Abstract
Objectives
To characterize the genetic basis of azithromycin resistance in Escherichia coli and Salmonella collected within the EU harmonized antimicrobial resistance (AMR) surveillance programme in 2014–18 and the Danish AMR surveillance programme in 2016–19.
Methods
WGS data of 1007 E. coli [165 azithromycin resistant (MIC > 16 mg/L)] and 269 Salmonella [29 azithromycin resistant (MIC > 16 mg/L)] were screened for acquired macrolide resistance genes and mutations in rplDV, 23S rRNA and acrB genes using ResFinder v4.0, AMRFinder Plus and custom scripts. Genotype–phenotype concordance was determined for all isolates. Transferability of mef(C)-mph(G)-carrying plasmids was assessed by conjugation experiments.
Results
mph(A), mph(B), mef(B), erm(B) and mef(C)-mph(G) were detected in E. coli and Salmonella, whereas erm(C), erm(42), ere(A) and mph(E)-msr(E) were detected in E. coli only. The presence of macrolide resistance genes, alone or in combination, was concordant with the azithromycin-resistant phenotype in 69% of isolates. Distinct mph(A) operon structures were observed in azithromycin-susceptible (n = 50) and -resistant (n = 136) isolates. mef(C)-mph(G) were detected in porcine and bovine E. coli and in porcine Salmonella enterica serovar Derby and Salmonella enterica 1,4, [5],12:i:-, flanked downstream by ISCR2 or TnAs1 and associated with IncIγ and IncFII plasmids.
Conclusions
Diverse azithromycin resistance genes were detected in E. coli and Salmonella from food-producing animals and meat in Europe. Azithromycin resistance genes mef(C)-mph(G) and erm(42) appear to be emerging primarily in porcine E. coli isolates. The identification of distinct mph(A) operon structures in susceptible and resistant isolates increases the predictive power of WGS-based methods for in silico detection of azithromycin resistance in Enterobacterales.
Introduction
The macrolide azithromycin is a critically important clinical antimicrobial,1 increasingly used as an alternative when typical first-line antimicrobials (e.g. quinolones) are no longer effective in the treatment of severe cases of bacterial gastrointestinal infections.2–5 Considering that azithromycin is one of the few available options for treatment of MDR bacteria and that the majority of the azithromycin resistance genes are acquired, the spread of azithromycin resistance could seriously decrease the options to fight life-threatening infections. Among the azithromycin resistance genes,6 mph(A) and erm genes, encoding macrolide 2′-phosphotransferase and rRNA methylases, respectively, are the two main mechanisms involved in high-level azithromycin resistance.7,8 Another recently identified resistance mechanism in Escherichia coli conferred by tandemly arranged plasmid-borne genes mef(C)-mph(G), encoding an efflux pump and a phosphorylase, respectively, has been described to mediate high-level azithromycin resistance.9 Additionally, substitutions in the 50S ribosomal subunit proteins L4 (rplD) and L22 (rplV), in 23S rRNA (rrlH) and in the efflux pump AcrB (R717Q/L) also can lead to increased macrolide resistance.2,6 However, genes encoding efflux pumps, e.g. msr(A), msr(D), mef(A), mef(B), ere genes encoding macrolide esterases, and mph(B) encoding a macrolide phosphorylase appear to have no role or only marginal roles in azithromycin resistance in Enterobacterales.6–8
Due to the transmission of antimicrobial-resistant bacteria between animals and humans, the EU has implemented harmonized monitoring and reporting of antimicrobial resistance (AMR) in zoonotic and commensal bacteria from food-producing animals and food since 2014.10,11 In this monitoring programme, azithromycin susceptibility of E. coli and Salmonella enterica is tested phenotypically by broth microdilution.
Phenotypic azithromycin susceptibility testing is technically challenging and presents reproducibility issues in classifying isolates consistently as susceptible or resistant, which could be overcome by using WGS methods. However, these methods perform with high accuracy only for well-studied AMR determinants. Therefore, elucidating AMR gene patterns to increase the accuracy and effectiveness of WGS approaches is of great importance.
Although the prevalence of azithromycin-resistant E. coli and Salmonella in the EU has generally been reported to be low depending on the country and isolation source,12 a proper assessment of the risk to humans posed by azithromycin-resistant bacteria from food animals and food requires knowledge of resistance determinants. Therefore, the objectives of this study were to elucidate the genetic basis of azithromycin resistance in E. coli and Salmonella from food-producing animals and meat from 27 European countries and explore the azithromycin genotype–phenotype correspondence. Furthermore, this study aimed to resolve the molecular basis of genotype–phenotype discordance previously observed for mph(A), which is the most common azithromycin resistance gene in E. coli, and to characterize the genetic context and transferability of mef(C)-mph(G) genes, which are emerging in Enterobacterales.
Methods
Bacterial isolates
A total of 1276 isolates were examined in this study. These isolates comprised 1007 E. coli and 269 Salmonella enterica subsp. enterica serovars (Table S1, available as Supplementary data at JAC Online) collected within the EU harmonized monitoring of AMR in zoonotic and indicator bacteria from food-producing animals and food in 2014–18,10,13 and the Danish Programme for surveillance of antimicrobial consumption and resistance in bacteria from food animals and food (www.danmap.org) in 2017–19. The EU harmonized monitoring system involved biannual surveillance of different animal types. Isolates from 2014, 2016 and 2018 originated from poultry caeca and meat, whereas isolates from 2015 and 2017 and from the Danish AMR surveillance programme were recovered from porcine and bovine caeca and meat. For the Danish collection, only isolates with azithromycin MIC > 16 mg/L were included. Antimicrobial susceptibility testing was performed by broth microdilution using Sensititre MIC susceptibility plates (EUVSEC1 and EUVSEC2, Thermo Fisher Scientific) in duplicate, by the originating laboratory and at the EU Reference Laboratory – Antimicrobial Resistance (EURL-AR). In cases of more than one 2-fold dilution difference, the tests were repeated a third time. The azithromycin MIC values were interpreted in accordance with The European Food Safety Authority (EFSA)-defined surveillance breakpoint. Isolates with MIC ≤ 16 mg/L were classified as WT and isolates with MIC > 16 mg/L as non-WT.13
Identification of genes and chromosomal mutations associated with macrolide resistance
WGS of the isolates was performed using Illumina paired-end sequencing on MiSeq, HiSeq or NextSeq platforms (Illumina, Inc., San Diego, CA, USA). Sequencing reads were trimmed using Trimmomatic v0.3814 and assembled by metaSPAdes v3.13.0.15 Assemblies were evaluated by Quast16 and genomes with ≤500 contigs were included in further analysis (Table S1). The ResFinder database v4.017 incorporated into ABRicate v0.9.8 (https://github.com/tseemann/abricate) was used to detect acquired macrolide resistance genes with minimum identity and coverage thresholds of 90% and 60%, respectively. Chromosomal mutations in acrB and 23S rRNA genes were screened for using AMRFinder (https://github.com/ncbi/amr/wiki). The sequences of chromosomal rplDV genes were screened for mutations mediating azithromycin resistance7,18 using custom scripts. Sequence alignments of rplDV genes were performed using MAFFT in Geneious Prime v2020.0.4 (https://www.geneious.com) and compared with rplDV of E. coli ATCC 25922 (NC_000913.3) and Salmonella enterica serovar Typhimurium LT2 (NC_003197) as reference genes. MLST was performed using mlst v2.19.0 according to the Achtman schemes (https://github.com/tseemann/mlst). Salmonella serovar determination was carried out by SeqSero2 v1.1.1.19 Sequences from isolates positive for mph(A) and mef(C)-mph(G) were screened for plasmid replicon genes using PlasmidFinder (database version February 2020)20 with minimum identity and coverage thresholds as above. IS and Tn were identified using ISfinder (https://www-is.biotoul.fr)21 and TnCentral (https://tncentral.ncc.unesp.br/index.html). Visualization of the genetic contexts of the macrolide resistance genes was performed by pyGenomeViz v0.4.3 (https://github.com/moshi4/pyGenomeViz). Graphs were created using ‘ggplot2’ and ‘networkD3’ packages in R v4.1.0.22,23 Raw sequence data were submitted to the European Nucleotide Archive (ENA) at EMBL-EBI under accession numbers PRJEB18618, PRJEB21546, PRJEB33169, PRJEB43436, PRJEB43584, PRJEB63535 and PRJEB63683 (Table S1).
