Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma and is characterized by high clinicopathologic heterogeneity.1 This has fueled several efforts at subclassification aiming to identify prognostic and clinically relevant subgroups. Currently, the most widely used classification is based on gene expression profiles, stratifying DLBCL according to the cell-of-origin (COO) into activated B-cell (ABC) and germinal center B-cell (GCB) subtypes.2-4 Patients with ABC-DLBCL have been reported to have less favorable responses to standard therapy than those with GCB-DLBCL. Unfortunately, clinical studies based on this classification have repeatedly failed to improve the outcome of DLBCL patients, suggesting that this simplified scheme is not sufficiently effective for clinical trial design.5 To address this problem, several recent studies have utilized high-resolution genomic analysis to subcategorize patients based on genetic alteration profiles.6-8 This approach has uncovered broader biological heterogeneity than the ABC/GCB paradigm. In a recent National Cancer Institute (NCI) study, observations have been applied to develop a probabilistic classification algorithm, LymphGen, that enables the subclassification of DLBCL into 6 molecularly distinct groups: MCD, BN2, EZB, ST2, A53, and N1, with prognostic correlations.9,10 Biomolecular clusters defined by other studies have overlapped with those of the NCI study.11 However, all large-scale studies have primarily been performed using data derived by whole genome, whole exome sequencing, or a large comprehensive target next-generation sequencing (NGS) panel carried out as part of wider research endeavors. In contrast, in routine clinical practice, sequencing is necessarily targeted to limited sets of genes with a key role for immediate clinical management. In this study, therefore, we aimed to apply the published LymphGen algorithm to a validated clinical NGS assay: the Memorial Sloan Kettering Cancer Center’s (MSKCC) Integrated Mutation Profiling of Actionable Cancer Targets for Hematologic malignancies NGS panel (MSK-IMPACT HEME, herein referred to as IMPACT), which is currently utilized in the management of patients with DLBCL in the clinical setting.
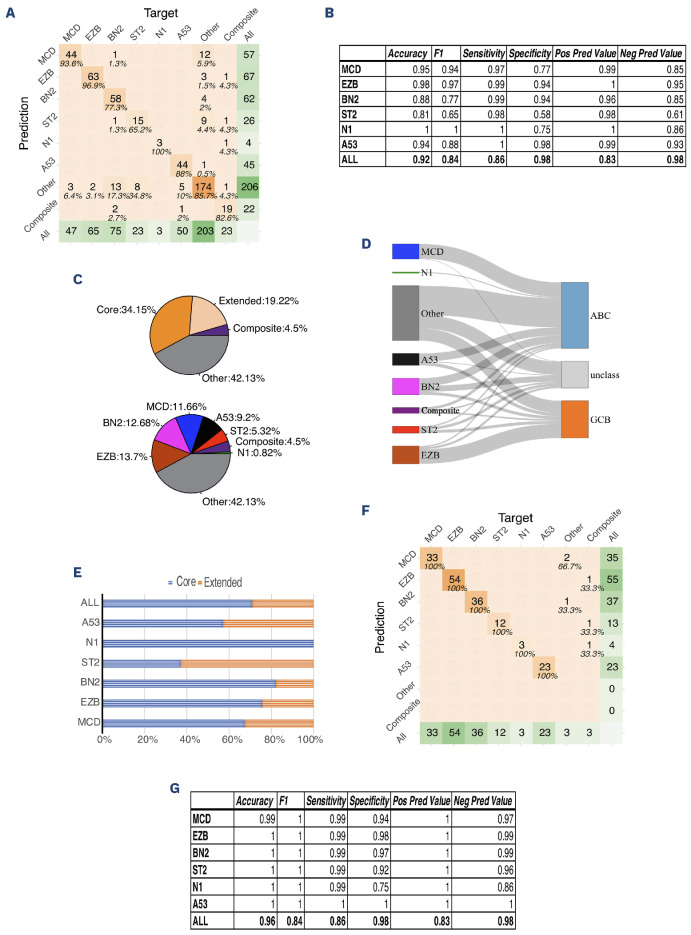
IMPACT is a clinical validated hybridization capture-based assay designed to detect genetic alterations, including single nucleotide variants (SNV), small insertions and deletions (INDEL), and copy number alterations (CNA), implicated in the oncogenesis of hematopoietic malignancies (Online Supplementary Figure S1). The custom designed DNA probes target all protein-coding exons and the adjacent 20 bp of intronic sequence of 400 key oncogenes and tumor suppressor genes. IMPACT uses either saliva or nail clippings as a source of germline DNA to confidently identify somatic mutations in hematologic tumor cells. Details of the methodology and analytical code have been published previously.12,13 Published data from the NCI DLBCL cohort was used to evaluate the performance of LymphGen with only the genes in the IMPACT panel. Applying the LymphGen cluster allocation to the limited set of 400 genes on cases from the NCI cohort by filtering the NCI variant calling data into genes on the IMPACT panel demonstrated an overall accuracy of 92%, with 86% sensitivity and 98% specificity, using cluster allocation based on the original panel as ground truth (Figure 1A, B). In total, 58% of cases were ultimately classified into one of the 6 subtypes (or composite subtypes), which was only marginally below the 63% reported with the use of comprehensive methods, including whole exon, deep amplicon and RNA-sequencing in the original NCI report. The cases that were successfully classified included 11.7% MCD type, 12.7% BN2 type, 13.7% EZB type, 9.2% A53 type, 5.3% ST2 type, and 0.8% N1 type, compared to the original NCI LymphGen study which included 13.9% MCD type, 16.1% BN2 type, 13.2% EZB type, 6.6% A53 type, 4.7% ST2 type, and 2.8% N1 type (Figure 1C). Misclassification primarily encompassed cases deemed as unclassified (“Other”) by either the comprehensive NCI panel or the IMPACT panel, as shown in the confusion matrix (Figure 1A).
