NST-628 is a potent and CNS penetrant pan-RAF-MEK molecular glue with broad efficacy in RAS- and RAF-driven models.
Abstract
Alterations in the RAS–MAPK signaling cascade are common across multiple solid tumor types and are a driver for many cancers. NST-628 is a potent pan-RAF–MEK molecular glue that prevents the phosphorylation and activation of MEK by RAF, overcoming the limitations of traditional RAS–MAPK inhibitors and leading to deep durable inhibition of the pathway. Cellular, biochemical, and structural analyses of RAF–MEK complexes show that NST-628 engages all isoforms of RAF and prevents the formation of BRAF–CRAF heterodimers, a differentiated mechanism from all current RAF inhibitors. With a potent and durable inhibition of the RAF–MEK signaling complex as well as high intrinsic permeability into the brain, NST-628 demonstrates broad efficacy in cellular and patient-derived tumor models harboring diverse MAPK pathway alterations, including orthotopic intracranial models. Given its functional and pharmacokinetic mechanisms that are differentiated from previous therapies, NST-628 is positioned to make an impact clinically in areas of unmet patient need.
Significance: This study introduces NST-628, a molecular glue having differentiated mechanism and drug-like properties. NST-628 treatment leads to broad efficacy with high tolerability and central nervous system activity across multiple RAS- and RAF-driven tumor models. NST-628 has the potential to provide transformative clinical benefits as both monotherapy and vertical combination anchor.
See first author Meagan B. Ryan discuss this research article, published simultaneously at the AACR Annual Meeting 2024: https://vimeo.com/932606757/4d0bd5aa98
Introduction
Dysregulation of RAS–MAPK (RAS–RAF–MEK–ERK) pathway signaling is one of the most frequently occurring events in tumor development and has presented a promising and challenging clinical target in precision oncology. RAS (KRAS, NRAS, and HRAS) mutations occur in approximately 27% of cancers, with KRAS mutations occurring most frequently in pancreatic (95%), colorectal (50%), and lung tumors (30%), with RAF (ARAF, BRAF, and CRAF) mutations most frequently occurring in BRAF in 7% of tumors, primarily in melanoma (50%; refs. 1–3). Additional activating or loss-of-function mutations, such as mutation or loss of neurofibromin 1 (NF1) or mutations in receptor tyrosine kinases (RTK), also frequently occur in cancer and contribute to aberrant activation and dependency on the RAS–MAPK pathway for tumor growth and maintenance. Selective and potent inhibitors have been developed for every node of the RAS–RAF–MEK–ERK pathway, with varying degrees of success related to tolerability and durability of response clinically.
Highly potent and selective ATP noncompetitive inhibitors of MEK1/2 and monomer selective (type I RAF) inhibitors of BRAF with potency in BRAF V600X (BRAF class I mutants) are some of the best clinically characterized inhibitors of the RAS–MAPK pathway. MEK inhibitors, such as trametinib, are limited clinically due to adverse side effects requiring dose reductions or disruptions and, despite efficacy in BRAF class I mutant cancers, are limited in efficacy in other mutational backgrounds (4, 5). Even in patients with clinical response to MEK inhibitors, the durability of response remains a hurdle due to multiple mechanisms of resistance to therapy often converging on reactivation of signaling through the RAS–MAPK pathway. One of the best characterized mechanisms of resistance to both MEK inhibitors and type I RAF inhibitors is CRAF-mediated bypass due to loss of negative feedback on the RAF node or activation of RTKs upstream leading to the formation of BRAF–CRAF heterodimers to maintain signaling through the RAS–MAPK pathway (6, 7). Type II RAF inhibitors, which selectively target BRAF–CRAF heterodimers, and the “paradox breaker” plixorafenib, which disrupts the formation of BRAF dimers, both were developed in an attempt to target CRAF-mediated bypass to other classes of RAF and MEK inhibitors, whereas sparing ARAF activity (8–11). However, the type II RAF inhibitors naporafenib and belvarafenib are limited by ARAF-mediated bypass, and response to single-agent inhibitors has been a disappointment clinically.
Successful targeting approaches to the RAS–MAPK pathway have been limited to single agents, and vertical combination strategies targeting multiple nodes of the RAS–RAF–MEK–ERK cascade are currently under investigation. Type I RAF and MEK inhibitor combinations have been successful clinically in BRAFV600-mutant melanoma and have more limited efficacy in other BRAFV600-mutant tumors, such as colorectal cancer, where triplet therapy with the addition of EGFR monoclonal antibodies deepens therapeutic response (12–14). Type II RAF inhibitor combinations are also under clinical evaluation in NRAS-mutant cancers; however, their promise is limited by tolerability to the treatment regimens (15, 16). Recent advances in targeting KRASG12C-mutant cancers have also been limited by the rapid onset of therapeutic resistance clinically, with proposed vertical combination strategies under evaluation as a mitigation strategy for resistance mechanisms converging on reactivation of the RAS–MAPK pathway (17–19).
NST-628 was developed as a nondegrading molecular glue of the RAF–MEK node of the RAS–MAPK pathway with activity on both active and inactive conformations of the proteins. With potent activity on both A/B/CRAF and MEK1/2, we show that NST-628 overcomes limitations of prior MEK and RAF inhibitors by blocking CRAF-mediated bypass and has broad efficacy in NRAS-, KRAS-, and BRAF-mutant tumor models. With a half-life optimized for daily dosing, balanced metabolic profile, and efficient blood–brain barrier penetrance, NST-628 is active in central nervous system (CNS) models and shows greater tolerability than other MEK inhibitors, such as trametinib. With superior pharmacokinetic and pharmacodynamic properties, our data show that NST-628 is positioned as a novel, potent, and tolerable single-agent inhibitor of both RAF and MEK and is an ideal combination partner in vertical combination strategies for RAS- and RAF-mutant cancers.
Results
NST-628 Is a Potent Pan-RAF–MEK Nondegrading Molecular Glue
NST-628 (Fig. 1A) was discovered by the deliberate optimization of potent dual inhibition of phospho-MEK and phospho-ERK in AlphaLISA and HTRF assays (Supplementary Fig. S1A), CNS penetration, and balanced ADME/DMPK properties suitable for an optimal predicted clinical half-life. The off-target kinase binding potential of NST-628 was determined using KINOMEscan profiling (Supplementary Table S1; ref. 20). NST-628 at a concentration of 1 μmol/L was screened against 97 human kinases and disease-relevant mutant variants to determine kinase activity. Following NST-628 administration, MEK1 and MEK2 activity was 0% and 0.05% of control, as expected by the pharmacologic action of the compound. RAF1 activity was 24% of control, whereas c-Jun N-terminal kinase (JNK) 1, JNK2, and JNK3 were 76%, 20%, and 17% of control, respectively. No other kinases displayed significantly decreased activity in the presence of NST-628, demonstrating the selectivity of the compound.
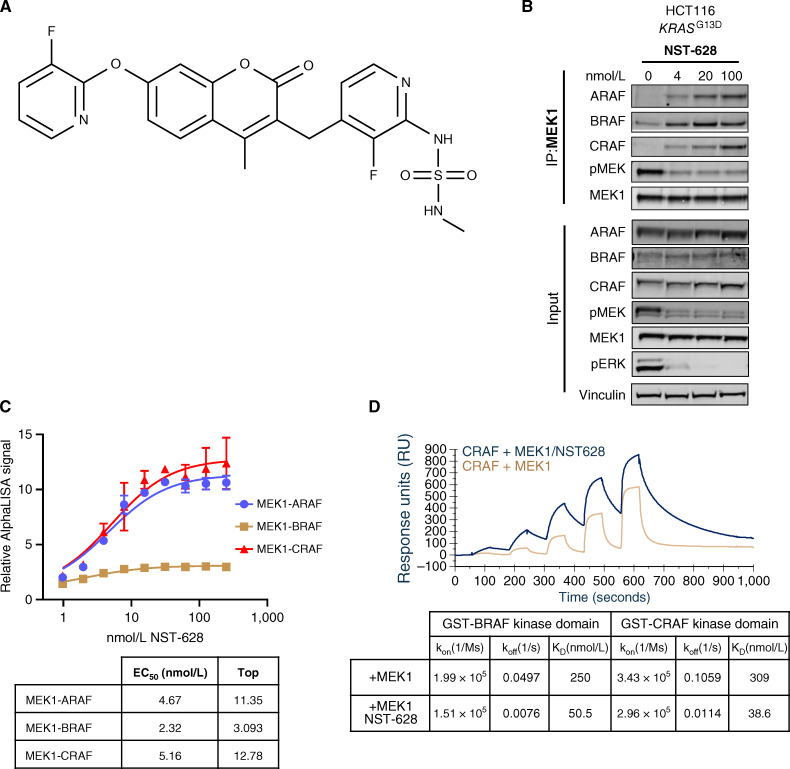
Figure 1.
NST-628 is a pan-RAF–MEK nondegrading molecular glue. A, Chemical structure of NST-628. B, MEK1 immunoprecipitation in HCT116 cell treated with indicated concentrations of NST-628 (4-100 nmol/L) for 2 hours, and blot analysis was performed for ARAF, BRAF, CRAF, phospho-MEK, MEK1, phospho-ERK, and vinculin as a loading control. C, MEK1–RAF complex formation monitored by AlphaLISA protein–protein interaction assays after treatment with various concentrations of NST-628 for 30 minutes at RT. D, Table of binding constants from SPR-based ternary complex assays where MEK1 is titrated with immobilized GST-BRAF or GST-CRAF in the presence and absence of 3 μmol/L NST-628.
In an endogenous MEK1 immunoprecipitation assay adapted from Khan and colleagues (21) in the KRASG13D-mutant HCT116 cell line, NST-628 engaged ARAF, BRAF, and CRAF in a complex with MEK1 (Fig. 1B). Increasing concentrations of NST-628 induced a dose-dependent increase in the interaction between MEK1–ARAF, MEK1–BRAF, and MEK1–CRAF and increased gluing of the MEK1–RAF complex correlated with a decrease in the levels of both phospho-MEK1 and phospho-ERK. To further confirm the stabilization of MEK1–RAF paralog complexes in the presence of NST-628, biochemical AlphaLISA protein–protein interaction (PPI) assays were performed with recombinant purified MEK1–RAF complexes (Fig. 1C). In an AlphaLISA PPI assay, ARAF, BRAF, or CRAF is captured on the glutathione donor beads, and MEK1 is captured on the nickel chelating acceptor beads. When the two proteins interact, the donor bead is brought into proximity of the acceptor bead, and excitation of the donor bead will result in signal generation on the acceptor bead. The AlphaLISA signal is proportional to the amount of complex in the assay condition. The addition of NST-628 showed a dose-dependent increase in signal for all MEK1–RAF complexes, suggesting that NST-628 promoted the complex formation between MEK1 and RAFs by acting as a molecular glue. In the MEK1–RAF AlphaLISA assays, NST-628 showed singledigit nanomolar EC50 values with a maximal relative signal of 11-fold, 3-fold, and 13-fold for MEK1–ARAF, MEK1–BRAF, and MEK1–CRAF, respectively (Fig. 1C). To further elucidate the molecular glue mechanism, surface plasmon resonance (SPR)–based ternary complex assays were performed by immobilizing GST-BRAF or GST-CRAF on the SPR chip surface (Fig. 1D) and titrating MEK1 in the presence and absence of 3 μmol/L of NST-628. The binding affinities between MEK1–BRAF and MEK1–CRAF were determined to be 250 and 310 nmol/L, respectively. These affinities are consistent with the previously reported numbers from SPR measurements (6). The addition of NST-628 increased the affinity between MEK1–BRAF from 250 to 50 nmol/L and increased the affinity between MEK1–CRAF from 309 to 39 nmol/L. The increase of affinities for both systems was solely driven by the decrease of koff suggesting that the presence of NST-628 slowed the dissociation between MEK1 and RAFs (Fig. 1D). NST-628 displayed a distinct profile compared with orthosteric MEK1 inhibitor trametinib which disrupted the PPI between MEK and RAFs (Supplementary Fig. S1B). By inhibiting the activity of MEK1 and enhancing the affinity between MEK1 and RAF, NST-628 can act as a potent molecular glue driving inhibition of the RAF and MEK1 nodes of the RAS–MAPK pathway.
