Abstract
Background
Ovarian cancer (OC) ranks as the fifth most prevalent neoplasm in women and exhibits an unfavorable prognosis. To improve the OC patient's prognosis, a pioneering risk signature was formulated by amalgamating disulfidptosis-related genes.
Methods
A comparative analysis of OC tissues and normal tissues was carried out, and differentially expressed disulfidptosis-related genes (DRGs) were found using the criteria of |log2 (fold change) | > 0.585 and adjusted P-value <0.05. Subsequently, the TCGA training set was utilized to create a prognostic risk signature, which was validated by employing both the TCGA testing set and the GEO dataset. Moreover, the immune cell infiltration, mutational load, response to chemotherapy, and response to immunotherapy were analyzed. To further validate these findings, QRT-PCR analysis was conducted on ovarian tumor cell lines.
Results
A risk signature was created using fourteen differentially expressed genes (DEGs) associated with disulfidptosis, enabling the classification of ovarian cancer (OC) patients into high-risk group (HRG) and low-risk group (LRG). The HRG exhibited a lower overall survival (OS) compared to the LRG. In addition, the risk score remained an independent predictor even after incorporating clinical factors. Furthermore, the LRG displayed lower stromal, immune, and estimated scores compared to the HRG, suggesting a possible connection between the risk signature, immune cell infiltration, and mutational load. Finally, the QRT-PCR experiments revealed that eight genes were upregulated in the human OC cell line SKOV3 compared with the human normal OC line IOSE80, while six genes were down-regulated.
Conclusions
A fourteen-biomarker signature composed of disulfidptosis-related genes could serve as a valuable risk stratification tool in OC, facilitating the identification of patients who may benefit from individualized treatment and follow-up management.
Keywords: Ovarian cancer, Immunity, Disulfidptosis, Bioinformatics, Disulfidptosis-related genes (DRGs)
1. Introduction
Ovarian cancer (OC) ranks as the fifth most prevalent malignancy among women, with an estimated annual global mortality rate of 125,000 individuals. In China, the prevalence of OC has gradually increased due to the aging population [1]. OC ranks third in the incidence of gynecological cancer and second in terms of mortality rate among women globally [2]. This disease primarily encompasses three types: germ cell and sex cord-stromal tumors, as well as epithelial OCs. The latter constitutes 85%–90 % of all OCs and represents the most aggressive subtype. Epithelial OC can be further classified into serous, mucous, endometrioid, and non-transparent cell types [3]. In comparison to other subtypes, epithelial OC exhibits accelerated progression, a higher rate of metastasis, unfavorable clinical outcomes, and elevated mortality rates [4]. Currently, the etiology of OC remains unelucidated, with hormones, genetics, environment, and lifestyle potentially serving as primary risk factors for its onset [5].
Cell death refers to the irreversible termination of vital processes and the cessation of life, whereas regulated cell death (RCD) is a specific kind of cell death that is governed by specific molecular pathways, which may be influenced by genetic or pharmacological interventions [6]. Multiple modes of cell death have been discovered, encompassing apoptosis, pyroptosis, ferroptosis, copper death, and other mechanisms [7]. The facilitation of glutathione biosynthesis and inhibition of oxidative stress plays a crucial role in ferroptosis, and cystine uptake is mediated by solute carrier family 7 member 11 (SLC7A11) [8]. The precise mechanism underlying the regulatory effects of SLC7A11 on ferroptosis requires further investigation; however, inhibiting SLC7A11 expression was found to lead to intracellular cysteine depletion, increasing cell susceptibility to oxidative stress and ferroptotic cell death [[9], [10], [11]]. Furthermore, a report from 2017 indicated that SLC7A11 plays a significant role in promoting cell death under glucose starvation. Recent studies have demonstrated that the conversion of cystine to cysteine, facilitated by SLC7A11, is heavily reliant on the reduced NADPH produced through the glucose-pentose phosphate pathway. This particular form of cell death has been termed disulfidptosis [12]. In recent years, multiple studies have applied advanced RNA sequencing and microarray technologies for bioinformatic research, aiding in the identification of potential cancer biomarkers. A common and widely used approach involves the comparison of gene expression levels in samples from various groups, particularly between cancer and normal groups, as a method to explore differentially expressed genes (DEGs) [13,14].
We proposed to create a gene signature associated with disulfidptosis to anticipate the OC patient's prognosis. Moreover, an investigation was conducted into potential signaling pathways. Our outcomes manifest that disulfide death may contribute to the occurrence and development of OC, which provides an important idea for treating OC.
2. Methods
2.1. Data collection, processing, and analyses
The gene transcriptome data of 469 OC patients were acquired from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Additionally, the clinical data of 300 patients and the gene mutation data of 196 patients were also gathered. The clinical data of the individuals under examination are documented in Table S1. Overall survival (OS) was established as the duration from the initiation of therapy till death from any cause. Only cases with comprehensive data on OS were incorporated into the research. Additional analysis was performed on these specimens. The transcriptome data was first measured in fragments per kilobase million (FPKM) and then converted to transcripts per million (TPM). Subsequently, one sample with incomplete survival data was excluded from the analysis. Thereafter, the GSE53963 dataset was downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), and tissue samples were retained to merge with the TCGA dataset. The UCSC Xena server (https://xena.ucsc.edu/) was used to download digital focal-level copy number variation (CNV) from the GDC TCGA OC project.
2.2. Analyzing differential expression to identify prognostic-related DRGs
The differential analysis of DRGs between OC samples and normal samples utilized the Wilcoxon rank-sum test. The DRGs associated with prognosis were further determined using Kaplan-Meier (KM) analysis and univariate Cox regression with a P-value <0.05 [15,16].
2.3. Analyses of consensus clustering for the DRGs GSVA and ssGSEA
Consensus clustering was conducted using the k-means algorithm to uncover unique patterns associated with the expression of disulfidptosis regulators in relation to disulfidptosis. The consensus clustering approach, implemented using the "ConsensuClusterPlus" package, was used to estimate the number of clusters and their stability. During this procedure, we conducted 1000 iterations to ensure the consistency of our categorization [17]. Furthermore, the appropriate number of categories was ascertained by doing consensus clustering analysis, cumulative distribution function (CDF), and consensus matrix. Analyzed using the "GGalluvial" R package (v.4.1.0), the study examined the different forms of OS and risk ratings. Furthermore, the "GSVA" R packages were used to conduct GSVA enrichment analysis, which was then shown in a heatmap [18]. Afterward, the file "c2.cp.kegg.v6.2.symbols" was obtained from the MSigDB database for the purpose of conducting GSVA analysis. A statistically significant difference was determined while comparing various subgroups using the "limma" software, with an adjusted P-value of less than 0.05. The ssGSEA (single-sample gene-set enrichment analysis) technique was used to measure the proportional abundance of each cell infiltrate in the OC TME. A PPI network was constructed employing the STRING database, with common genes as input (https://string-db.org/). The confidence for the required interaction score was established at 0.4, while all other parameters were left at their default values. The MCODE plug-in, which is a part of Cytoscape, was used to selectively extract the essential functional modules. The settings were left at their default values.
2.4. A selection Hub gene network based on genes and enrichment analysis
Data from the TCGA OC study has been integrated with GSE53963 cancer data. Statistical significance was ascertained by a log2 (fold change) more than 0.585 and an adjusted P-value lower than 0.05. Subsequently, the Gene Ontology GO enrichment studies were conducted using the "org.Hs.eg.db" and "enrichplot" packages to investigate biological functions and structures. Additionally, the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were employed to ascertain the associated pathways.
