8+ T cells to tumor tissue and stimulates anticancer immunity, thereby preventing c-MYC/Mcl 1 hepatocellular carcinoma (HCC) initiation in vivo [6]. For melanoma, mRNA vaccines designed based on individual mutations of patients have an exciting effect on preventing tumor recurrence after surgery [[7], [8], [9]]. Cancer vaccines allow immune cells to precisely recognize antigens within tumor cells, offering advantages such as a broad therapeutic window, minimal nonspecific effects, and persistent immunological memory. Cancer vaccines can overcome the medication resistance, unpleasant effects, limited therapeutic efficacy, and high costs associated with conventional chemotherapy and immunotherapy. However, mRNA vaccines that recognize COAD antigens have not yet been developed, and the subpopulation of patients suitable for mRNA vaccines has not been determined.
Necroptosis is a novel type of programmed death mediated by receptor interacting protein kinase 1 (RIPK1), RIPK3, and mixed lineage kinase domain-like protein (MLKL). Previous studies have shown that tumor cell necrosis created a favorable environment for tumor growth and metastasis; however, necroptosis of tumor cells has been found to play a role in suppressing tumor processes. In 2016, Aaes et al. first reported that necroptosis in tumor tissues is a type of immunogenic cell death, increasing the immunogenicity of tumor cells, and then they developed a vaccine from necroptotic tumor cells and demonstrated its efficacy in repressing tumor growth in an animal model [10]. Similarly, necroptosis of tumor cells could promote the maturation of dendritic cells, cross-priming of cytotoxic T cells, and the production of IFN-γ. Therefore, necroptosis is a promising therapeutic option, particularly in drug-resistant cancers. Recently, necroptosis is discovered to be strongly correlated with tumor immunity. The expression of necroptosis-related genes (NRGs) is closely associated with immune cell infiltrations and has potential value in predicting the prognosis of patients with various types of tumors [11,12]. Therefore, necroptosis combined with mRNA vaccines represents a potentially effective treatment for COAD.
In this study, we screened two hub NRGs that might be suitable for mRNA vaccine development and identified two necroptosis subtypes using the data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). Subsequently, we depicted the immune characterization, necroptosis landscape, and drug sensitivity of the two necroptosis subtypes to verify the close relationship between necroptosis and tumor immune microenvironment (TIME) and to propose a prognostic tool and immune infiltration indicator for COAD. These results will aid in the precision of immunotherapy and the development of mRNA vaccines.
2. Materials and methods
2.1. Data acquisition and processing
A COAD dataset with 512 samples was downloaded from the UCSC Xena database (https://xena.ucsc.edu/) [13]. The data types Count and FPKM were chosen, and “primary solid tumor (01A)” data in the dataset were extracted and then transformed into transcripts per million (TPM) format. The “Masked Somatic Mutation” data were selected as somatic mutation data for patients with COAD and the data were pre-processed using the VarScan software. Subsequently, the R package maftools was used to visualize the somatic mutation feature. Clinical information on patients, including age, tumor node metastasis (TNM) stage, survival time, and status, was downloaded. Patients with incomplete clinical information were excluded. Finally, 412 patients with survival status and 331 patients with other clinical information were included in the study. ChIP data for the expression profiles of COAD samples were obtained from the following GEO datasets (https://www.ncbi.nlm.nih.gov/geo/): GSE87211 (363 COAD samples with clinical information of the patients, platform GPL13497), GSE73360 (86 COAD samples, platform GPL17586), and GSE44861 (111 COAD samples, platform GPL3921). ChIP data were normalized using the R package limma, and GEO datasets were used as validation sets. Information on the datasets is presented in Table 1.
Table 1.
Information of datasets from the GEO database.
| GSE number | Platform | Samples | Source types | Disease | Group |
|---|---|---|---|---|---|
| GSE87211 | GPL13497 | 203 patients and 160 controls | rectum | READ | Validation cohort |
| GSE73360 | GPL17586 | 55 patients and 31 controls | colon and rectum | COADREAD | Validation cohort |
| GSE44861 | GPL3921 | 56 patients and 55 controls | colon and rectum | COADREAD | Validation cohort |
| GSE20916 | GPL570 | 36 patients and 24 controls | colon and rectum | COADREAD | Validation cohort |
| GSE74602 | GPL6104 | 30 patients and 30 controls | colon | COAD | Validation cohort |
In addition, NRGs were obtained from the Gene Set Enrichment Analysis (GSEA) database (http://www.gsea-msigdb.org/gsea/index.jsp) [14] and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/) [15]. Eight necroptosis-related signaling pathways were obtained from the GSEA database, and one was obtained from the KEGG database. Finally, we combined the NRGs from the two databases to obtain 159 NRGs for further analysis (Table S1). The workflow of this study is illustrated in Fig. 1.
Fig. 1.
Workflow of data processing and analysis. WGCNA: Weighted gene co-expression network analysis. ssGSEA: single sample gene set enrichment analysis.
2.2. GEPIA and cBioPortal analysis
Raw data of RNA-seq from TCGA were generated using the UCSC Xena browser. Differential genes were identified using Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia2.cancer-pku.cn) [16], with the criteria of |log2FC|>1 and q value < 0.05. Survival analysis was conducted on a cohort of 412 patients with essential survival information (including survival time and status), based on clinical data obtained from the UCSC Xena database and the GEO database. The survival package (version 3.2–11) in R was utilized to perform survival analysis. The Kaplan-Meier (K-M) method was used to assess the differences in overall survival (OS) and progression-free survival (PFS) among patients in different groups based on the median gene expression. The log-rank test was conducted to evaluate the differences in the survival statuses between different groups, and p < 0.05 was considered statistically significant.
Similarly, to study gene alterations, we integrated raw RNA-seq data from TCGA using the cBio Cancer Genomics Portal (http://www.cbioportal.org) [17]. The microsatellite instability and tumor mutation burden (TMB) for patients with COAD were extracted from the altered genes, and p < 0.05 was considered statistically significant.
2.3. Correlations between tumor antigens and immune cell infiltration
Based on modules of gene expression, somatic mutations, clinical outcomes, and somatic copy number alterations in the Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) [18], we analyzed and visualized the correlation between enriched tumor-infiltrating immune cells (TIICs) and the expression of tumor antigens. Statistical significance was evaluated using Spearman's correlation adjusted for tumor purity, and p < 0.05 was deemed statistically significant.
In addition, we assessed the infiltration of immune and other stromal cells based on gene expression using the R package MCPCounter [19]. We used MCPCounter to calculate the enrichments of eight immune cell sub-clusters, including CD4+ cells, CD8+ cells, natural killer (NK) cells, B lymphocytes, monocytes, dendritic cells, neutrophils and cytotoxic lymphocytes. Similarly, the infiltration enrichment of two types of stromal cells (fibroblasts and endothelial cells) was also calculated using MCPCounter. Spearman correlation analysis was utilized to determine the correlations between MCPcounter enrichments and antigens expression, and p < 0.05 was considered statistically significant.
2.4. Identification and validation of necroptosis subtypes
The NRGs were clustered according to gene expression profiles using the R package “ConsensusClusterPlus” [20], and a consistency matrix was then constructed to identify corresponding necroptosis subtypes and gene modules. The algorithm of the median distance measured based on 1-pearson correlation was used for zoning with 1000 replications, and 80 % of the patients resampling each time. The value range of the cluster sets was 2–9, and the optimal partitions were defined according to the consensus matrix and cumulative distribution function. Subsequently, the same settings were used to validate necroptosis subtypes in the GSE87211 dataset.
2.5. Prognostic evaluation of necroptosis subtypes
To compare whether there were significant differences in survival curves between different necroptosis subtype patient groups, we conducted a Log-rank test. We firstly organized the clinical information of the patients, including key data such as age, TNM stage, survival time, and survival status, and excluded patient samples with missing major clinical information. For each patient enrolled, we recorded their survival time and key medical events, such as death or disease progression. Then, the patients were divided into the high expression and the low expression groups based on the median expression levels of each subtype. When performing the log-rank test, we used the survival package in the R package (version 3.2–11). The correlation between necroptosis subtypes and NRGs and cell features were determined using analysis of variance. The most frequently mutated gene was screened using Chi-square test. The immune enrichment score was calculated using the single-sample GSEA (ssGSEA) algorithm in the R package GSVA [21].
2.6. Gene co-expression network analysis
Modules of NRGs were screened using the R package “WGCNA” (version 1.70–3) [22]. The soft power threshold was analyzed using the “pickSoftThreshold” function, and the optimal soft threshold was “six”. Subsequently, a scale-free network was constructed according to the soft threshold, a topology matrix was constructed, hierarchical clustering was performed based on the topology matrix, and eigengenes were calculated. Correlations among the eigengenes modules were constructed, and hierarchical clustering was performed based on those correlations. Gene Ontology (GO) analysis is used to analyze the functional enrichment of large-scale genes, including biological processes (BP), molecular functions, (MF), and cellular components (CC) [23]. The KEGG is a database widely used to store the information on genomic and biological processes, diseases and drugs [24]. We performed GO and KEGG analysis of genes using the R package clusterProfiler [25] and screened the GO categories and pathways following the threshold of Benjamini-Hochberg-adjusted P-value <0.05 and FDR value q. value < 0.20.
2.7. Necroptosis landscape of the tumor microenvironment
To visualize the distribution of necroptosis subtypes among the patients, a graph learning-based dimensionality reduction analysis was conducted using the reduced dimension function of the Monocle package with a Gaussian distribution. We set a maximum of components to 4, and used discriminative dimensionality reduction with “DDRtree”. In addition, the necroptosis landscape was visualized using a function plot of the cell trajectory with color-coded necroptosis subtypes.
2.8. Drug sensitivity analysis
Alterations in the cancer genome have a strong effect on the clinical responses to therapy and are effective biomarkers of drug responses in many cases. Genomics of Drug Sensitivity in Cancer (GDSC: www.cancerRxgene.org) is a public database related to drug sensitivity in cancer cells and molecular markers of drug responses [26]. We screened the sensitive markers of cancer drug response using the GDSC database. The IC50 value was calculated based on the TPM and expression matrix from data sets of the TCGA-COAD datasets using the pRRophetic algorithm (version 0.5) [27]. We compared the differences in IC50 of anticancer drugs or small-molecule compounds between different necroptosis subtypes.
2.9. Cell culture and cell transfection
The SW480 cells was purchased from Cellverse (iCell) Bioscience Technology Co., Ltd (China). SW480 cells were cultivated in Leibovitz's L-15 Medium (Sangon) supplemented with 10 % fetal bovine serum (Gibco) and 1 % penicillin/streptomycin (Gibco) under standard conditions (5 % CO2, 37 °C).
BAX overexpression plasmid and IL1B overexpression plasmid were acquired from GenePharma Co., Ltd. (Shanghai, China). Lipofectamine 2000 reagent (Invitrogen) was utilized to transfect BAX overexpression plasmid and IL1B overexpression plasmid into SW480 cells according to manufacturer's manual.
2.10. Quantitative reverse transcription PCR (qRT-PCR)
Total RNA isolation from SW480 cells was conducted using Trizol reagent (Invitrogen) following the manufacturer's instruction. cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo). qRT-PCR was performed using ABI Q6 PCR System (Applied Biosystems Inc., USA) with a SYBR Green Master Mix (Roche). The 2−ΔΔCt method was applied for detecting the relative expression of BAX and IL1B. The primer sequences are shown in Table S2.