To better understand the pan-RAF–MEK glue activity of NST-628, we determined X-ray cocrystal structures of NST-628 bound to the MEK1–ARAF, MEK1–BRAF, and MEK1–CRAF complexes (Fig. 2A; Supplementary Table S2). To our knowledge, these are the first high-resolution crystal structures of MEK1–ARAF and MEK1–CRAF complexes. In all complexes, MEK1 and RAF kinase domains arrange in a face-to-face organization mediated via the NST-628 allosteric site in MEK and the αG-helix of RAF; this arrangement creates an interfacial pocket that would not be anticipated by the isolated MEK1 and RAF structures (21, 22). NST-628 engages the interfacial pocket, positioning the 2-fluoropyridine in a highly hydrophobic crevice formed by MEK1 residues Phe209, Leu118, Phe129, Met143, and Ile141. The coumarin core and 3-fluoropyridine interact with MEK1 residues Leu215, Ile216, and Met219, which are contributed by the activation loop helix, and the core carbonyl forms hydrogen bonds with the backbone amides of residues Val211 and Ser212. These interactions stabilize the helical conformation of the MEK1 activation loop and prevent RAF-mediated phosphorylation of Ser218 and Ser222. The N-methylsul-famide of NST-628 is hydrogen bonded to the side chain of MEK1 Arg234 and the backbone carbonyl of MEK1 Arg189, positioning it adjacent to the αG-helix of RAF. Despite the sequence similarity in the αG-helices of each RAF paralog, the characteristics of the interfacial pockets are different due to variability in a conserved arginine residue from RAF. ARAF Arg515 is positioned in the interfacial pocket directly adjacent to NST-628 and forms hydrogen bonds with the backbone carbonyl of MEK1 residue Met219 from the activation loop, resulting in a smaller interfacial pocket than other RAF paralog complexes. CRAF Arg554 is hydrogen bonded to CRAF Asp555 from the αG-helix, generating a larger interfacial pocket. BRAF Arg662 is in an intermediate conformation where it can interact with the backbone carbonyls of MEK1 residues Met219 and Ala222 (Ser222 in wild-type MEK1) from the activation loop. NST-628 can accommodate these differences in the interfacial pockets to promote pan-RAF–MEK interactions. Additionally, in the MEK1–ARAF and MEK1–BRAF complex structures, a network of ordered waters bridge NST-628 and the RAF αG-helix. Because the MEK1 allosteric pocket generates a similar interfacial binding site with KSR1, we determined the crystal structure of the MEK1–KSR1 complex with NST-628 (Supplementary Fig. S2A; Supplementary Table S3). Although we can observe binding of NST-628 consistent with the MEK1–RAF complex structures, we are unable to attribute biological significance to this interaction in the context of HCT116 tumor cells (Supplementary Fig. S2B and S2C). The increased affinities for MEK1 and RAF in the presence of NST-628, as well as the positioning of NST-628 in an interfacial pocket contacting both MEK1 and RAF, support NST-628 functioning as a pan-RAF–MEK molecular glue with the ability to inhibit multiple key nodes of the RAS–MAPK pathway.
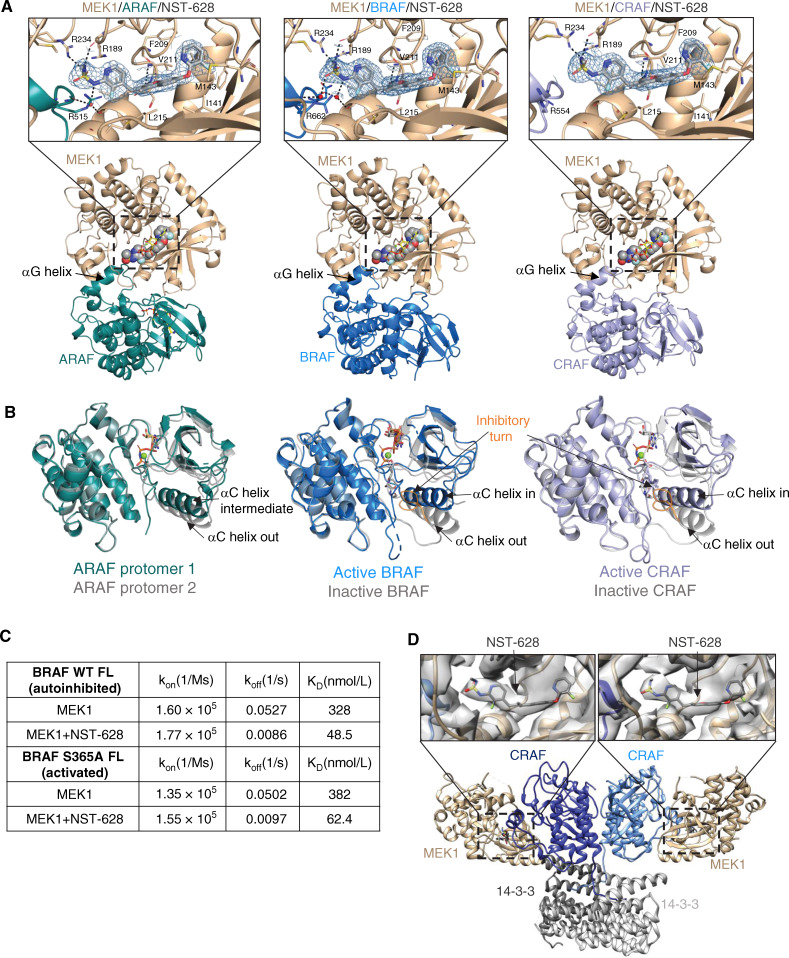
Figure 2.
NST-628 engages pan-RAF–MEK complexes with active and inactive RAF conformations. A, Overview of MEK1–RAF heterodimers in the crystal structures of MEK1–ARAF (2.42 Å resolution), MEK1–BRAF (2.07 Å resolution), and MEK1–CRAF (2.59 Å resolution) with NST-628 (shown as spheres) and active RAF. Insets show electron density for NST-628 (blue mesh) in the interfacial allosteric site with key interactions highlighted by black dashes and waters represented as red spheres. B, Overlay of the distinct RAF conformations observed in crystal structures. C, Table of binding constants from SPR-based ternary complex assays titrating MEK1 with immobilized full-length WT BRAF or full-length S365A BRAF in the presence or absence of 3 μmol/L NST-628. D, Overview of cryo-EM structure of MEK1–CRAF–14-3-3 with NST-628 (4.36 Å resolution). Insets show electron density for NST-628 and the interfacial allosteric pocket.
NST-628 Glues Both Active and Inactive Conformations of RAF
Signaling through the RAS–MAPK pathway is dynamic and tightly regulated in normal tissues; however, in RAS- or RAF-mutant cancers, equilibrium shifts to active conformations of protein (23). Previous efforts to target the RAF signaling node have been limited due to selectively inhibiting either active or inactive conformations of RAF (24). Through thorough structural and biophysical characterization, we demonstrate that NST-628 can engage MEK–RAF complexes with inactive and active RAF conformations. In the MEK1–ARAF, MEK1–BRAF, and MEK1–CRAF structures with NST-628, back-to-back active RAF dimers are observed, resulting in heterotetrameric RAF–MEK complexes (Supplementary Fig. S3A). In the MEK1–BRAF and MEK1–CRAF heterotetrameric complexes, the A-loops of both RAF protomers are in an extended conformation, supporting the αC-helix IN conformation and RAF dimerization. However, the organization of the ARAF dimer in the MEK1–ARAF heterotetramer complex is unique, with the αC-helices of the RAF protomers in an intermediate and OUT conformation. The ARAF protomer with the αC intermediate conformation has Glu354 directly stabilizing the DFG-motif, whereas the ARAF protomer with the αC out conformation has the DFG-motif IN despite no obvious stabilizing interactions through the αC-helix and a poorly ordered inhibitory turn. Although unexplored further in this work, the unusual ARAF dimer arrangement observed in the structure may underlie the unique pharmacology of ARAF, including toward type II RAF inhibitors (25).
To determine the structures of MEK–RAF complexes with NST-628 and inactive RAF, we generated alternative crystal forms using modified MEK1 constructs with different termini and loop modifications (Supplementary Fig. S3B; Supplementary Table S4). Despite the lack of a side-to-side RAF dimer in these structures, the overall architecture of the interface and the binding mode of NST-628 are similar to the previous structures in which RAF is in an active, dimeric arrangement. Notably, the RAF αC-helix adopts the outward conformation supported by the inhibitory turn of the A-loop, supporting that NST-628 can engage MEK–RAF complexes with inactive RAF as well as active RAF conformations (Fig. 2B). We confirmed these findings with SPR-based ternary complex assays using immobilized full-length wild-type BRAF-14-3-3 (autoinhibited conformation) and full length S365A BRAF-14-3-3 (active conformation; Fig. 2C). In both cases, NST-628 enhances the affinity of MEK1 to BRAF-14-3-3 complex by ∼6-fold, driven primarily by a dramatic decrease in off-rate. In a cellular assay, NST-628 is insensitive to upstream pathway flux driven by epidermal growth factor, maintaining pan-RAF–MEK complexes in a MEK1 immunoprecipitation assay, and inhibition as measured by both phospho-MEK and phospho-ERK is sustained (Supplementary Fig. S4). To further characterize the binding of NST-628 to active RAF signaling complexes, we determined a 4.36 Å resolution cryo-electron microscopy structure of MEK12–CRAF2–14-3-32 with NST-628 (Fig. 2D; Supplementary Fig. S5A–S5E; Supplementary Table S5). The overall architecture of the complex is consistent with the active BRAF2–14-3-32 complexes determined previously (26–28) and confirms the heterotetrameric arrangement of MEK1 with active CRAF that was observed in the crystal structure.