Figure 1.
LymphGen classification on National Cancer Institute cohort data filtered by IMPACT panel. (A) Confusion matrix of IMPACT panel compared to National Cancer Institute (NCI) panel (all cases). The target represents the classification by NCI panel; the prediction represents the classification by IMPACT panel. The center number shows the number of cases; the percentage represents the proportion of cases classified by the IMPACT panel for a certain subtype in the cases classified by the NCI panel for the same subtype (considered as ground truth in this comparison). (B) Performance of the classification. (C) Cases from the NCI cohort filtered into the IMPACT panel were classified by LymphGen into 6 subclasses (MCD, EZB, BN2, N1, ST2, A53). The cases with more than one assigned subclass are indicated as “Composite”. The cases that cannot be classified into these subclasses are labeled as “Other”. Regarding categories of the subclassification: tumors with subtype probabilities of >90% or 50-90% were defined as ‘‘Core’’ or ‘‘Extended’’ subtype members, respectively. (D) Correlation of LymphGen classification and cell-of-origin by Han’s algorithm. (E) Composition of “Core” and “Extended” group in each subtypes. (F) Confusion matrix of the IMPACT panel compared to the NCI panel for “Core” predicted group only. The target represents the classification by the NCI panel; the prediction represents the classification by the IMPACT panel. The center number shows the number of cases; the percentage represents the proportion of cases classified by IMPACT for a certain subtype in the cases classified by the NCI panel for the same subtype (considered as ground truth in this comparison). (G) Performance of the IMPACT panel compared to the NCI panel within the “Core” group. ABC: activated B-cell subtype; GCB: germinal center B-cell subtype.
Similar to the NCI study, most cases of EZB subtypes were of the GCB COO; most cases of MCD and N1 subtypes were of ABC COO; and BN2 and A53 subtypes carried both GCB and ABC types, while cases in the ST2 subtype contained more GCB type than ABC type (Figure 1D). In our classification using the genes on the targeted IMPACT panel approach, within 58% of classified cases, 34% of cases were called with high confidence (“Core” group, >90% probability), while 19% of cases were called with lower confidence (“Extended” group, 50-90% probability) (Figure 1C). Compared to the original NCI allocation (47% “Core” and 10% “Extended” group cases), there is a slight reduction in calling confidence from the narrower IMPACT panel. The percentage of cases being classified in the “Core” group varies in subtypes, with the highest in N1 subtype and the lowest in ST2, which represents the confidence of classification in these subtypes (Figure 1E). In the “Core” group, classification accuracy by the IMPACT gene panel increased to 96%; sensitivity and specificity were 86% and 98%, respectively. These results suggested that we could confidently classify cases into subtypes if defined by the “Core” (Figure 1F, G).