2.11. Isolation of human CD8+T cells and co-culture of CD8+T cells with SW480 cells
CD8+ T cells were separated from whole blood of healthy individuals using EasySep™ Direct Human CD8+T cell Isolation Kit following the manufacturer's instructions. CD8+T cells were cultured in T cell serum-free medium (RC-003, STEMERY) containing 10 % FBS. CD8+T cells were co-cultured with SW480 cells at 1:1 ratio at 37 °C for 48 h.
2.12. Western blot
SW480 cells were lysed with RIPA lysate buffer following the manufacturer's manual. After testing the protein concentrations by BCA protein assay, samples were separated by 10 % SDS-PAGE gel, transferred on polyvinylidene fluoride (PVDF) membranes, and blocked with 5 % skim milk. Subsequently, the membranes were incubated with primary antibodies anti-RIPK1 (1:4000, 29932-1-AP, Proteintech), anti-RIPK3 (1:2000, 17563-1-AP, Proteintech), anti-MLKL (1:1500, 21066-1-AP, Proteintech), anti–IFN–γ (1:1000, 8455, CST), anti-TNF-α (1:1000, ab1793, Abcam) and anti-GAPDH (1:15000, 60004-1-lg, Proteintech) overnight at 4 °C, and then incubated with HRP-conjugated Goat Anti-Rabbit IgG (H + L) secondary antibody (1:5000, SA00001-2, Proteintech) or HRP-conjugated Goat Anti-mouse IgG (H + L) secondary antibody (1:5000, SA00001-1, Proteintech) at room temperature for 2 h. The protein bands were exposed with enhanced chemiluminescence (ECL) kit. Finally, the image was captured using an electrochemiluminescence imaging system.
2.13. Flow cytometry
SW480 cells were seeded into six-well plates and harvested 48 h after the transfection with BAX overexpression vector, IL1B overexpression vector, and empty vector, or after co-culture with CD8+T cells. Afterwards, cells were washed in PBS and stained with 5 μL PI (Vazyme Biotech) and 5 μL FITC-conjugated Annexin V for 10 min at room temperature in the dark. Samples were detected on a flow cytometer (BD FACSVerse, USA). Annexin V (+) and PI (+) cells in the upper right quadrant were regarded as necroptosis.
2.14. Statistical analysis
R software (version 4.1.2) was used for data processing and analysis. For continuous variables of the two groups, an independent Student's t-test was used to compare differences in normally distributed variables and the Mann-Whitney U test was used to compare the differences in non-normally distributed variables. The Kruskal-Wallis test was used to compare the differences between more than three groups. The Chi-squared test or a Fisher's exact test was used to compare the categorical variables. Correlations between different molecules were calculated using Spearman's correlation coefficient. The R package survival (version 3.2–11) was used for the survival analysis. K-M survival curves were plotted to show survival differences, and the log-rank test was used to compare the difference in survival time between the two groups. The “WGCNA” R package (version 1.70–3) was used to screen for immune-related gene modules. For functional enrichment analysis, we employed the “clusterProfiler” R package (version 4.0). All p values were two-sided, with a p < 0.05 considered to be statistically significant.
3. Results
3.1. Identification of potential necroptosis related antigens from COAD
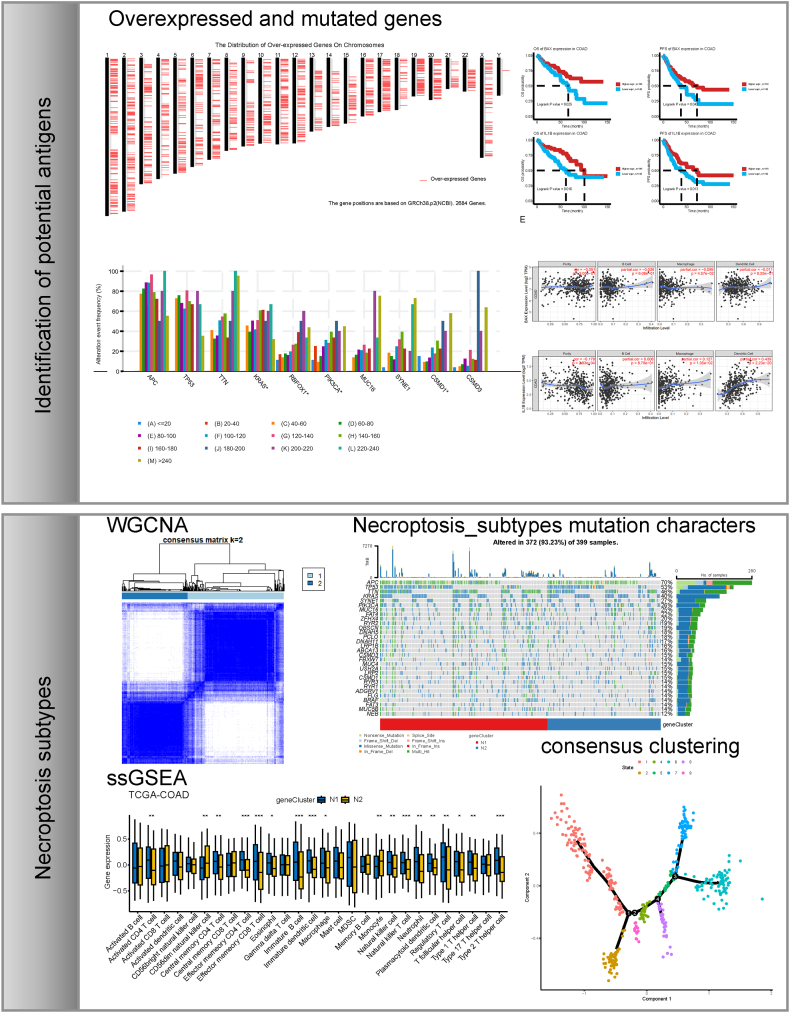
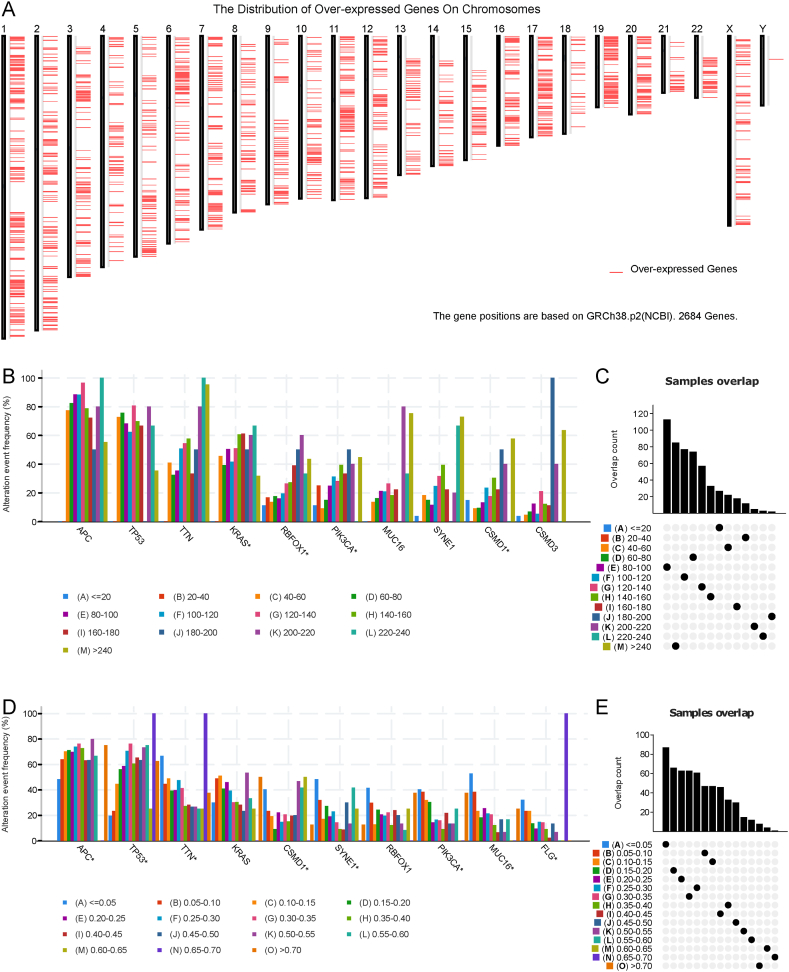
The workflow of data processing and analysis is outlined in Fig. 1. To identify potential mRNA vaccines for COAD, we first screened differentially expressed genes and obtained 5352 differentially expressed genes, among which 2684 coding genes were overexpressed and considered as potential tumor-related antigens (Fig. 2A). Next, we analyzed the mutated gene fragments that potentially encoded specific tumor antigens and calculated their mutation counts. In total, 18,276 mutated gene fragments were screened (Fig. 2C and E). Mutational analysis revealed that APC was the most frequently mutated gene in terms of both mutated gene fragments and mutational counts (Fig. 2B and D). In addition, our analysis showed that both the mutation counts and mutation frequency of APC, TP53, TTN, and KRAS were altered. Integrating the results of mutated genes and overexpressed genes, we screened 2058 frequently mutated and overexpressed tumor-related genes that were potential tumor antigens.
Fig. 2.
Identification of potential tumor antigens of COAD. A Distribution of upregulated genes in chromosomes. B-E Identification of potential specific tumor antigen based on TCGA-COAD mutation data. Samples overlapped in alterations of genomic fragments (B) and mutation counts (D). The highest number of genes in the altered genomic fragments (C) and mutation counts (E). TCGA: The Cancer Genome Atlas; COAD: colon adenocarcinoma.
