Despite significant advances, outcomes for children with Down syndrome (DS, trisomy 21) who develop acute lymphoblastic leukemia (ALL) remain poor. Reports of large DS-ALL cohorts have shown that children with DS have inferior event-free survival (EFS) and overall survival (OS) compared to children without DS.1-3
Children with DS also exhibit increased treatment-related mortality due to infections and toxicities following chemotherapy, higher cumulative risk of relapse and inferior outcomes following relapse.4 This situation highlights the dire need for the development of more potent and targeted therapies to improve the survival and quality of care for these vulnerable children, who often have additional co-morbidities linked to trisomy 21 that complicates their clinical management. Targeted approaches and immunotherapies have shown promising results for pediatric leukemia.5 Hence, the development of new models of DS-ALL are needed to rapidly advance drug discovery and refine existing treatment strategies. We recently established DS-ALL patient-derived xenografts (PDX) and demonstrated that targeting somatic alterations found in DS-ALL using MEK inhibitors combined with conventional treatment has the potential to improve outcomes for these children.6 Targeting the dosage-sensitive mechanisms resulting from the extra copy of chromosome 21 is also an area of intense investigation.7-9 As such, inhibition of the chromosome 21 kinase DYRK1A using EHT1610 or of its direct targets FOXO1 and STAT3, has shown promising cytotoxic effects both in vitro and in vivo.8
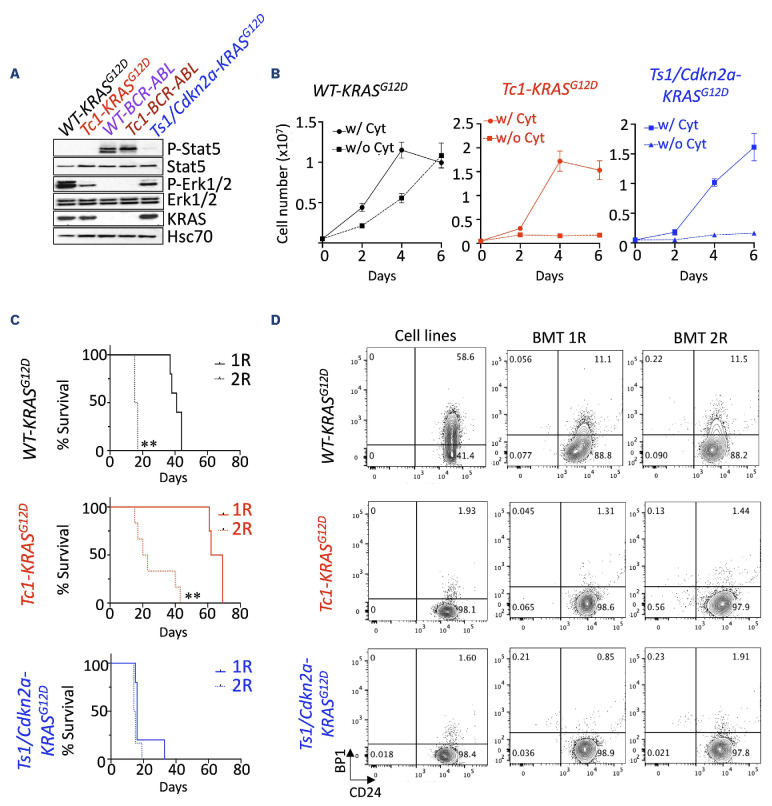
In this study, we developed novel clinically relevant models of DS-ALL to facilitate the assessment of new therapeutic agents. First, we modeled oncogenic cooperation seen in DS-ALL in vitro.10 To this end, we transduced wild-type (WT) and trisomic (Tc1) 8-10-week-old bone marrow cells with retroviruses encoding the frequently observed mutant KRASG12D or the more rarely seen BCR-ABL fusion11 (animal experiments were approved by institutional ethics committee and followed Australian guidelines for the care and use of animals). Of note, the bone marrow stroma and hematopoietic stem cell and progenitor compartment composition did not significantly differ between disomic and trisomic mice, except from an increased proportion of multipotent progenitors MPP2 at the expense of the less committed MPP1 (Online Supplementary Figure S1A-C). Trisomic Tc1 progenitors exhibited increased capacity to form colony-forming unit (CFU) pre-B colonies in vitro compared to WT (Online Supplementary Figure S1D), as seen previously in the partially trisomic Ts1Rhr (Ts1) model.7 Ectopic expression of KRASG12D or BCR-ABL enhanced the number and replating capacity of both WT and Tc1 CFU pre-B colonies compared to the empty vector MIC (MSCV-IRES-mCherry) (Online Supplementary Figure S1E, F). Next, we established two murine DS-ALL cell lines (Tc1-KRASG12D and Tc1-BCR-ABL), disomic controls (WT-KRASG12D and WT-BCR-ABL), as well as an independent Ts1/Cdkn2a-KRASG12D cell line (established from triple transgenic Ts1Rhr, Mb1-Cre, Cdkn2afl/fl donor mice); attempts to develop CRLF2-rearranged/JAK2 mutant murine DS-ALL cell lines were unsuccessful. Interestingly, although KRASG12D led to a constitutive phosphorylation of Erk1/2, the highest levels of Erk1/2 phosphorylation