Abstract
Heterogeneity and high plasticity are common features of cells from the mononuclear phagocyte system: monocytes (MOs), macrophages, and dendritic cells (DCs). Upon activation by microbial agents, MO can differentiate into MO-derived DCs (MODCs). In previous work, we have shown that during acute infection with Plasmodium berghei ANKA (PbA), MODCs become, transiently, the main CD11b+ myeloid population in the spleen (SP) and once recruited to the brain play an important role in the development of experimental cerebral malaria (ECM). Here, we isolated 4 cell populations: bone marrow (BM) MOs (BM-MOs) and SP-MOs from uninfected mice; BM inflammatory MOs (BM-iMOs) and SP-MODCs from PbA-infected mice and used a system biology approach to a holistic transcriptomic comparison and provide an interactome analysis by integrating differentially expressed miRNAs (DEMs) and their differentially expressed gene targets (DEGs) data. The Jaccard index (JI) was used for gauging the similarity and diversity among these cell populations. Whereas BM-MOs, BM-iMOs, and SP-MOs presented high similarity of DEGs, SP-MODCs distinguished by showing a greater number of DEGs. Moreover, functional analysis identified an enrichment in canonical pathways, such as DC maturation, neuroinflammation, and IFN signaling. Upstream regulator analysis identified IFNγ as the potential upstream molecule that can explain the observed DEMs–Target DEGs intersections in SP-MODCs. Finally, directed target analysis and in vivo/ex vivo assays indicate that SP-MODCs differentiate in the SP and IFNγ is a main driver of this process.
Keywords: Plasmodium, MODC, cell differentiation, miRNA, systems biology
1 |. INTRODUCTION
The mononuclear phagocyte system comprises monocytes (MOs), macrophages (MAs), and dendritic cells (DCs) with bone marrow (BM) origin. These cells have high plasticity and share many functional features in the immune defense as well as homeostasis maintenance, for example, inflammation-induced migration, cytokine and chemokine secretion, clearance of microorganisms and apoptotic cells, antigen processing and presentation.1 During inflammatory response and/or infection, activated MOs migrate from blood to tissues, produce inflammatory cytokines, perform antimicrobial functions, and may differentiate into MAs or DCs.2,3 These differentiated cells are main players of innate immune response and host resistance to malaria.4 Infection with Plasmodium vivax or P. falciparum induces significant increase in circulating MOs, including an inflammatory MO (iMOs) subset (CD14+CD16+), and changes of their distribution in different organs, such as spleen (SP), liver, and lungs.5 iMOs recruitment from BM to blood is mediated by the chemokine CCL2 and its receptor, CCR2.6 These cells are highly efficient in phagocytizing Plasmodium-infected erythrocytes and produce ROS,7,8 thus, the expanded iMO population play a pivotal role in resistance to both human and experimental rodent malaria.9,10
Previous studies from our group have shown that during acute rodent malaria, another subset of MO, the MO-derived DCs (MODCs) emerge as a main myeloid population in the SP and non-lymphoid organs.11,12 We have also shown that the development of experimental cerebral malaria (ECM) during P. berghei ANKA (PbA) infection is dependent of splenic MODC infiltration in the brain.11 These cells promote neuroinflammation by producing critical chemokines (CXCL9 and CXCL10) involved in T cell recruitment to the brain.13 We also observed that infection of CCR2−/− mice with PbA compromises the emergence of MODCs in the SP, whereas the migration of MODCs to the brain and the development of ECM are not affected.11 These findings suggested that splenic MODCs differentiate locally and independent from iMOs recruited from BMs. The omic technologies have been applied to reveal new factors or markers that allow us to better understand the heterogeneity of these MO subsets, but these key molecules are far from being fully characterized.