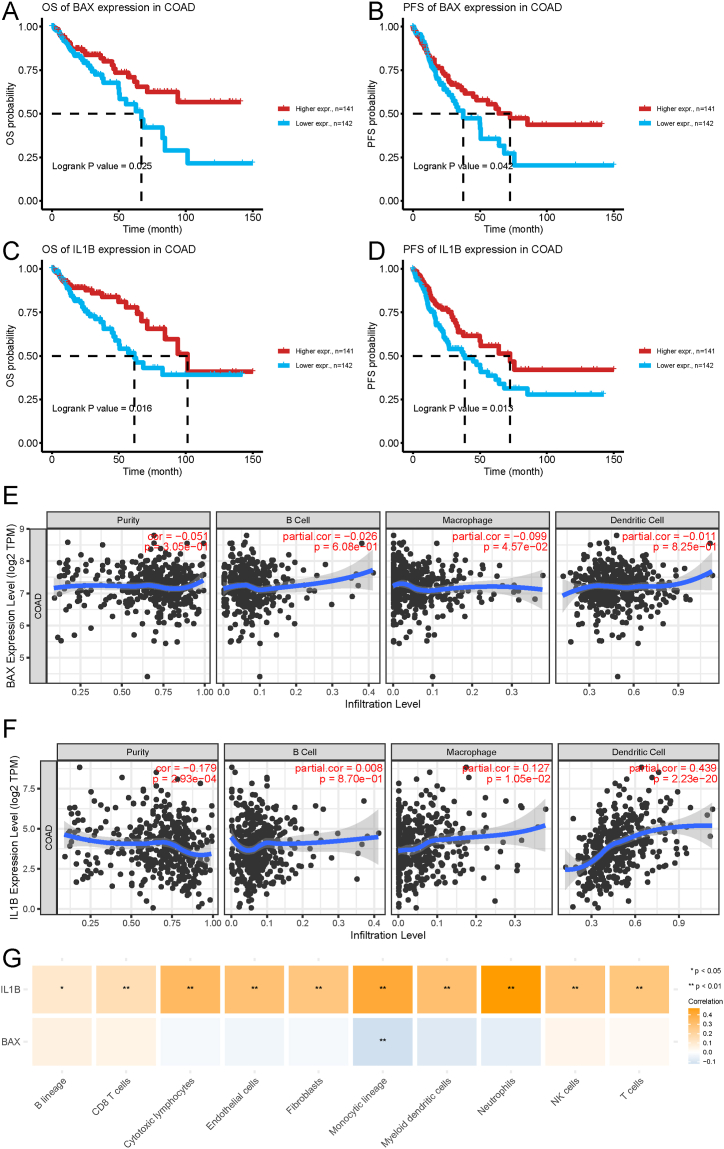
Subsequently, we focused on NRGs as potential candidates for mRNA vaccine development. Based on the survival analysis results for OS and PFS, we identified two NRGs, BLC-2-associated X protein (BAX) and interleukin 1 beta (IL1B), from the 159 potential antigens. As shown in Fig. 3A–D, patients in the low NRGs expression group had a poor prognosis, as manifested by reduced OS and PFS. Considering the interaction between tumor antigens and antigen-presenting cells (APCs), we analyzed the correlations between potential antigens and APCs using the MCPcounter algorithm integrated with the TIMER database. According to the MCPcounter algorithm, IL1B expression was positively correlated with various types of APCs, while BAX expression level was negatively correlated with macrophages (Fig. 3G). These results were validated using the TIMER database (Fig. 3E and F). Subsequently, GSEA was used to analyze the function of BAX and IL1B. Results showed that BAX was mainly enriched in Glycolysis/Gluconeogenesis, Biosynthesis of amino acids, and Ribosome (Fig. S1A). IL1B was mainly involved in Ribosome biogenesis in eukaryotes, mRNA surveillance pathway, and RNA polymerase (Fig. S1B).
Fig. 3.
Identification of tumor antigens associated with the prognosis of COAD patients and APCs. A-D Kaplan-Meier curves displaying OS and PFS of COAD patients based on median BAX (A and B) and IL1B (C and D) expression levels. E Association of BAX expression level with tumor purity, infiltrations of B cells, macrophages and dendritic cells. F Association of IL1B expression level with tumor purity and infiltration of B cells, macrophages, and dendritic cells. G Association of immune cell and stromal cell infiltrations with expression levels of BAX and IL1B. *p < 0.05, **p < 0.01. COAD: colon adenocarcinoma; APCs: antigen-presenting cells; OS: overall survivals; PFS: progression-free survival.
3.2. Identification of potential necroptosis subtypes of COAD
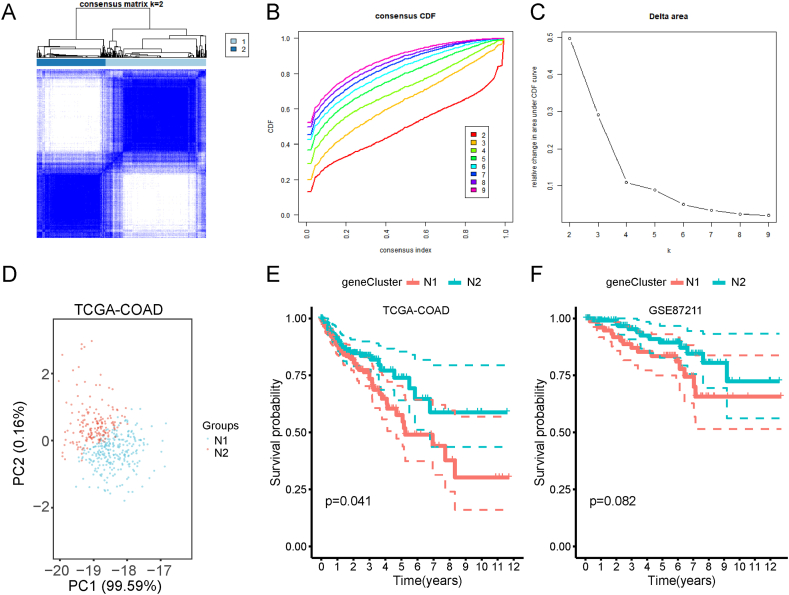
Recent studies have shown that necroptosis plays a vital role in killing tumor cells and is closely related to tumor immunity. Necroptosis subtypes that reflect the necroptosis status in tumor tissues and the tumor microenvironment could help to select suitable patients for vaccination. Therefore, we analyzed the expression profiles of 159 NRGs from the TCGA-COAD cohort to construct a consensus cluster. Based on their cumulative distribution function and delta area, K = 2 was chosen because the NRGs appeared to cluster stably (Fig. 4A–C), and two necroptosis subtypes were obtained, namely N1 and N2. Principal component analysis (PCA) showed that the two necroptosis subtypes could be obviously differentiated (Fig. 4D). Survival analysis revealed that the patients from the TCGA-COAD in the N2 cohort had a better prognosis than those in the N1 cohort (Fig. 4E). In the validated cohort GSE87211, patients in the N1 cohort had a significantly poorer prognosis than those in the N2 cohort (Fig. 4F).
Fig. 4.
Identification of potential necroptosis subtypes in the COAD cohort. A Clustered heat-map of TCGA-COAD samples using the threshold K = 2. B–C Cumulative distribution function curve (B) and δ area (C) of necroptosis subtype in the TCGA-COAD cohort. D Principal component analysis of necroptosis subtypes in the TCGA-COAD cohort. E-F Kaplan-Meier curves display the overall survival (OS) of necroptosis subtypes in the TCGA-COAD and GSE39582 cohorts. TCGA: The Cancer Genome Atlas; COAD: colon adenocarcinoma.
3.3. Association between necroptosis subtypes and immune mutational status
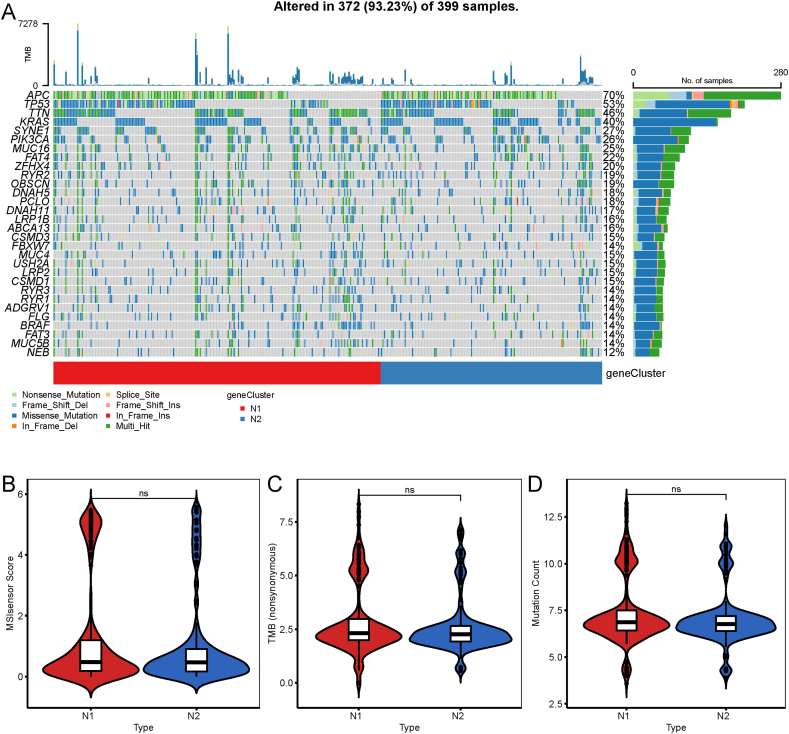
Higher TMB and somatic mutation rate are associated with stronger anticancer immune response. We observed that 30 genes, including APC, TP53, TTN, KRAS, SYNE1, and PIK3CA, had different mutational statuses in the two necroptosis subtypes (Fig. 5A). Subsequently, we calculated the TMB, MSI, and total mutation counts of each patient according to the mutational datasets from TCGA-COAD cohort and compared the differences between the two necroptosis subtypes. As shown in Fig. 5B–D, there were no differences in MSI, TMB, and total mutational counts between the two necroptosis subtypes.
Fig. 5.
Association of necroptosis subtypes with TMB and mutations. A Waterfall plot displaying the characteristic mutation genes in the N1 and N2 subtypes. B-D Association of the necroptosis subtypes with mutation count (B), TMB (C), and MSI (D). n.s. p > 0.05. TMB: tumor mutation burden.
3.4. Association between necroptosis subtypes of COAD and immune modulators
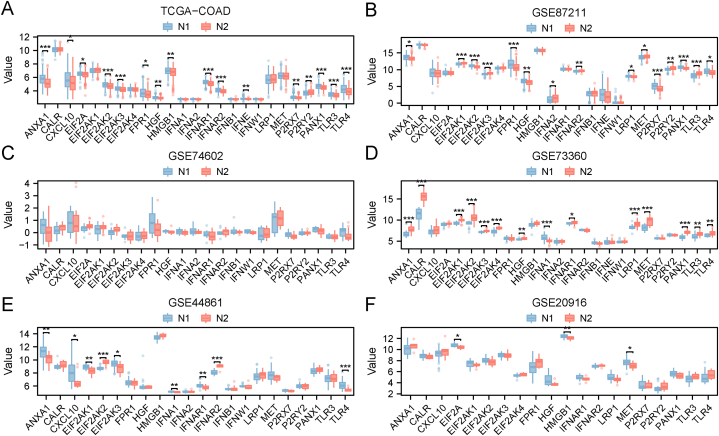
Previous studies have demonstrated the significance of immunological checkpoints (ICPs) and immune cell death (ICD) modulators in anticancer immune responses and their impact on mRNA vaccine efficacy [28,29]. Necroptosis is closely associated with immune regulation. Therefore, we assessed the differences in ICPs and ICD between the two necroptosis subtypes. We performed consistent cluster analyses on the datasets GSE20916, GSE44861, GSE73360, and GSE74602, and obtained two necroptosis subtypes (N1 and N2). In the TCGA-COAD cohort, we detected 25 ICD genes, of which 16 were significantly differentially expressed between the two subtypes (Fig. 6A). In the GSE87211 cohort, we detected 24 ICD genes, of which 16 were significantly differentially expressed between the two necroptosis subtypes (Fig. 6B). In the GSE74602 cohort, we detected 23 ICD genes, and none of them showed significant difference between the two subtypes (Fig. 6C). In the GSE73360 cohort, we detected 25 ICD genes, of which 14 were significantly differentially expressed between the two necroptosis subtypes (Fig. 6D). In the GSE44681 cohort, we detected 22 ICD genes, of which 9 were significantly differentially expressed between the two subtypes (Fig. 6E). In the GSE20916 cohort, we detected 20 ICD genes, of which 3 were significantly differentially expressed between the two subtypes (Fig. 6F).
Fig. 6.
Association of necroptosis subtypes with ICD modulators. A-F Differences in the expression of ICD modulators between the two necroptosis subtypes N1 and N2 in flowing cohorts: TCGA-COAD (A), GSE87211 (B), GSE74602 (C), GSE73360 (D), GSE44861 (E), and GSE20916 (F). *p < 0.05, **p < 0.01, ***p < 0.001. ICD: immune cell death; TCGA: The Cancer Genome Atlas; COAD: colon adenocarcinoma.