Conjugation experiments and characterization of mef(C)-mph(G)-harbouring contigs
Isolates harbouring mef(C)-mph(G) (Table 1) were used as donors in filter-mating experiments with a rifampicin-resistant, lactose-negative E. coli J62-2 strain as recipient.24 Transconjugants (TCs) were selected on MacConkey agar (Sigma, Denmark) supplemented with 16 mg/L azithromycin (Sigma, Denmark) and 50 mg/L rifampicin (Sigma, Denmark). TCs were initially verified by MIC determination as described for the original isolates and by colony PCR targeting mef(C)-mph(G) using primers F3220-5′-ATTGGCGGTGTCATCCTGAG-3′ and R3221-5′-CGCTGACTTGTGCAGTTGAC-3′. Plasmid DNA from TCs with azithromycin MIC > 16 mg/L and mef(C)-mph(G) positive by PCR was extracted using the QIAGEN Plasmid Midi kit (QIAGEN, Germany), prepared for sequencing using the Nextera XT DNA Library Preparation Kit and sequenced with a NextSeq 500/550 Mid Output v2.5 Kit (300 cycles) on a NextSeq 500 platform (Illumina). Raw plasmid sequence data were subjected to quality checking using FastQC v0.11.5,25 trimmed with BBTools v.36.4926 and assembled by SPAdes v3.15.3.27 Assemblies were analysed by ResFinder and PlasmidFinder with thresholds as described above. Due to the common occurrence of co-extraction of chromosomal DNA during plasmid DNA extraction, seven-gene MLST analysis was successfully performed to distinguish between true TCs (having ST of the recipient strain) and mutated donor strains (having ST of the respective donor strain).
Table 1.
Characteristics of E. coli and Salmonella sp. isolates carrying mef(C)-mph(G) genes
| Strain ID | Species | ENA run accession number | Host | Country of origin | Source of isolation | Year of isolation | MLST ST | Azithromycin MIC (mg/L) |
Conjugative transfer of mef(C)-mph(G) |
Plasmid repliconsa | Other AMR genesa |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NRS-2015 ESBL-08-69 | E. coli | ERR2019198 | Porcine | Netherlands | Caecal content | 2015 | 453 | 32 | Yes | IncIFIB, IncX1, IncIγ, IncFII, IncQ1, Col8282 | tet(A), sul3, sul2, ant(3′)-Ia, cmlA1, aph(3′)-Ib, aph(6)-Id, dfrA5, blaTEM-52 |
| ZTA15:00420EB1 | E. coli | ERR2019249 | Porcine | Spain | Caecal content | 2015 | 7456 | 32 | No | ColE10, Col156, IncFII(pRSB107), Col(MG828), IncIγ, IncX4, IncHI2; IncHI2A, IncQ1 | tet(M), mcr-4.1, aph(4)-Ia, aac(3)-Ia, tet(A), blaCTX-M-14, blaTEM-1B, aph(3′)-Ia, mcr-1.1, mph(B), sul1, ant(3′)-Ia, drfA1, aph(6)-Id, aph(3′)-Ib, sul2 |
| HP-6957 | E. coli | ERR3393110 | Porcine | Czechia | Caecal content | 2017 | 57 | 64 | Yes | IncFIB, IncFIC(FII), IncIγ | aadA1 |
| ECO NRS 187.57 | E. coli | ERR3393279 | Porcine | Netherlands | Caecal content | 2017 | 88 | >64 | Yes | IncFIB, IncFIC(FII), IncIγ | — |
| 17-AB00518_0 | E. coli | ERR3393142 | Bovine | Germany | Caecal content | 2017 | 48 | 64 | Yes | IncIγ, Col(pHAD28) | sul2 , blaCTX-M-1 |
| 10047105/3 | E. coli | ERR3393194 | Porcine | Hungary | Meat | 2017 | 453 | >64 | Yes | IncFII, IncIγ, IncY | sul2 , blaCTX-M-1, tet(A), dfrA17, aadA5 |
| HP36996 | E. coli | ERR3393109 | Porcine | Czechia | Caecal content | 2017 | 165 | >64 | No | IncFII, IncIγ, Col(MG828) | — |
| 17-AB01235_0 | E. coli | ERR3393151 | Bovine | Germany | Caecal content | 2017 | NTb | 32 | No | IncFIA, IncFIB, IncFIC, IncIγ, IncX4 | sul2, tet(B), blaCTX-M-1, fosA7 |
| 17-AB02375_0 | E. coli | ERR3393160 | Porcine | Germany | Caecal content | 2017 | 57 | 64 | No | IncFIA(HI1), IncFIB, IncFIC(FII), IncIγ, IncHI1A, IncHI1B | sul2, sul3, blaCTX-M-1, tet(A), tet(B), dfrA12, dfrA1, qnrS1, aadA1, aadA2, aadA5, cmlA, floR, aph(3′)-Ib, aph(3′)-Ia, aph(6)-Id, blaTEM-1B, ant(3′)-1, tet(M) |
| 10849-2GR | E. coli | ERR3393337 | Porcine | Romania | Caecal content | 2017 | 101 | 64 | No | Col156, ColRNAI, IncFIB, IncFIC(FII), IncIγ, IncFII, Col(pHAD28) | sul2, blaCTX-M-1, tet(A), aadA5, aadA1, dfrA17, dfrA1, ant(3′)-Ia |
| Ec151 | E. coli | ERR11629170 | Bovine | Denmark | Caecal content | 2019 | 8863 | >64 | ND | IncIα | sul2, aadA5, dfrA17, bla CTX-M-1 |
| S308 | Salmonella 1,4,[5],12:i:-c | ERR11628334 | Porcine | Denmark | Clinical | 2017 | 19 | >64 | No | IncFIB, IncFII | sul1, sul2, blaTEM-1B, aadA1, aac(6′)-Ia; tet(A), dfrA1 |
| S514 | Salmonella Derbyd | ERR11628347 | Porcine | Denmark | Clinical | 2018 | 40 | >16 | No | ColpVC, IncQ1 | sul2, aac(6′)-Ia; aph(6)-Id, aph(3′)-Ib, fosA7, dfrA17 |
aIn bold, plasmid replicons and AMR genes detected in TCs.