and cytokine independence for cell proliferation were only observed in WT-KRASG12D cells (Figure 1A, B). All KRASG12D-expressing murine cell lines engrafted in sub-lethally irradiated C57Bl/6J mice in primary and secondary recipients and ultimately succumbed to leukemia with complete penetrance, with recipients displaying mCherry-positive cells in the peripheral blood, bone marrow, and in the spleen (data not shown). Engrafted recipient mice exhibited splenomegaly associated with a leukemia-driven disorganized architecture (Online Supplementary Figure S1G, H). We also confirmed that the engrafted cell lines exhibit a phenotype similar to the cell lines cultured in vitro; although we noted clonal selection of a CD24+/BP1- pro-B population from the WT-KRASG12D cells in vivo, which was retained in secondary recipients (Figure 1C, D; and data not shown). Altogether, we developed novel murine models of DS-ALL, providing a unique platform suitable for testing targeted therapies.
Figure 1.
Establishment of novel Down syndrome acute lymphoblastic leukemia models for preclinical testing. (A) Constitutive phosphorylation of Stat5 and Erk1/2 in KRASG12D and BCR-ABL-expressing murine cells (starved for 6 hours). (B) Growth of murine wild-type (WT)-KRASG12D, Tc1-KRASG12D and Ts1/Cdkn2a-KRASG12D cells with or without cytokines (Il-7, Scf and Flt3-L, 10 ng/mL) over 6 days. (C) Kaplan-Meier analysis comparing survival of primary (1R) and secondary (2R) sub-lethally irradiated recipient mice engrafted with 1-2x106 Tc1-KRASG12D (1R N=4, 2R N=6), WT-KRASG12D (1R N=5, 2R N=6) and Ts1Rhr/Cdkn2a-KRASG12D (1R N=5, 2R N=6) cell lines; **P<0.01. (D) Phenotype of the murine cell lines assessing surface expression of BP1 and CD24 (phenotype of the WT-BCR-ABL and Tc1-BCR-ABL cell lines are in the Online Supplementary Figure 1F), and representative flow plots showing phenotype of mCherry-positive cell lines in primary and secondary recipients. w/: with; w/o: without. Cyt: cytokine.
In order to validate the clinical relevance of these murine cells, we focused on the therapeutically targetable chromosome 21 kinase DYRK1A, as recent reports have emphasized its role in childhood leukemia, regardless of Down syndrome.8 First, we used short hairpin RNA (shRNA) interference to show that all five cell lines, disomic (N=2) or trisomic (N=3) and expressing either KRASG12D or BCR-ABL oncogenes, were sensitive to Dyrk1a knock-down (KD) (Figure 2A, B; Online Supplementary Figure S1I). We next assessed the efficacy of new potent DYRK1A inhibitors using a DYRK1A-focused library which included EHT1610 used as control, Leucettinib-21 and its inactive isomer (compounds inspired by Leucettines and Leucettamine B, a natural substance produced by the marine sponge Leucetta microraphis),12,13 and three additional DYRK1A inhibitors whose chemical structure is based on the 7-azaindole scaffold, AM28, AM30 and AM45.14 In dose-response experiments, we showed that AM30 and Leucettinib-21 were cytotoxic in both WT-KRASG12D and Tc1-KRASG12D cell lines (Figure 2C). We also observed that Leucettinib-21, AM30 and AM45 were more potent than EHT1610 in decreasing cellular growth in all cell lines tested, and that Tc1-KRASG12D cells always exhibited lower half-maximal inhibitory concentration (IC50) values than its disomic counterpart WT-KRASG12D (Figure 2D, E; Online Supplementary Figure S2A).