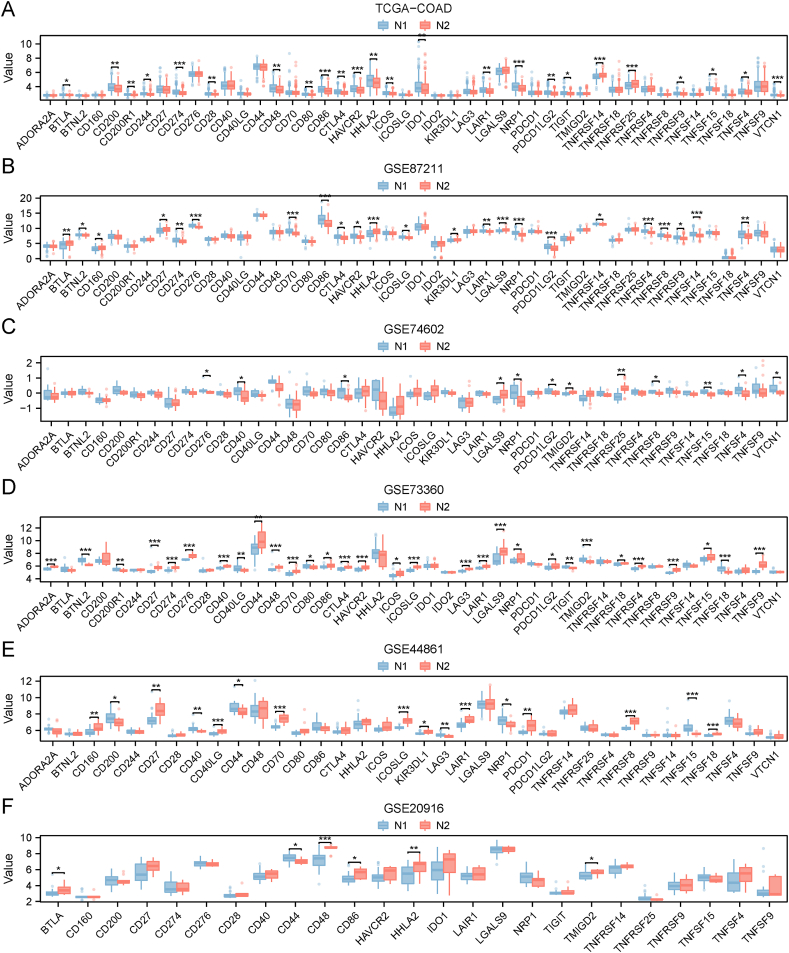
In the TCGA-COAD cohort, we detected 46 ICPs, of which 24 were significantly differentially expressed between the two subtypes (Fig. 7A). In the GSE87211 cohort, we detected 46 ICPs, of which 23 were significantly differentially expressed between the two subtypes (Fig. 7B). In the GSE74602 cohort, we detected 43 ICPs, of which 12 were significantly differentially expressed between the two necroptosis subtypes (Fig. 7C). In the GSE73360 cohort, we detected 43 ICPs, of which 30 were significantly differentially expressed between the two subtypes (Fig. 7D). In the GSE44861 cohort, we detected 36 ICPs, of which 16 were significantly differentially expressed between the two subtypes (Fig. 7E). In the GSE2096 cohort, we detected 25 ICPs, of which 6 were significantly differentially expressed between the two subtypes (Fig. 7F).
Fig. 7.
Association of necroptosis subtypes with immune checkpoint (ICP) modulators. A-F Differences in the expression of ICP modulators between the two necroptosis subtypes N1 and N2 in the flowing cohorts: TCGA-COAD (A), GSE87211 (B), GSE74602 (C), GSE73360 (D), GSE44861 (E), and GSE20916 (F). *p < 0.05, **p < 0.01, ***p < 0.001. ICP: immune checkpoints; TCGA: The Cancer Genome Atlas; COAD: colon adenocarcinoma.
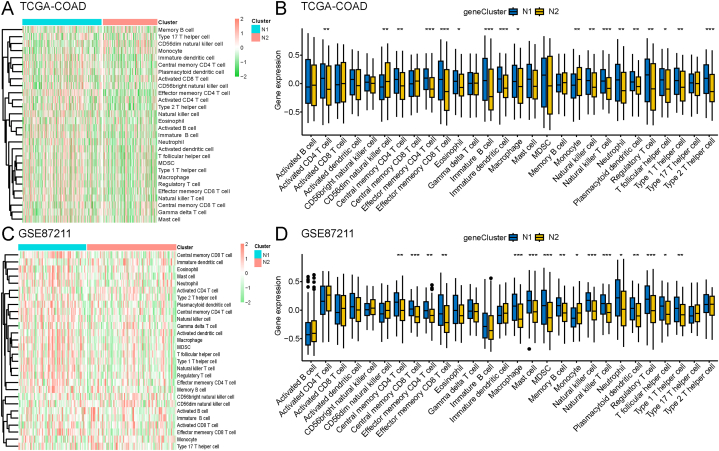
3.5. Cellular and molecular characterization of necroptosis subtypes
We scored 28 previously reported signature genes in the TCGA-COAD and GSE87211 cohorts using ssGSEA to characterize the immune cell components between the two necroptosis subtypes. As shown in Fig. 8A and C, the immune cell components were divided into two clusters and showed remarkable differences between the two subtypes. Immune cell enrichment scores revealed that immune cells mostly infiltrated N1 subtype patients (Fig. 8B and D). Next, we analyzed the correlation between the expression of BAX and IL1B and 28 N1- and N2-related immune cell types. Results showed that BAX expression was negatively correlated with regulatory T cells, immature dendritic cells, and effector memeory CD4+ T cells, whereas IL1B expression was positively correlated with activated CD4+ T cells, central memory CD8+ T cells, and regulatory T cells (Fig. S2).
Fig. 8.
Twenty-eight immune cell signatures of the necroptosis subtypes based on ssGSEA. A-B Differential enrichment scores of 28 immune cell signatures between the two necroptosis subtypes in the TCGA-COAD (A) and GSE 87211 (B) cohorts displayed using heat-maps. C Differences in enrichment scores of the 28 immune cell signatures between the two necroptosis subtypes in the TCGA-COAD cohort. D Differences in enrichment scores of the 28 immune cell signatures between the two necroptosis subtypes in GSE87211. *p < 0.05, **p < 0.01, ***p < 0.001. ssGSEA: single-sample GSEA; TCGA: The Cancer Genome Atlas; COAD: colon adenocarcinoma.
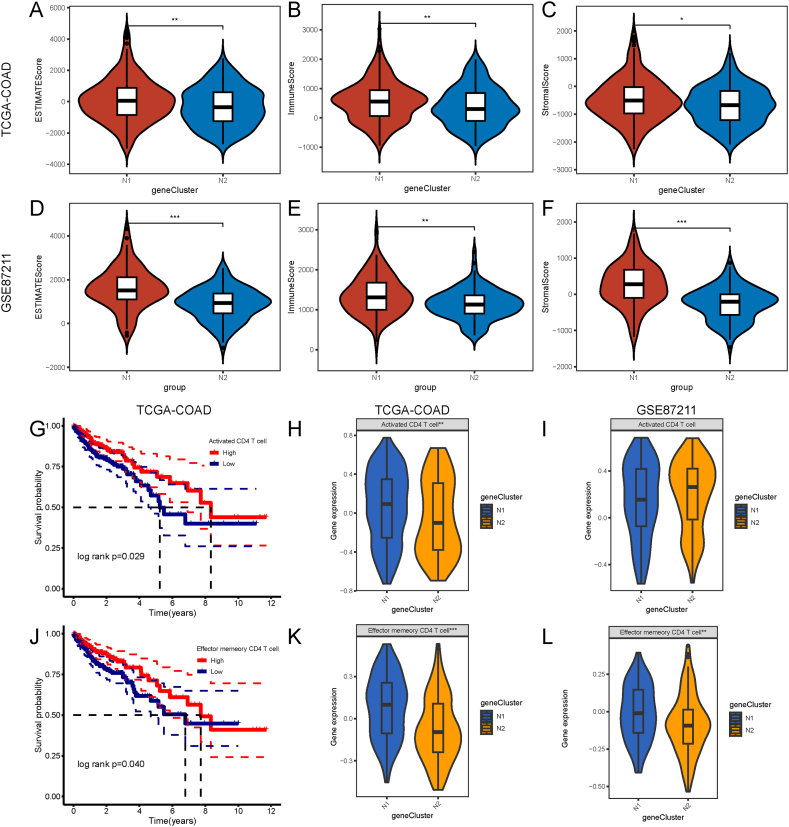
The tumor immune status affects the response to mRNA vaccines. According to the ESTIAMTE algorithm, we validated that the immune, stromal, and ESTIAMTE scores were remarkably higher in the N1 subtype than in the N2 subtype for patients in both TCGA-COAD and GSE87211 cohorts (Fig. 9A–F). The K-M curves showed that of the 22 types of immune cells, significant differences occurred in the prognosis of activated CD4+ T cells and effector memory CD4+ T cells, with low scores correlated with a poorer COAD prognosis (Fig. 9G–J). A comparison of activated CD4+ T cells and effector memory CD4+ T cells revealed significant differences in enrichment scores between the two necroptosis subtypes from the TCGA-COAD cohort. In the GSE87211 dataset, the differences were consistent with those in the TCGA-COAD cohort (Fig. 9H-L). Therefore, N1 represents an immune “hot” and immunosuppressive phenotype, while N2 represents an immune “cold” phenotype.
Fig. 9.
Cellular and molecular characterization of necroptosis subtypes. A-C Differences in the ESTIMATE, immune and stromal score between the two necroptosis subtypes in the TCGA-COAD cohort. D-F Differences in the ESTIMATE, immune and stromal score between the two necroptosis subtypes in the GSE87211 cohort. G Prognostic difference between high and low -scores of activated CD4+ T-cells in the TCGA-COAD cohort. J Prognostic differences between high and low -scores of effector memory CD4+ T-cells. H–I Significant differences in activated CD4+ T-cell scores between the N1 and N2 subtypes in the TCGA-COAD (H) and GSE87211 cohorts (I). K-L Significant differences in effector memory CD4+ T-cell scores between the N1 and N2 subtypes in the TCGA-COAD (K) and GSE87211 cohorts (L). *p < 0.05, **p < 0.01, ***p < 0.001. TCGA: The Cancer Genome Atlas; COAD: colon adenocarcinoma.