bNot typeable.
c S. enterica subsp. enterica serovar 1,4, [5],12:i:-.
d S. enterica subsp. enterica serovar Derby.
Results and discussion
Macrolide resistance genes in E. coli and Salmonella from food-producing animals and meat in the EU
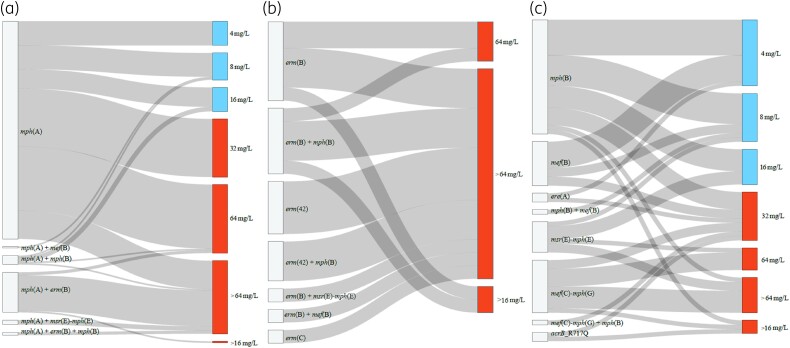
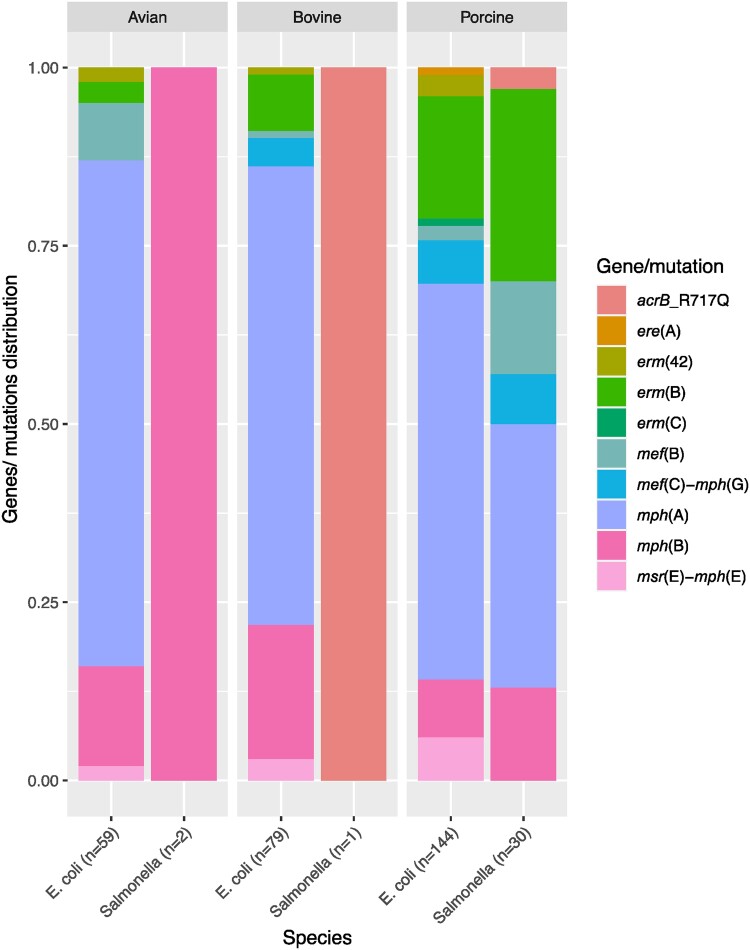
We screened commensal E. coli and Salmonella isolates from food-producing animals and meat in Europe for the presence of macrolide resistance determinants and compared these genotypes with their respective azithromycin phenotypes. Eleven different macrolide resistance genes and five non-synonymous rplDV mutations were detected in 248 E. coli and 26 Salmonella (enterica serovars S. Rissen, S. Blockley, S. Typhimurium, S. Paratyphi-B-var-Java, S. Bredeney, S. Infantis, S. 1,4, [5],12:i:-, S. Derby and S. Dublin) isolates (Tables S4, S5 and S6). The presence of one or more macrolide resistance gene(s) or known mutations was associated with the azithromycin resistance phenotype in 159 (66%) E. coli and 24 (92%) Salmonella isolates (Figure 1 and Table S4). No known genes or mutations mediating azithromycin resistance were detected in the remaining 1002 isolates, although 10 of these isolates exhibited azithromycin MIC > 16 mg/L (Table S4). Six genes, namely mph(A), mph(B), mef(B), erm(B) and mef(C)-mph(G), were detected in both E. coli and Salmonella, whereas erm(C), erm(42), ere(A) and msr(E)-mph(E) were detected in E. coli only (Table S4). Seven of all detected macrolide resistance genes [mph(A), mph(B), mef(B), erm(B), erm(42), msr(E)-mph(E)] were found in isolates from all animal sources, while mef(C)-mph(G) were detected in isolates of bovine and porcine origin only, and ere(A) and erm(C) were harboured by isolates of porcine origin only (Figure 2, Table S4).
Figure 1.
Sankey plots showing macrolide resistance genes/mutations and azithromycin MICs in the E. coli and Salmonella isolates in this study. mph(A) gene and its combinations with other macrolide resistance genes (a), erm genes and their combinations with other macrolide resistance genes (b), other macrolide resistance genes (c). The rplDV non-synonymous mutations detected in seven susceptible isolates were not included in the graph (Table S4). It is important to note that in resistant isolates harbouring mph(A) in combination with mph(B) or mef(B), the full mph(A) operon was present, and the promoter region was complete in all cases (Table S4). The 10 resistant isolates (MIC > 16 mg/L) without known azithromycin resistance mechanisms are not included in the graph. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Figure 2.