Although the resolution is modest, clear density for NST-628 is visible in the allosteric site of MEK1, further supporting that NST-628 can engage the activated forms of RAF signaling complexes that promote oncogenic growth. NST-628 can also bind MEK1 with monomeric RAF in an autoinhibited conformation, leading to greater efficacy in cancers with diverse RAS–MAPK pathway alterations.
NST-628 Does Not Drive RAF Heterodimer Formation and Is Active in Tumor Models Dependent on RAF Heterodimer Signaling
NST-628 potently inhibits wild-type ARAF, BRAF, and CRAF complexes with MEK. However, there are many tumor types harboring mutations at the MEK1–BRAF interface, with BRAF non-V600E mutations comprising approximately 35% of BRAF-mutant cancers. BRAF class II/III mutations are dimerization dependent and respond at lower rates to MAPK targeting agents in the clinic than BRAF class I mutations (29, 30). The BRAF G466A and G469A mutations were identified for further characterization due to their proximity to the MEK1–BRAF interface and their confirmed status as class II/III driver mutations insensitive to treatment by classic, orthosteric BRAF inhibitors (29). The binding of BRAF class II/III mutants G466A and G469A on MEK1 was characterized by SPR and a modest decrease in affinity for MEK1 with BRAF G466A mutant and a roughly 2-fold decrease in affinity for MEK1 with BRAF G469A mutant was seen (Fig. 3A). These affinity changes are driven primarily by reduced on-rates predicted to result from changes in P-loop dynamics that directly impair BRAF P-loop/MEK1 interactions or BRAF dimerization-dependent MEK binding (31). In the presence of NST-628, off-rates of MEK1 from both class II mutants are decreased ∼5-fold, resulting in ∼3- to 4-fold higher affinities. The enhanced affinity between MEK1 and class II BRAF mutants in the presence of NST-628 confirmed its ability to act as a potent molecular glue on the mutants and the potential for efficacy in tumor types harboring these detrimental mutations.
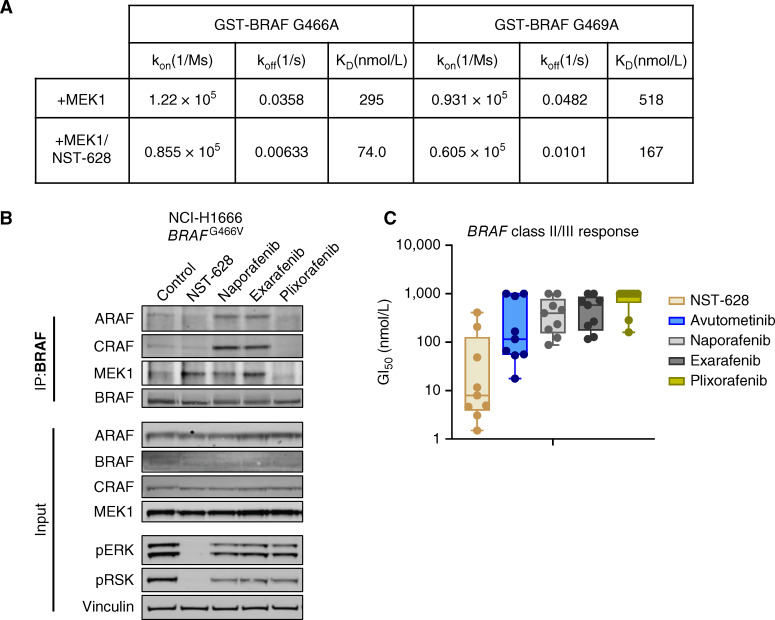
Figure 3.
NST-628 inhibits BRAF class II/III mutants and does not induce the formation of RAF heterodimers. A, Table of binding constants from SPR-based ternary complex assays titrating MEK1 with immobilized GST-BRAF G466A or GST-BRAF G469A in the presence and absence of 3 μmol/L NST-628. B, BRAF immunoprecipitation in the NCI-H1666 cell line treated with 100 nmol/L of the indicated inhibitors for 2 hours, and blot analysis was performed for ARAF, BRAF, CRAF, MEK1, phospho-ERK, and vinculin as a loading control. C, BRAF class II/III mutant cell line panel (OV90, NCI-H1666, NCI-H1755, NCI-H2405, WM1963, WM3629, WM3670, WM3912, and WM3928) was treated for 72 hours with a dose titration of NST-628, avutometinib, naporafenib, exarafenib, or plixorafenib, and viability was measured by CellTiter-Glo.
The activity of NST-628 on the BRAF G466A and G469A mutant proteins in a biophysical assay also translates to activity on the endogenous BRAF G466V mutant protein in the NCI-H1666 cell line. In a BRAF immunoprecipitation assay in the NCI-H1666 cell line, NST-628 treatment does not induce the heterodimerization of BRAF and CRAF after 2 hours of treatment (Fig. 3B). In contrast, the type II RAF inhibitors naporafenib and exarafenib induce the formation of BRAF–CRAF heterodimers, as well as BRAF-ARAF heterodimers, whereas the RAF dimer inhibitor plixorafenib disrupts all BRAF heterodimer formation without impacting pathway activity. This mechanistic difference leads to greater RAS–MAPK suppression as measured by phospho-ERK and phospho-RSK with NST-628. In the HCT116 KRASG13D mutant cell line, NST-628 drives the formation of pan-RAF–MEK complexes in a MEK1 immunoprecipitation assay while in a BRAF immunoprecipitation assay, BRAF heterodimer formation is observed only with type II RAF inhibitor treatment (Supplementary Fig. S6A). No heterodimer formation is also observed using a CRAF IP and CRAF IP/MS approach in the HCT116 cell line after 2 hours of NST-628 treatment, which contrasts with CRAF-BRAF heterodimer formation driven by naporafenib treatment (Supplementary Fig. S6B–S6D).
NST-628 activity on BRAF class II and III mutations translates into potent antiproliferative activity in models harboring BRAF class II and III mutations. In a proliferation assay in a panel of BRAF class II and BRAF class III mutant models, NST-628 demonstrated greater potency than both type II RAF inhibitors naporafenib and exarafenib and the BRAF dimer breaker plixorafenib (Fig. 3C). NST-628 is also more potent than the RAF–MEK dual inhibitor avutometinib (VS-6766) in the same panel of BRAF class II and III mutant cell lines. Clinically, avutometinib is limited due to modest activity as a single agent and toxicity with daily dosing and is currently under investigation with intermittent dosing in combination with the focal adhesion kinase inhibitor defactinib (32, 33).
NST-628 Is Active across Models Driven by RAS- and RAF Mutations
With a differentiated mechanism from non-ATP competitive MEK inhibitors and type II RAF inhibitors, NST-628 was also broadly efficacious in an unbiased cell line panel screen. We performed an unbiased 553 cell line OMNI screen to determine a biomarker profile and identify tumor models with sensitivity to NST-628 treatment (Fig. 4A; Supplementary Fig. S7A; Supplementary Table S6). In a cell proliferation assay, sensitivity to NST-628 treatment (GI50 ≤100 nmol/L) was 66%, 56%, 47%, and 38% for NRAS-mutant, KRAS-mutant, BRAF-mutant, and HRAS-mutant models, respectively, whereas sensitivity in NF1-mutant models was 24% and RAS/RAF wild-type models was 23% (Fig. 4A; Supplementary Fig. S6A and S6B). Among BRAF-mutant models, BRAF class I responded at a higher rate than other BRAF-mutant models (57% vs. 32%; Supplementary Fig. S7A and S7B).
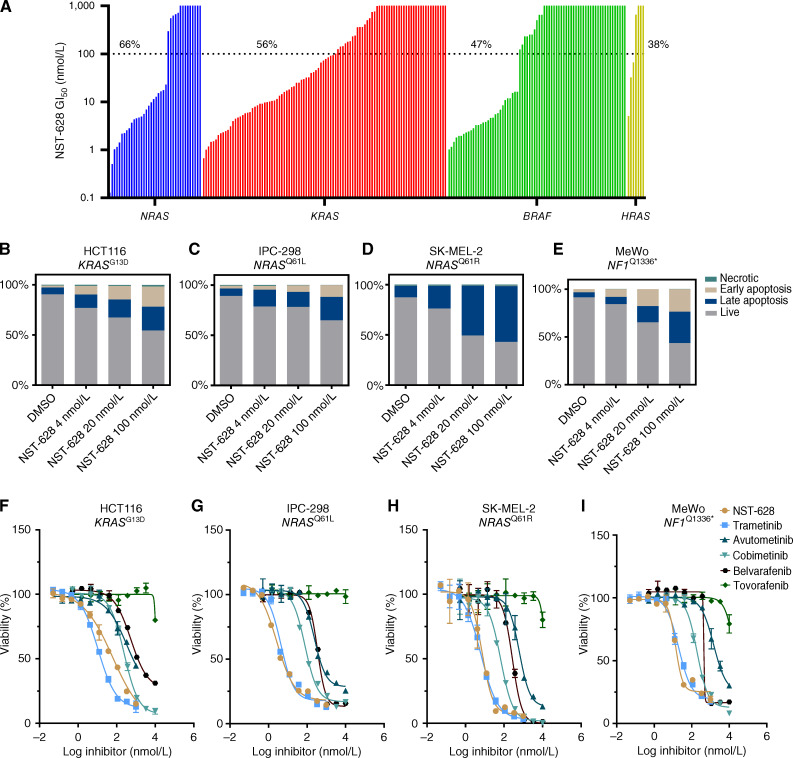
Figure 4.
NST-628 inhibits the growth of RAS- and RAF-driven cancers. A, OMNI cell line panel was treated with a dose-response of NST-628 for between 3 and 7 days, and viability was measured by CellTiter-Glo. Response rates in each mutational background were calculated for models with a GI50 of ≤100 nmol/L. B, HCT116 (C) IPC-298, (D) SK-MEL-2, and (E) MeWo cell lines were treated with 4, 20, or 100 nmol/L NST-628 for 48 hours and were stained with Annexin V and DAPI, and live, early apoptosis, late apoptosis, and necrotic cells were analyzed by flow cytometry. F, HCT116 (G), IPC-298 (H), SK-MEL-2, and (I) MeWo cells were treated with a dose response of NST-628, trametinib, avutometinib, cobimetinib, belvarafenib, or tovorafenib for 72 hours, and viability was assessed by CellTiter-Glo.
Next, we determined the mechanism of reduced proliferation by looking at cytotoxic effects of NST-628. In the NRAS-mutant IPC-298 and SK-MEL-2, NF1-mutant MeWo, and KRAS-mutant HCT116 cell lines, NST-628 treatment increased levels of early and late apoptotic cells and reduced live cells in a dose-dependent fashion. Across all cellular models, 100 nmol/L NST-628 induced the greatest level of apoptosis (Fig. 4B–E). In the same cell lines, NST-628 showed strong antiproliferative activity and greater potency than other MAPK targeting therapies. NST-628 is equipotent to the MEK inhibitor trametinib and demonstrates greater potency than the MEK inhibitor cobimetinib, the RAF–MEK inhibitor avutometinib, and the type II RAF inhibitors belvarafenib and tovorafenib (Fig. 4F–I).