3.6. Necroptosis landscape of COAD
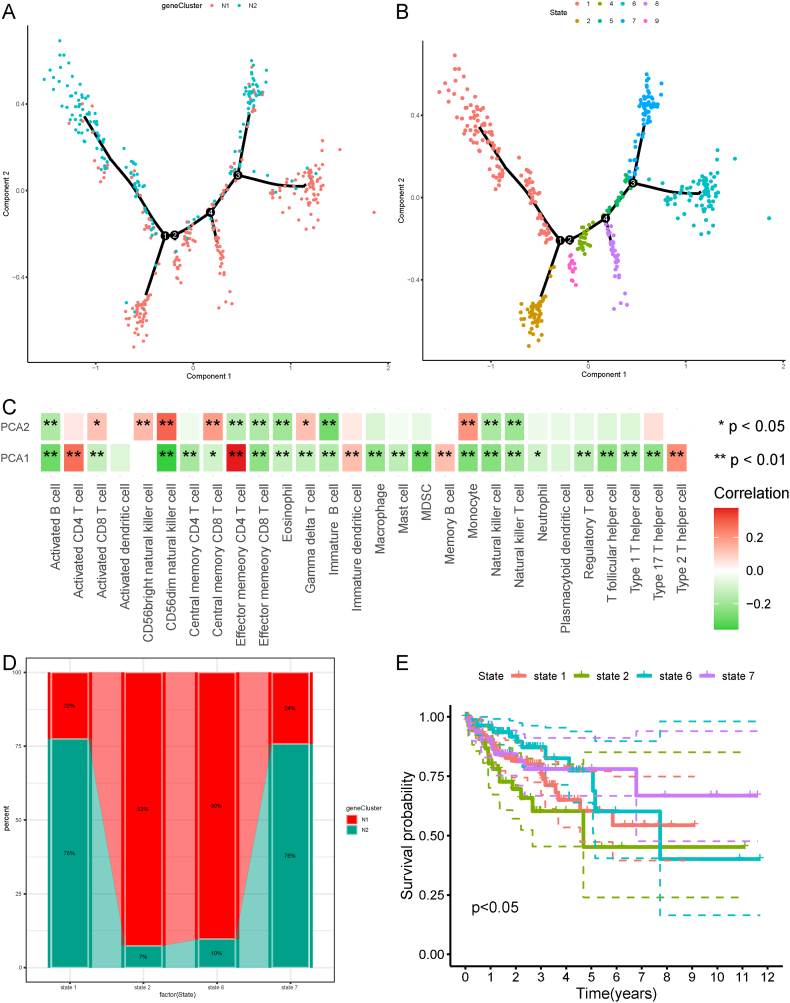
The NRG expression profile of each patient was used to construct a necroptotic landscape of COAD (Fig. 10A). As shown in Fig. 10C, the X-axis (PCA1) was significantly correlated with many types of immune cells and positively correlated with activated CD4 T cells, effector memory CD4+ T cells, immature dendritic cells, memory B cells, and type 2 T helper cells. Similarly, the Y-axis (PCA2) was positively correlated with activated CD8+ T cells, CD56bright natural killer cells, CD56dim natural killer cells, and central memory CD8+ T cells (Fig. 10C). We observed obvious inter-cluster heterogeneity within the necroptosis subtypes. Based on the locations of the two necroptosis subtypes in the necroptosis landscape, we further divided the patients into eight stages (Fig. 10B). Based on the eight types of stages and their positions, we selected the stages at the endpoints, including stages 1, 2, 6, and 7, for further analysis. The proportions of the two necroptosis subtypes at the four stages were shown in Fig. 10D. Survival analysis revealed that significant differences in the prognosis of the four stages, with stage 2 having the poorest prognosis (Fig. 10E). Taken together, the necroptosis landscape based on the necroptosis subtype could not identify the necroptosis status and prognosis of each patient with COAD; thus, additional samples are required for validation in subsequent studies.
Fig. 10.
Necroptosis landscape of COAD. A The location of individual patients in the necroptosis landscape, with the color corresponding to the necroptosis subtype identified above, which represents the overall characteristics of the TIME. B Further stratification of N1 and N2 based on their location on the necroptosis landscape. C Correlation between PCA1/2 and immune modules. D Proportion of necroptosis subtypes for the patients located in end point stages. E Different stages in the necroptosis landscape associated with different prognosis, with stage 2 showing the poorest prognosis. *p < 0.05, **p < 0.01, ***p < 0.001. COAD: colon adenocarcinoma; TIME: tumor immune microenvironment; PCA: principal components analysis. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.7. Identification of necroptosis gene co-expression modules and necroptosis hub genes of COAD
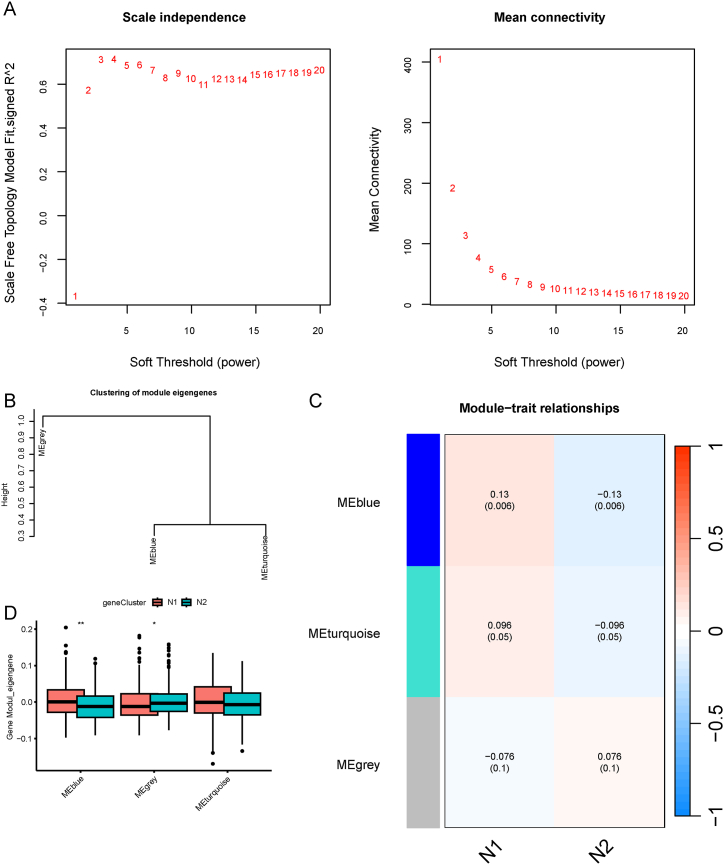
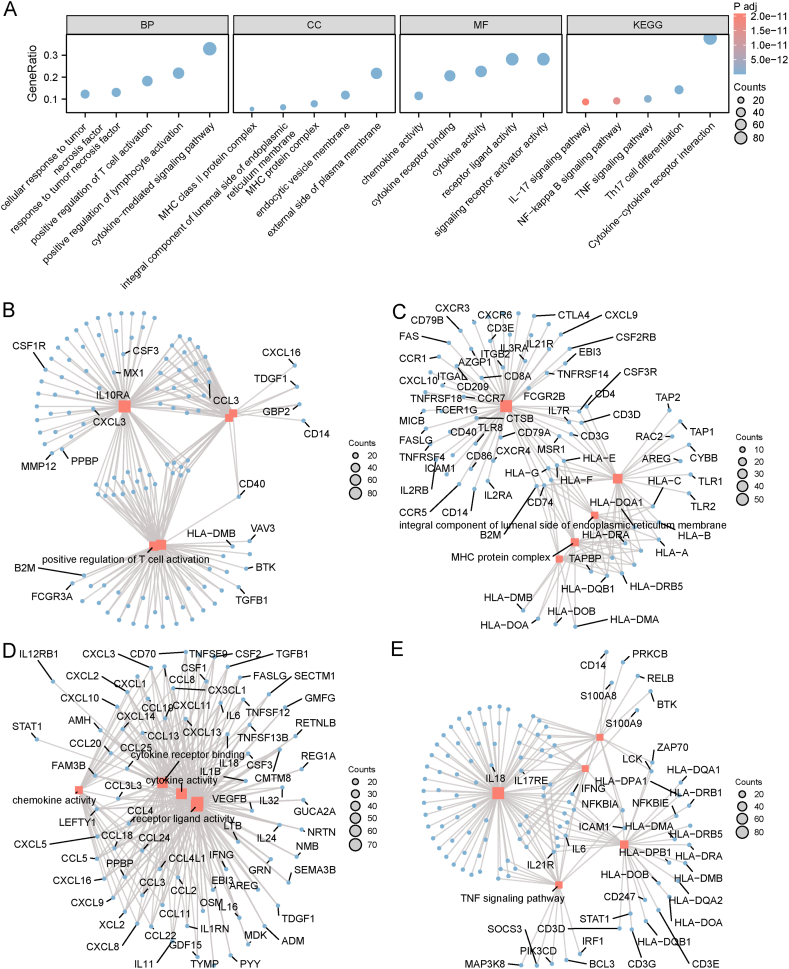
Hub NRGs can be used to screen patients who are suitable for mRNA vaccines because the expression of NRGs significantly influences the effectiveness of mRNA vaccines. To identify hub genes, we constructed a WGCNA of the NRGs. The soft threshold was set to 3 in the scale-free network (Fig. 11A). The gene matrix was transformed into adjacent and topological matrices and the number of genes in each module was set to at least 20. Next, we calculated the signature genes of each module and merged the similar modules. Finally, we obtained three gene modules, and the grey module represented unassigned genes (Fig. 11B and C). When comparing the eigengene score of each module between the two subtypes, we discovered that the eigengene score was obviously higher in N1 than in N2, excepted in the grey module (Fig. 11D). Subsequently, we selected the blue module that was most closely correlated was necroptosis subtypes for further study and performed functional enrichment analysis for the genes in the module (Fig. 12A). The genes were enriched in BP categories, including cytokine-mediated signaling pathway, positive regulation of lymphocyte activation, positive regulation of T cell activation, response to tumor necrosis factor, and cellular response to tumor necrosis factor (Fig. 12B); in CC categories, including external side of plasma membrane, MHC protein complex, MHC class II protein complex, endocytic vesicle membrane, and integral component of luminal side of endoplasmic reticulum membrane (Fig. 12C); in MF categories, including cytokine activity, receptor ligand activity, signaling receptor activator activity, cytokine receptor binding, and chemokine activity (Fig. 12D). In KEGG pathway enrichment, genes in blue module participated in cytokine-cytokine receptor interaction, Th17 cell differentiation, TNF signaling pathway, NF-kappa B signaling pathway, and IL-17 signaling pathway (Fig. 12E). Detailed information regarding the enrichment analysis is presented in Table 2.
Fig. 11.
Identification of necroptosis related hub genes of COAD. A Best soft threshold value was three. B–C Three modules were obtained according to the WGCNA. D Comparison of the eigengene score of different modules between the two necroptosis subtypes. *p < 0.05, **p < 0.01. COAD: colon adenocarcinoma; WGCNA: weighted gene co-expression network analysis.
Fig. 12.