2.5. Validation and generation of signatures
The DEGs obtained from various DRG clusters were first normalized across all OC specimens, and the genes that were common to all clusters were obtained. The patients were categorized into many groups employing an unsupervised clustering technique, and overlapping DEGs were discovered. The consensus clustering approach was deployed to ascertain both the quantity and stability of gene groups. Subsequently, the prognostic analysis of each gene in the signature was conducted employing the univariate Cox regression model. The genes that exhibited statistical significance in the prognostic analysis were selected for further study. Afterward, principal component analysis (PCA) was deployed to create the gene profile associated with disulfidptosis. Principal components 1 and 2 were chosen as signature scores. This method has the benefit of concentrating the score on the group that contains the most closely correlated (or anti-correlated) genes while mitigating the gene's influence that does not align with the rest of the group. Subsequently, the DRGscore was determined with a methodology that closely resembles the GGI:
Where i is the expression of disulfidptosis-related genes [19,20].
2.6. Analyzing the tumor microenvironment, immune infiltration, and tumor mutation burden
The immunological, stromal, and ESTIMATE scores were computed using the ESTIMATE method. An algorithm successfully determined the numbers of stromal and immune cells in malignant tumor tissues by examining gene expression patterns. The ESTIMATE algorithm was constructed with the R "estimate" package, which may be found at the following link: https://sourceforge.net/projects/estimateproject/. TMB scores were computed for each OC patient in the TCGA cohort based on somatic mutation analysis. A waterfall plot depicting the high-risk group (HRG) and low-risk groups (LRG) was created employing the R package "maftools."
2.7. Cell culture and Real-time quantitative PCR (QRT-PCR)
The SKOV3 cell line (Homo sapiens, human; RRID: CVCL_0532) and IOSE80 cell line (Homo sapiens, human; RRID: CVCL_5546) were obtained from the Cell Bank of the Chinese Academy of Sciences in Shanghai, China. The cell lines underwent verification by STR profiling. The cells were cultured in DMEM/F12 medium enriched with 10 % fetal bovine serum at 37 °C and a CO2 concentration of 5 %. The SKOV3 and IOSE80 cell lines were used to extract total RNA using the RNA simple total RNA kit (Invitrogen, Shanghai, China). The RNA was converted into complementary DNA (cDNA) using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Shanghai, China). In addition, QRT-PCR was performed employing an ABI 7500 Real-Time PCR System (Applied Biosystems). The PCR primers are specified in Table S2, and they were formulated and produced by GENEWIZ. The mRNA expression was quantified using the 2−△△Ct technique with reference to GAPDH.
3. Results
3.1. Differential expression and genetic alterations of DRGs
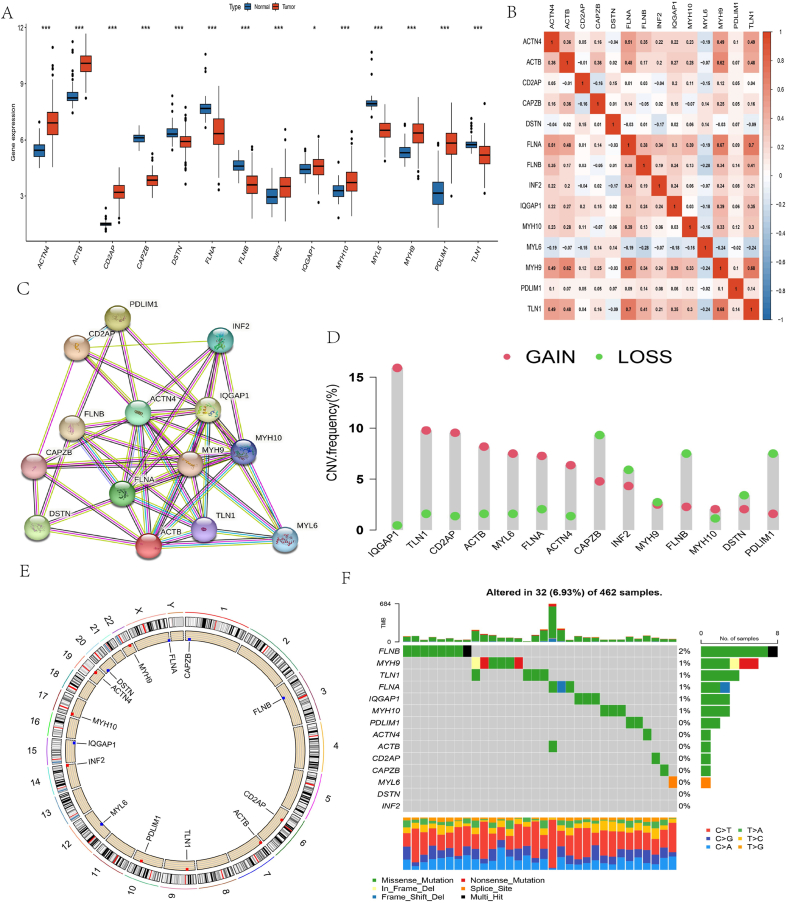
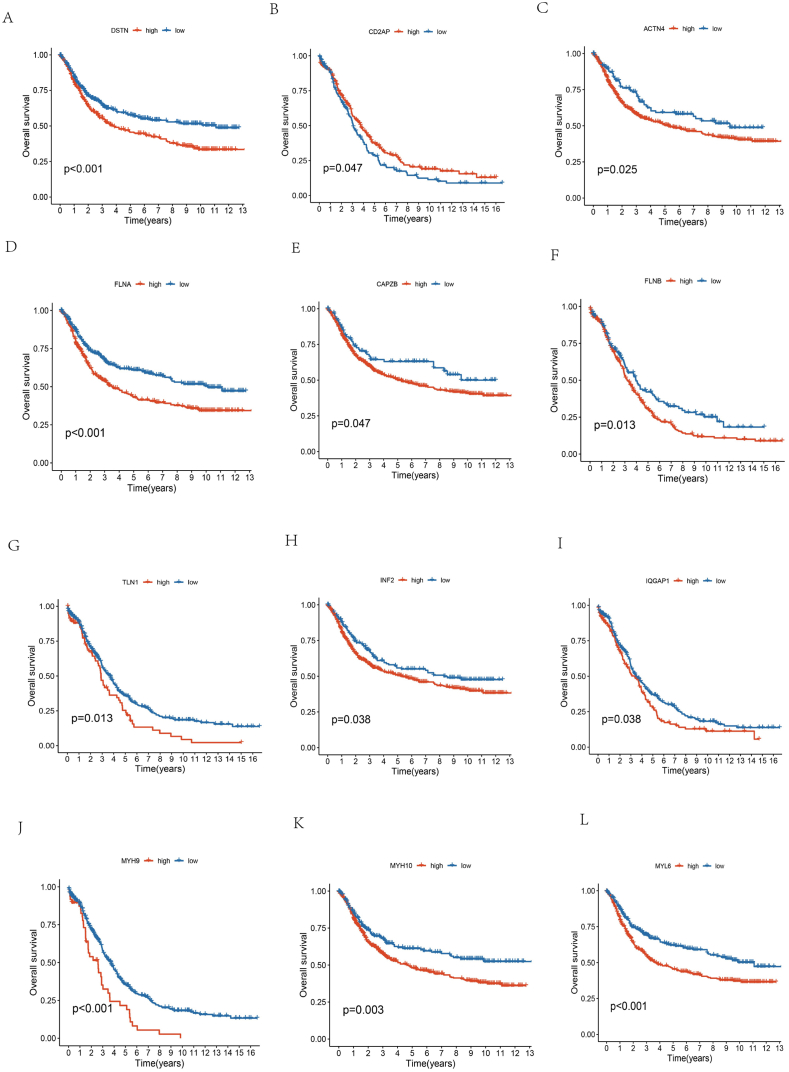