Finally, we sought to characterize the durability of RAS–MAPK pathway inhibition in driving the antiproliferative activity of NST-628 in RAS- and RAF-mutant cancers. In SK-MEL-2 (NRAS-Q61R mutant) and MeWo (NF1-Q1336* mutant) cell lines, NST-628 treatment leads to a durable downregulation of the canonical MAPK transcriptional targets DUSP6 and SPRY4 over a 72-hour time course (Supplementary Fig. S8A; Supplementary Table S7). Minimal phospho-ERK rebound is observed in HCT116 cells over a 72-hour time course of NST-628 treatment (Supplementary Fig. S8B and S8C). MAPK inhibition has been previously demonstrated to enhance the efficacy of immunotherapy in multiple tumor types by modulating the tumor microenvironment via tumor-intrinsic induction of interferon-stimulated genes (34, 35). In all models tested, there is evidence of an upregulation of immune-related genes, including transcription of genes associated with interferon stimulation (ISG) and antigen presentation machinery (Supplementary Fig. S8A and S8B). Global proteomics in the HCT116 cell line confirmed broad upregulation of ISGs/antigen presentation and RTKs while MAPK targets remained downregulated over time out to 72 hours of NST-628 treatment (Supplementary Fig. S8D–S8G; Supplementary Table S8). We also characterized long-term resistance mechanisms to NST-628 in the SK-MEL-2 and MeWo cell line models by selecting resistant populations emerging after >3 months of NST-628 treatment. After confirming resistance to NST-628, significant upregulation of phospho-AKT and modest upregulation of phospho-ERK were observed (Supplementary Fig. S9A–S9C). Signaling mechanisms that potentially drive activation of parallel pathways include activation of receptor tyrosine kinases (EGFR) and genomic alteration of additional nodes of the RAS network (KRASG12C mutation, ARAF amplification, MAPK1 amplification, TSC2A460T mutation; Supplementary Fig. S9D and S9E; Supplementary Table S9). No mutations predicted to affect the binding of NST-628 to either MEK or RAF paralogs were detected. Collectively, short-term “adaptive” and long-term “acquired” resistance mechanisms strongly converge on the activation of parallel oncogenic pathways and modulation of tumor-intrinsic immune signatures.
NST-628 Displays Antitumor Properties In Vivo
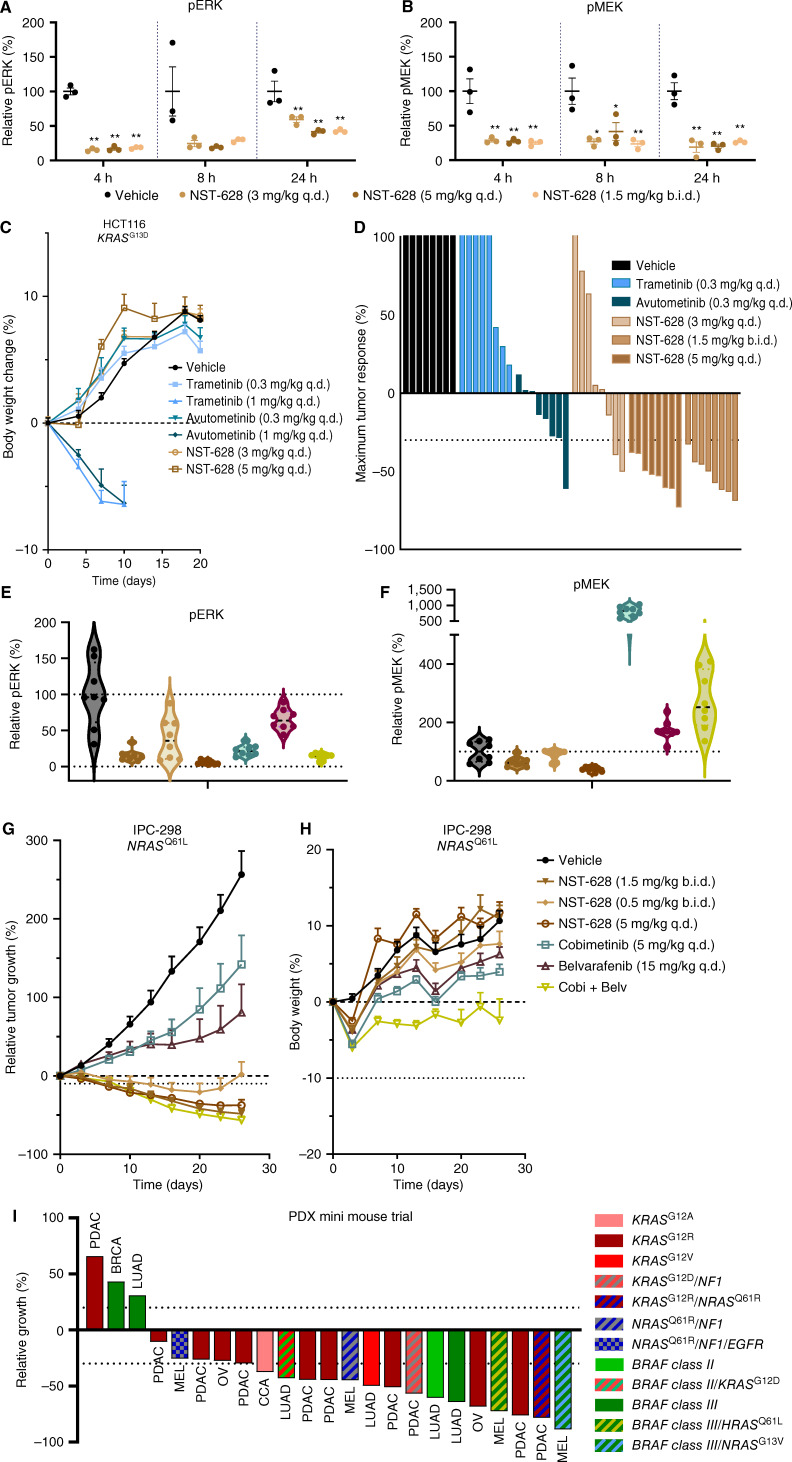
After demonstrating the potency of NST-628 across cell line models of diverse tumor types and genetic backgrounds with dysregulation of the RAS–MAPK in vitro and characterizing both acute and long-term signaling modulation by NST-628, we next assessed the therapeutic potential of NST-628 in vivo. In the HCT116 KRASG13D-mutant tumor model, a single oral dose of NST-628 at 3 mg/kg qd, 5 mg/kg qd, and 1.5 mg/kg b.i.d. reduces both phospho-MEK and phospho-ERK at 4, 8, and 24 hours (Fig. 5A and B). In the same tumor model, NST-628 is tolerable with weight gain comparable with vehicle-treated animals, and no evidence of skin keratinization was observed across all treatment groups, up to 5 mg/kg qd. However, at a 1 mg/kg dose, both trametinib and avutometinib are not tolerable, with marked weight loss seen and discontinuation of treatment (Fig. 5C). NST-628 reduced tumor growth in all tumors in the 5 mg/kg qd and 1.5 mg/kg b.i.d. treatment arms, and controlled growth of tumors in the 3 mg/kg arms with regressions in 3/8 tumors at maximum tumor response (day 10; Fig. 5D; Supplementary Fig. S10A and S10B).
Figure 5.
NST-628 displays potent antitumor activity in KRAS- and NRAS-mutant models. HCT116 tumors treated with a single dose of 3 mg/kg qd, 5 mg/kg qd, or 1.5 mg/kg b.i.d. NST-628 and assessed for (A) phospho-ERK or (B) phospho-MEK by immunoblot 4, 8, or 24 hours after treatment. C, Body weights of HCT116 tumor-bearing mice treated with 0.3 or 1 mg/kg qd trametinib, 0.3 or 1 mg/kg qd avutometinib, or 3 or 5 mg/kg qd NST-628. D, Day 10 tumor volume of HCT116 tumors treated with 0.3 mg/kg qd trametinib, 0.3 mg/kg qd avutometinib, 3 mg/kg qd, 5 mg/kg qd, or 1.5 mg/kg b.i.d. NST-628; tumors are normalized to D0 starting volume. IPC-298 tumors treated with 0.5 mg/kg b.i.d., 1.5 mg/kg b.i.d., or 5 mg/kg qd NST-628, cobimetinib (5 mg/kg qd), belvarafenib (15 mg/kg qd), or a combination of cobimetinib (5 mg/kg daily) and belvarafenib (15 mg/kg) and immunoblotted for (E) phospho-ERK or (F) phospho-MEK 4 hours after treatment. G, IPC-298 tumor volume and (H) body weights of tumor-bearing mice treated as in E and F. Tumor volume and body weights are normalized to D0. I, CTG-0723, CTG-1684, CTG-1351, CTG-0308, CTG-3063, CTG-0889, CTG-1086, CTG-1375, CTG-0941, CTG-1612, CTG-0302, CTG-0881, CTG-1471, CTG-1358, CTG-0381, CTG-0291, CTG-1441, CTG-2841, CTG-0964, CTG-3059, CTG-0314, CTG-1068, and CTG-1501 (left–right) PDX tumors treated with 3 mg/kg qd NST-628. Data are represented as day 13/14 or maximum response tumor volume, normalized to D0.
NST-628 also showed strong activity in a model of NRAS-mutant melanoma. NRAS-mutant melanomas are dependent on signaling through the RAF node, yet these tumors are treatment-refractory to MAPK targeting agents and also do not respond to MEK inhibitors as single agents (36, 37). The combination of the type II RAF inhibitor belvarafenib and the MEK inhibitor cobimetinib has been shown to overcome ARAF-driven resistance to belvarafenib as a single agent and is currently under clinical investigation in NRAS-mutant melanoma (NCT04835805; refs. 9, 16). We investigated if NST-628 can show equivalent efficacy and superior tolerability to the loose RAF and MEK inhibitor combination of belvarafenib and cobimetinib in the NRASQ61L-mutant model IPC-298. NST-628 at 5 mg/kg qd or 1.5 mg/kg b.i.d. dosing effectively inhibited phospho-ERK and phospho-MEK to a greater degree than belvarafenib or cobimetinib alone or in combination in the IPC-298 model (Fig. 5E and F). Superior pathway inhibition of both phospho-MEK (biomarker for pathway reactivation) and phospho-ERK (biomarker for efficacy) led to a deep inhibition and regression of the IPC-298 model treated with NST-628 at the top two doses. Although the combination of belvarafenib and cobimetinib also induced tumor regressions in this sensitive xenograft tumor model, the combination had a much higher overall drug burden and limited tolerability as measured by body weight loss in comparison with NST-628 (Fig. 5G and H; Supplementary Fig. S10C).