MicroRNAs (miRNAs) are a class of noncoding RNAs capable of promoting gene expression regulation by inducing mRNA decay or translation repression. The miRNA–mRNA (target) pairing is determined by the called seed region of the miRNA, a sequence of 8 nucleotides located at the 5´ end of the mature miRNA.14,15 Because of the great number of possible pair-wise miRNA–-target interactions, computation models, databases, and tools are essential in the prediction of biologic function of the miRNAs targets.16 Although miRNA importance has already been extensively demonstrated, their participation on overall gene expression control in mechanisms of differentiation and plasticity of different cell populations during Plasmodium infection is still poorly explored.
Here, we analyzed the interactome of both differentially expressed miRNAs (DEMs) and their differentially expressed gene targets (DEGs) to depict the heterogeneity and differentiation of MOs isolated from uninfected mice BM (BM-MOs) and SP (SP-MOs), and the newly differentiated inflammatory MOs (BM-iMOs) and SP-MODCs from PbA-infected mice. Strikingly, SP-MODCs present unique DEMs and DEGs profile with a DC signature. In addition, the combination of functional in vivo and in vitro assays with an in silico mRNA and miRNA network predictions we show that MODCs differentiate in the SP from SP-MO, not needing to be recruited from the BM and that IFNγ is the main driver of this process.
2 |. MATERIALS AND METHODS
2.1 |. Mice
All animals used in this work were 8–12-weeks old. C57BL/6 were obtained from either Animal Facility of the UFMG or initially purchased from Jackson Laboratory (Bar Harbor, ME). IL-4−/−, IL-12−/−, IL-17−/−, and IFNγ−−/− mice were obtained from Fiocruz-Minas (Belo Horizonte, MG, Brazil). The REX3 mouse lineage was kindly provided by Andrew D. Luster from Massachusetts General Hospital (MGH)17 and the GREAT (IFNγ YFP-reporter) mice by John E. Harris from UMMS (Worcester, MA, USA). These mice were bred and reared at the animal facility of University of Massachusetts Medical School (UMMS). All animal procedures were performed in accordance with the guidelines of the American Association for Laboratory Animal Science (AALAS) and approved by the Institutional Animal Care and Use Committee of the UMMS (ID A-2371–18) as well as the Institutional Ethical Committee for Animal Experimentation at Fiocruz (CEUA LW 15–14).
2.2 |. Infection
The PbA strain was stored in liquid nitrogen, thawed, and maintained in C57BL/6 mice by weekly passages for up to 12 weeks. For experimental infection, mice were injected intraperitoneally (i.p.) with 105 infected RBCs. These mice were observed daily and parasitemia was estimated by counting Giemsa-stained thin blood smears. ECM signs were evaluated by different parameters that included ruffled fur, abnormal postural responses, reduced reflexes, reduced grip strength, coma, and convulsions. Mice that demonstrated complete disability in all parameters or died between days 7 and 9 postinfection were considered as having ECM.18
2.3 |. SP and BM cell cultures
Spleens and BMs from C57BL/6 mice uninfected and at 6 days postinfection with PbA were harvested, suspended in RPMI medium supplemented with 10% of FBS at 10 × 106 cells/ml. Cells were cultured with medium alone or with LPS (100 ng/ml) for 24 h with addition of Brefeldin A 6 h prior the end time and processed for flow cytometry to detect intracellular cytokines. For the main IFNγ source in PbA-infected mice, we used splenocytes and BM cells from GREAT (IFNγ YFP-reporter) mice. Samples were acquired using a LSRII (BD Biosciences, Franklin Lakes, NJ, USA) cytometer and analyzed with FlowJo software.