Functional enrichment analysis of the genes in blue module based on the WGCNA. A Functional enrichment of genes displayed using a bar graph. B-E Network of BP (B), CC (C), MF (D), and KEGG pathway (E). GO: Gene Ontology, BP: biological process, CC: cellular component, MF: molecular function, KEGG: Kyoto Encyclopedia of Genes and Genomes; WGCNA: weighted gene co-expression network analysis. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Detailed information of GO and KEGG enrichment analysis.
| ONTOLOGY | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue |
|---|---|---|---|---|---|---|---|
| BP | GO:0019221 | cytokine-mediated signaling pathway | 83/252 | 486/18800 | 3.84E-69 | 1.23E-65 | 7.75E-66 |
| BP | GO:0051251 | positive regulation of lymphocyte activation | 55/252 | 371/18800 | 1.63E-41 | 3.06E-39 | 1.94E-39 |
| BP | GO:0050870 | positive regulation of T cell activation | 46/252 | 223/18800 | 1.79E-41 | 3.18E-39 | 2.01E-39 |
| BP | GO:0034612 | response to tumor necrosis factor | 33/252 | 249/18800 | 2.28E-23 | 1.03E-21 | 6.48E-22 |
| BP | GO:0071356 | cellular response to tumor necrosis factor | 31/252 | 229/18800 | 2.79E-22 | 1.1E-20 | 6.94E-21 |
| CC | GO:0009897 | external side of plasma membrane | 55/253 | 455/19594 | 1.73E-37 | 3.84E-35 | 2.5E-35 |
| CC | GO:0042611 | MHC protein complex | 20/253 | 25/19594 | 3.89E-34 | 4.32E-32 | 2.8E-32 |
| CC | GO:0042613 | MHC class II protein complex | 14/253 | 17/19594 | 1.64E-24 | 1.21E-22 | 7.88E-23 |
| CC | GO:0030666 | endocytic vesicle membrane | 30/253 | 194/19594 | 8.97E-24 | 4.98E-22 | 3.23E-22 |
| CC | GO:0071556 | integral component of lumenal side of endoplasmic reticulum membrane | 16/253 | 29/19594 | 2.16E-23 | 8.01E-22 | 5.2E-22 |
| MF | GO:0005125 | cytokine activity | 57/252 | 235/18410 | 2.17E-55 | 8.75E-53 | 6.54E-53 |
| MF | GO:0048018 | receptor ligand activity | 71/252 | 489/18410 | 7.67E-53 | 1.55E-50 | 1.16E-50 |
| MF | GO:0030546 | signaling receptor activator activity | 71/252 | 496/18410 | 2.12E-52 | 2.86E-50 | 2.14E-50 |
| MF | GO:0005126 | cytokine receptor binding | 52/252 | 272/18410 | 1.14E-44 | 1.15E-42 | 8.59E-43 |
| MF | GO:0008009 | chemokine activity | 29/252 | 49/18410 | 3.85E-42 | 3.11E-40 | 2.32E-40 |
| KEGG | hsa 04060 | Cytokine-cytokine receptor interaction | 82/217 | 295/8164 | 1.53E-64 | 3.28E-62 | 2.15E-62 |
| KEGG | hsa 04659 | Th17 cell differentiation | 31/217 | 108/8164 | 3.17E-24 | 5.69E-23 | 3.73E-23 |
| KEGG | hsa 04668 | TNF signaling pathway | 22/217 | 112/8164 | 1.09E-13 | 7.57E-13 | 4.97E-13 |
| KEGG | hsa 04064 | NF-kappa B signaling pathway | 20/217 | 104/8164 | 2.31E-12 | 1.5E-11 | 9.86E-12 |
| KEGG | hsa 04657 | IL-17 signaling pathway | 19/217 | 94/8164 | 3.29E-12 | 2.08E-11 | 1.37E-11 |
3.8. Drug sensitivity analysis
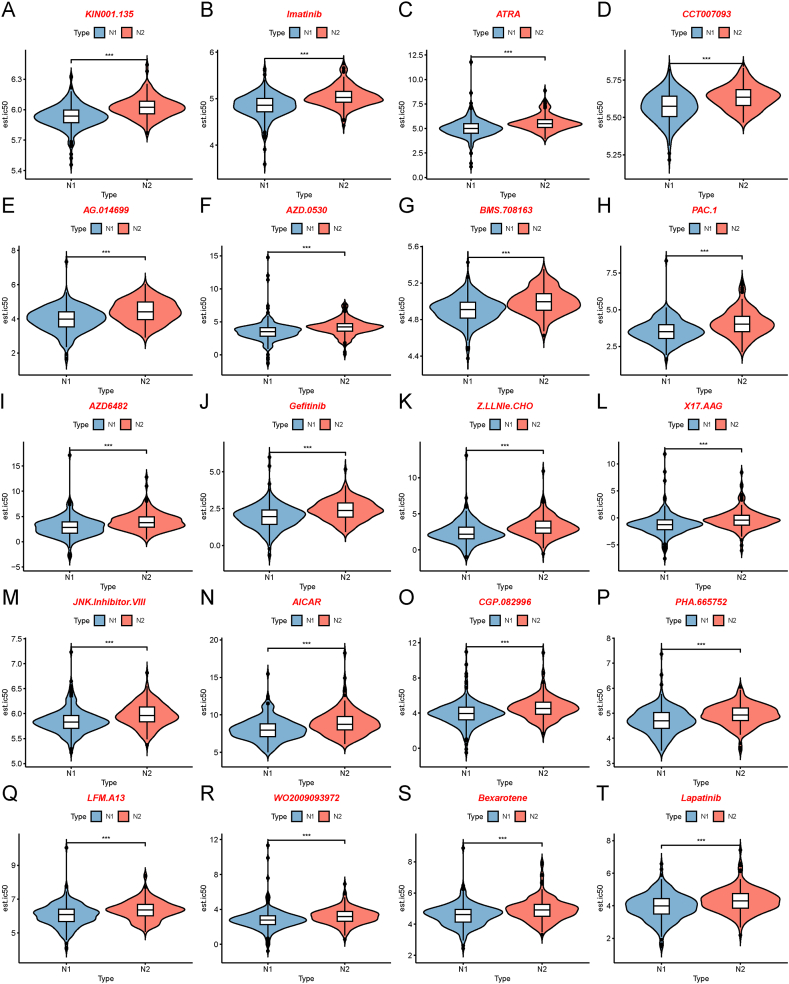
To probe the therapeutic strategies of mRNA vaccine for patients with different necroptosis subtypes, we chose drug sensitivity data from GDSC as the training set to predict the sensitivities of common anticancer drugs for patients with different necroptosis subtypes, based on the TCGA-COAD datasets. We determined the differences in drug sensitivity between the two necroptosis subtypes using the Wilcoxon rank sum (Mann-Whitney U) test, and the top 20 drugs with the most significant differences were displayed using violin plots in Fig. 13A-T. The top 20 drugs were as follows: KIN001.135, imatinib, ATRA, CCT007093, AG.014699, AZD.0530, BMS.708163, PAC.1, AZD6482, gefitinib, Z.LLNle.CHO, X17.AAG, JNK.Inhibitor.VIII, AICAR, CGP.082996, PHA.665752, LFM.A13, WO2009093972, bexarotene, and lapatinib. Our drug sensitivity analysis suggested that patients with the N1 subtype had higher drug sensitivity than those with the N2 subtypes, which stressed the importance of individual therapies for patients with tumors.
Fig. 13.
Differences in drug sensitivity between the two necroptosis subtypes. A-T Sensitivity of patients in the two necroptosis subtypes to the drugs KIN001.135 (A), imatinib (B), ATRA (C), CCT007093 (D), AG.014699 (E), AZD.0530 (F), BMS.708163 (G), PAC.1 (H), AZD6482 (I) gefitinib (J), Z.LLNle.CHO (K), X17.AAG (L), JNK.Inhibitor.VIII (M), AICAR (N), CGP.082996 (O), PHA.665752 (P), LFM.A13 (Q), WO2009093972 (R), bexarotene (S), and lapatinib (T) based on the data from Genomics of Drug Sensitivity in Cancer (GDSC). ***p < 0.001.
3.9. Overexpression of both BAX and IL1B induces necroptosis of COAD cells and enhances the cytotoxicity of CD8+ T cells
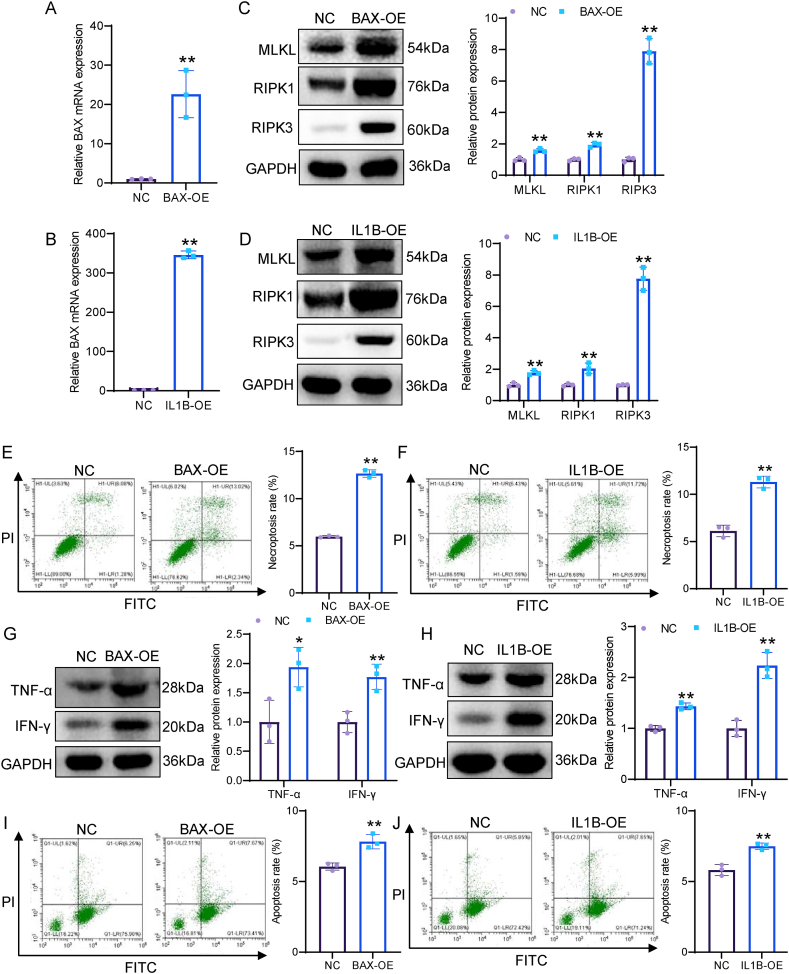
To investigate the role of BAX and IL1B in COAD cell necroptosis, we overexpressed BAX and IL-1β in SW480 cells. qRT-PCR assay confirmed that the expression of BAX and IL1B was significantly increased in SW480 cells transfected with BAX and IL1B overexpression plasmids (Fig. 14A and B). Subsequently, we detected the expression of necroptosis markers (MLKL, RIPK1, and RIPK3). Western blot assay indicated that both BAX overexpression and IL1B overexpression significantly increased the expression of MLKL, RIPK1, and RIPK3 in SW480 cells (Fig. 14C and D). Flow cytometry results showed that both BAX overexpression and IL1B overexpression remarkably enhanced the proportion of necroptotic cells (Fig. 14E and F). Next, to investigate the role of BAX and IL1B in immune response, we co-cultured BAX- and IL1B-overexpressed SW480 cells with CD8+ T cells. Western blot showed that both BAX overexpression and IL1B overexpression in SW480 cells markedly enhanced the expression of TNF-α and INF-γ in cell supernatants (Fig. 14G and H). Flow cytometry analysis showed that both BAX overexpression and IL1B overexpression in SW480 cells obviously enhanced the cytotoxicity of CD8+T cells (Fig. 14I and J). Overall, both BAX and IL1B promoted necroptosis of COAD cells and enhanced the cytotoxicity of CD8+ T cells.
Fig. 14.
BAX overexpression and IL1B overexpression both induce necroptosis of COAD cells and enhance the cytotoxicity of CD8+T cells. A, B The overexpression efficiency of BAX and IL-1β was detected by qRT-PCR. C, D The expression of RIPK1, RIPK3, and MLKL was determined by Western blot. E, F Necroptotic proportions of SW480 cells was examined by flow cytometry. G, H The expression of INF-γ and TNF-α in cell supernatants were measured by Western blot. I, J The cytotoxicity of CD8+T cells upon co-cultured with SW480 cells. *p < 0.05; **p < 0.01.