Figure 2.
Genetic and pharmacological inhibition of DYRK1A decreases growth of Down syndrome acute lymphoblastic leukemia cells. (A) Ratio of wild-type (WT)-KRASG12D and Tc1-KRASG12D transduced with GFP-expressing Banshee vectors encoding two shDyrk1a compared to empty Banshee-U6 counterparts over 9 days (N=4 replicates); ***P<0.001. (B) Validation of Dyrk1a knockdown at the protein level 48 hours after transduction (GFP-sorted). Dyrk1a band intensities were quantified and normalized as a ratio of shDyrk1a-transduced to control U6-transduced WT-KRASG12D cells. (C) Cytotoxic effect of increasing doses (in μM) of the DYRK1A inhibitors EHT1610, AM30, Leucettinib-21 (LCTB-21) and its inactive isomer iso-Leucettinib-21 (Iso LCTB-21) at 48 hours in murine WT-KRASG12D and Tc1-KRASG12D cells assessed by flow cytometry (Annexin V staining). (D) Dose-response curves assessing efficacy of EHT1610, LCTB-21 and Iso LCTB-21 at 72 hours by alamarBlue cell viability assay in murine WT-KRASG12D and Tc1-KRASG12D cells. (E) Heatmap integrating relative half-maximal inhibitory concentration (IC50) values obtained for the DYRK1A inhibitors tested in our murine cell lines.
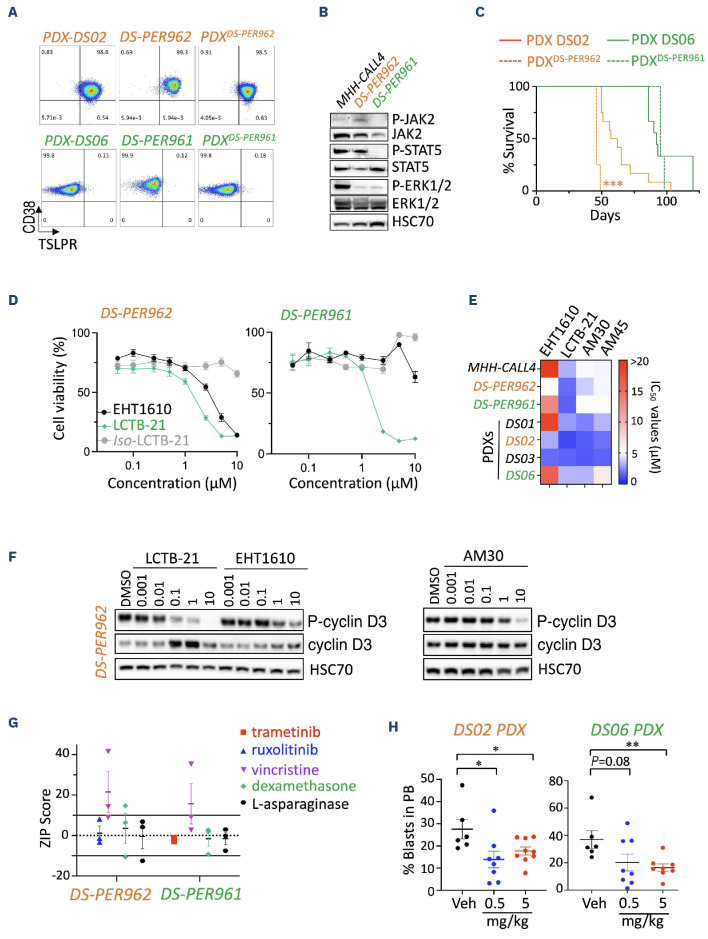
In order to expand our observations, we next established two human DS-ALL cell lines: DS-PER961 and DS-PER962 from our previously reported DS06 (KRASG12S-positive) and DS02 (CRLF2-rearranged/JAK2I682F-positive) PDX.6 These unique models represent the first human cell lines for DS-ALL, with comprehensive characterization at the genomic, transcriptomic and phenotypic levels, confirming their resemblance to the PDX models from which they originated (Figure 3A; Online Supplementary Figure S3A-E). Western blot analyses confirmed phosphorylation of ERK1/2 and STAT5 downstream of KRASG12S and JAK2I682F mutants in DS-PER961 and DS-PER962 respectively (Figure 3B). Using NOD-SCID-γc-/- (NSG) mice, we also showed that both DS-ALL cell lines engrafted into immunocompromised recipients with DS-PER962 cells being more aggressive than their PDX counterparts (Figure 3C). Using these human cells, we confirmed that Leucettinib-21, AM30 and AM45 were potent inhibitors in human DS-ALL cells and in MHH-CALL4 (a non-DS CRLF2-rearranged and JAK2 mutant ALL cell line known to be sensitive to DYRK1A inhibition) (Figure 3D, E; Online Supplementary Figure S2B-D).8 No significant effect was seen for AM28 nor for the inactive isomer of Leucettinib-21 (Figure 3D; Online Supplementary Figure S2B, C). Importantly, efficacy of these DYRK1A inhibitors was demonstrated in DS-ALL blasts freshly harvested from four DS-ALL PDX models (described in 6), validating the suitability of our DS-ALL cells to assess efficacy of new therapies (Figure 3E). Compared to EHT1610 and AM30, Leucettinib-21 was the most potent compound in inhibiting phosphorylation of the known DYR-K1A target cyclin D3 in a dose-dependent manner shown in DS-PER962 (Figure 3F), DS-PER961 and in murine cells (Online Supplementary Figure S2E), but had limited effect on FOXO1-phosphorylation (Online Supplementary Figure S2F-I). Next, we evaluated in vitro drug combinations of Leucettinib-21 with targeted and conventional therapies and identified synergy with vincristine and an additive effect with dexamethasone and L-asparaginase (Figure 3G; Online Supplementary Figure S2J). An additive effect between Leucettinib-21 and the targeted therapies ruxolitinib (a JAK1/2 inhibitor) and trametinib (a MEK1/2 inhibitor) was also seen in DS-PER962 (CRLF2-positive/JAK2I682F mutant) and in the DS-PER961 (KRASG12S) cell lines, respectively (Figure 3G). Finally, we assessed the efficacy of Leucettinib-21 in vivo in the DS06 and DS02 PDX models and observed that in vivo treatment with Leucettinib-21 decreased leukemia burden but did not fully eradicate leukemia (Figure 3H). Together, this data demonstrates the suitability and clinical relevance of the novel murine and human models we have established and emphasizes the key role of DYRK1A in DS-ALL.
Figure 3.
Efficacy of DYRK1A inhibition in novel human Down syndrome acute lymphoblastic leukemia cell lines and patient-derived xenograft models. (A) Representative flow plots assessing CD38 and TSLPR expression of the human DS-PER962 and DS-PER961 Down syndrome acute lymphoblastic leukemia (DS-ALL) cell lines compared to their corresponding DS06 and DS02 patient-derived xenografts (PDX), and to NSG recipients engrafted with 1x106 DS-ALL cell lines (PDXcells). (B) Western blots assessing the constitutive phosphorylation of JAK2, STAT5 and ERK1/2 in DS-PER962 and DS-PER961 cell lines following a 6-hour starvation; the non-DS MHH-CALL4 (CRLF2-rearranged/JAK2 mutant) ALL cell line was used as control. (C) Kaplan-Meier curves comparing the survival of the DS-PER962 and DS-PER961 PDX (N=3-4) to their corresponding DS02 and DS06 PDX (N=9-12); ***P<0.001. (D) Efficacy of EHT1610, Leucettinib-21 (LCTB-21) and its inactive isomer iso-Leucettinib-21 (Iso LCTB-21) in the human DS-PER962 and DS-PER961 cell lines (72 hours). (E) Heatmap representing the relative half-maximal inhibitory concentration (IC50) values obtained in the non-DS-ALL, DS-ALL human cell lines and DS-ALL cells freshly harvested from PDX models. (F) Western blot comparing the effect of EHT1610, LCTB-21 and AM30 on phospho-cyclin D3 and total cyclin D3 stability after 6 hours of treatment in the DS-PER962 cell line. (G) ZIP scores obtained from combining Leucettinib-21 (10 doses from 0.01 to 10 μM) with trametinib (0.0001-20 μM), ruxolitinib (0.0001-20 μM), vincristine (0.0005-0.5 μM), dexamethasone (0.00025-20 μM) and L-asparaginase (0.1-20 μM) in the human DS-ALL cell lines. ZIP scores <-10 =antagonism; -10 to 10 =additive; >10 =synergy. (H) In vivo effect of Leucettinib-21 on leukemia burden in the peripheral blood (PB) of the DS02 and DS06 PDX models (oral gavage, 2 weeks and 4 weeks of treatment respectively); *P<0.05, **P<0.01. Veh: vehicle.