First, 14 DRGs (ACTN4, ACTB, CD2AP, CAPZB, DSTN, FLNA, FLNB, INF2, IQGAP1, MYH10, MYL6, MYH9, PDLIM1, TLN1) were identified from previous studies. Next, the expression of these 14 DRGs between tumor and normal tissues was analyzed in OC patients from the TCGA database. The results revealed significant variations in these 14 DRG expression levels between normal tissues and OC samples (P-value <0.05). ACTN4, ACTB, CD2AP, INF2, IQGAP1, MYH10, MYH9, and PDLIM1 exhibited significantly elevated expression in tumor tissues relative to normal tissues. Conversely, CAPZB, DSTN, FLNA, FLNB, MYL6, and TLN1 exhibited greater expression in normal tissues (Fig. 1A). Furthermore, FLNA and PDLIM1 expression exhibited a strong positive correlation (Fig. 1B). Next, the protein-protein interaction (PPI) networks of the 14 DEGs were created by String (https://string-db.org/) (Fig. 1C). After analyzing the copy number variation (CNV) in DRGs, PDLIM1, FLNB, INF2, and CAPZB demonstrated a deletion frequency of over 5 % of copies, revealing a significantly higher occurrence compared to the frequency of amplification. Conversely, copies of IQGAP1, TLN1, ACTB, MYL6, FLNA, ACTN4, and CD2AP demonstrated a higher frequency of increase, exceeding 5 %. (Fig. 1D–E). Additionally, out of 462 OC samples, 322 somatic cells (6.93 %) exhibited mutations, with MYH10 (8 %) displaying a higher mutation frequency (Fig. 1F). Following that, KM analysis and Cox analysis were conducted on the TCGA and GSE53963 datasets to estimate the prognostic importance of DRGs in patients with OC. The KM analysis revealed that 14 genes linked to disulfidptosis were significantly correlated with the prognosis of OS. Among these, DSTN, FLNA, MYH9, and MYL6 showed the most significant differences in survival outcomes, with a P-value less than 0.001. (Fig. 2A–L, Table S3). To assess the predictive specificity and sensitivity of 14 disulfidptosis-related genes, receiver operating characteristic (ROC) and the area under the ROC curve (AUC) were presented from the TCGA database. (Fig. S1).
Fig. 1.
Genetic and transcriptional alterations of DRGs in OC. (A) Distributions of 14 DRGs that differ in expression between the normal ovary and OC tissues (*p < 0.05, ***p < 0.001); (B) Expression correlation between 14 DRGs; (C) PPI network showing the interactions of the DRGs; (D) Frequencies of CNV gain, loss, and non-CNV among DRGs; (E) Locations of CNV alterations in DRGs on 23 chromosomes; (F) Mutation frequencies of 14 DRGs in OC patients from the TCGA cohort.
Fig. 2.
The prognosis significance of DRGs of OV patients. (A) Kaplan–Meier survival curves (KMSC) indicated that OC patients with high DSTN mRNA expression had a shorter OS; (B) KMSC manifested that OC patients with low CD2AP mRNA expression had a shorter OS; (C–L) KMSC manifested that OC patients with high ACTN4, FLNA, CAPZB, FLNB, TLN1, INF2, IQGAP1, MYH9, MYH10, and MYL6 mRNA expression had a shorter OS.
3.2. Cluster analysis of the division subtypes, the GSVA and the ssGSEA
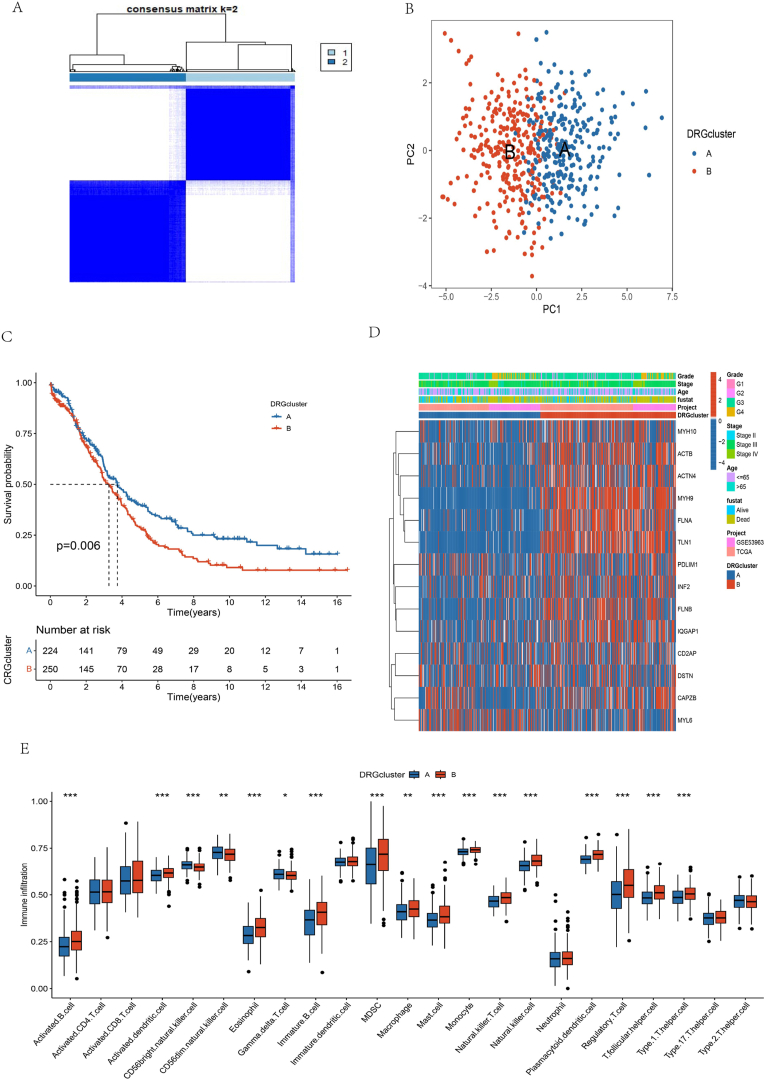
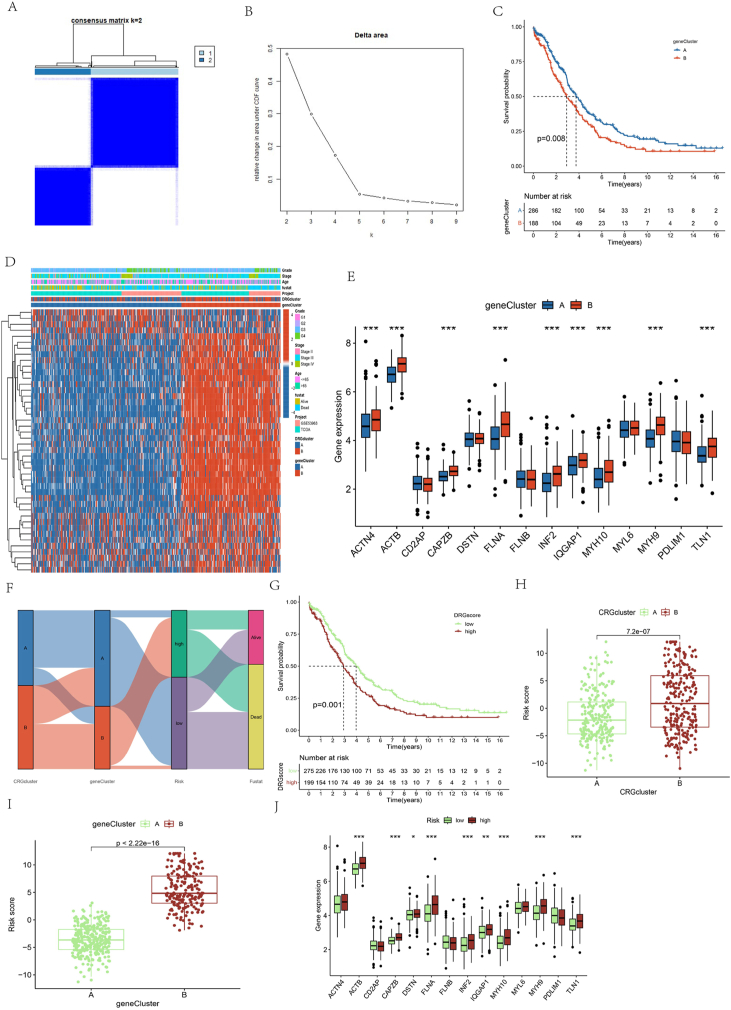