2.4 |. Flow cytometry
The single cell suspensions of BM or SPs were incubated with anti-FcγIII/II and fluorochrome-conjugated antibodies against surface markers in PBS containing 1% FBS for 15 min at room temperature. CD11b (PE-Cy7, clone: M1/70), F4/80 (PE-Cy5, clone: BM8), CD11c (Alexa 700, clone: N418), MHCII (APC eFluor-780, clone: M5/114.15.2), DC-SIGN (APC e-fluor 660, clone: MMD3), and Ly6C (e-fluor 450, clone: HK1.4) were purchased from eBioscience (San Diego, CA, USA). CD80 (FITC, clone: 16–10A1) and CD86 (PE, clone: GL1, BD) were purchased from BD. CD40 (FITC, clone: 3/23) was purchased from Biolegend (San Diego, CA, USA) and CCR2 (FITC, clone 475301) and CCR5 (APC, clone CTC5) were from R&D (Minneapolis, MN, USA). In REX3 mice, endogenous RFP (CXCL9) and BFP (CXCL10) were read in PE and Pacific Blue channels, respectively. Sample acquisition was performed by using a Fortessa or LRSII Cytometers (BD) and analyzed using FlowJo software. For detection of IFNγ-YFP, cells were gated on CD3+CD4+, CD3+CD8+, or CD3−NK1.1+. The monoclonal antibodies specific for CD3 (FITC, clone 145–2C11), CD4 (APC, clone RM4–5), CD8 (APC-Cy7, clone 53–6.7), and NK1.1 (BV421, clone PK136) were purchased from eBioscience and used to gate YFP+ cells. For intracellular detection of cytokines, after surface staining, cells were fixed and permeabilized using the Cytofix/Cytoperm™ Kit (BD). Prior to fixation. Live/Dead Fixable Aqua Cell Stain Kit (Invitrogen, Carlsbad, CA, USA) was used to exclude dead cells. Cells were stained for 30 min at 4C with TNF-α (PE, clone MP6-XT22), IL-12p40 (PE, clone C17.8), IL-10 (FITC, clone JES5–16E3) and NOS2 (PE, clone CXNFT), purchased from eBioscience. Cells were gated on CD11b+F4/80+ and then on DC-SIGN+MHCII+ for detection of cytokines on MODCs.19
2.5 |. Ex vivo differentiation of Ly6Chi MOs with IFNγ
Ly6Chi MOs FACS sorted from the SP or the BM of uninfected C57BL/6 mice were cultured with IFNγ (100 ng/ml). After 24 h, dead cells were excluded using Live/Dead Fixable Aqua Cell Stain Kit (Invitrogen) and then stained with fluorochrome-conjugated antibodies for CD11b, F4/80, MHCII, DC-SIGN, and Ly6C as described previously.
2.6 |. In vivo administration of IFNγ
C57BL/6 mice received i.p. injection containing 5 μg of recombinant carrier-free IFNγ (Biolegend) in 200 μl sterile PBS once each day for 3 days.20 After 16 h of the last injection, mice were euthanized for SP and BM harvested for detection of MODCs by flow cytometry.
2.7 |. Cell sorting
Splenic CD11b+ cells were enriched using magnetically labeled microbeads (Miltenyi, Bergisch Gladbach, Germany), stained with Ly6G (PE), CD11b (PE-Cy7), F4/80 (PE-Cy5), CD11c (FITC), MHCII (APC eFluor 780), and DC-SIGN (APC e-fluor 660), and then purified by using a cell sorting ARIA (BD). The cells were first gated on SSC-A/FSC-A followed by FSC-H/FSC-A, to avoid doublets, and then on Ly6G+ cells to exclude neutrophils (Supplemental Fig. 2). Sorted SP-MOs and BM-MOs (Ly6G−CD11b+ F4/80+DC-SIGN−MHCII−) from uninfected mice, as well as BM-iMOs (Ly6G−CD11b+ F4/80+DC-SIGNloMHCII−) and SP-MODCs (Ly6G−CD11b+F4/80+DC-SIGNhiMHCIIhi) from infected mice were counted, resuspended in RNAlater and stored at −20°C up to RNA extraction.19
2.8 |. Adoptive cell transfer
Briefly, splenocytes from uninfected CD45.2 donor mice were incubated with a mix of PE-labeled antibodies: CD4 (clone GK1.5), CD8 (clone 53–6.7), TCR-b (clone H57–597), CD19 (clone 1D3), Ly6G (clone 1A8), Ly49 (clone YLI-90), and CD11c (clone N418), and MOs were enriched up to 40–60% using the PE-multisort kit (Miltenyi Biotec) by negative selection. Enriched MOs (CD11b+F4/80+DC-SIGN−MHCII− cells) were injected intravenously (i.v.) into 6-week-old, nonirradiated CD45.1 recipient mice uninfected or 3 days postinfection with PbA. After 48 h, SP and BM cells were obtained and analyzed by flow cytometry for detection of MoDCs from CD45.2+ or CD45.1+ cells.