Compared to other children, higher sensitivity to treatment-related toxicity in children with DS-ALL remains a major clinical challenge. This has significantly limited the development of novel targeted therapies for this patient population, ultimately delaying translation into clinical trials. Recently, integration of immunotherapeutic approaches has offered promise in reducing toxicity. Indeed, Laetsch et al. reported comparable outcomes between DS- and non-DS children with relapsed/refractory B-ALL who received CD19-directed chimeric antigen receptor T-cell therapy, including similar rates of CD19-negative relapse,15 and an ongoing clinical trial is currently assessing whether blinatumomab can replace two blocks of consolidation chemotherapy for treatment of de novo DS-ALL (clinicaltrial gov. Identifier: NCT03911128). As an alternative approach, identifying key vulnerabilities in DS-ALL blasts could provide the molecular basis for development of novel targeted therapies. While resources such as the recent in-depth characterization of the genetic landscape of DS-ALL can provide such insight,10 we have developed novel models of DS-ALL and shown that inhibiting dosage-sensitive mechanisms altered by trisomy 21 may also represent a new avenue to integrate agents with low toxicity and ultimately improve outcomes and quality of care for children with DS-ALL. We demonstrated that a reduction in DYRK1A expression is sufficient to decrease the growth of DS-ALL cell lines, confirmed the sensitivity of human and murine cells to DYRK1A inhibition and showed that the leading candidate, Leucettinib-21, potentiates the cytotoxic effect of other chemotherapeutic and targeted agents and delayed leukemia expansion in vivo, with no detectable toxicity identified in the peripheral blood (Online Supplementary Figure S2K). Leucettinib-21 has recently completed regulatory preclinical safety studies and is primed for early phase clinical assessment. Strikingly, studies have demonstrated the preclinical impact of targeting DYRK1A activity in different subtypes of childhood leukemia,8 further emphasizing the potential benefit for investigating Leucettinib-21 or other new potent DYRK1A inhibitors in clinical trials for ALL.
Altogether, this study has established and comprehensively characterized the first DS-ALL cell lines, providing suitable and clinically relevant cellular models to identify new molecular weaknesses in DS-ALL and test the efficacy of novel targeted therapies (as exemplified here with DYRK1A inhibition), alone or in combination with standard of care, to ultimately develop new, less toxic treatments to improve the outcome for children with DS-ALL.
Supplementary Material
Acknowledgments
We thank L. Munoz, T Johns, G. Chua, P. Kumar, T Lassmann, and staff members of the Bioresources facility at Telethon Kids Institute for their support in obtaining reagents, processing samples and housing/monitoring animals related to this study.
Funding Statement
Funding: This research was supported by Australian Government Research Training Program (RTP) Scholarships (to SLCS and KP). CAB is supported by a Fellowship from the Jerome Lejeune and Sisley-d’Ornano Foundations. TB is supported by the University of Rouen Normandy, INSA Rouen Normandy, the Centre National de la Recherche Scientifique (CNRS), European Regional Development Fund (ERDF), Labex SynOrg (ANR-11-LABX-0029), Carnot Institute I2C, the XL-Chem Graduate School of Research (ANR-18-EURE-0020 XL CHEM), and by Region Normandie. APM is supported by a postdoctoral research fellowship from the University of Sydney’s Drug Discovery Initiative. MK is a recipient of a National Health and Medical Research Council Principal Research Fellowship (APP1154692). LM is supported by grants from the Jerome Lejeune Foundation, l’Agence Nationale de la Recherche (ANR) (DYRK-DOWN), BpiFrance (i-Nov, vague 9), the European Union’s Horizon 2020 research and innovation programme (Grant #848077) (GO-DS21), and the European Innovation Council (EIC) Accelerator Down-Autonomy project (190138295). SM is supported by a fellowship from the Cancer Council Western Australia (CCWA, Grant #877). This study was supported by project grants from the Child Cancer Research Foundation (RSK, LCC and SM), Cancer Council Western Australia (Grant #1068) and the Jerome Lejeune Foundation (Grant #1806).