Distribution of macrolide resistance genes in E. coli and Salmonella isolates in this study according to their origin. Twenty-one percent (n = 51) of the isolates carried more than one macrolide resistance gene and are included under each of the genes they carry. The total number of genes in each species or from each source is given in parentheses. Two mph(A)- and two mph(B)-harbouring E. coli isolates for which isolation sources are not available are not included. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Regarding chromosomal mutations, five non-synonymous rplDV mutations were detected in seven E. coli isolates with MIC ≤ 16 mg/L (Table S4), while no rplDV mutations were detected in Salmonella. Mutations in 23S rRNA genes were not detected in any of the analysed genomes. Two azithromycin-resistant Salmonella isolates (serovars 1,4, [5],12:i:- and Dublin) with no azithromycin resistance gene harboured the AcrB-R717Q substitution. The 10 isolates with no identified mechanism of azithromycin resistance were screened for additional non-synonymous mutations in the acrR and acrB genes, and either no mutations were found, or the detected amino acid substitutions/deletions were also observed in susceptible isolates (Table S4).
Mph(A) operon structures in azithromycin-resistant and -susceptible isolates
One of the established macrolide resistance genes that confers high-level azithromycin resistance in Enterobacterales is mph(A).5,6,28 Here, mph(A) was detected in 15% of the isolates (n = 186). The E. coli mph(A)-harbouring isolates (n = 175) were genetically diverse, belonging to 77 STs, with ST744, ST1011, ST10 and ST410 being the most represented. The mph(A)-carrying Salmonella isolates (n = 11) belonged to four STs (ST469, ST34, ST19 and ST52) (Table S4). The isolates harbouring mph(A) displayed a wide range of azithromycin MIC values ranging from 4 to >64 mg/L (Figure 2 and Table S4). Of these isolates, 27% (n = 50) had a susceptible phenotype (MIC ≤ 16 mg/L). Previous studies have also shown that mph(A) can be present in azithromycin-susceptible isolates. For instance, Gomes et al.7 reported that 7% of the mph(A)-harbouring E. coli isolates in their study were susceptible, which suggests that the mph(A) gene does not always confer a resistant phenotype in Enterobacterales.
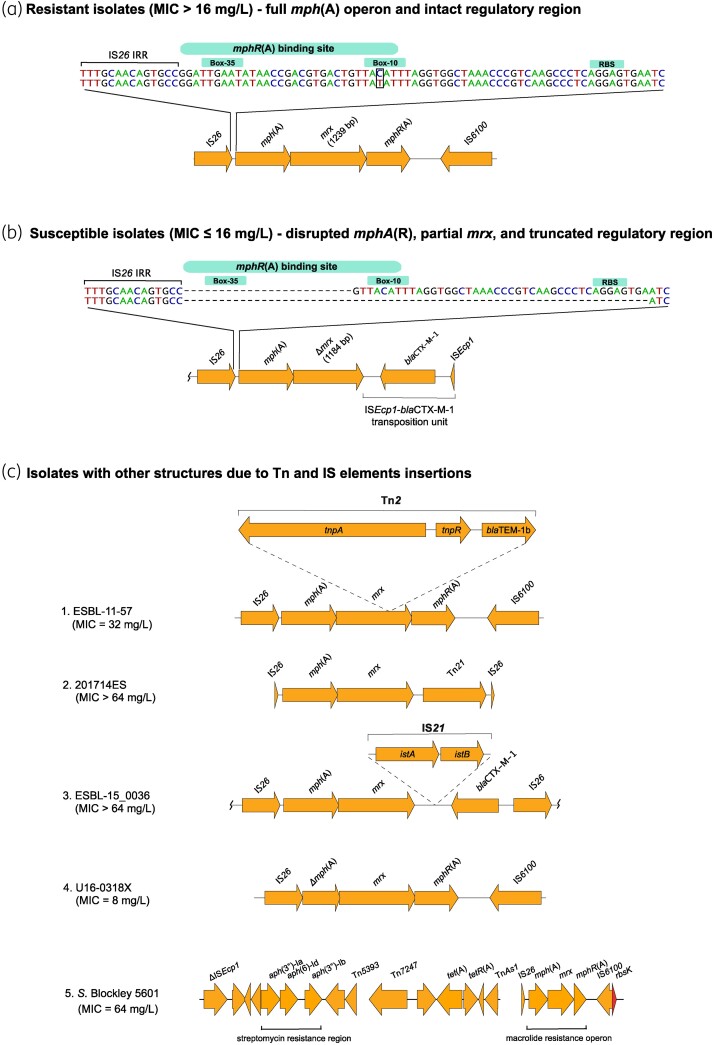
The mph(A) gene encoding macrolide 2′-phosphotransferase I is part of an operon, mph(A)-mrx-mphR(A), in which the downstream genes mrx and mphR(A) encode a protein with unknown function and a repressor that controls the inducible expression of mph(A), respectively.29 In this study, annotation of the complete mph(A) operon and analysis of the mph(A) promoter region, which overlaps with the mphR(A) binding site, revealed differences between azithromycin-susceptible and -resistant isolates. In general, resistant isolates (n = 136; 73%) had an intact mph(A) operon (Figure 3a), while mph(A)-harbouring susceptible isolates (n = 50; 27%) had altered operon structure (Figure 3b). Most commonly, susceptible isolates lacked the mphR(A) repressor gene and part of the mrx gene, disrupted by an ISEcp1-blaCTX-M-1 transposition unit. Additionally, the susceptible isolates (n = 50) had either a 24 bp (n = 23) or a 66 bp (n = 14) deletion spanning the mphR(A) binding site and/or the transcription start site and the ribosomal binding site (RBS). Because it has been previously suggested that bacterial cells sense the presence of macrolides through the MphR(A) repressor by forming a macrolide–repressor complex,29 our interpretation of these results is that mph(A) is not expressed in susceptible isolates due to the lack of the macrolide–repressor complex.
Figure 3.