2.9 |. RNA-Seq and miRNA sample preparation
For RNA-seq and miRNA analysis, biologic replicates (3 samples) of MOs (SP-MOs) and (BM-MO) from uninfected mice and inflammatory MOs (BM-iMOs), and SP-MODCs from PbA-infected mice were obtained by cell sorting. RNA-seq libraries were prepared using the TruSeq Stranded mRNA Kit (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. The library was submitted for sequencing on the Illumina NextSeq 500 (Bauer Core Facility Harvard University). The RNAseq data is available at Gene Expression Omnibus, access: GSE126381, platform GPL19057. miRNAs were isolated from total RNA using the mirVana™ miRNA Isolation Kit (Thermofisher, Whaltam, MA, USA), following the manufacturers’ protocol. Expression profiling of 641 miRNAs was performed using a multiplexed RT reaction (Megaplex™ RT Primers, Rodent Pool Set v3.0 kit) to produce cDNA. The cDNA sample and master mix (TaqMan Universal Master Mix II, no UNG- Thermofisher) were loaded into preprinted TaqMan Low Density Arrays (TLDA) microfluidic cards (Rodent Card A + B v3, format 384 each). Raw data files (.EDS) were processed using automatic baseline corrections and manually checked for each assay if threshold cycle (Ct) value corresponded to midpoint of logarithmic amplification curve using the software Connect (Thermofisher).
2.10 |. RNA-Seq and miRNA statistical and bioinformatics analysis
For RNA-seq, reads previously trimmed with Trimmomatic 21 were mapped to the Genome Reference Consortium Mouse Build 38 patch release 5 (GRCm38.p5) using STAR aligner 22 and the Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values were calculated with CUFFLINKS 23 to the following contrasts: BM-MOs x BM-iMOs, SP-MOs x SP-MODCs, BM-MOs x SP-MOs, BM-MOs x SP-MODCs and BM-iMOs x SP-MODCs. MicroRNA statistical analysis was carried using the Linear Model for microarray data (Limma) with comparisons p-values submitted to false discovery rate (FDR) adjustment according to the Benjamini–Hochberg method (https://www.jstor.org/stable/2346101).24 The miRNAs and genes were considered differentially expressed if adjusted P ≤ 0.05 and absolute fold change FC ≥ 1.5) (Supplemental Table 1). The comparative threshold cycle method was used to calculate the relative miRNA expression (ΔCt) after global normalization. The samples hierarchical clustering was performed using the squared Euclidean distance of measure method and z-score normalization for the top 2000 most variable genes (FPKM values) and 351 miRNAs (ΔCt values). The principal component analysis (PCA) of gene and miRNA expression was performed for all samples and the same set of miRNAs and genes used in the hierarchical clustering by using a median centering of the data set. The x-axis corresponds to principal component 1 (PC1) and y-axis to the principal component 2 (PC2) and the percentages of variance in both. Both hierarchical clustering and PCA were built using ClustVis web tool.25
2.11 |. DEGs-DEMs integration, functional, and network analysis
MiRNA target prediction, canonical pathways, network and upstream regulator analysis were performed with ingenuity pathway analysis (IPA; Qiagen, Hilden, Germany). The miRNAs and gene expression profiles were integrated to identify putative targets within the differentially expressed genes of the differentially expressed miRNAs using the IPA target filter tool which relies on 3 popular algorithms (TargetScan, TarBase, and miRecords) enabling prioritization of experimentally validated and high predicted mRNA targets based on the content of date 2019–12.
IPA was also used to identify significantly enriched canonical pathways within the list of DEMs and their target DEGs and to build DEMs-DEGs (considering fold change inverse pairing) networks. The significance of the association between each list and the pathway was measured by Fisher’s exact test. As a result, a P value was obtained, determining the probability that the association between the genes in our data set and the networks generated can be explained by chance alone.