Data-sharing statement
RNA-sequencing files are available via the Gene Expression Omnibus (GEO) database under the accession number GSE245056.
References
- 1.Buitenkamp TD, Izraeli S, Zimmermann M, et al. Acute lymphoblastic leukemia in children with Down syndrome: a retrospective analysis from the Ponte di Legno study group. Blood. 2014;123(1):70-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ceppi F, Stephens D, den Hollander BS, et al. Clinical presentation and risk factors of serious infections in children with Down syndrome treated for acute lymphoblastic leukemia. Pediatr Blood Cancer. 2016;63(11):1949-1953. [DOI] [PubMed] [Google Scholar]
- 3.Rabin KR, Devidas M, Chen Z, et al. Outcomes in children, adolescents, and young adults with Down syndrome and ALL: a report from the Children’s Oncology Group. J Clin Oncol. 2024;42(2):218-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyr F, Escherich G, Mann G, et al. Outcomes of treatment for relapsed acute lymphoblastic leukaemia in children with Down syndrome. Br J Haematol. 2013;162(1):98-106. [DOI] [PubMed] [Google Scholar]
- 5.Rafei H, Kantarjian HM, Jabbour EJ. Targeted therapy paves the way for the cure of acute lymphoblastic leukaemia. Br J Haematol. 2020;188(2):207-223. [DOI] [PubMed] [Google Scholar]
- 6.Laurent AP, Siret A, Ignacimouttou C, et al. Constitutive activation of RAS/MAPK pathway cooperates with trisomy 21 and is therapeutically exploitable in Down syndrome B-cell leukemia. Clin Cancer Res. 2020;26(13):3307-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lane AA, Chapuy B, Lin CY, et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat Genet. 2014;46(6):618-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhansali RS, Rammohan M, Lee P, et al. DYRK1A regulates B cell acute lymphoblastic leukemia through phosphorylation of FOXO1 and STAT3. J Clin Invest. 2021;131(1):e135937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Page EC, Heatley SL, Eadie LN, et al. HMGN1 plays a significant role in CRLF2 driven Down Syndrome leukemia and provides a potential therapeutic target in this high-risk cohort. Oncogene. 2022;41(6):797-808. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Chang TC, Junco JJ, et al. Genomic landscape of Down syndrome-associated acute lymphoblastic leukemia. Blood. 2023;142(2):172-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Doherty A, Ruf S, Mulligan C, et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309(5743):2033-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tahtouh T, Durieu E, Villiers B, et al. Structure-activity relationship in the Leucettine family of kinase inhibitors. J Med Chem. 2022;65(2):1396-1417. [DOI] [PubMed] [Google Scholar]
- 13.Lindberg MF, Deau E, Miege F, et al. Chemical, biochemical, cellular, and physiological characterization of Leucettinib-21, a Down syndrome and Alzheimer’s disease drug candidate. J Med Chem. 2023;66(23):15648-15670. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Q, Phoa AF, Abbassi RH, et al. Structural optimization and pharmacological evaluation of inhibitors targeting dual-specificity tyrosine phosphorylation-regulated kinases (DYRK) and CDC-like kinases (CLK) in glioblastoma. J Med Chem. 2017;60(5):2052-2070. [DOI] [PubMed] [Google Scholar]
- 15.Laetsch TW, Maude SL, Balduzzi A, et al. Tisagenlecleucel in pediatric and young adult patients with Down syndrome-associated relapsed/refractory acute lymphoblastic leukemia. Leukemia. 2022;36(6):1508-1515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing files are available via the Gene Expression Omnibus (GEO) database under the accession number GSE245056.