Given the prevalence of cancers with RAS–MAPK alterations across diverse histologies, we next investigated the efficacy of NST-628 in patient-derived xenograft (PDX) models. In a murine efficacy trial, lung adenocarcinoma, pancreatic adenocarcinoma (PDAC), melanoma (MEL), ovarian, breast cancer, and cholangiocarcinoma (CCA) PDX models with NRAS, KRAS, or BRAF class II/III mutations, NST-628 was administered at an intermediate dose of 3 mg/kg qd for a range of 27 to 60 days (Fig. 5I; Supplementary Table S10). The overall response rate (maximum response ≤ −30%) was 69.5% and the disease control rate (maximum response ≤20%) was 87%. Regressions were observed in models with KRASG12V, KRASG12R, NRASQ61x, and BRAF class II/III mutations, and the responses were durable, frequently up to 60 days of treatment with NST-628. Collectively, the activity of NST-628 in cell line xenografts and PDX models justifies clinical investigation of the compound in solid tumors with NRAS, KRAS, and BRAF class II/III mutations.
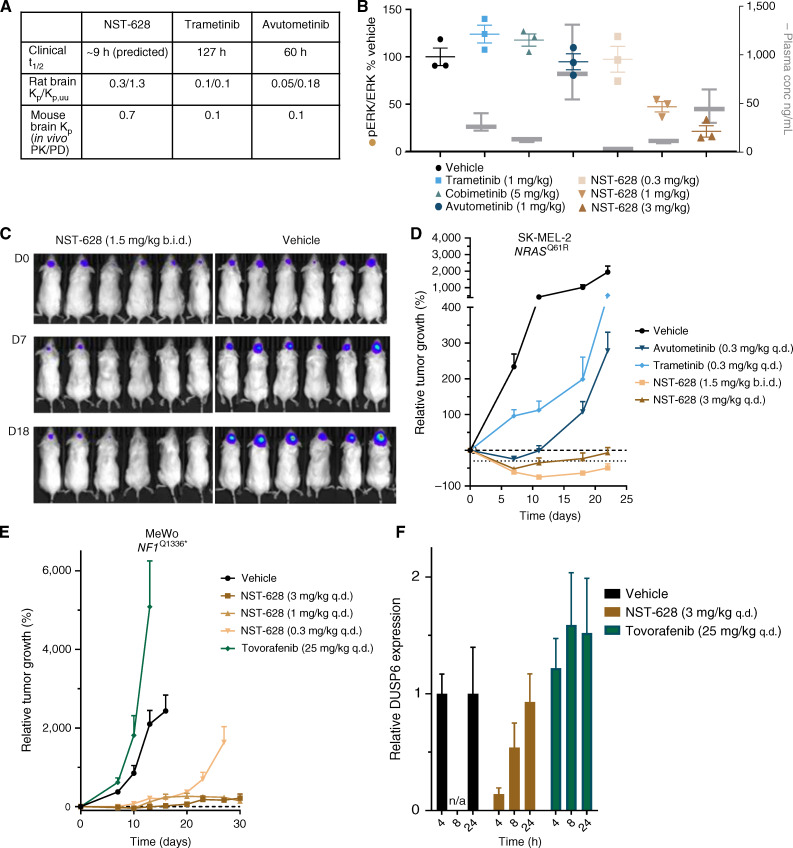
NST-628 Is a Potent and Fully Brain-Penetrant Inhibitor
NST-628 was designed with a favorable pharmacokinetic profile in mind to overcome the dosing limitations of other MEK inhibitors. In contrast to other RAS–MAPK pathway therapeutics, NST-628 has a best-in-class metabolic profile consisting of a short predicted effective half-life of 9 to 10 hours in humans, optimized tissue accumulation, and low risk for drug–drug interactions. As such, NST-628 is compatible with once-daily dosing with a target coverage and modeled plasma concentration maintained above tumor stasis concentration (Fig. 6A; Supplementary Fig. S11A and S11B; ref. 38). Additionally, NST-628 is fully brain penetrant, with a Kp/KP,uu ratio of 0.3/1.3, in contrast with trametinib and avutometinib, which have a KP,uu value of 0.1 and 0.18, respectively (Fig. 6A). In normal mouse brains, with intact blood–brain barrier, only NST-628 was able to inhibit the RAS–MAPK pathway as measured by basal levels of phospho-ERK in a dose-dependent fashion, despite equivalent levels of drug for other MEK inhibitors, cobimetinib, trametinib, and avutometinib, in plasma (Fig. 6B). Next, we investigated whether the superior blood–brain barrier penetrance would translate into antitumor activity in CNS models with RAS–MAPK alterations, representing patient populations with either brain metastases or primary CNS cancers. Brain metastases occur in approximately 40% of patients presenting with metastatic cancer and often occur in tumor types driven by RAS–MAPK pathway dysregulation, including melanoma, where response rates are low in intracranial disease (39, 40). To model melanoma brain metastases, we used a luciferase-tagged model system for the study of intracranial disease response rates to NST-628. In the SK-MEL-2-luc intracranial xenograft, harboring an NRASQ61R mutation, NST-628 at 3 mg/kg qd and 1.5 mg/kg b.i.d. dosing led to tumor regressions as measured by bioluminescence imaging. In comparison, trametinib and avutometinib only modestly delayed SK-MEL-2 intracranial tumor growth compared with the vehicle control (Fig. 6C and D; Supplementary Fig. S11C and S11D). In addition to strong antitumor activity in the SK-MEL-luc model, NST-628 also showed potent antitumor activity in the MeWo-luc intracranial model, a melanoma cell line harboring an NF1 mutation (Fig. 6E; Supplementary Fig. S11E and S11F). Tovorafenib, a type II RAF inhibitor that has reported CNS activity in BRAF-altered pediatric low-grade glioma (pLGG), had no activity in the MeWo-luc model and accelerated tumor growth. Antitumor activity was due to on-target RAS–MAPK pathway inhibition by NST-628 as measured by DUSP6 transcript as a tumor-specific pharmacodynamic readout (Fig. 6F).
Figure 6.
NST-628 is a fully brain-penetrant inhibitor. A,In vivo half-life, rat brain Kp, Kp, uu, and mouse Kp values for NST-628, trametinib, and avutometinib. B, Plasma concentration compared with phospho-ERK levels 4 hours after dose in normal mouse brains of mice treated with 1 mg/kg trametinib, 1 mg/kg avutometinib, 5 mg/kg cobimetinib, or 0.3, 1, or 3 mg/kg NST-628. C, Bioluminescent imaging of SK-MEL-2-luc intracranial tumors treated with vehicle or 1.5 mg/kg b.i.d. NST-628 at D0, D7, and D18 of the study. D, Intracranial tumor volume measured by bioluminescent imaging of SK-MEL-2-luc tumors treated with 0.3 mg/kg qd trametinib, 0.3 mg/kg qd avutometinib, 3 mg/kg qd or 1.5 mg/kg b.i.d. NST-628. E, Tumor volume as measured by bioluminescent imaging of intracranial MeWo-luc tumors treated with 25 mg/kg qd tovorafenib or 0.3, 1, 3 mg/kg qd NST-628. F, DUSP6 transcript levels in intracranial MeWo-luc tumors treated with 25 mg/kg tovorafenib or 3 mg/kg NST-628 at 4, 8, and 24 hours after single dose of inhibitor.
NST-628 Enhances the Activity of KRAS Inhibitors
Clinically, direct KRAS inhibitors have changed the treatment paradigm for patients with KRAS-altered tumors. However, limitations remain for KRAS inhibitors as single agents clinically, with initial response and durability of response limited for KRASG12C inhibitors (17, 18, 41). Adaptive reactivation of RAS–MAPK signaling due to multiple mechanisms underscores the need for the identification of convergent signaling nodes upstream and downstream to enhance the efficacy of KRAS inhibitors in the clinic by a vertical treatment strategy (19, 42). NST-628 represents a convergent signaling node downstream of KRAS inhibition and we asked if it could enhance the efficacy of FDA-approved KRASG12C inhibitors.
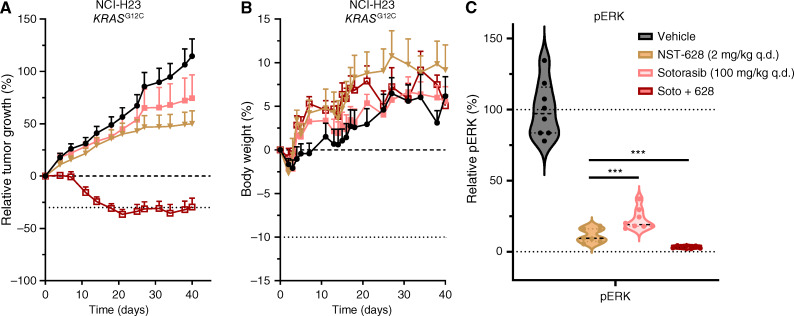
We observed that NST-628 enhances the efficacy of both adagrasib and sotorasib in vitro in the KRASG12C-mutant model NCI-H23 and in multiple models of KRASG12C-mutant cancer, the same combinations displayed an additive effect (Supplementary Fig. S12A–S12D). The combinatorial activity of NST-628 with KRAS mutant–selective inhibition was also confirmed by assessing signaling in two KRASG12C-mutant cell line models. Decreased phospho-ERK was observed with the combination of NST-628 and sotorasib, thereby suggesting a convergent mechanism on the RAS–MAPK pathway as well as inhibition of compensatory signaling through the PI3K–AKT pathway (Supplementary Fig. S12E and S12F). In vivo, low-dose NST-628 (2 mg/kg qd) and high-dose sotorasib (100 mg/kg qd) slowed the growth of NCI-H23 tumors, but only the combination of NST-628 and sotorasib led to deep tumor regressions that were durable to the end of the 40-day study (Fig. 7A). The combination of NST-628 and sotorasib was also well tolerated, with all mice gaining weight throughout the treatment duration (Fig. 7B). Mechanistically, the combination of NST-628 and sotorasib led to a significant decrease in the levels of phospho-ERK, indicating a convergent mechanism on inhibition of the RAS–MAPK pathway (Fig. 7C). Collectively our data support the clinical investigation of NST-628 as a potent and tolerable inhibitor of a convergent signaling node in KRAS inhibitor anchored treatment regimens.
Figure 7.
NST-628 enhances the efficacy of KRASG12C inhibition. A, Tumor volume of NCI-H23 tumors and (B) body weights of NCI-H23 tumor-bearing mice treated with 2 mg/kg NST-628, 100 mg/kg qd sotorasib, or a combination of NST-628 and sotorasib. C, Phospho-ERK levels in NCI-H23 endpoint tumors from A and B collected 4 hours after last dose.