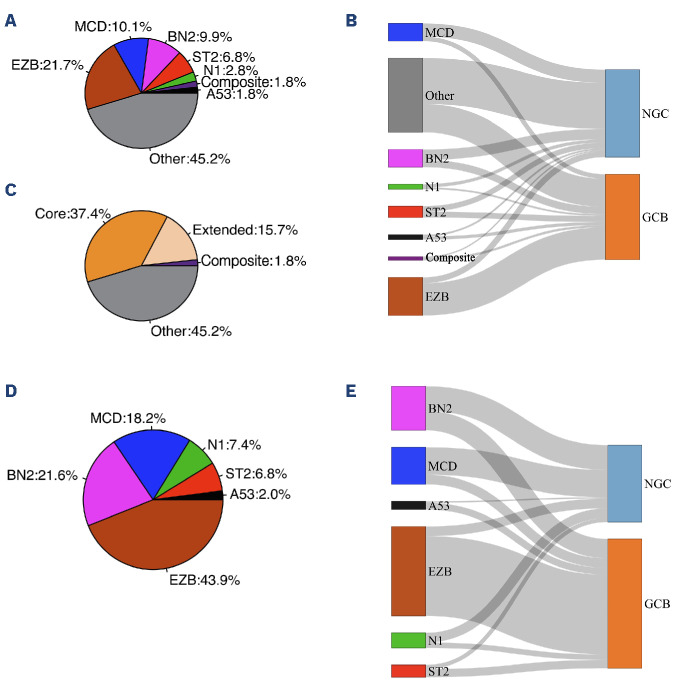
Copy number alteration was included in LymphGen input data to use the full algorithm. However, in the clinical setting, the copy number calling accuracy might be compromised by poor sample quality or low tumor content. To explore the accuracy of classification using the IMPACT gene panel by LymphGen when CNA is not available, we applied only SNV, small INDEL, and fluorescence in situ hybridization (FISH) for BCL2 and BCL6 translocation information to the algorithm. The overall balanced accuracy was 81%, with 83% sensitivity and 89% specificity overall (Online Supplementary Figure S2A). The A53 subtype was omitted as expected, because the classification of the A53 subtype mainly relied on CNA. Twenty-three cases (4.7%) were misclassified into the “Other” (unclassified) group, suggesting that the lack of CNA reduces the algorithm’s ability to classify cases into subclasses (Online Supplementary Figure S2B). These results showed that the optimized CNA information is essential for classifying the A53 subtype and further helps increase the overall accuracy of the classification method. We then applied LymphGen to the MSK DLBCL clinical cohort of 396 cases (320 cases of the cohort have both BCL2 and BCL6 clinical cytogenetic results available) sequenced by IMPACT (data available at https://www.cbioportal.org/ study/summary?id=mbn_msk_2024). When the LymphGen algorithm was applied to the MSK DLBCL cohort, 55% of the cases were assigned into one of the subtypes (MCD: 10%, EZB: 22%, BN2: 10%, N1: 3%, ST2: 7%, A53: 2%, composite: 2%) (Figure 2A). Within these cases, about two-thirds were in the “Core” group, and one-third were in the “Extended” group (Figure 2C). The correlation of genetic subtypes and COO based on Hans’ algorithm was similar to the NCI cohort. Most EZB subtypes were GCB type, while most MCD and N1 subtypes were non-GCB type; BN2, ST2, and A53 subtypes carry a mixture of GCB and non-GCB types (Figure 3B). We further investigated the composition of each subclass and its relationship with COO classification in the “Core” group; 148 (37.4% of total cases) cases classified as “Core” group were assigned into one of the 6 classes (MCD: 18.2%, EZB: 44%, BN2: 22%, N1: 7%, ST2: 7%, A53: 2%). The correlation of genetics subtypes and COO was similar to that observed in all cases. (Figure 2D, E). Essential genes in the IMPACT panel which significantly distinguish each subtype were identified by Benjamini-Hochberg procedure and χ2 test (Online Supplementary Figure S3A).
Figure 2.
LymphGen classification on the Memorial Sloan Kettering cohort. (A) 396 diffuse large B-cell lymphoma cases sequenced by IMPACT were classified by LymphGen into 6 subclasses: MCD, EZB, BN2, N1, ST2, A53. Cases with more than one assigned subclass are indicated as “Composite”. Cases that cannot be classified into these subclasses are labeled as “Other”. (B) Correlation of LymphGen classification and cell-of-origin (COO) by Han’s algorithm. (C) Categories of the subclassification: tumors with subtype probabilities of >90% or 50-90% were defined as ‘‘Core’’ or ‘‘Extended’’ subtype members, respectively. (D) Composition of the subtypes in the cases from the “Core” group (classified based on probability >90%). (E) Correlation of LymphGen classification and COO by Han’s algorithm in the cases from the “Core” group.
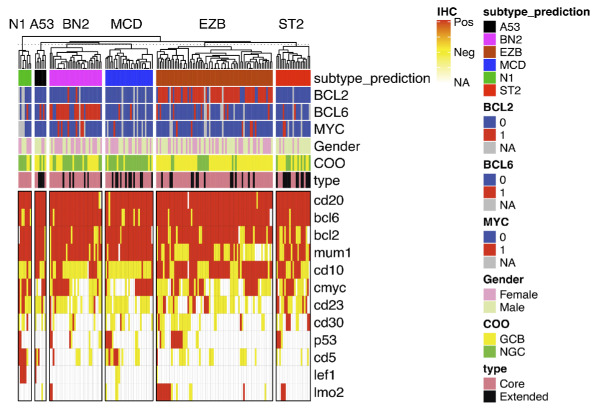
Figure 3.