OC patients were categorized into two primary subtypes, namely cluster A and cluster B, as K = 2 yielded the most optimal consistency matrix for the subtypes (Fig. 3A). Afterward, t-distributed stochastic neighbor embedding (t-SNE) was deployed to distinguish the two clusters by analyzing the expression levels of DRGs (Fig. 3B). Furthermore, the survival study demonstrated a significant disparity in survival rates between the two clusters (P-value of 0.006), as evidenced by the KM curve. The results suggest a more favorable prognosis for OC patients in cluster B compared to cluster A (Fig. 3C). Subsequently, a sophisticated heat map was created employing a cluster-based approach, utilizing the clinical stage, gender, and age of OC patients from the TCGA dataset (Fig. 3D). Combined with the outcomes of the survival analysis, samples in Cluster A corresponding to favorable survival outcomes showed abundant infiltration by CD56dim natural killer cell, Gamma delta T cell, and Type 2 T helper cell. In contrast, samples in Cluster B corresponding to an unfavorable clinical prognosis manifested the infiltration of activated CD8 T cells, eosinophils, MDSC, regulatory T cells, and natural killer T cells (Fig. 3E).
Fig. 3.
Landscape of the DRGs and biological characteristics of disulfidptosis subtypes in ovarian cancer. (A) Consensus matrix of OC patients, k = 2, using the unsupervised consensus clustering method; (B) Principal component analysis of 14 DRGs in all OC cohorts found two distinct subtypes; (C) Kaplan–Meier curves for overall survival of all OC patients; (D) Variations in clinicopathologic features and expression levels of 14 DRGs in all OC cohorts among the two distinct subtypes. Tumor stage, age, survival status, and cluster were used as patient annotations. Red and blue represent high and low expression of disulfidptosis genes, respectively; (E) Comparison of the ssGSEA scores for immune cells in the two OC subtypes. The line in the box represents the median value (*p < 0.05, **p < 0.01, ***p < 0.001).
3.3. Analysis of intersection genes and enrichment
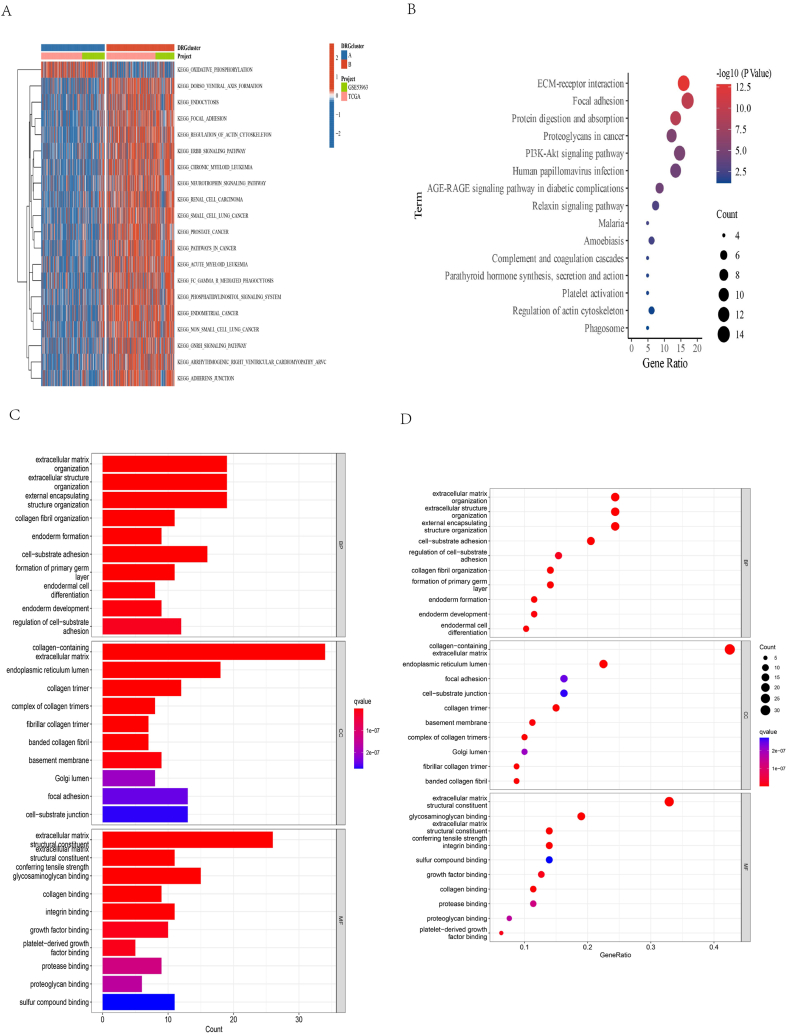
KEGG and GO enrichment analyses were conducted to determine potential pathways and functions of differentially regulated genes (DRGs). Based on the GSVA heat map, two distinct clusters exhibited disparate pathways (Fig. 4A). The KEGG enrichment analysis revealed the involvement of DRGs in focal adhesion (Fig. 4B). The GO analysis indicated a close association between DRGs and the plasma membrane in the Cellular Component (CC) category. In the Biological Process (BP) and Molecular Function (MF) categories, DRGs were predominantly linked to the plasma membrane and protein binding, respectively (Fig. 4C–D).
Fig. 4.
Correlations of TME and biological characteristics in two OC subtypes. (A) a GSVA of KEGG biological pathways in two disulfidptosis subtypes; (B) KEGG enrichment analysis of DRGs; (C–D) GO enrichment analysis of DRGs.
3.4. Analysis of consensus clustering for partition subtypes and signatures construction
To categorize OC patients into distinct subtypes, the optimal k-value was determined based on the highest correlation coefficient, thereby identifying clusters A and B (Fig. 5A–B). The KM survival analysis manifested a significant variation, with a P-value of 0.008 showing a significant difference. These findings indicated a more favorable survival outcome in cluster A compared to cluster B (Fig. 5C).
Fig. 5.