Discussion
RAS–MAPK targeted therapies have had mixed results clinically as single agents and are limited by tolerability, depth of response, and durability. NST-628 was designed to overcome the limitations of previous generations of RAF and MEK inhibitors. NST-628 engages both inactive and active conformations of pan-RAF–MEK complexes, leading to broad efficacy across RAS- and RAF-driven cancers both in vitro and in vivo. Additionally, NST-628 addresses patient populations with no currently approved targeted therapies, including BRAF class II/III-mutant, NRAS-mutant, and KRAS–non-G12C-mutant cancers as well as patients presenting with CNS disease, both primary and metastatic.
We leveraged structural and biochemical insights of the RAF–MEK interface to design a compound that potently binds both RAF and MEK, with a pan-RAF activity. This nondegrading molecular glue does not induce the formation of RAF heterodimers and also lacks the ARAF-sparing Achilles heel of type II RAF inhibitors (8, 43). NST-628 binds both active and inactive conformations of RAF and is insensitive to upstream RAS–MAPK pathway flux, leading to deep and durable pathway inhibition. In comparison, RAF and MEK inhibitor combinations currently under clinical investigation are anchored with either type I or II RAF inhibitors and show highly variable response rates depending on both tumor type and tolerability of the combination regimen (44). NST-628 represents a chemical class with the properties of both an RAF inhibitor and MEK inhibitor, and we demonstrated significant efficacy and greater tolerability than the combination of type II RAF inhibitor belvarafenib and MEK inhibitor cobimetinib in NRAS-mutant melanoma (16). Given that MEK inhibitors as a class have had minimal efficacy in NRAS-mutant tumors as monotherapy, this represents a RAS–MAPK-dependent patient class with unmet therapeutic need and potential for transformative benefit and accelerated approval (45–47).
Beyond demonstrated efficacy in NRAS-mutant cancer models, NST-628 was broadly efficacious in RAS- and RAF-driven cancer models across diverse tumor types, including melanoma, lung, and pancreatic histologies. Mechanistically, NST-628 inhibits multiple BRAF mutants, including BRAF class II/III mutations that sit at the interface between BRAF and MEK, while not driving the formation of RAF heterodimers, rather inhibiting A/B/CRAF–MEK complexes in an inactive conformation. The differentiated mechanism compared with the type II RAF inhibitors, which drive heterodimer formation, more effectively inhibits CRAF-mediated bypass signaling, especially in BRAF class II/III mutant models. Direct CRAF inhibition in the absence of targeting other RAF isoforms leads to paradoxical activation of RAS–MAPK signaling, emphasizing the importance of targeting all RAF isoforms therapeutically rather than targeting CRAF alone (48). Given its best-in-class PK profile, consisting of optimized half-life, volume of distribution, and blood–brain barrier penetrance, NST-628 also overcomes the limitations of MEK inhibitors such as trametinib and the RAF–MEK inhibitor avutometinib that have had toxicities requiring dose reductions and intermittent dosing schedule clinically while expanding the impact on the underserved patient population with RAS-driven CNS malignancies.
With emerging therapeutic strategies directly targeting RAS mutations and diverse, yet convergent mechanisms of resistance involving the RAS–MAPK pathway, NST-628 is positioned to become an ideal combination partner for KRAS inhibitors by effectively inhibiting a convergent signaling node (49). NST-628 effectively combines with the KRASG12C inhibitor sotorasib in an NSCLC tumor model that is refractory to KRASG12C-directed monotherapy and is effective as a single agent in other KRAS-mutant cancers. Our study also showed RTK modulation, parallel PI3K–AKT involvement, and upregulation of tumor-intrinsic immune pathways in short-term adaptive responses and/or resistance to NST-628. These findings identify new strategies for potentially efficacious inhibitor combinations in future preclinical and clinical investigation of NST-628. Collectively, this study shows that with a differentiated mechanism, a balanced metabolic profile, and a potent and tolerable antitumor activity, the nondegrading pan-RAF–MEK glue NST-628 provides a promising therapeutic impact for patients with RAS- and RAF-driven tumors. NST-628 is entering first-in-human clinical trials for the monotherapy treatment of solid tumors with RAS- or RAF mutations.
Methods
Reagents and Cell Lines
NST-628 was synthesized at Wuxi AppTec (Shanghai, CN; WO2023211812, compound 35). NST-628 in powder form was stored at room temperature and protected from light. NST-628 was formulated in 100% DMSO for in vitro assays and aliquoted for long-term storage at −20°C. Trametinib, avutometinib (VS-6766), cobimetinib, naporafenib (LXH254), belvarafenib, exarafenib (KIN-2787), tovorafenib, plixorafenib (PLX8394), and sotorasib were purchased from Selleckchem. Cell lines were obtained directly from ATCC (HCT116, MeWo, NCI-H1666, NCI-H1755, NCI-H2405, OV90), DSMZ (IPC-298), and the Wistar Institute (WM1963, WM3629, WM3670, WM3912 WM3928) and maintained in either McCoy’s 5A, DMEM, or RPMI (Gibco) supplemented with 10% FBS (Gibco) and were not cultured longer than 6 months after receipt from cell banks. Cell lines are regularly tested for Mycoplasma and profiled by STR.
Inhibitor Treatment Assays
Short-term sensitivity to compound was determined by CellTiter-Glo (Promega). Briefly, cell lines were seeded at 1–5 × 103 cells/well of a 96-well plate, and 24 hours after seeding, a serial dilution of compound was added to cells. After 72 hours of inhibitor treatment, plates were developed with CellTiter-Glo and luminescence read on a plate reader (Envision). For the OMNI screen run at Pharmaron, cell lines were seeded in 384-well plates (Corning) overnight and the next day a titration of NST-628 was prepared in DMSO using a TECAN (EVO200) liquid handler, and the compound was added to plates using and Echo 655. Plates were incubated for 3–7 days at 37°C and on the final day of the assay, CellTiter-Glo was added to plates and luminescence read on a plate reader (Envision).
Immunoprecipitation and Western Blot
HCT116 or NCI-H1666 cells were seeded at 10 × 106 cells in a 10-cm dish. The next day, cells were treated with inhibitors for 2 hours and then either a MEK1 or BRAF immunoprecipitation was performed. Briefly, cell lysates were collected in IP lysis buffer (Thermo Fisher) and subjected to overnight immunoprecipitation with MEK1 antibody (Cell Signaling Technology, #2352) or BRAF antibody (Santa Cruz Biotechnology, #sc-5284). The next day protein complexes were incubated with Protein G Dynabeads (Thermo Fisher) and then collected. Immunoprecipitation samples and input samples were resolved on 4% to 12% Bis-Tris Gels, and Western blotting was performed using antibodies against ARAF (Abcam,# ab200653), BRAF (Santa Cruz Biotechnology, #sc-5284), CRAF (BD #610152), MEK1 (Cell Signaling Technology, #2352), phospho-MEK (S217/221; Cell Signaling Technology, #9154), phospho-ERK (T402/Y404; Cell Signaling Technology, #4370), phospho-p90 RSK (Abcam,# ab32413), and vinculin (Millipore Sigma, #V9131).
MEK–RAFs AlphaLISA Protein–Protein Interaction Assay
Protein working solution was prepared by diluting MEK–ARAF, MEK–BRAF or MEK–CRAF to 20 nmol/L into dilution buffer containing 50 mmol/L HEPES pH 7.5, 100 mmol/L NaCl, 0.1% BSA, 2 mmol/L TCEP, 0.05% Tween-20. On a 384-well assay plate (Greiner 781280), compounds were dispensed by a MosquitoHTS liquid handler (SPT Labtech) with a top concentration of 1 μmol/L, 2-fold dilution, and 10-point. 10 μL of protein working solution was transferred onto the assay plate, and the plate was mixed and incubated at 25°C for 30 minutes. After incubation, 5 μL of 80 μg/mL nickel chelate acceptor beads (Revvity, AL108M) in dilution buffer was added and incubated at 25°C for 60 minutes. Finally, 5 μL of 80 μg/mL glutathione donor beads (Revvity, 6765301) in dilution buffer was added to initiate the signal generation. The plate was incubated at 25°C for 120 minutes to stabilize the signal before read on EnVision platereader (PerkinElmer) with the following setting: Mirror: AlphaScreen (444); emission filter: Europium 615 (203); excitation time: 0.18 seconds; emission time: 0.37 seconds. The AlphaLISA signals were normalized to DMSO control, and the normalized data were plotted and fitted in GraphPad Prism 8.0 by using the [Agonist] versus response (three parameters) analysis.
MEK–RAF SPR Experiment
All experiments were performed using a Biacore T200 instrument (Cytiva) at 15°C. WT BRAF, BRAF mutants (S365A, G466A, and G469A) and CRAF were immobilized on a CM5 sensor chip (Cytiva) by amine coupling according to the manufacturer’s instructions. The buffer conditions were 10 mmol/L HEPES pH 7.5, 150 mmol/L NaCl, 1 mmol/L MgCl2, 1 mmol/L DTT, 0.05% Tween-20, and 0.01% DMSO. MEK was injected in single-cycle experiments at concentrations of 0.0156, 0.0625, 0.25, and 1 μmol/L for 60 seconds at a flow rate of 30 μL/minute, followed by a dissociation phase up to 600 seconds. The effects of NST-628 on the BRAF– or CRAF–MEK interactions were determined using the same procedure by injecting premixed MEK protein and 3 μmol/L of NST-628. The resulting sensorgrams were double-referenced, DMSO-calibrated prior to global analysis using a 1:1 interaction model including a linear drift parameter by Biacore Insight Evaluation Software (Cytiva).
Recombinant Protein Expression and Purification
All recombinant protein expression and purification were performed using standard methods. For detailed information about the constructs used in each experiment, see the Supplementary Methods.
Structure Determination
Crystal structures and cryo-EM structures were determined using standard methods. Data collection and refinement statistics can be found in Supplementary Tables S2–S4. All figures were made using Pymol or UCSF Chimera (50), and cryo-EM image processing software was managed through the SBgrid Consortium software collection (51). More detailed information about the determination of each structure can be found in the Supplementary Methods.
Apoptosis Assays
HCT116, IPC-298, SK-MEL-2, and MeWo cells were seeded at 2.5 × 104 cells in a 24-well plate and the next day were treated with 4, 20, 100 nmol/L of NST-628. After 48 hours of treatment, cells were collected and stained for annexin and DAPI using a kit (Thermo Fisher #V13245). Samples were run on an Attune Flow Cytometer (Thermo Fisher) and analyzed using FlowJo software.