4. Discussion
Recently, an increasing number of patients with early-stage colon cancer has been screened owing to the popularity of colonoscopy, thereby the long-term survival rate has greatly improved. However, the therapeutic outcomes for patients in the advanced stage are still very unsatisfactory with very high mortality because therapeutic methods are limited [30]. The initiation and development of colon cancer are very complex and are associated with various physiological and pathological processes. Necroptosis plays an important role in cellular activities and its dysregulation can result in disease states, including tumor progression. Necroptosis plays a dual role in carcinogenesis and colon cancer development because it is associated with cancer cell death and immune escape. Zhang et al. [31] reported that PRMT1 inhibited immune escape of colon cancer cells by enhancing RIP3 methylation. In addition, previous studies have shown that many NRGs had potential value in predicting the prognosis of COAD patients [32,33]. Therefore, studying the correlation between necroptosis and immune regulation in COAD can provide new therapeutic strategies for patients with COAD.
The development of mRNA vaccine technology in recent years has provided new ideas and solutions for cancer treatment. mRNA vaccines are nucleic acid-based vaccines that introduce artificially synthesized mRNA molecules into human cells to induce the production of vaccine antigen proteins, thereby eliciting an immune response against specific pathogens. Compared to traditional vaccines, mRNA vaccines have a shorter development cycle, lower research costs, and higher immune protection and safety [34].
Although mRNA vaccine technology has shown excellent performance in COVID-19 prevention and control, its potential in other diseases has not been fully explored. For example, the treatment of a specific disease might require the selection of suitable targets and antigens based on its pathogenic mechanism; however, the screening process for these targets and antigens is complex and challenging. In addition, mRNA vaccine technology confronts certain challenges, such as stability, delivery efficiency, and toxic side effects [[35], [36], [37]]. Therefore, further research and development of new mRNA vaccine technologies are needed to overcome the challenges of various diseases. For unexplored applications of mRNA in a particular disease, researchers could utilize high-throughput technologies and bioinformatics methods to find suitable targets and antigens and continuously optimize mRNA vaccine technology to improve its efficacy and safety [38].
This study aims to identify potential antigens for developing mRNA vaccines to treat patients with COAD. Therefore, we constructed a landscape of aberrant gene expression and genomic alterations to identify potential antigenic targets. To verify the clinical signature of the candidate antigens, we conducted prognostic and immunity analyses. Candidate antigens were identified based on their association with poor prognosis and high APC infiltration. As COAD develops and progresses, these antigens play an important role in their direct presentation to CD8+ T cells to induce an immune response upon lymphocyte infiltration. The potential of these candidate genes for mRNA vaccine development is supported by previous reports, although they have yet to be functionally validated [39]. This study is the first to screen for the potential antigens IL1B and BAX for the development of mRNA vaccines to treat patients with COAD.
This study revealed aberrant expression and genomic alterations of various genes in different cancer subtypes, thus revealing potential targets for developing therapeutic interventions. For example, IL1B and BAX have been identified as promising mRNA vaccine candidates for colon cancer, and their upregulation has been found to be associated with better prognosis. Additionally, BAX has been identified as a potential target for the development of therapeutic interventions for COAD and BAX promotes mitochondrial apoptosis process in cells. Abnormal BAX expression is involved in the initiation and development of COAD [30,40]. In our study, BAX was down-regulated in COAD patients, which was correlated with the improved prognosis of patients with COAD. Furthermore, BAX interacts with and opens the mitochondrial voltage-dependent anion channel (VDAC), leading to the loss of membrane potential and the release of various mitochondrial components, which might also generate tumor antigens, and deserves more in-depth study for future tumor antigen identification [41]. Similarly, IL1B is a type of cytokine that plays an important role in mediating the inflammatory response [42]. In this study, we discovered that patients with COAD with high IL1B expression had a better prognosis, and further immune infiltration analysis showed that IL1B was positively correlated to many types of antigen-presenting cells. In vitro experiments demonstrated that BAX and IL1B overexpression promoted necroptosis of COAD cells and enhanced the cytotoxicity of CD8+T cells. Therefore, BAX and IL1B are potential candidates for mRNA vaccine for patients with COAD.
This article discusses the identification of necroptosis subtypes in patients with COAD, which could aid in the selection of the appropriate populations for mRNA vaccine therapy. Two necroptosis subtypes (N1 and N2) were identified based on necroptosis gene expression profiles, and each subtype exhibited different molecular, cellular, and clinical features. Compared to patients with the N1 subtype, those with the N2 subtype had a better prognosis in the TCGA-COAD cohort, which was validated in the GEO dataset GSE87211. Although there were 30 NRGs with differential mutation status between the two necroptosis subtypes, there were no significant differences in TMB, total number of MS, somatic mutations. However, other immune characteristics presented significant differences, such as the expression level of ICPs and ICD modulators. Since the therapeutic benefits of mRNA vaccines are limited to a specific fraction of patients, mRNA vaccines could be more effective in patients who had upregulated ICD modulators but not suitable for patients with upregulated ICPs [8,43]. In our study, we found that some ICD modulators were highly expressed in the N2 subtype both in TCGA-COAD and GEO cohorts, and most ICPs were expressed at low levels in the N2 subtype. Therefore, patients with the N2 subtype may be more suitable for mRNA vaccines.
For patients with COAD, mRNA vaccines are not commonly used because of the heterogeneity and complex tumor immune microenvironment (TIME). In the present study, the two necroptosis subtypes showed clear differences in the TIME. The N1 subtype presented an immune “hot” phenotype and displayed increased immune cell infiltration, thus representing an excessive inflammatory environment in tumor tissues. On the contrary, the N2 subtype showed the opposite immune status and displayed an immune desert phenotype, thereby representing a non-inflamed environment in tumor tissues. Although the N1 subtype showed higher immune infiltration, the patients had a poor prognosis and potential mechanism is associated with the immunosuppressive environment in tumor tissues. A previous study demonstrates that SPRY1 recruited neutrophils and macrophages to tumor tissues, thereby promoting pancreatic cancer progression, and showed that the mechanism was associated with cancer-related inflammation [44]. The N2 subtype was characterized as an immune “cold” phenotype because of lack of the immune cell infiltration. However, N2 subtype may be suitable for mRNA vaccination because of its non-suppressive immune environment. A previous study reveals that COAD cells escape from immune surveillance by downregulating MHC-1 expression at the cell surface and hindering immune cell infiltration [45]. In our study, APC infiltration was attenuated in the N2 subtype, and the potential tumor antigens IL1B and BAX could recruit APCs to tumor tissues, thereby increasing immune infiltration and reinvigorating the immune response in the patients. Therefore, IL1B and BAX are potential mRNA vaccine targets for patients with COAD. Further construction of the necroptosis landscape revealed intra-cluster heterogeneity between the two necroptosis subtypes. Based on the position of the patients in the trajectories, eight stages were identified. The prognosis of patients among the stages was significantly different, with stage 2 showing the poorest prognosis and the highest proportion of the N1 subtype compared to the other stages. Therefore, our necroptosis subtype analysis integrated with the necroptosis landscape could aid in the development of individualized therapeutic strategies for patients with COAD.
Gene co-expression network analysis can help us to understand the potential functions of NRGs. We obtained three modules based on the WGCNA. The genes in the MEblue modulate were closely related to necroptosis subtypes, and significantly enriched in immune regulation. For example, positive regulation of lymphocyte activation in the BP category is involved in the tumor immune response, and CD8+ T-cells are considered the major effector immune cells in anticancer immune processes. β-catenin suppresses CD8+ T-cell infiltration in COAD tissues, resulting in poor prognosis of patients with COAD [46]. Therefore, our study revealed that necroptosis subtypes were closely related to immune responses and the potential mechanism may be associated with immune regulation in tumor tissues. Furthermore, the drug sensitivity analysis revealed that the two necroptosis subtypes showed significant differences in anticancer drug reactions and patients with N1 subtypes were more sensitive to most anticancer drugs, including imatinib, gefitinib, lapatinib. Overall, our necroptosis subtype analysis provides a guide for individualized therapy using anticancer drugs in patients with COAD.
mRNA vaccines have offered a hopeful approach for cancer immunotherapy. Nevertheless, only a few mRNA vaccines targeting COAD have been developed [47]. mRNA vaccine development is a significant challenge. The tumors of different patients exhibit varying genetic and molecular characteristics, which complicates the development of vaccines targeting specific tumor antigens [39]. Immune suppressive factors within the tumor microenvironment may affect the efficacy of mRNA vaccines [48]. The application strategies of mRNA vaccines in cancer treatment include antigen presentation, immune activation, introduction of antigen receptors, and expression of immune regulatory proteins [49]. These strategies aim to combat cancer cells by activating the immune system. In the development and application of tumor mRNA vaccines, data-driven healthcare plays a crucial role in personalized treatment, efficacy monitoring, and disease prediction and management [50]. Data-driven healthcare offers strong support for the development and application of mRNA vaccines. In this study, we identified BAX and IL1B as the potential candidates for mRNA vaccines against COAD, and confirmed patients with the N2 subtype were appropriate for vaccination. In the future, we plan to optimize the design of mRNA vaccines and develop more effective delivery systems for the identified targets, and to test the safety and efficacy of the vaccines in preclinical models.
This study has some limitations. First, our analysis based on bioinformatics methods and data obtained from public databases requires further validation using multicenter clinical data. Second, the differences in RNA and protein expression of the candidate antigens BAX and IL1B need to be detected using clinical samples. Third, the anti-tumor effects of mRNA vaccines developed based on the antigens BAX and IL1B are still unclear; therefore, cell and animal experimental validations are imperative.
5. Conclusion
In conclusion, we identified two potential antigens BAX and IL1B, which were associated with better prognosis in COAD. Moreover, two necroptosis subtypes were identified, including N1 and N2. Our findings provide a theoretical basis for developing anti-cancer mRNA vaccines, predicting patient prognosis, and selecting patients for vaccination.
Abbreviations: COAD, colon adenocarcinoma; TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; NRG, necroptosis-related gene; APC, antigen-presenting cell; ICP, immune checkpoint protein; ICD, immunogenetic cell death; AFP, α-fetoprotein; HCC, hepatocellular carcinoma; GEPIA, Gene Expression Profiling Interactive Analysis; TIMER, Tumor Immune Estimation Resource; TIIC, tumor-infiltrating immune cells; GSEA, Gene Set Enrichment Analysis; OS, overall survival; PFS, progression-free survival; TMB, tumor mutation burden; ssGSEA, single-sample GSEA; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; WGCNA, weighted gene co-expression network analysis; BAX, BLC2-associated X protein; VDAC, voltage-dependent anion channel.
Ethics approval and consent to participate
Ethical review and approval were waived for this study due to the fact that the data are fully deidentified and no intervention on patients was performed.
Consent for publication
Not applicable.
Availability of data and materials
All data here are publicly available in the UCSC Xena database (https://xena.ucsc.edu/) and GEO datasets (https://www.ncbi.nlm.nih.gov/geo/).
Competing interests
The authors declare that they have no competing interests.