2.12 |. Venn diagrams and Jaccard similarity index
To accomplish testing following 2 hypothesis studied here, we performed pairwise comparisons between the lists of DEGs from all cell types involved in each possible differentiation route (P value ≤ 0.05 and FC ≥ 1.5): Hypothesis 1: BM-MOs x BM-iMOs, BM-MO x SP-MODC, BM-iMO x SP-MODC; and Hypothesis 2: BM-MOs x SP-MODCs, BM-MOs x SP-MO, and SP-MOs x SP-MODCs. Using the overlap DEGs numbers between the Venn diagrams, we measured the similarity between sets using the Jaccard index.
2.13 |. Flow cytometry data statistical analysis
All flow cytometry data represented with bar graphs was analyzed using Graphpad Prism 8.0 Software. The difference between 3 or more groups was verified using 2-way ANOVA test via Bonferroni’s test for multiple comparisons. The differences between 2 groups were verified using the t-test for parametric data. A P ≤ 0.05 value was considered statistically significant.
3 |. RESULTS
3.1 |. MO populations dynamics and phenotypic heterogeneity in the SP and BM of PbA-infected mice
Here, we characterize MO subsets harvested from BM and SP of uninfected and C57BL/6 mice at 6 days postinfection with PbA. During PbA infection, splenic MODCs, that represents over 60% of total MOs, are CD11b+F4/80+DC-SIGNhiMHCIIhi and express different levels of Ly6C, whereas iMOs (approximately 25% of total MOs) are Ly6Chi and CD11b+F4/80+DC-SIGNintMHCII− cells. We have also identified in the BM of PbA-infected mice newly differentiated inflammatory MOs (iMOs) and a MODC-like population that share some markers with MODCs, such as DC-SIGN and MHCII (Figs. 1A and B). Thus, to determine the equivalence between BM MODC-like and SP-MODCs, we evaluated the expression of previously described markers.4 SP-MODCs express CD11c and other activation markers, such as, CD80 and CD40; the chemokines CXCL9 or CXCL10 and most importantly the receptor CCR5 which is essential for migration of MODCs to the brain and development of ECM.11 In contrast, BM MODC-like cells lack in the expression of these markers (Fig. 1C). In addition, LPS-activated MODCs, but not the MODC-like cells, express high levels of TNFα, IL-10, and iNOS 26 and Plasmodium12 infection. Hence, BM MODC-like cells are not mature MODCs (Supplemental Figs. 1A and B).
FIGURE 1. Monocyte and MODC populations in the spleen and bone marrow of PbA-infected mice.
Spleens and BMs were harvested 6 days after PbA infection. (A) Splenocytes or BM cells were first gated for CD11b+F4/80+ cells and then for DC-SIGN and MHCII expression. (B) Expression of Ly6C and DC-SIGN was evaluated in CD11b+F4/80+DC-SIGN+MHCII+ cells. The data shown are representative of 3 independent experiments. (C) Splenic and BM CD11b+F4/80+DC-SIGN+MHCII+ cells were evaluated for the expression of DC/MODC markers and chemokines, such as, CD11c, CD40, CD80, CXCL9, CXCL10, and CCR5. Dotted lines show cells from uninfected mice and full lines from infected mice. Numbers indicate MFI for each marker on infected samples. (D) Emergency of DC-SIGN+MHCII+ cells in the spleen and BM follows the increase in of parasitemia in PbA-infected mice. Line graphs shows the frequency of each MO population within CD11b+F4/80+ cells. (E) Parasitemia was estimated by counting Giemsa-stained thin blood smears and is expressed as percentage of infected RBCs. Data shown are representative of 2 independent experiments. Results are expressed as average ± S.E.M. **P ≤ 0.001
Inflammation or infection can induce iMOs differentiation into MODCs 6,27 and egress of these cells from BM is CCR2-dependent.28 However, a previous study from our group has shown that during ECM, the emergence of splenic MODCs is only partially compromised in PbA-infected CCR2−/− mice.11 Here, we observed that emergence of MODCs in the SP parallels the parasitemia, whereas MO and iMOs decrease overtime. On the other hand, in the BM there is a significant enrichment of iMOs with infection, which on day 7 postinfection represents the predominant population among total CD11b+F4/80+ MOs (∼50%) (Figs. 1D and E and Supplemental Fig. 1C). In addition, BM iMOs express intermediary levels of DC-SIGN, suggesting that this population could be an intermediate differentiation state and contribute for the PbA-induced splenic MODCs.