Heatmap of immunophenotype and cytogenetic features in subtypes. All cases classified into one of the 6 classes are clustered by the subclass. The immunohistochemistry status of each maker is shown in a color block (red: positive; yellow: negative; white: not available / not performed). Fluorescence in situ hybridization results for BCL2, BCL6, and MYC translocations are shown in the color block as indicated here. Cell-of-origin (COO) and classification type (“Core” vs. “Extended”) are also annotated by color block.
In the MSK DLBCL cohort, cytogenetics and immunophenotypes observed in each genetic subtype were similar to those identified in the NCI cohort. As expected, BCL2 translocations were enriched in the EZB subtype (54 out of 67 cases, 81%), and BCL6 translocations were enriched in the BN2 subtype (24 out of 31 cases, 77%). MYC translocations were seen more frequently (6 out of 39 cases, 15.3%) in BN2 subtypes when co-existing with BCL6 translocation. MYC translocations were also seen in a small proportion of EZB (7 out of 86 cases, 8.1%), MCD (2 out of 40 cases, 5.0%), and ST2 (2 out of 27 cases, 7.4%) subtypes. Immunophenotypes were evaluated by immunohistochemical stains in all cases as part of the diagnostic work-up. Consistent with the germinal center phenotype, CD10, CD23, and LMO2 were more frequently found expressed in the EZB and ST2 subtypes, while MUM1 was seen more frequently in the BN2, MCD, N1, and A53 subtypes. Interestingly, 50% (4 out of 8 cases) of N1 subtypes expressed CD5 and LEF1 by immunohistochemistry, indicating that some of the N1 subtype cases might represent Richter’s transformation of chronic lymphocytic leukemia. Only 2 cases with BCL2 translocation fell in the unclassified group, suggesting that BCL2 translocation is a robust classifier for the EZB subtype (Figure 3A). Furthermore, survival analyses within the MSK cohort have demonstrated trends consistent with those observed in other studies. However, statistical significance was not achieved, likely due to the inherent heterogeneity of the cohort and the relatively short duration of follow-up (data not shown).
In conclusion, in this study, we validated genetic classification for DLBCL by LymphGen on our clinically validated 400-gene targeted NGS panel, IMPACT, with high sensitivity and specificity and showed that the classification can be applied to routine clinical practice. To note, in our practice, over 99% of DLBCL cases were successfully sequenced, thanks to detailed pre-analytical checks that identified and excluded inadequate samples. Success rates, however, may differ with other workflows. The findings also indicate the accuracy of the classification can be further enhanced if detection of CNA can be improved. Our study supports the view that genomic classifications of DLBCL using high complexity testing such as LymphGen can be readily translated into routine clinical care using well-designed smaller targeted NGS panels such as IMPACT. Such panels offer opportunities for clinical decision-making and selection of patients for risk-adapted and biomarker selected clinical trials.
Supplementary Material
Funding Statement
Funding: We thank the MSK Cancer Center for their support (Grant/Core Grant: P30 CA008748). ARD was funded by a grant from Fundación Española de Hematología y Hemoterapia.
Data-sharing statement
NGS sequencing data of this study is available on https://www. cbioportal.org
References
- 1.Ennishi D, Hsi ED, Steidl C, Scott DW. Toward a new molecular taxonomy of diffuse large B-cell lymphoma. Cancer Discov. 2020;10(9):1267-1281. [DOI] [PubMed] [Google Scholar]
- 2.Boltezar L, Prevodnik VK, Perme MP, Gasljevic G, Novakovic BJ. Comparison of the algorithms classifying the ABC and GCB subtypes in diffuse large B-cell lymphoma. Oncol Lett. 2018;15(5):6903-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275-282. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937-1947. [DOI] [PubMed] [Google Scholar]
- 5.Sehn LH, Gascoyne RD. Diffuse large B-cell lymphoma: optimizing outcome in the context of clinical and biologic heterogeneity. Blood. 2015;125(1):22-32. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacy SE, Barrans SL, Beer PA, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood. 2020;135(20):1759-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Runge HFP, Lacy S, Barrans S, et al. Application of the LymphGen classification tool to 928 clinically and genetically-characterised cases of diffuse large B cell lymphoma (DLBCL). Br J Haematol. 2021;192(1):216-220. [DOI] [PubMed] [Google Scholar]
- 10.Wright GW, Huang DW, Phelan JD, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37(4):551-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morin RD, Arthur SE, Hodson DJ. Molecular profiling in diffuse large B-cell lymphoma: why so many types of subtypes? Br J Haematol. 2022;196(4):814-829. [DOI] [PubMed] [Google Scholar]
- 12.Ptashkin RN, Ewalt MD, Jayakumaran G, et al. Enhanced clinical assessment of hematologic malignancies through routine paired tumor and normal sequencing. Nat Commun. 2023;14(1):6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS sequencing data of this study is available on https://www. cbioportal.org