Funding
This work was funded in part by the following: Guangdong Basic and Applied Basic Research Foundation (2022A1515220075, to Y. Q. Luo). The Scientific Research Projects of Medical and Health Institutions of Shenzhen Longhua District, Shenzhen (2022049, to Y.Q. Luo). Guangzhou Basic and Applied Basic Research Foundation (202201020509 to Y.W. Huang). Science and Technology Planning Project of Nansha District (2021MS004 to W.H. Liao). Shenzhen Science and Technology Program (JCYJ20220531092607017 to Y.Q. Luo).
CRediT authorship contribution statement
Yuqi Luo: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing. Caijie Lu: Data curation, Methodology, Supervision, Writing – original draft. Yiwen Huang: Data curation, Formal analysis, Supervision, Writing – original draft. Weihua Liao: Methodology. Yaoxing Huang: Conceptualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e32531.
Contributor Information
Yuqi Luo, Email: luoyuqi2004@tom.com.
Yaoxing Huang, Email: huangyaoxing@sina.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. 2021. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics. CA: a cancer journal for clinicians. 2023;73(1):17–48. doi: 10.3322/caac.21763. 2023. [DOI] [PubMed] [Google Scholar]
- 3.Zhao H., Ming T., Tang S., Ren S., Yang H., Liu M., Tao Q., Xu H. Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol. Cancer. 2022;21(1):144. doi: 10.1186/s12943-022-01616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Requena D.O., Garcia-Buitrago M. Molecular insights into colorectal carcinoma. Arch. Med. Res. 2020;51(8):839–844. doi: 10.1016/j.arcmed.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Reck M., Rodriguez-Abreu D., Robinson A.G., Hui R., Csoszi T., Fulop A., Gottfried M., Peled N., Tafreshi A., Cuffe S., O'Brien M., Rao S., Hotta K., Leal T.A., Riess J.W., Jensen E., Zhao B., Pietanza M.C., Brahmer J.R. Five-year outcomes with pembrolizumab versus chemotherapy for metastatic non-small-cell lung cancer with PD-L1 tumor proportion score >/= 50. J. Clin. Oncol. 2021;39(21):2339–2349. doi: 10.1200/JCO.21.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu X., Deng S., Xu J., Green B.L., Zhang H., Cui G., Zhou Y., Zhang Y., Xu H., Zhang F., Mao R., Zhong S., Cramer T., Evert M., Calvisi D.F., He Y., Liu C., Chen X. Combination of AFP vaccine and immune checkpoint inhibitors slows hepatocellular carcinoma progression in preclinical models. J. Clin. Invest. 2023;33(11) doi: 10.1172/JCI163291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H., Yuan Y., Xue H., Yu R., Jin X., Wu X., Huang H. Reprogramming mitochondrial metabolism of macrophages by miRNA-released microporous coatings to prevent peri-implantitis. J. Nanobiotechnol. 2023;21(1):485. doi: 10.1186/s12951-023-02244-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorentzen C.L., Haanen J.B., Met O., Svane I.M. Clinical advances and ongoing trials on mRNA vaccines for cancer treatment. Lancet Oncol. 2022;23(10):e450–e458. doi: 10.1016/S1470-2045(22)00372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prieto P.A., Mannava K., Sahasrabudhe D.M. COVID-19 mRNA vaccine-related adenopathy mimicking metastatic melanoma. Lancet Oncol. 2021;22(6):e281. doi: 10.1016/S1470-2045(21)00197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aaes T.L., Kaczmarek A., Delvaeye T., De Craene B., De Koker S., Heyndrickx L., Delrue I., Taminau J., Wiernicki B., De Groote P., Garg A.D., Leybaert L., Grooten J., Bertrand M.J., Agostinis P., Berx G., Declercq W., Vandenabeele P., Krysko D.V. Vaccination with necroptotic cancer cells induces efficient anti-tumor immunity. Cell Rep. 2016;15(2):274–287. doi: 10.1016/j.celrep.2016.03.037. [DOI] [PubMed] [Google Scholar]
- 11.Song Y., Zhang J., Fang L., Liu W. Prognostic necroptosis-related gene signature aids immunotherapy in lung adenocarcinoma. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.1027741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L., Zhang X., Zhang Q., Zhang T., Xie J., Wei W., Wang Y., Yu H., Zhou H. A necroptosis related prognostic model of pancreatic cancer based on single cell sequencing analysis and transcriptome analysis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1022420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman M.J., Craft B., Hastie M., Repecka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N., Zhu J., Haussler D. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47(D1):D590–d595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269) doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X., Xiong X., Zhang M., Wang K., Chen Y., Zhou J., Mao Y., Lv J., Yi D., Chen X.W., Wang C., Qian S.B., Yi C. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell. 2017;68(5):993–1005 e9. doi: 10.1016/j.molcel.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becht E., de Reynies A., Giraldo N.A., Pilati C., Buttard B., Lacroix L., Selves J., Sautes-Fridman C., Laurent-Puig P., Fridman W.H. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin. Cancer Res. 2016;22(16):4057–4066. doi: 10.1158/1078-0432.CCR-15-2879. [DOI] [PubMed] [Google Scholar]
- 20.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G. Gene Ontology semantic similarity analysis using GOSemSim. Methods Mol. Biol. 2020;2117:207–215. doi: 10.1007/978-1-0716-0301-7_11. [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson I.R., Ramaswamy S., Futreal P.A., Haber D.A., Stratton M.R., Benes C., McDermott U., Garnett M.J. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batista-Duharte A., Hassouneh F., Alvarez-Heredia P., Pera A., Solana R. Immune checkpoint inhibitors for vaccine improvements: current status and new approaches. Pharmaceutics. 2022;14(8) doi: 10.3390/pharmaceutics14081721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng X., Xu H., Yi X., Zhang T., Wei Q., Li H., Ai J. Tumor-antigens and immune landscapes identification for prostate adenocarcinoma mRNA vaccine. Mol. Cancer. 2021;20(1):160. doi: 10.1186/s12943-021-01452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biller L.H., Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. 2021;325(7):669–685. doi: 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L., He Y., Jiang Y., Wu Q., Liu Y., Xie Q., Zou Y., Wu J., Zhang C., Zhou Z., Bian X.W., Jin G. PRMT1 reverts the immune escape of necroptotic colon cancer through RIP3 methylation. Cell Death Dis. 2023;14(4):233. doi: 10.1038/s41419-023-05752-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W., Lu S., Peng L., Zhang Z., Zhang Y., Guo D., Ma F., Hua Y., Chen X. Integrated analysis of necroptosis-related genes for evaluating immune infiltration and colon cancer prognosis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1085038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Lin M.G., Meng L., Chen Z.M., Wei Z.J., Ying S.C., Xu A. Identification of necroptosis-related genes for predicting prognosis and exploring immune infiltration landscape in colon adenocarcinoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.941156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gote V., Bolla P.K., Kommineni N., Butreddy A., Nukala P.K., Palakurthi S.S., Khan W. A comprehensive review of mRNA vaccines. Int. J. Mol. Sci. 2023;24(3) doi: 10.3390/ijms24032700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alharbi N., Skwarczynski M., Toth I. The influence of component structural arrangement on peptide vaccine immunogenicity. Biotechnol. Adv. 2022;60 doi: 10.1016/j.biotechadv.2022.108029. [DOI] [PubMed] [Google Scholar]
- 36.Mirtaleb M.S., Falak R., Heshmatnia J., Bakhshandeh B., Taheri R.A., Soleimanjahi H., Zolfaghari Emameh R. An insight overview on COVID-19 mRNA vaccines: advantageous, pharmacology, mechanism of action, and prospective considerations. Int. Immunopharm. 2023;117 doi: 10.1016/j.intimp.2023.109934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Son S., Lee K. Development of mRNA vaccines/therapeutics and their delivery system. Mol Cells. 2023;46(1):41–47. doi: 10.14348/molcells.2023.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei S., Sun Q., Chen J., Li X., Hu Z. Bioinformatics analyses for the identification of tumor antigens and immune subtypes of gastric adenocarcinoma. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.1068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan H., Yu T., Liu C., Wang Y., Jing F., Ding Z., Liu J., Shi H. Identifying tumor antigens and immuno-subtyping in colon adenocarcinoma to facilitate the development of mRNA vaccine. Cancer Med. 2022;11(23):4656–4672. doi: 10.1002/cam4.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X., Feng H., Wang Y., Wu Y., Guo Q., Feng Y., Ma M., Guo W., Song X., Zhang Y., Han S., Cao L. Septin 4 promotes cell death in human colon cancer cells by interacting with BAX. Int. J. Biol. Sci. 2020;16(11):1917–1928. doi: 10.7150/ijbs.44429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf P., Schoeniger A., Edlich F. Pro-apoptotic complexes of BAX and BAK on the outer mitochondrial membrane. Biochim. Biophys. Acta Mol. Cell Res. 2022;1869(10) doi: 10.1016/j.bbamcr.2022.119317. [DOI] [PubMed] [Google Scholar]
- 42.Rebe C., Ghiringhelli F. Interleukin-1 beta and cancer. Cancers. 2020;12(7) doi: 10.3390/cancers12071791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miao L., Zhang Y., Huang L. mRNA vaccine for cancer immunotherapy. Mol. Cancer. 2021;20(1):41. doi: 10.1186/s12943-021-01335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi T., Li X., Zheng J., Duan Z., Ooi Y.Y., Gao Y., Wang Q., Yang J., Wang L., Yao L. Increased SPRY1 expression activates NF-kappaB signaling and promotes pancreatic cancer progression by recruiting neutrophils and macrophages through CXCL12-CXCR4 axis. Cell. Oncol. 2023 doi: 10.1007/s13402-023-00791-z. [DOI] [PubMed] [Google Scholar]
- 45.Zeestraten E.C., Reimers M.S., Saadatmand S., Goossens-Beumer I.J., Dekker J.W., Liefers G.J., van den Elsen P.J., van de Velde C.J., Kuppen P.J. Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br. J. Cancer. 2014;110(2):459–468. doi: 10.1038/bjc.2013.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue J., Yu X., Xue L., Ge X., Zhao W., Peng W. Intrinsic beta-catenin signaling suppresses CD8(+) T-cell infiltration in colorectal cancer. Biomed. Pharmacother. 2019;115 doi: 10.1016/j.biopha.2019.108921. [DOI] [PubMed] [Google Scholar]
- 47.Lesterhuis W.J., De Vries I.J., Schreibelt G., Schuurhuis D.H., Aarntzen E.H., De Boer A., Scharenborg N.M., Van De Rakt M., Hesselink E.J., Figdor C.G., Adema G.J., Punt C.J. Immunogenicity of dendritic cells pulsed with CEA peptide or transfected with CEA mRNA for vaccination of colorectal cancer patients. Anticancer research. 2010;30(12):5091–5097. [PubMed] [Google Scholar]
- 48.Wang B., Pei J., Xu S., Liu J., Yu J. Recent advances in mRNA cancer vaccines: meeting challenges and embracing opportunities. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1246682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D., Liu C., Li Y., Tenchov R., Sasso J.M., Zhang D., Li D., Zou L., Wang X., Zhou Q. Messenger RNA-based therapeutics and vaccines: what's beyond COVID-19? ACS Pharmacol. Transl. Sci. 2023;6(7):943–969. doi: 10.1021/acsptsci.3c00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amri M.M., Abed S.A. The data-driven future of healthcare: a review. Mesopotamian Journal of Big Data. 2023;2023:68–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data here are publicly available in the UCSC Xena database (https://xena.ucsc.edu/) and GEO datasets (https://www.ncbi.nlm.nih.gov/geo/).