mph(A) operon structure in azithromycin-resistant isolates (MIC > 16 mg/L) (a), azithromycin-susceptible isolates (MIC ≤ 16 mg/L) (b), and in isolates with Tn and IS insertions (c). S. Blockley 5601 (c5) carries the mph(A) operon as part of an MDR region, consisting of the streptomycin resistance cluster aph(3′)-Ib–aph(6)-Id–aph(3′)-Ia and the tet(A) gene. NCBI blastn revealed 99.24%–100% identity and 100% coverage between the whole streptomycin and azithromycin resistance genomic region of S. Blockley isolate 5601 and S. Blockley strain 159383 (GenBank accession number CP043662.1: 4326383-4352831). The MDR region was reconstructed using Bandage (http://github.com/rrwick/Bandage). istA—IS21-like element IS21 family transposase IstA, istB—IS21-like element IS21 family helper ATPase IstB. In (a), the difference between the two sequences is T instead of C in the −10 region. Wavy lines at the end(s) of diagrams show where only part of the contig is included. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Among the resistant mph(A)-harbouring isolates (n = 136), 117 followed the common mph(A) structure for resistant isolates [intact regulatory region and full mph(A) operon] (Figure 3a). One isolate (16037780A201X5, MIC = 32 mg/L) had a truncated regulatory region (Table S4) and three isolates had disruptions into the mph(A) operon due to Tn insertions (Figure 3c, 1–3). Additionally, four isolates had too short contigs to assess the completeness of the regulatory regions (Table S4). In S. Blockley 5601, the complete mph(A) operon was chromosomally integrated (Figure 3c, 5) as previously reported for S. Blockley strain 159383.30
Exceptions to the mph(A) operon structure that is typical in the susceptible isolates [truncated regulatory region and lack of mphR(A)] were also observed. The regulatory region was intact in 13 isolates, and in 5 of them mphR(A) was absent, whereas in 7 of them mphR(A) was present but mrx had a deletion mutation at nucleotide position 576 leading to a premature stop codon (PMSC) and truncated Mrx. However, previous cloning experiments in minicells carrying mrx with a non-sense mutation showed that Mph(A) production was still enhanced in presence of erythromycin as in minicells carrying non-mutated (WT) mrx gene.29 Therefore, the deletion mutation in mrx in the seven isolates in our study might not be the explanation for the observed susceptible phenotype. Additionally, in isolate NRS_2017_ESBL_19.14 (MIC = 8 mg/L), mphR(A) was present and the regulatory region intact, but mrx was located in two contigs. In isolate U16-0318X (MIC = 8 mg/L) mph(A) had a deletion (Figure 3c, 4).
Figure 4.
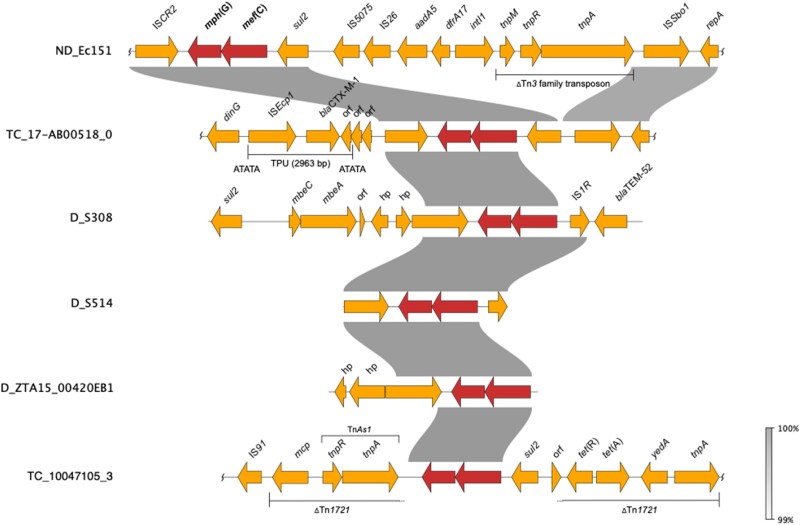
Comparative analyses of contigs harbouring mef(C)-mph(G) in E. coli and Salmonella. D, donor (included for isolates that did not yield TCs), ND, conjugation not performed. Wavy lines at the end(s) of diagrams show where only part of the contig is included. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Despite exceptions to each of the two most common mph(A) operon structures, the size of the dataset analysed and the high concordance between mph(A) operon structures and observed phenotypes suggest that in addition to the mph(A) gene, the mphR(A) repressor gene is also required for azithromycin resistance. Nevertheless, in vitro studies are needed to elucidate the association between the absence of mphR(A), truncation in the regulatory region and susceptibility to azithromycin.
mef(C)-mph(G) genes
The mef(C)-mph(G) tandem genes, conferring high-level azithromycin resistance, were first identified in Photobacterium damselae subsp. damselae31 on an MDR plasmid pAQU1 and subsequently found in other marine and enteric bacteria from fish intestines.31,32 Since their first report in Enterobacterales, in a Shiga toxin-producing E. coli (STEC) isolate in 2021,9 mef(C)-mph(G) have been detected in E. coli from various origins and geographical locations.33–36
We first detected mef(C)-mph(G) in E. coli and Salmonella isolates included in the 2018 EU harmonized monitoring of AMR and the Danish Programme for surveillance of antimicrobial consumption and resistance (results included in this study). Here, screening of all E. coli and Salmonella isolates collected between 2014 and 2018 identified mef(C)-mph(G) in 11 E. coli and two Salmonella isolates from bovine and porcine origin. In all isolates, mef(C)-mph(G) were associated with an azithromycin-resistant phenotype (MIC > 16 mg/L). No other known macrolide resistance mechanisms were detected in these isolates, except mph(B) in isolate ZTA15:00420EB1 (Tables 1 and S4). However, other AMR genes were found in the mef(C)-mph(G)-harbouring isolates, i.e. tet(A), sul2, blaTEM-52 and blaCTX-M-1, in all cases located on the same contig as mef(C)-mph(G) (Figure 4). The E. coli isolates carrying mef(C)-mph(G) belonged to eight different STs, and the two Salmonella isolates, S. Derby and S. 1,4, [5],12:i:-, belonged to ST40 and ST19, both of which have frequently been associated with the pork production chain (Table 1).37,38
Since mef(C)-mph(G) genes were previously described to be located on plasmids and associated with various mobile genetic elements, including plasmids of various sizes and integrative conjugative elements,9,32 we carried out conjugation experiments to assess their transferability. The conjugation performed for 10 E. coli and two Salmonella isolates showed that mef(C)-mph(G) could be located on transferrable plasmids, as in 5 of the E. coli isolates the genes were successfully transferred to the E. coli J62-2 recipient (Table 1). Unequivocal association between mef(C)-mph(G) and a plasmid replicon gene could not be made using short-read sequencing data. However, in all TCs, except TC_NRS-2015 ESBL-08-69, only one plasmid replicon was detected, which allowed us to associate mef(C)-mph(G) with IncFII in isolate TC_10047105-3 and IncIγ in isolates TC_17-AB00518-0, TC_HP-6957 and TC_ECO-NRS-187-57 (Table 1). Analyses of the genetic contexts of mef(C)-mph(G) in the donor strains and TCs revealed that the genes were associated with truncated TnAs1 consisting of tnpR and tnpA genes in IncFII plasmids and ISCR2 in IncIγ plasmids, with both elements located downstream of mef(C)-mph(G) (Figure 4). The ISCR2 is known to be associated with dissemination of multiple resistance genes in various species.39
Other azithromycin resistance genes
The relevance of the plasmid-borne erm genes in mediating high-level azithromycin resistance was confirmed in this study as previously described.7,8,35,40 The presence of erm(C), erm(B) and erm(42), alone or in combination with other macrolide resistance genes, was associated with a resistant phenotype (MIC ≥ 64 mg/L) in all cases. Association of erm genes with high azithromycin MIC levels has been reported in E. coli even in the presence of efflux pump inhibitors.6,41,42
The mph(B) gene, the second most detected gene in our collection, encodes macrolide 2′-phosphotransferase II, which phosphorylates 14-membered (e.g. erythromycin) and 16-membered (e.g. tylosin) macrolides.43,44 Nevertheless, 6 out of 26 isolates harbouring mph(B) as the sole macrolide resistance gene in our collection had azithromycin MIC > 16 mg/L (Table S4). The only mph(B)-harbouring isolate in the study of Gomes et al.7 exhibited an MIC of 16 mg/L in the absence of the efflux pump inhibitor PaβN. Previous cloning and expression analysis of mph(B) in E. coli also showed that mph(B) did not confer resistance to azithromycin.44
In addition, we detected mef(B), msr(E)-mph(E) or ere(A) as the only macrolide resistance gene in isolates with azithromycin MIC > 16 mg/L. While azithromycin-resistant ere(A)-carrying E. coli, even in the presence of PaβN, has been observed,7 there is no description of msr(E)-mph(E) or mef(B) conferring azithromycin resistance in E. coli or Salmonella. Generally, the decreased azithromycin susceptibility in isolates with MIC = 32 mg/L without a known mechanism of azithromycin resistance could be explained by the one 2-fold dilution MIC variation from the cut-off for resistance. However, the occurrences of high-level azithromycin resistance (MIC ≥ 64 mg/L) that cannot be explained by genes detected in this study suggest that unknown mechanism(s) could be involved in the reduced azithromycin susceptibility. For instance, a recent study using hidden Markov models identified and experimentally validated five novel macrolide resistance genes that increased the azithromycin resistance when cloned into E. coli.45
Study limitations
A limitation in our study was that we could not unequivocally determine whether the detected macrolide resistance genes were located on the chromosome or on plasmids due to the nature of the Illumina short-read sequencing technology. Furthermore, the association of AMR genes with IS or Tn elements is not entirely certain, due to the possibility of mis-assemblies, although studies suggest that these tend to be rare for high-coverage data. Both limitations could be overcome by long-read sequencing technologies.