Identification of gene subtypes based on the DEGs of Disulfidptosis-related clusters. (A–B) Identification of gene subtypes based on prognostic DEGs among TWO disulfidptosis subtypes in OC cohort. (C) Kaplan–Meier curves for overall survival of all OC patients with two gene subtypes. (D) Heat map showing the relationships between clinicopathologic features and the two gene subtypes. (E) Differences in the expression of 14 DRGs among the two gene clusters. (F) An alluvial diagram of the distribution of disulfidptosis cluster, gene cluster in two risk groups, as well as survival outcomes. (G) The Kaplan–Meier survival curves were stratified by disulfidptosis cluster and risk subgroup. (H) Difference of risk score among two cuproptosis clusters. (I) Difference of risk score among two gene clusters. (J) The differential analysis of DRGs expression in the collection file (*p < 0.05, **p < 0.01, ***p < 0.001).
Subsequently, a comprehensive cluster-based heat map was generated, incorporating the clinical stage, grade, and age of OC patients (Fig. 5D). Additionally, DEGs associated with DRGs were analyzed. Notably, the boxplot revealed that ACTB and FLNA displayed the highest expression levels in cluster B (Fig. 5E). Except for the genes CD2AP, DSTN, MYL6, FLNB, and PDLIM1, all other genes manifested significant variations in expression between the two clusters, with a P-value less than 0.001. The patient distribution in the two clusters of genes related to disulfidptosis, two gene clusters, and two groups based on disulfidptosis-related genes_score groups is displayed in Fig. 5F. The KM survival curve illustrates that patients with a low score signature are associated with a significantly more favorable survival outcome compared to those with a high score (P-value = 0.001) (Fig. 5G). In the boxplots below, the DRG signatures are shown differently among the clusters of genes and DRGs. The results indicated that the HRG was located in subgroup B of the DRGCluster and subgroup B of geneCluster. Additionally, previous studies have shown that the HRG has poor prognostic outcomes. Concurrently, poor prognostic outcomes can be observed in subgroup B of the DRGCluster and subgroup B of geneCluster (Fig. 5H–I). In the collection file, a boxplot showed the differential expression of DRGs. The outcomes manifested that the vast majority of genes emerged with a significantly greater expression in the HRG compared to the LRG (P-value <0.05) (Fig. 5J).
3.5. Immune cell infiltration and mutational load are associated with the signature
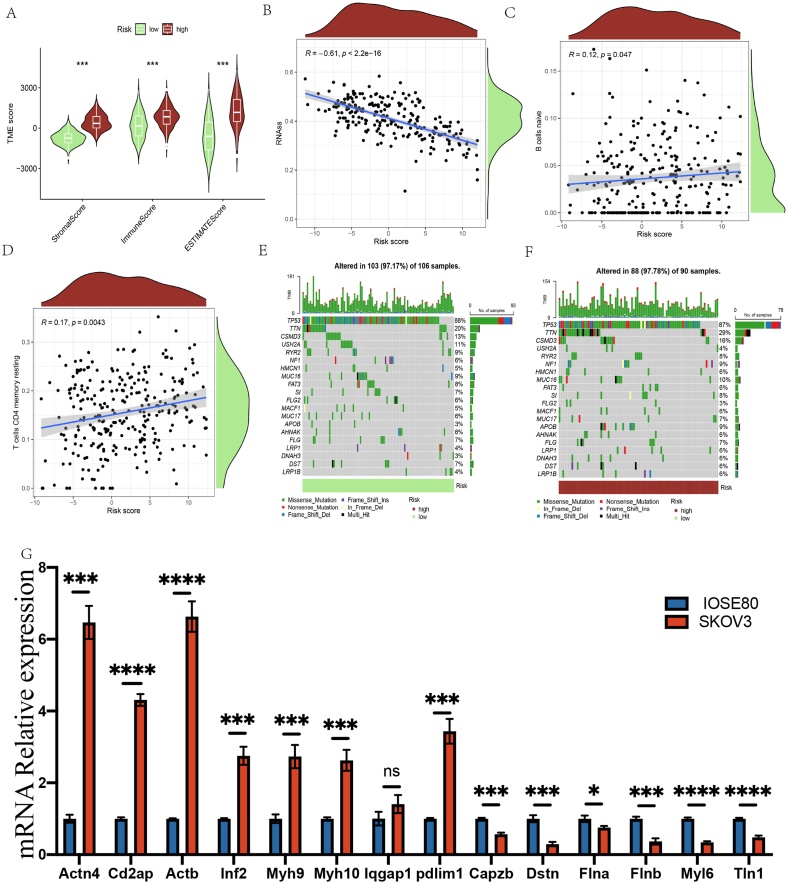
An extensive analysis was executed to ascertain the connection between signatures and immunity. The outcomes emerged that the stromal, immune, and estimated scores of the LRG were significantly lower compared to the HRG (Fig. 6A). The stem cell concentration and risk score showed a significant negative correlation (R = −0.61, P-value <2.2e-16) (Fig. 6B). Different immune cells were associated with different risk scores. Specifically, naive B cells (R = 0.12, P-value = 0.047) and resting memory CD4 T cells (R = 0.17, P-value = 0.0043) showed significant correlations (Fig. 6C–D). As shown in Fig. 6E–F, the HRG presented a more extensive tumor mutation burden than the LRG. Moreover, the poor prognostic outcomes in the HRG can be partially attributed to TMB.
Fig. 6.
The landscape of immune microenvironment with prognostic signature.
(A) Correlations between risk score and immune score, stromal score, and ESTIMATE score. (B) Correlation between risk score and stem cell content (RNAss). (C–D) Correlations between risk score and different immune cells. (E–F) Mutation frequencies of the high- and low-risk groups. (G) The mRNA expression of fourteen genes in cell line SKOV3 and IOSE80 was measured by QRT-PCR (****P < 0.0001; ***P < 0.001; *P < 0.05; ns, not statistically different, Error bars are ± SEM).
3.6. Expression validation of the DRGs by QRT-PCR
In order to provide a more detailed analysis of the DRG expression levels in both normal and OC cells, we grew the SKOV3 human OC and the IOSE80 human normal ovarian cell lines. The QRT-PCR experiments exhibited that eight genes were upregulated in the human OC cell line SKOV3 compared with the human normal ovarian cell line IOSE80, while five genes were down-regulated. Compared with normal samples, the expression levels of ACTN4, ACTB, CD2AP, INF2, IQGAP1, MYH10, MYH9, and PDLIM1 were significantly upregulated in OC specimens, while CAPZB, DSTN, FLNA, FLNB, MYL6, and TLN1were significantly downregulated (Fig. 6G).