Xenograft Studies
For the HCT116 studies, 6- to 8-week-old female BALB/c nude mice (GemPharmatech Co., Ltd) were inoculated with 5 × 106 cells subcutaneously in the flank. Treatment of NST-628 (3, 5 mg/kg qd, 1.5 mg/kg b.i.d.), trametinib (0.3, 1 mg/kg qd), or avutometinib (0.3, 1 mg/kg qd) by oral gavage was initiated when tumor size reached 125 mm3 and tumor size was assessed by caliper measurements for 22 days. For the IPC-298 studies, 6- to 8-week-old female BALB/c nude mice (GemPharmatech Co., Ltd) were inoculated with 5 × 106 cells subcutaneously in the flank. Treatment with NST-628 (5 mg/kg qd; 0.5, 1.5 mg/kg b.i.d.), cobimetinib (5 mg/kg qd), belvarafenib (15 mg/kg qd), or a combination of cobimetinib (5 mg/kg daily) and belvarafenib (15 mg/kg) by oral gavage began when tumors reached 130 mm3 in size and tumor size was assessed by caliper measurements for 26 days. For the NCI-H23 study, 6- to 8-week-old NOG mice (Beijing Vital River Laboratory Animal Technology Co., Ltd) were inoculated with 1.5 × 107 cells in an RPMI/Matrigel mixture subcutaneously in the flank. Treatment with NST-628 (2 mg/kg qd), sotorasib (100 mg/kg QD), or a combination of NST-628 (2 mg/kg qd) and sotorasib (100 mg/kg qd) by oral gavage began when tumors reached 170 mm3 and tumor size was assessed by caliper measurements for 40 days. All xenograft studies were performed at Pharmaron Inc. All the procedures related to animal handling, care, and treatment in this study were performed according to guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of Pharmaron following the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Intracranial Xenograft Studies
For the SK-MEL-2-luc and MeWo-luc xenograft studies, 6- to 8-week-old female NOD SCID mice (Beijing AniKeeper Biotech Co., Ltd) were inoculated intracranially with a luciferase tagged SK-MEL-2 cell line (SK-MEL-2-luc) or the luciferase tagged MeWo cell line (MeWo-luc) with the following procedure. Animals were anesthetized by i.m. injection of ZoletilTM 50 (Virbac S.A.) and xylazine hydrochloride. The skin over the coronal and sagittal sutures of anesthetized mice were sterilized with iodine followed by alcohol. An incision of 0.5 cm was made along the skin over the midline to expose coronal and sagittal suture junctions. Animals were placed on the stereotaxic instrument (Stoelting) for intracranial injection. 2 × 105 luciferase-expressing SK-MEL-2-luc tumor cells suspended in 2 μL EMEM media were injected into the right forebrain by positioning the needle at 2.0 mm lateral to the sagittal suture, 0.5 to 1.0 mm anterior to coronal suture with the injection depth precisely controlled at 3.0 mm. The injection was slowly proceeded over a one-minute period. Upon completing the injection, the needle was retained for another minute. After the needle removal, the hole was sealed with bone wax, and the incision was closed. The tumor growth was monitored by image analysis. Mice were then randomized into each group based on bioluminescent signals of tumors and body weights around days 18 to 20 after tumor cells inoculation. SK-MEL-2-luc mice were treated with NST-628 (3 mg/kg qd, 1.5 mg/kg b.i.d.), trametinib (0.3 mg/kg qd), or avutometinib (0.3 mg/kg qd) by oral gavage. MeWo-luc mice were treated with NST-628 (0.3, 1, 3 mg/kg qd) or tovorafenib (25 mg/kg qd) by oral gavage. Mice were injected intraperitoneally with 15 mg/mL (at 5 μL/g bw) of D-luciferin (PerkinElmer) and anesthetized with 1% to 2% isoflurane inhalation. Ten minutes after the luciferin injection, the mice were imaged using IVIS Lumina III (PerkinElmer) once per week. Living Image software (PerkinElmer) was used to compute regions of interest (ROI) and integrate the total bioluminescence signal in each ROI from the brain. All the procedures relating to animal handling, care, and treatment in this study were performed according to guidelines approved by the IACUC of Pharmaron following the guidance of the Association for AAALAC.
PDX Mini Mouse Trial
Low-passage Champions TumorGraft models (Champions Oncology) were implanted subcutaneously into the left flank of 6- to 8-week-old female athymic nude mice (Envigo) with tumor fragments from each model (CTG-0723, CTG-1684, CTG-1351, CTG-0308, CTG-3063, CTG-0889, CTG-1086, CTG-1375, CTG-0941, CTG-1612, CTG-0302, CTG-0881, CTG-1471, CTG-1358, CTG-0381, CTG-0291, CTG-1441, CTG-2841, CTG-0964, CTG-3059, CTG-0314, CTG-1068, and CTG-1501). After the tumors reached 150 to 300 mm³, mice (n = 2/group) were treated with the vehicle control or NST-628 at 3 mg/kg daily by oral gavage for 28 to 60 days, and tumor volume was assessed by caliper measurements. Vehicle and NST-628 were formulated in 5% DMSO and 95% (20% hydroxypropyl-beta-cyclodextrin (HP-β-CD) in sterile saline. All experimental procedures were approved by Champions Oncology’s IACUC.
Data Availability
Crystal and cryo-EM structures described in this study can be accessed at https://www.rcsb.org under accession codes 9AXM (MEK1–ARAF with NST-628), 9AXX (MEK1–BRAF active with NST-628), 9AYA (MEK1–CRAF active with NST-628), 9AXY (MEK1–BRAF inactive with NST-628), 9AY7 (MEK1–CRAF inactive with NST-628), 9AXH (MEK1–KSR1 with NST-628), 9AXC (MEK1–CRAF with NST-628 focused refinement), and 9AXA (MEK1–CRAF–14-3-3 with NST-628 consensus refinement). Cryo-EM maps described in this study can be accessed at https://www.ebi.ac.uk/emdb/ under accession codes EMDB-43932 (MEK1–CRAF focused refinement) and EMDB-43931 (MEK1–CRAF–14-3-3 consensus refinement). RNA-seq and whole-exome sequencing data sets can be accessed at https://www.ncbi.nlm.nih.gov/sra/PRJNA1087369, submission SUB14270352. Other data sets are available upon reasonable request from the corresponding author.
Supplementary Material
Supplementary methods with detailed protein production, mass spectrometry, in vivo, and in vitro methodology
KINOMESCAN data
Data collection and refinement statistics for crystal structures; Cryo-EM data collection, processing, and model statistics
OMNI screen panel data with NST-628
RNAseq DEG data
Global proteomics data
WES results
PDX model patient characteristics
Supplementary Figure S1 supports Figure 1 and contains cellular properties of NST-628 and a comparison with tramentib Supplementary Figure S2 supports Figure 2 and contains KSR1 data Supplementary Figure S3 supports Figure 2 and contains additional RAF-MEK crystal structures Supplementary Figure S4 supports Figure 2 and contains EGF stimulation experiment Supplementary Figure S5 supports Figure 2 and contains CRAF-MEK cryo-EM Supplementary Figure S6 supports Figure 3 and contains BRAF and MEK IPs, and CRAF IP/MS Supplementary Figure S7 supports Figure 4 and contains the full OMNI screen panel Supplementary Figure S8 supports Figure 4 and contains NST-628 RNAseq, signaling and proteomics studies Supplementary Figure S9 supports Figure 4 and contains NST-628 resistance studies Supplementary Figure S10 supports Figure 5 and contains HCT116 and IPC298 in vivo data Supplementary Figure S11 supports Figure 6 and contains NST-628 PK as well as SK-MEL-2 and MeWo in vivo data Supplementary Figure S12 Supports Figure 7 and contains KRAS G12C inhibitor combination studies
Acknowledgments
We thank our colleagues at Nested Therapeutics for their thoughtful discussions and constructive feedback throughout the NST-628 program and for the critical review of the manuscript. We also thank the scientists at Pharmaron and Wuxi AppTec for their excellent in vivo study and medicinal chemistry support, respectively, as well as additional research support at Viva Biotech, Beactica, HD Bio, and Champions Oncology. Finally, we thank the patients and their families for their participation in the phase I clinical trial with NST-628. This study was funded by Nested Therapeutics.
Footnotes
Note Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
M.B. Ryan reports other support from Nested Therapeutics during the conduct of the study. B. Quade reports other support from Nested Therapeutics, Inc. during the conduct of the study. S.E. Cohen reports that he is an employee of and shareholder in Nested Therapeutics. A. Ozen reports being an employee with equity interest in Nested Therapeutics. C. Ye reports other support from Nested Therapeutics during the conduct of the study. A.C. Dar reports personal fees and other support from Nested Therapeutics during the conduct of the study; and ACD is a cofounder, consultant, shareholder, and advisory board member to Nested Therapeutics. K.P. Hoeflich reports personal fees from Turbine AI outside the submitted work. M. Hale reports a patent for WO/2023/211812 pending. No disclosures were reported by the other authors.
Authors’ Contributions
M.B. Ryan: Conceptualization, data curation, formal analysis, writing–original draft, writing–review and editing. B. Quade: Data curation, formal analysis, visualization, methodology, writing–original draft, writing–review and editing. N. Schenk: Data curation, formal analysis, visualization, methodology, writing–review and editing. Z. Fang: Data curation, formal analysis, methodology, writing–original draft, writing–review and editing. M. Zingg: Data curation, formal analysis, visualization, methodology, writing–review and editing. S.E. Cohen: Data curation, methodology, writing–original draft, writing–review and editing. B.M. Swalm: Data curation, formal analysis, methodology, writing–review and editing. C. Li: Formal analysis, visualization, methodology, writing–review and editing. A. Ozen: Data curation, formal analysis, visualization, methodology, writing–review and editing. C. Ye: Software, formal analysis, visualization, methodology, writing–review and editing. M. Ritorto: Data curation, formal analysis, supervision, methodology, writing–review and editing. X. Huang: Formal analysis, supervision, writing–review and editing. A.C. Dar: Conceptualization, writing–review and editing. Y. Han: Conceptualization, supervision, writing–review and editing. K.P. Hoeflich: Conceptualization, resources, supervision, project administration, writing–review and editing. M. Hale: Conceptualization, formal analysis, supervision, methodology, writing–review and editing. M. Hagel: Conceptualization, methodology, writing–review and editing.