Conclusions
We investigated azithromycin resistance determinants and phenotype–genotype correlation in a collection of 1276 E. coli and Salmonella isolates from food-producing animals and meat in Europe. Eleven different genes previously associated with azithromycin resistance were detected. Our study highlights that the interpretation of mph(A)-mediated phenotypes in Enterobacterales requires consideration of the entire mph(A) operon and its regulatory region. Generally, azithromycin-susceptible isolates lacked the mphR(A) repressor gene and had a truncated mphR(A) binding site, while azithromycin-resistant isolates had an intact mph(A) operon and complete mphR(A) binding site. We observed elevated azithromycin MIC values (≥64 mg/L) in all cases when an erm gene was found alone or associated with another macrolide resistance determinant. Moreover, our study provides additional insight into the genetic context and transferability of mef(C)-mph(G) genes, which are emerging in E. coli and Salmonella. These genes were found to be associated with various transposable elements, often harboured on conjugative plasmids and containing additional AMR genes, thus highlighting concerns for co-selection and spread.
Supplementary Material
Acknowledgements
We gratefully acknowledge Birthe S. Rosenqvist Lund, Inge Marianne Hansen, Christina Aaby Svendsen, Jacob Dyring Jensen and Gunhild Larsen for performing antimicrobial susceptibility testing and WGS. The Department of Animal Health and Antimicrobial Strategies (Uppsala, Sweden) is acknowledged for sharing isolates for this study. We thank Veterinary Services (Nicosia, Cyprus), Istituto Zooprofilattico Sperimentale del Lazio e della Toscana ‘M. Aleandri’ (Rome, Italy), Laboratoire de Médecine Vétérinaire de l’État (LMVE, Dudelange, Luxembourg) and National Veterinary Research Institute (Pulawy, Poland) for the WGS of selected isolates for this study. We would also like to acknowledge the anonymous reviewers for their expertise to improve the manuscript.
Contributor Information
Mirena Ivanova, European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR), Research Group for Global Capacity Building, Technical University of Denmark, Kongens Lyngby, Denmark.
Armen Ovsepian, European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR), Research Group for Global Capacity Building, Technical University of Denmark, Kongens Lyngby, Denmark; DIANA-Lab, Dept. of Computer Science and Biomedical Informatics, University of Thessaly, Lamia, Greece.
Pimlapas Leekitcharoenphon, Research Group for Genomic Epidemiology, Technical University of Denmark, Kongens Lyngby, Denmark.
Anne Mette Seyfarth, European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR), Research Group for Global Capacity Building, Technical University of Denmark, Kongens Lyngby, Denmark.
Hanne Mordhorst, Research Group for Genomic Epidemiology, Technical University of Denmark, Kongens Lyngby, Denmark.
Saria Otani, Research Group for Genomic Epidemiology, Technical University of Denmark, Kongens Lyngby, Denmark.
Sandra Koeberl-Jelovcan, Austrian Agency for Health and Food Safety, Graz, Austria.
Mihail Milanov, National Diagnostic and Research Veterinary Institute, Sofia, Bulgaria.
Gordan Kompes, Croatian Veterinary Institute, Zagreb, Croatia.
Maria Liapi, Bacteriology Serology Laboratory, Veterinary Services, Cyprus.
Tomáš Černý, State Veterinary Institute, Prague, Czech Republic.
Camilla Thougaard Vester, Danish Veterinary and Food Administration, Ringsted, Denmark.
Agnès Perrin-Guyomard, French Agency for Food, Environmental and Occupational Health & Safety, Maisons-Alfort, France.
Jens A Hammerl, German Federal Institute for Risk Assessment, Berlin, Germany.
Mirjam Grobbel, German Federal Institute for Risk Assessment, Berlin, Germany.
Eleni Valkanou, Veterinary Laboratory of Chalkis, Chalkis, Greece.
Szilárd Jánosi, National Food Chain Safety Office, Veterinary Diagnostic Directorate, Budapest, Hungary.
Rosemarie Slowey, Central Veterinary Research Laboratory, Kildare, Ireland.
Patricia Alba, Istituto Zooprofilattico Sperimentale del Lazio e della Toscana ‘M. Aleandri’, Rome, Italy.
Virginia Carfora, Istituto Zooprofilattico Sperimentale del Lazio e della Toscana ‘M. Aleandri’, Rome, Italy.
Jelena Avsejenko, Institute of Food Safety, Animal Health and Environment BIOR, Riga, Latvia.
Asta Pereckiene, National Food and Veterinary Risk Assessment Institute, Vilnius, Lithuania.
Dominique Claude, Laboratoire de Médecine Vétérinaire de l’État, Dudelange, Luxembourg.
Renato Zerafa, Public Health Laboratory, Valletta, Malta.
Kees T Veldman, Wageningen Bioveterinary Research, Part of Wageningen University & Research, Lelystad, Netherlands.