3.2 |. Unsupervised and interactome analyses of differentially expressed genes and miRNAs in MO subsets during PbA infection
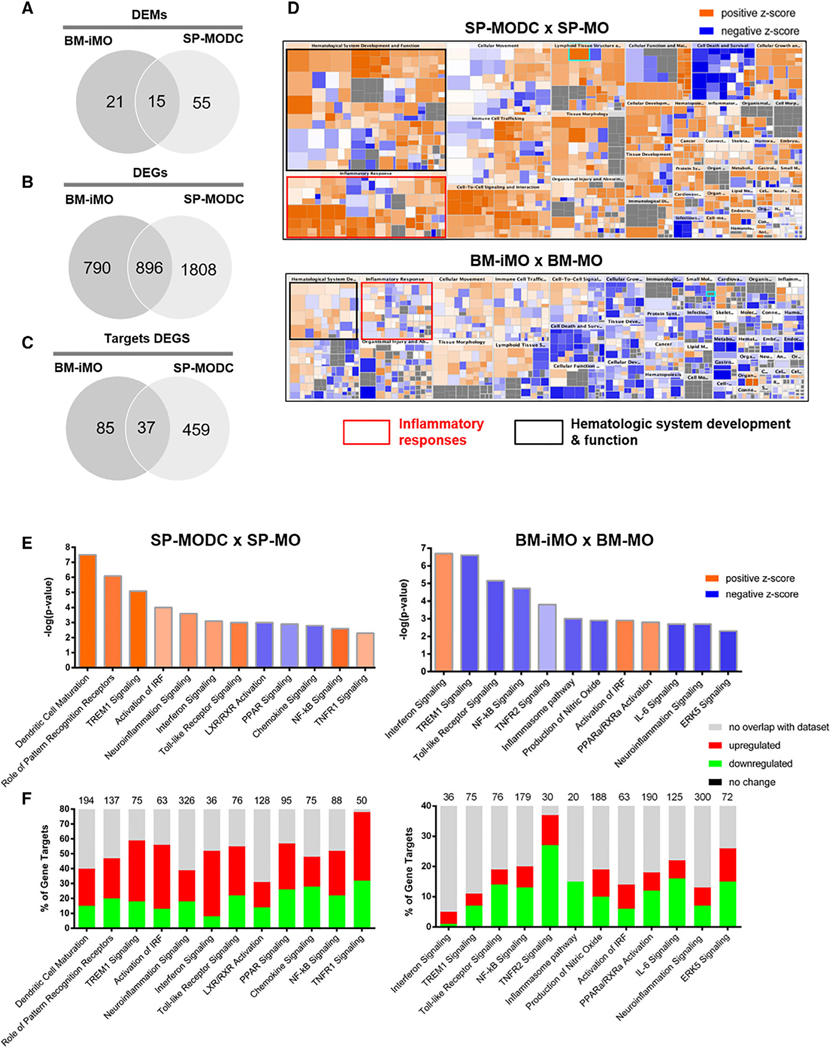
In order to determine the similarities and heterogeneities of SP and BM MO populations, we purified 4 cell populations (Supplemental Fig. 2): SP-MO and BM-MO cells (Ly6G−CD11b+F4/80+DC-SIGN−MHCII−) from uninfected mice and BM-iMOs (Ly6G−CD11b+F4/80+DC-SIGNintMHCII−) and SP-MODCs (Ly6G−CD11b+ F4/80+ DC-SIGNhiMHCIIhi) from PbA-infected mice. In Fig. 2A, we summarized the data processing and analysis of gene and miRNA expression profiles as a workflow. Briefly, statistical analysis and identification of DEGs and DEMs were performed and both transcriptome data were submitted to unsupervised analyses based on PCA, heatmap representation of hierarchical clustering and Jaccard index (JI) analysis, in order to observe grouping and/or segregation of the 4 MO populations. The list of DEGs and DEMs, were also used for integration, network, miRNA target, upstream regulator, and functional analyses. As shown in Fig. 2B, unsupervised analysis based on the expression of 2000 genes and 351 miRNAs with the higher expression variance showed segregation of the 4 groups and intergroup clustering both in PCA and hierarchical clustering indicating that gene and miRNA expression profiles were specific to each cell population. Both PCA and hierarchical clustering showed that BM-MOs (blue dots) and SP-MOs (green dots) samples grouped together, indicating similarities in transcriptome profiling. SP-MODCs (red dots) and BM-iMOs (magenta dots) clustered apart, indicating that these populations are unique and present individual DEGs and DEMs profile.