References
- 1. Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol 2018;15:709–20. [DOI] [PubMed] [Google Scholar]
- 2. Prior IA, Hood FE, Hartley JL. The frequency of Ras mutations in cancer. Cancer Res 2020;80:2969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Papke B, Der CJ. Drugging RAS: know the enemy. Science 2017;355:1158–63. [DOI] [PubMed] [Google Scholar]
- 4. Kun E, Tsang YTM, Ng CW, Gershenson DM, Wong KK. MEK inhibitor resistance mechanisms and recent developments in combination trials. Cancer Treat Rev 2021;92:102137. [DOI] [PubMed] [Google Scholar]
- 5. Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nat Rev Clin Oncol 2014;11:385–400. [DOI] [PubMed] [Google Scholar]
- 6. Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 2014;25:697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wildtype BRAF. Nature 2010;464:427–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Monaco K-A, Delach S, Yuan J, Mishina Y, Fordjour P, Labrot E, et al. LXH254, a potent and selective ARAF-sparing inhibitor of BRAF and CRAF for the treatment of MAPK-driven tumors. Clin Cancer Res 2021;27:2061–73. [DOI] [PubMed] [Google Scholar]
- 9. Yen I, Shanahan F, Lee J, Hong YS, Shin SJ, Moore AR, et al. ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma. Nature 2021;594:418–23. [DOI] [PubMed] [Google Scholar]
- 10. Zhang C, Spevak W, Zhang Y, Burton EA, Ma Y, Habets G, et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature 2015;526:583–6. [DOI] [PubMed] [Google Scholar]
- 11. Yao Z, Gao Y, Su W, Yaeger R, Tao J, Na N, et al. RAF inhibitor PLX8394 selectively disrupts BRAF dimers and RAS-independent BRAF-mutant-driven signaling. Nat Med 2019;25:284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367:1694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, et al. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov 2015;5:358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corcoran RB, Andre T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF(V600E)-mutant colorectal cancer. Cancer Discov 2018;8:428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Braud F, Dooms C, Heist RS, Lebbe C, Wermke M, Gazzah A, et al. Initial evidence for the efficacy of naporafenib in combination with trametinib in NRAS-mutant melanoma: results from the expansion arm of a phase Ib, open-label study. J Clin Oncol 2023;41:2651–60. [DOI] [PubMed] [Google Scholar]
- 16. Shin SJ, Lee J, Kim TM, Kim J-S, Kim YJ, Hong YS, et al. A phase Ib trial of belvarafenib in combination with cobimetinib in patients with advanced solid tumors: interim results of dose-escalation and patients with NRAS-mutant melanoma of dose-expansion. J Clin Oncol 39, 2021. (suppl 15; abstr 3007). [Google Scholar]
- 17. Awad MM, Liu S, Rybkin II, Arbour KC, Dilly J, Zhu VW, et al. Acquired resistance to KRAS(G12C) inhibition in cancer. N Engl J Med 2021;384:2382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanaka N, Lin JJ, Li C, Ryan MB, Zhang J, Kiedrowski LA, et al. Clinical acquired resistance to KRASG12C inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS–MAPK reactivation. Cancer Discov 2021;11:1913–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ryan MB, Fece de la Cruz F, Phat S, Myers DT, Wong E, Shahzade HA, et al. Vertical pathway inhibition overcomes adaptive feedback resistance to KRASG12C inhibition. Clin Cancer Res 2020;26:1633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fabian MA, Biggs WH, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, et al. A small molecule–kinase interaction map for clinical kinase inhibitors. Nat Biotechnol 2005;23:329–36. [DOI] [PubMed] [Google Scholar]
- 21. Khan ZM, Real AM, Marsiglia WM, Chow A, Duffy ME, Yerabolu JR, et al. Structural basis for the action of the drug trametinib at KSR-bound MEK. Nature 2020;588:509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gonzalez-Del Pino GL, Li K, Park E, Schmoker AM, Ha BH, Eck MJ. Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation. Proc Natl Acad Sci U S A 2021;118:e2107207118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yaeger R, Corcoran RB. Targeting alterations in the RAF–MEK pathway. Cancer Discov 2019;9:329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karoulia Z, Gavathiotis E, Poulikakos PI. New perspectives for targeting RAF kinase in human cancer. Nat Rev Cancer 2017;17:676–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vasta JD, Michaud A, Zimprich CA, Beck MT, Swiatnicki MR, Zegzouti H, et al. Protomer selectivity of type II RAF inhibitors within the RAS/RAF complex. Cell Chem Biol 2023;30:1354–65. [DOI] [PubMed] [Google Scholar]
- 26. Liau NPD, Wendorff TJ, Quinn JG, Steffek M, Phung W, Liu P, et al. Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization. Nat Struct Mol Biol 2020;27:134–41. [DOI] [PubMed] [Google Scholar]
- 27. Park E, Rawson S, Li K, Kim B-W, Ficarro SB, Pino GG-D, et al. Architecture of autoinhibited and active BRAF–MEK1–14-3-3 complexes. Nature 2019;575:545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kondo Y, Ognjenović J, Banerjee S, Karandur D, Merk A, Kulhanek K, et al. Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases. Science 2019;366:109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dankner M, Wang Y, Fazelzad R, Johnson B, Nebhan CA, Dagogo-Jack I, et al. Clinical activity of mitogen-activated protein kinase–targeted therapies in patients with non–V600 BRAF-mutant tumors. JCO Precis Oncol 2022;6:e2200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haling JR, Sudhamsu J, Yen I, Sideris S, Sandoval W, Phung W, et al. Structure of the BRAF-MEK complex reveals a kinase activity independent role for BRAF in MAPK signaling. Cancer Cell 2014;26:402–13. [DOI] [PubMed] [Google Scholar]
- 32. Guo C, Chénard-Poirier M, Roda D, de Miguel M, Harris SJ, Candilejo IM, et al. Intermittent schedules of the oral RAF-MEK inhibitor CH5126766/VS-6766 in patients with RAS/RAF-mutant solid tumours and multiple myeloma: a single-centre, open-label, phase 1 dose-escalation and basket dose-expansion study. Lancet Oncol 2020;21:1478–88. [DOI] [PubMed] [Google Scholar]
- 33. Banerjee SN, Ring KL, Nieuwenhuysen EV, Fabbro M, Aghajanian C, Oaknin A, et al. Initial efficacy and safety results from ENGOT-ov60/GOG-3052/RAMP 201: A phase 2 study of avutometinib (VS-6766) ± defactinib in recurrent low-grade serous ovarian cancer (LGSOC). J Clin Oncol 41, 2023. (suppl 16; asbtr 5515). [Google Scholar]
- 34. Tian J, Chen JH, Chao SX, Pelka K, Giannakis M, Hess J, et al. Combined PD-1, BRAF and MEK inhibition in BRAF(V600E) colorectal cancer: a phase 2 trial. Nat Med 2023;29:458–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ribas A, Lawrence D, Atkinson V, Agarwal S, Miller WH, Carlino MS, et al. Combined BRAF and MEK inhibition with PD-1 blockade immunotherapy in BRAF-mutant melanoma. Nat Med 2019;25:936–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murphy BM, Terrell EM, Chirasani VR, Weiss TJ, Lew RE, Holderbaum AM, et al. Enhanced BRAF engagement by NRAS mutants capable of promoting melanoma initiation. Nat Commun 2022;13:3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Garcia-Alvarez A, Ortiz C, Muñoz-Couselo E. Current perspectives and novel strategies of NRAS-mutant melanoma. Oncotargets Ther 2021;14:3709–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wong H, Choo EF, Alicke B, Ding X, La H, McNamara E, et al. Anti-tumor activity of targeted and cytotoxic agents in murine subcutaneous tumor models correlates with clinical response. Clin Cancer Res 2012;18:3846–55. [DOI] [PubMed] [Google Scholar]
- 39. Kim AE, Nieblas-Bedolla E, de Sauvage MA, Brastianos PK. Leveraging translational insights toward precision medicine approaches for brain metastases. Nat Cancer 2023;4:955–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tawbi HA, Boutros C, Kok D, Robert C, McArthur G. New Era in the management of melanoma brain metastases. Am Soc Clin Oncol Educ Book 2018:741–50. [DOI] [PubMed] [Google Scholar]
- 41. Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N Engl J Med 2020;383:1207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov 2020;19:533–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yen I, Shanahan F, Lee J, Hong YS, Shin SJ, Moore AR, et al. ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma. Nature 2021;594:418–23. [DOI] [PubMed] [Google Scholar]
- 44. Poulikakos PI, Sullivan RJ, Yaeger R. Molecular pathways and mechanisms of BRAF in cancer therapy. Clin Cancer Res 2022;28:4618–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moschos SJ. War against NRAS-mutant melanoma using targeted therapies remains challenging. Clin Cancer Res 2022;28:2977–9. [DOI] [PubMed] [Google Scholar]
- 46. Johnson DB, Puzanov I. Treatment of NRAS-mutant melanoma. Curr Treat Options Oncol 2015;16:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murphy BM, Terrell EM, Chirasani VR, Weiss TJ, Lew RE, Holderbaum AM, et al. Enhanced BRAF engagement by NRAS mutants capable of promoting melanoma initiation. Nat Commun 2022;13:3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Morgan CW, Dale IL, Thomas AP, Hunt J, Chin JW. Selective CRAF inhibition elicits transactivation. J Am Chem Soc 2021;143:4600–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sattler M, Mohanty A, Kulkarni P, Salgia R. Precision oncology provides opportunities for targeting KRAS-inhibitor resistance. Trends Cancer 2023;9:42–54. [DOI] [PubMed] [Google Scholar]
- 50. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF chimera: a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605–12. [DOI] [PubMed] [Google Scholar]
- 51. Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, et al. Collaboration gets the most out of software. eLife 2013;2:e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods with detailed protein production, mass spectrometry, in vivo, and in vitro methodology
KINOMESCAN data
Data collection and refinement statistics for crystal structures; Cryo-EM data collection, processing, and model statistics
OMNI screen panel data with NST-628
RNAseq DEG data
Global proteomics data
WES results
PDX model patient characteristics
Supplementary Figure S1 supports Figure 1 and contains cellular properties of NST-628 and a comparison with tramentib Supplementary Figure S2 supports Figure 2 and contains KSR1 data Supplementary Figure S3 supports Figure 2 and contains additional RAF-MEK crystal structures Supplementary Figure S4 supports Figure 2 and contains EGF stimulation experiment Supplementary Figure S5 supports Figure 2 and contains CRAF-MEK cryo-EM Supplementary Figure S6 supports Figure 3 and contains BRAF and MEK IPs, and CRAF IP/MS Supplementary Figure S7 supports Figure 4 and contains the full OMNI screen panel Supplementary Figure S8 supports Figure 4 and contains NST-628 RNAseq, signaling and proteomics studies Supplementary Figure S9 supports Figure 4 and contains NST-628 resistance studies Supplementary Figure S10 supports Figure 5 and contains HCT116 and IPC298 in vivo data Supplementary Figure S11 supports Figure 6 and contains NST-628 PK as well as SK-MEL-2 and MeWo in vivo data Supplementary Figure S12 Supports Figure 7 and contains KRAS G12C inhibitor combination studies
Data Availability Statement
Crystal and cryo-EM structures described in this study can be accessed at https://www.rcsb.org under accession codes 9AXM (MEK1–ARAF with NST-628), 9AXX (MEK1–BRAF active with NST-628), 9AYA (MEK1–CRAF active with NST-628), 9AXY (MEK1–BRAF inactive with NST-628), 9AY7 (MEK1–CRAF inactive with NST-628), 9AXH (MEK1–KSR1 with NST-628), 9AXC (MEK1–CRAF with NST-628 focused refinement), and 9AXA (MEK1–CRAF–14-3-3 with NST-628 consensus refinement). Cryo-EM maps described in this study can be accessed at https://www.ebi.ac.uk/emdb/ under accession codes EMDB-43932 (MEK1–CRAF focused refinement) and EMDB-43931 (MEK1–CRAF–14-3-3 consensus refinement). RNA-seq and whole-exome sequencing data sets can be accessed at https://www.ncbi.nlm.nih.gov/sra/PRJNA1087369, submission SUB14270352. Other data sets are available upon reasonable request from the corresponding author.