Cécile Boland, Sciensano, Brussels, Belgium.
Cristina Garcia-Graells, Sciensano, Brussels, Belgium.
Pierre Wattiau, Sciensano, Brussels, Belgium.
Patrick Butaye, Department of Pathobiology, Ghent University, Merelbeke, Belgium; Jockey Club College of Veterinary Medicine and Life Sciences, Kowloon, Hong Kong.
Magdalena Zając, National Veterinary Research Institute, Pulawy, Poland.
Ana Amaro, Instituto Nacional de Investigação Agrária e Veterinária, Oeiras, Portugal.
Lurdes Clemente, Instituto Nacional de Investigação Agrária e Veterinária, Oeiras, Portugal.
Angela M Vaduva, Institute for Hygiene and Veterinary Public Health, Bucharest, Romania.
Luminita-Maria Romascu, Institute for Diagnosis and Animal Health, Bucharest, Romania.
Nicoleta-Manuela Milita, Institute for Diagnosis and Animal Health, Bucharest, Romania.
Andrea Mojžišová, State Veterinary and Food Institute, Dolny Kubin, Slovakia.
Irena Zdovc, Institute for Microbiology and Parasitology, Ljubljana, Slovenia.
Maria Jesús Zamora Escribano, Spanish Agency for Food Safety and Nutrition, Madrid, Spain.
Cristina De Frutos Escobar, Spanish Agency for Food Safety and Nutrition, Madrid, Spain.
Gudrun Overesch, Vetsuisse Faculty, Institute of Veterinary Bacteriology, University of Bern, Bern, Switzerland.
Christopher Teale, Animal & Plant Health Agency, Weybridge, UK.
Guy H Loneragan, School of Veterinary Medicine, Texas Tech University, Amarillo, TX, USA.
Beatriz Guerra, European Food Safety Authority, Parma, Italy.
Pierre Alexandre Beloeil, European Food Safety Authority, Parma, Italy.
Amanda M V Brown, Department of Biological Sciences, Texas Tech University, Lubbock, TX, USA.
Rene S Hendriksen, European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR), Research Group for Global Capacity Building, Technical University of Denmark, Kongens Lyngby, Denmark.
Valeria Bortolaia, European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR), Research Group for Global Capacity Building, Technical University of Denmark, Kongens Lyngby, Denmark; Statens Serum Institut, Copenhagen, Denmark.
Jette Sejer Kjeldgaard, European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR), Research Group for Global Capacity Building, Technical University of Denmark, Kongens Lyngby, Denmark.
Funding
This work was supported by general funding to the European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR) (http://www.eurl-ar.eu/).
Transparency declarations
B.G. and P.A.B. are currently employed by the European Food Safety Authority (EFSA) in its BIOHAW Unit that provides scientific and administrative support to EFSA’s scientific activities in the area of microbial risk assessment. The positions and opinions presented in this article are those of the authors alone and are not intended to represent the views or scientific works of EFSA. All other authors: none to declare.
Author contributions
M.I. and V.B. designed the experiments. M.I., A.O. and H.M. carried out the conjugation experiments. M.I., A.O., A.M.S., R.S.H., V.B. and J.S.K. analysed the data at the EURL-AR. B.G. and P.A.B. analysed the data and coordinated the isolate selection at EFSA. M.I. performed the bioinformatic analyses, data visualizations, wrote the draft manuscript and addressed reviewers comments during revisions. V.B., J.S.K. and A.M.V.B. revised the draft manuscript. J.S.K. and A.M.V.B. helped with addressing reviewers’ comments. R.S.H., A.M.V.B. and G.H.L. supervised the project. All additional authors contributed to sample preparation, data analysis in the originating countries and revision of the draft manuscript.
Supplementary data
Tables S1–S6 are available as Supplementary data at JAC Online.
References
- 1. WHO . The WHO list of Critically Important Antimicrobials. 2019. https://www.who.int/groups/advisory-group-on-the-who-list-of-critically-important-antimicrobials.
- 2. Hooda Y, Sajib MS, Rahman H et al. Molecular mechanism of azithromycin resistance among typhoidal Salmonella strains in Bangladesh identified through passive pediatric surveillance. PLoS Negl Trop Dis 2019; 13: e0007868. 10.1371/journal.pntd.0007868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lübbert C. Antimicrobial therapy of acute diarrhoea: a clinical review. Expert Rev Anti Infect Ther 2016; 14: 193–206. 10.1586/14787210.2016.1128824 [DOI] [PubMed] [Google Scholar]
- 4. Darton TC, Tuyen HT, The HC et al. Azithromycin resistance in Shigella spp. in Southeast Asia. Antimicrob Agents Chemother 2018; 62: e01748-17. 10.1128/AAC.01748-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Timmermans M, Latour S, Ceyssens P-J et al. The AMR-ARRAY: a modular bead array detecting β-lactam, (fluoro) quinolone, colistin, aminoglycoside and macrolide resistance determinants in Gram-negative bacteria. J Microbiol Methods 2022; 196: 106472. 10.1016/j.mimet.2022.106472 [DOI] [PubMed] [Google Scholar]
- 6. Gomes C, Martínez-Puchol S, Palma N et al. Macrolide resistance mechanisms in Enterobacteriaceae: focus on azithromycin. Crit Rev Microbiol 2017; 43: 1–30. 10.3109/1040841X.2015.1136261 [DOI] [PubMed] [Google Scholar]
- 7. Gomes C, Ruiz-Roldán L, Mateu J et al. Azithromycin resistance levels and mechanisms in Escherichia coli. Sci Rep 2019; 9: 6089. 10.1038/s41598-019-42423-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nguyen MCP, Woerther PL, Bouvet M et al. Escherichia coli as reservoir for macrolide resistance genes. Emerg Infect Dis 2009; 15: 1648–50. 10.3201/eid1510.090696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bizot E, Cointe A, Bidet P et al. Azithromycin resistance in Shiga-toxin producing Escherichia coli in France between 2004 and 2020 and detection of mef(C)- mph(G) genes. Antimicrob Agents Chemother 2021; 66: e0194921. 10.1128/AAC.01949-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. European Commission . Commission implementing decision of 12 November 2013 on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria (2013/652/EU). https://eur-lex.europa.eu/eli/dec_impl/2013/652/oj. [Google Scholar]
- 11. European Commission . Commission implementing decision (EU) 2020/1729 of 17 November 2020 on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria and repealing implementing decision 2013/652/EU. (2020/1729/EU). https://eur-lex.europa.eu/eli/dec_impl/2020/1729/oj. [Google Scholar]
- 12. European Food Safety Authority, ECDC . The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J 2023; 21: e07867. 10.2903/j.efsa.2023.7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. European Food Safety Authority, Aerts M, Battisti A et al. Technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food-producing animals and food. EFSA J 2019; 17: 5709. 10.2903/j.efsa.2019.5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nurk S, Meleshko D, Korobeynikov A et al. metaSPAdes: a new versatile metagenomic assembler. Genome Res 2017; 27: 824–34. 10.1101/gr.213959.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gurevich A, Saveliev V, Vyahhi N et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013; 29: 1072–5. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bortolaia V, Kaas RS, Ruppe E et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 2020; 75: 3491–500. 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gunell M, Kotilainen P, Jalava J et al. In vitro activity of azithromycin against nontyphoidal Salmonella enterica. Antimicrob Agents Chemother 2010; 54: 3498–501. 10.1128/AAC.01678-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang S, den Bakker HC, Li S et al. SeqSero2: rapid and improved Salmonella serotype determination using whole-genome sequencing data. Appl Environ Microbiol 2019; 85: e01746-19. 10.1128/AEM.01746-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carattoli A, Zankari E, García-Fernández A et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58: 3895–903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siguier P. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 2006; 34: D32–6. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wickham H. Ggplot2: Elegant Graphics for Data Analysis. 2nd edn. Springer International Publishing, 2016. [Google Scholar]
- 23. Allaire JJ, Gandrud C, Russell K et al. NetworkD3: D3 JavaScript Network Graphs from R. 2017. https://CRAN.R-project.org/package=networkD3.