4. Discussion
Notably, OC is recognized as one of the most aggressive solid tumors [21,22]. According to data provided by the World Health Organization, OC shows a high prevalence, with a majority of cases being diagnosed at an advanced stage [[23], [24], [25]]. Despite notable advancements in surgical and chemotherapeutic treatment approaches for OC, researchers have discovered that following the conventional histological classification as a basis for anti-tumor therapy has not effectively enhanced the OS rate of OC patients. At present, the 5-year survival rate remains below 45 % [26,27]. Therefore, the precise determination of molecular subtypes of OC and the development of predictive models are crucial in guiding personalized therapy [[28], [29], [30]]. Such models could enable the accurate identification of molecular subtypes of OC, thereby facilitating personalized therapy [31,32]. Disulfidptosis is a recently proposed form of cell death. SLC7A11 is widely recognized for its primary role in facilitating the transportation of amino acids across the plasma membrane, providing a crucial pathway for the sustenance of cancer cells. Gan et al. demonstrated that in the absence of glucose, cancer cells exhibiting elevated levels of SLC7A11 exhibited rapid depletion of NADPH, resulting in abnormal accumulation of disulfide molecules, notably cystine. This abnormal accumulation triggers an interaction between actin cytoskeleton proteins. The aberrant disulfide bonding hampers the organization of the actin network, eventually resulting in the breakdown of the network and subsequent cellular apoptosis [33]. Considering that numerous cancer treatments exert their effects by inducing apoptosis, further research should be conducted on the potential of targeting disulfide death as a viable cancer therapy. Following advances in high-throughput sequencing technology, various public databases and bioinformatics algorithms have been developed, offering valuable resources and methodologies to explore the intricate interplay between cell death and comprehensive immunity [[34], [35], [36]]. Nevertheless, the correlation between OC and disulfidptosis remains ambiguous, as no investigation has ascertained the combined implications of immunity and disulfidptosis on OC prognosis. Therefore, the present study explored the combined effect of disulfidptosis-related genes in OC. In addition, a gene signature combining immunity and disulfidptosis was created to forecast the subtype, prognosis, and implication of immunotherapy in OC.
In this study, a set of 14 genes associated with disulfidptosis (DRGs) was initially gathered from previously published articles. Through an intersection analysis with the differential genes in the TCGA-OC database, these 14 DRGs were found to be present in both OC samples and normal tissues. Furthermore, these genes were differentially expressed and were linked to the OC patient's prognosis. Consensus classification manifested that OC patients could be classified into two distinct subtypes depending on these DRG expression levels, showing significant variations in immune cell infiltration between the two clusters. Based on this characteristic, the patients were categorized into the HRG and LRG. Our outcomes validate the ability of the risk score to be an autonomous determinant for predicting the outcome of OC. The HRG exhibited a notable enrichment in various KEGG signaling pathways. Moreover, significant variations in immune infiltration were found between the HRG and LRG. The high-risk patients displayed indications of immunosuppression within the tumor microenvironment, leading to an unfavorable prognosis. Conversely, the LRG may benefit more from immunotherapy. Consequently, this model offers an improved means of assessing the prognosis of OC, which can assist in guiding the clinical treatment of OC patients. Finally, the efficacy of chemotherapy and immunotherapy for OC patients was also predicted based on this feature.
Recent investigations have manifested that cancer progression is impacted by various molecular channels and mechanisms, with intricate internal interactions among different molecules and signaling pathways [[37], [38], [39]]. Therefore, multi-gene signatures have been investigated to predict cancer prognosis prediction, presenting reliable and accurate results [[40], [41], [42], [43]]. This study has identified two distinct subtypes of OC on the basis of the expression levels of 14 genes associated with disulfidptosis. Notably, these two subtypes demonstrated a significant difference in prognosis. To investigate the underlying causes, GSVA and ssGSEA enrichment analyses were performed. The findings clearly indicated that cluster B was enriched in diverse signaling pathways, including oxidative phosphorylation, focal adhesion, actin cytoskeleton, regulation of ERBB signaling pathways, neurotrophic factor signaling pathways, adhesion molecule junctions, and other signaling pathways. Furthermore, GO analysis further revealed the enrichment of cluster B in cell adhesion, regulation of actin cytoskeleton, and the PI3K-Akt signaling pathway. Given that disulfidptosis induces cancer cell death by interfering with the actin cytoskeleton, the above enrichment outcomes emerged, and the clustering model based on DRGs effectively distinguished different subtypes of OC. Additionally, our findings revealed that ACTB and FLNA had the highest expression levels in cluster B. Previous studies have reported the close association of both ACTB [44,45] and FLNA [46,47] with the progression of various cancers. ACTB, or beta-actin, is a prevalent and strongly preserved protein that forms part of the cytoskeleton structure in all eukaryotic cells. ACTB may undergo fast assembly and disassembly to create filaments in response to cellular needs. These filaments play a crucial role in many biological processes, including cell migration and division [48]. While ACTB has traditionally been regarded as a housekeeping gene [49], recent research has revealed notable variations in ACTB expression levels under specific cancer conditions. Notably, upregulated levels of ACTB have been observed in lung and prostate cancer; in HHC, significantly elevated ACTB expression has been associated with more aggressive tumor characteristics. In the context of invasive melanoma cells, the mRNA expression of ACTB was observed to be twice as high as that in non-invasive melanoma cells [50]. Dysregulation of ACTB protein has also been noted in renal cancer cells and tissues, although further validation is required to confirm this finding [51]. Additionally, a comparative genetics study revealed that ACTB was significantly upregulated in serous OC specimens during the screening for potential prognostic molecular markers against OC [52]. FLNA, a regulator of the cytoskeleton, has been widely investigated, and numerous reports have demonstrated its role as a tumor-promoting protein in cancer progression [53]. For instance, Leung et al. conducted proteomic investigations and discovered a significantly elevated expression of FLNA in carboplatin-resistant OVCAR5 cells compared to carboplatin-sensitive OVCAR5 cells. Additionally, OC patients with high FLNA expression demonstrated a poorer prognosis [54]. Nonetheless, FLNA also showed the potential to inhibit tumor progression under specific circumstances [53]. Notably, in melanoma, the full-length nuclear variant of FLNA was found to suppress the activated transcription of FOXC1 [55]. In this study, OC patients with high FLNA expression exhibited a more favorable prognosis, implying the potential role of FLNA in OC patients.
Research on various cancers revealed a correlation between cell death and the immune system, exemplified by ferroptosis, cuproptosis, and other mechanisms [[40], [41], [42], [43]]. Consequently, this investigation aimed to examine the connection between disulfidptosis and the immune system in OC to establish a theoretical foundation for clinical immunotherapy. Our findings revealed that the HRG exhibits significantly elevated stromal, immune, and estimated scores in comparison to the LRG, suggesting a distinct connection between the HRG and infiltration of immune cells. Subsequently, a correlation analysis was performed on B and CD4+ T cells, indicating that the HRG exhibited higher immune infiltration of these cells. Disulfidptosis may have an impact on the function and activation of B cells and CD4+ T cells in OC patients. Notably, this study is the first to establish a connection between disulfidptosis and OC prognosis, thereby offering novel insights into the molecular mechanisms of OC. Nevertheless, the limitations of our study should be acknowledged. Firstly, a substantial quantity of OC samples is required to validate the stability of the typing. Secondly, further experimental verification is necessary to confirm the relationship between disulfidptosis and immunity. Thirdly, conducting overexpression and deletion studies on specific genes would enhance the persuasiveness of our results. Lastly, the mechanism underlying the prognostic genes in OC progression and experimentation should be explored using clinical tissue samples.
5. Conclusions
In conclusion, a robust correlation exists between the prognosis of OC patients and a signature comprising fourteen genes associated with disulfidptosis. This signature holds the potential to offer risk stratification value, making the fourteen genes viable candidates as prognostic biomarkers in OC patients. Consequently, these biomarkers can aid in disulfidptosis-targeted drug therapy, facilitate individualized therapeutic decision-making, and enhance follow-up management.
Funding sources
The Nature Science Foundation of Zhejiang Province (LY23H160015).
Data availability statement
The dataset used in this study, the GSE53963 dataset, was downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).
CRediT authorship contribution statement
Miaojia Jin: Conceptualization. Dan Ni: Data curation. Jianshu Cai: Formal analysis. Jianhua Yang: Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would also like to thank Home for Researchers (www.home-for-researchers.com) for this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e32273.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Hong S., Fu N., Sang S., Ma X., Sun F., Zhang X. Identification and validation of irf6 related to ovarian cancer and biological function and prognostic value. J. Ovarian Res. 2024;17(1):64. doi: 10.1186/s13048-024-01386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Pineros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: globocan sources and methods. Int. J. Cancer. 2019;144(8):1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 3.Lee E., Lokman N.A., Oehler M.K., Ricciardelli C., Grutzner F. A comprehensive molecular and clinical analysis of the pirna pathway genes in ovarian cancer. Cancers. 2020;13(1) doi: 10.3390/cancers13010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akashi K., Nagashima Y., Tabata T., Oda H. Immunochemical analysis of iron transporters and m2 macrophages in ovarian endometrioma and clear cell adenocarcinoma. Mol Clin Oncol. 2021;15(2):159. doi: 10.3892/mco.2021.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spyrou G., Appelgren D., Rosen A., Ingelsson B. Sizing up extracellular DNA: instant chromatin discharge from cells when placed in serum-free conditions. Front. Cell Dev. Biol. 2020;8(634) doi: 10.3389/fcell.2020.00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong L., Huang D., Shi Y., Liang Z., Bu H. Regulated cell death in cancer: from pathogenesis to treatment. Chin Med J (Engl) 2023;136(6):653–665. doi: 10.1097/CM9.0000000000002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Z.S., Wu Y., Zhao H.G., Liu C.X., Cai H.Y., Guo B.Z., Xie Y.A., Shi H.R. Association between the rs11614913 variant of mirna-196a-2 and the risk of epithelial ovarian cancer. Oncol. Lett. 2016;11(1):194–200. doi: 10.3892/ol.2015.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., Morrison B. 3rd, Stockwell BR: ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin C.S., Mishra P., Watrous J.D., Carelli V., D'Aurelio M., Jain M., Chan D.C. The glutamate/cystine xct antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat. Commun. 2017;8 doi: 10.1038/ncomms15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koppula P., Zhang Y., Shi J., Li W., Gan B. The glutamate/cystine antiporter slc7a11/xct enhances cancer cell dependency on glucose by exporting glutamate. J. Biol. Chem. 2017;292(34):14240–14249. doi: 10.1074/jbc.M117.798405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goji T., Takahara K., Negishi M., Katoh H. Cystine uptake through the cystine/glutamate antiporter xct triggers glioblastoma cell death under glucose deprivation. J. Biol. Chem. 2017;292(48):19721–19732. doi: 10.1074/jbc.M117.814392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Olszewski K., Zhang Y., Lim E.W., Shi J., Zhang X., Zhang J., Lee H., Koppula P., Lei G., Zhuang L., You M.J., Fang B., Li W., Metallo C.M., Poyurovsky M.V., Gan B. Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat. Cell Biol. 2020;22(4):476–486. doi: 10.1038/s41556-020-0496-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang K., Li X., Peng Y., Zhou Y. Comprehensive analysis of disulfidptosis-related lncrnas in molecular classification, immune microenvironment characterization and prognosis of gastric cancer. Biomedicines. 2023;11(12) doi: 10.3390/biomedicines11123165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Zhou Q., Wu Z., Lu X. Identification of novel key genes associated with uterine corpus endometrial carcinoma progression and prognosis. Ann. Transl. Med. 2023;11(2):100. doi: 10.21037/atm-22-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh C.T., Liao G.Y., Emura T. Sensitivity analysis for survival prognostic prediction with gene selection: a copula method for dependent censoring. Biomedicines. 2023;11(3) doi: 10.3390/biomedicines11030797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emura T., Matsui S., Chen H.Y. Compound.Cox: univariate feature selection and compound covariate for predicting survival. Comput Methods Programs Biomed. 2019;168:21–37. doi: 10.1016/j.cmpb.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Wilkerson M.D., Hayes D.N. Consensusclusterplus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanzelmann S., Castelo R., Guinney J. Gsva: gene set variation analysis for microarray and rna-seq data. BMC Bioinf. 2013;14(7) doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., Nordgren H., Farmer P., Praz V., Haibe-Kains B., Desmedt C., Larsimont D., Cardoso F., Peterse H., Nuyten D., Buyse M., Van de Vijver M.J., Bergh J., Piccart M., Delorenzi M. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 20.Zeng D., Li M., Zhou R., Zhang J., Sun H., Shi M., Bin J., Liao Y., Rao J., Liao W. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol. Res. 2019;7(5):737–750. doi: 10.1158/2326-6066.CIR-18-0436. [DOI] [PubMed] [Google Scholar]
- 21.Odunsi K. Immunotherapy in ovarian cancer. Ann. Oncol. 2017;28(suppl_8):viii1–viii7. doi: 10.1093/annonc/mdx444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaFargue C.J., Dal Molin G.Z., Sood A.K., Coleman R.L. Exploring and comparing adverse events between parp inhibitors. Lancet Oncol. 2019;20(1):e15–e28. doi: 10.1016/S1470-2045(18)30786-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bray F., Ren J.S., Masuyer E., Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int. J. Cancer. 2013;132(5):1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 24.Ferlay J., Steliarova-Foucher E., Lortet-Tieulent J., Rosso S., Coebergh J.W., Comber H., Forman D., Bray F. Cancer incidence and mortality patterns in europe: estimates for 40 countries in 2012. Eur. J. Cancer. 2013;49(6):1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 25.Siegel R., Naishadham D., Jemal A. Cancer statistics. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. 2013. [DOI] [PubMed] [Google Scholar]
- 26.Eisenhauer E.A. Real-world evidence in the treatment of ovarian cancer. Ann. Oncol. 2017;28(suppl_8):viii61–viii65. doi: 10.1093/annonc/mdx443. [DOI] [PubMed] [Google Scholar]
- 27.Kurta M.L., Edwards R.P., Moysich K.B., McDonough K., Bertolet M., Weissfeld J.L., Catov J.M., Modugno F., Bunker C.H., Ness R.B., Diergaarde B. Prognosis and conditional disease-free survival among patients with ovarian cancer. J. Clin. Oncol. 2014;32(36):4102–4112. doi: 10.1200/JCO.2014.55.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olbrecht S., Busschaert P., Qian J., Vanderstichele A., Loverix L., Van Gorp T., Van Nieuwenhuysen E., Han S., Van den Broeck A., Coosemans A., Van Rompuy A.S., Lambrechts D., Vergote I. High-grade serous tubo-ovarian cancer refined with single-cell rna sequencing: specific cell subtypes influence survival and determine molecular subtype classification. Genome Med. 2021;13(1):111. doi: 10.1186/s13073-021-00922-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kossai M., Leary A., Scoazec J.Y., Genestie C. Ovarian cancer: a heterogeneous disease. Pathobiology. 2018;85(1–2):41–49. doi: 10.1159/000479006. [DOI] [PubMed] [Google Scholar]
- 30.Cook D.P., Vanderhyden B.C. Ovarian cancer and the evolution of subtype classifications using transcriptional profilingdagger. Biol. Reprod. 2019;101(3):645–658. doi: 10.1093/biolre/ioz099. [DOI] [PubMed] [Google Scholar]