- 24. Hansen KH, Bortolaia V, Nielsen CA et al. Host-specific patterns of genetic diversity among IncI1-Iγ and IncK plasmids encoding CMY-2 β-lactamase in Escherichia coli isolates from humans, poultry meat, poultry, and dogs in Denmark. Appl Environ Microbiol 2016; 82: 4705–14. 10.1128/AEM.00495-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 26. Bushnell B, Rood J, Singer E. BBMerge—accurate paired shotgun read merging via overlap. PLoS One 2017; 12: e0185056. 10.1371/journal.pone.0185056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bankevich A, Nurk S, Antipov D et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiang Y, Wu F, Chai Y et al. A new plasmid carrying mphA causes prevalence of azithromycin resistance in enterotoxigenic Escherichia coli serogroup O6. BMC Microbiol 2020; 20: 247. 10.1186/s12866-020-01927-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noguchi N, Takada K, Katayama J et al. Regulation of transcription of the mph(A) gene for macrolide 2-phosphotransferase I in Escherichia coli: characterization of the regulatory gene mphR(A). J Bacteriol 2000; 182: 5052–8. 10.1128/JB.182.18.5052-5058.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nair S, Ashton P, Doumith M et al. WGS for surveillance of antimicrobial resistance: a pilot study to detect the prevalence and mechanism of resistance to azithromycin in a UK population of non-typhoidal Salmonella. J Antimicrob Chemother 2016; 71: 3400–8. 10.1093/jac/dkw318 [DOI] [PubMed] [Google Scholar]
- 31. Nonaka L, Maruyama F, Suzuki S et al. Novel macrolide-resistance genes, mef(C) and mph(G), carried by plasmids from Vibrio and Photobacterium isolated from sediment and seawater of a coastal aquaculture site. Lett Appl Microbiol 2015; 61: 1–6. 10.1111/lam.12414 [DOI] [PubMed] [Google Scholar]
- 32. Sugimoto Y, Suzuki S, Nonaka L et al. The novel mef(C)–mph(G) macrolide resistance genes are conveyed in the environment on various vectors. J Glob Antimicrob Resist 2017; 10: 47–53. 10.1016/j.jgar.2017.03.015 [DOI] [PubMed] [Google Scholar]
- 33. González-Santamarina B, Weber M, Menge C et al. Comparative genomic analysis of antimicrobial-resistant Escherichia coli from South American camelids in Central Germany. Microorganisms 2022; 10: 1697. 10.3390/microorganisms10091697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Irrgang A, Tenhagen B-A, Pauly N et al. Characterization of VIM-1-producing E. coli isolated from a German fattening pig farm by an improved isolation procedure. Front Microbiol 2019; 10: 2256. 10.3389/fmicb.2019.02256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma Y, Pirolo M, Subramani P et al. Macrolide resistance and in vitro potentiation by peptidomimetics in porcine clinical Escherichia coli. mSphere 2022; 7: e00402-22. 10.1128/msphere.00402-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suzuki S, Kadoya A, Masuda N et al. Macrolide resistance genes and mobile genetic elements in waterways from pig farms to the sea in Taiwan. J Glob Antimicrob Resist 2022; 29: 360–70. 10.1016/j.jgar.2022.04.024 [DOI] [PubMed] [Google Scholar]
- 37. Cai Y, Tao J, Jiao Y et al. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int J Food Microbiol 2016; 222: 56–64. 10.1016/j.ijfoodmicro.2016.01.020 [DOI] [PubMed] [Google Scholar]
- 38. Zhou Z, Li J, Zheng H et al. Diversity of Salmonella isolates and their distribution in a pig slaughterhouse in Huaian, China. Food Control 2017; 78: 238–46. 10.1016/j.foodcont.2017.02.064 [DOI] [Google Scholar]
- 39. Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol 2006; 70: 296–316. 10.1128/MMBR.00048-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu X, Yang X, Ye L et al. Genetic characterization of a conjugative plasmid that encodes azithromycin resistance in Enterobacteriaceae. Microbiol Spectr 2022; 10: e00788-22. 10.1128/spectrum.00788-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harmer CJ, Holt KE, Hall RM. A type 2 A/C2 plasmid carrying the aacC4 apramycin resistance gene and the erm(42) erythromycin resistance gene recovered from two Salmonella enterica serovars. J Antimicrob Chemother 2015; 70: 1021–5. 10.1093/jac/dku489 [DOI] [PubMed] [Google Scholar]
- 42. Chiou C-S, Hong Y-P, Wang Y-W et al. Antimicrobial resistance and mechanisms of azithromycin resistance in nontyphoidal Salmonella isolates in Taiwan, 2017 to 2018. Microbiol Spectr 2023; 11: e03364-22. 10.1128/spectrum.03364-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kono M. Purification and characterization of macrolide 2′-phosphotransferase type II from a strain of Escherichia coli highly resistant to macrolide antibiotics. FEMS Microbiol Lett 1992; 97: 89–94. 10.1016/0378-1097(92)90369-y [DOI] [PubMed] [Google Scholar]
- 44. Chesneau O, Tsvetkova K, Courvalin P. Resistance phenotypes conferred by macrolide phosphotransferases. FEMS Microbiol Lett 2007; 269: 317–22. 10.1111/j.1574-6968.2007.00643.x [DOI] [PubMed] [Google Scholar]
- 45. Lund D, Kieffer N, Parras-Moltó M et al. Large-scale characterization of the macrolide resistome reveals high diversity and several new pathogen-associated genes. Microb Genomics 2022; 8: 000770. 10.1099/mgen.0.000770 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.