- 31.Shen L., Fei X., Zhou Y., Wang J., Zhu Y., Zhuang Y. The effect of felt trust from patients among nurses on attitudes towards nursing service delivery. J. Adv. Nurs. 2022;78(2):404–413. doi: 10.1111/jan.14973. [DOI] [PubMed] [Google Scholar]
- 32.Zeng Y., Guan Q., Su Y., Huang Q., Zhao J., Wu M., Guo Q., Lyu Q., Zhuang Y., Cheng A.S. A self-administered immersive virtual reality tool for assessing cognitive impairment in patients with cancer. Asia Pac J Oncol Nurs. 2023;10(3) doi: 10.1016/j.apjon.2023.100205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Nie L., Zhang Y., Yan Y., Wang C., Colic M., Olszewski K., Horbath A., Chen X., Lei G., Mao C., Wu S., Zhuang L., Poyurovsky M.V., James You M., Hart T., Billadeau D.D., Chen J., Gan B. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat. Cell Biol. 2023;25(3):404–414. doi: 10.1038/s41556-023-01091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao R., Peng C., Song C., Zhao Q., Rong J., Wang H., Ding W., Wang F., Xie Y. Bicc1 as a novel prognostic biomarker in gastric cancer correlating with immune infiltrates. Int Immunopharmacol. 2020;87 doi: 10.1016/j.intimp.2020.106828. [DOI] [PubMed] [Google Scholar]
- 35.Dong C., Dang D., Zhao X., Wang Y., Wang Z., Zhang C. Integrative characterization of the role of il27 in melanoma using bioinformatics analysis. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.713001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng M., Sun S., Zhao R., Guan R., Zhang Z., Li S., Wei W., Guo R. The pyroptosis-related gene signature predicts prognosis and indicates immune activity in hepatocellular carcinoma. Mol Med. 2022;28(1):16. doi: 10.1186/s10020-022-00445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C., Liu N. Ferroptosis, necroptosis, and pyroptosis in the occurrence and development of ovarian cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.920059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian W., Lei N., Zhou J., Chen M., Guo R., Qin B., Li Y., Chang L. Extracellular vesicles in ovarian cancer chemoresistance, metastasis, and immune evasion. Cell Death Dis. 2022;13(1):64. doi: 10.1038/s41419-022-04510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong-Rolle A., Wei H.K., Zhao C., Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell. 2021;12(5):426–435. doi: 10.1007/s13238-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng P., Li N., Zhan X. Ovarian cancer subtypes based on the regulatory genes of rna modifications: novel prediction model of prognosis. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.972341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang L., Yu J., Li J., Li N., Liu J., Xiu L., Zeng J., Wang T., Wu L. Integration of scrna-seq and bulk rna-seq to analyse the heterogeneity of ovarian cancer immune cells and establish a molecular risk model. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.711020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang L., Li J., Yu J., Liu J., Xiu L., Zeng J., Wang T., Li N., Wu L. Establishment and validation of a novel invasion-related gene signature for predicting the prognosis of ovarian cancer. Cancer Cell Int. 2022;22(1):118. doi: 10.1186/s12935-022-02502-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang D., Lu W., Cui S., Mei H., Wu X., Zhuo Z. Establishment of an ovarian cancer omentum metastasis-related prognostic model by integrated analysis of scrna-seq and bulk rna-seq. J. Ovarian Res. 2022;15(1):123. doi: 10.1186/s13048-022-01059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Y., Tang S., Wang Z., Cai L., Lian H., Shen Y., Zhou Y. A pan-cancer analysis of the prognostic and immunological role of beta-actin (actb) in human cancers. Bioengineered. 2021;12(1):6166–6185. doi: 10.1080/21655979.2021.1973220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo C., Liu S., Wang J., Sun M.Z., Greenaway F.T. Actb in cancer. Clin. Chim. Acta. 2013;417:39–44. doi: 10.1016/j.cca.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Cheng L., Tong Q. Interaction of flna and anxa2 promotes gefitinib resistance by activating the wnt pathway in non-small-cell lung cancer. Mol. Cell. Biochem. 2021;476(10):3563–3575. doi: 10.1007/s11010-021-04179-1. [DOI] [PubMed] [Google Scholar]
- 47.Wang A., Liu L., Yuan M., Han S., You X., Zhang H., Lei F., Zhang Y. Role and mechanism of flna and ucp2 in the development of cervical cancer. Oncol. Rep. 2020;44(6):2656–2668. doi: 10.3892/or.2020.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuvertino S., Stuart H.M., Chandler K.E., Roberts N.A., Armstrong R., Bernardini L., Bhaskar S., Callewaert B., Clayton-Smith J., Davalillo C.H., Deshpande C., Devriendt K., Digilio M.C., Dixit A., Edwards M., Friedman J.M., Gonzalez-Meneses A., Joss S., Kerr B., Lampe A.K., Langlois S., Lennon R., Loget P., Ma D.Y.T., McGowan R., Des Medt M., O'Sullivan J., Odent S., Parker M.J., Pebrel-Richard C., Petit F., Stark Z., Stockler-Ipsiroglu S., Tinschert S., Vasudevan P., Villa O., White S.M., Zahir F.R., Study D.D.D., Woolf A.S., Banka S. Actb loss-of-function mutations result in a pleiotropic developmental disorder. Am. J. Hum. Genet. 2017;101(6):1021–1033. doi: 10.1016/j.ajhg.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song R., He S., Wu Y., Chen W., Song J., Zhu Y., Chen H., Wang Q., Wang S., Tan S., Tan S. Validation of reference genes for the normalization of the rt-qpcr in peripheral blood mononuclear cells of septic patients. Heliyon. 2023;9(4) doi: 10.1016/j.heliyon.2023.e15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goidin D., Mamessier A., Staquet M.J., Schmitt D., Berthier-Vergnes O. Ribosomal 18s rna prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mrna levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 2001;295(1):17–21. doi: 10.1006/abio.2001.5171. [DOI] [PubMed] [Google Scholar]
- 51.Ferguson R.E., Carroll H.P., Harris A., Maher E.R., Selby P.J., Banks R.E. Housekeeping proteins: a preliminary study illustrating some limitations as useful references in protein expression studies. Proteomics. 2005;5(2):566–571. doi: 10.1002/pmic.200400941. [DOI] [PubMed] [Google Scholar]
- 52.Zadeh Fakhar H.B., Zali H., Rezaie-Tavirani M., Darkhaneh R.F., Babaabasi B. Proteome profiling of low grade serous ovarian cancer. J. Ovarian Res. 2019;12(1):64. doi: 10.1186/s13048-019-0535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savoy R.M., Ghosh P.M. The dual role of filamin a in cancer: can't live with (too much of) it, can't live without it. Endocr. Relat. Cancer. 2013;20(6):R341–R356. doi: 10.1530/ERC-13-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leung D., Price Z.K., Lokman N.A., Wang W., Goonetilleke L., Kadife E., Oehler M.K., Ricciardelli C., Kannourakis G., Ahmed N. Platinum-resistance in epithelial ovarian cancer: an interplay of epithelial-mesenchymal transition interlinked with reprogrammed metabolism. J. Transl. Med. 2022;20(1):556. doi: 10.1186/s12967-022-03776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berry F.B., O'Neill M.A., Coca-Prados M., Walter M.A. Foxc1 transcriptional regulatory activity is impaired by pbx1 in a filamin a-mediated manner. Mol. Cell Biol. 2005;25(4):1415–1424. doi: 10.1128/MCB.25.4.1415-1424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset used in this study, the GSE53963 dataset, was downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).