FIGURE 2. Profiling of differentially expressed genes and miRNAs in MO populations during PbA infection.
(A) Workflow of data processing and analysis of gene and miRNA expression profiles BM and SP cells from C57BL/6 mice were used for FACS isolation of the specific cell populations: SP-MO and BM-MO cells (Ly6G−CD11b+F4/80+DC-SIGN−MHCII−) from uninfected mice; BM-iMO (Ly6G−CD11b+F4/80+DC-SIGNintMHCII−) and SP-MODC (Ly6G−CD11b+F4/80+DC-SIGNhiMHCIIhi) from mice 6 days post-infection with PbA. (B) Principal component analysis (PCA) based on the top 2000 differentially expressed genes (DEGs) and 500 differentially expressed miRNA (DEMs) was performed, by using a median centering of the data set (the percentage of the variance is indicated between brackets). Heatmap and hierarchical clustering was performed on all the samples using squared Euclidean distance measure and Ward’s method for linkage analysis and z-score normalization. Each row represents 1 mRNA (left panel) and miRNA (right panel) significantly regulated and each column represents 1 sample. Specific cell populations are designed by the following colors: BM-MO: blue, BM-iMO: magenta, SP-MO: green, and SP-MODC: red. The color-coded scale (blue: expression levels lower than the mean and red: expression level over the mean) illustrates the mRNA and miRNA fold change (log2ΔCt) after global normalization is indicated at the bottom of the figure. (C) Schematic representation of the mathematical test of 2 hypotheses on preferential MODC differentiation route. Hypothesis 1: assumes that BM-MOs differentiates on BM-iMOs during infection and then migrate to the spleen where they complete maturation to SP-MODCs. Hypothesis 2: assumes that resident SP-MO along with BM-MO that migrate to the spleen during infection with an unchanged phenotype will differentiate into SP-MODC. The Venn Diagram indicate all the possible virtual comparisons between the cells on the tested hypothesis. In these diagrams, we compare DEGs from each differential analysis. Yellow dots indicate the intersection where is expected the biggest change on gene expression, that is in agreement with the organ where the differentiation took place. Red and orange dots indicate those intersections where is expected few genes in common mainly because they make reference to the migration process and we expect that during migration the cells suffer a minor change on
Next, using the list of DEGs, we have tested 2 hypotheses regarding the possible routes of MODC differentiation: hypothesis 1, the MODC differentiate from BM-MOs into BM-iMOs and then migrate to the SP where they complete their differentiation; or hypothesis 2, BM-MOs migrate to SP where they compose the pool of SP-MOs, which differentiate into MODC upon PbA challenge (Fig. 2C). If the first hypothesis is true, we expected that the contrast BM-MO versus BM-iMOS would show a greater number of DEGs than BM-iMOs versus SP-MODCs. And, if the second hypothesis is correct, BM-MOs/BM-iMOS versus SP-MOs would show less DEGs, which would be more pronounced when comparing BM-MOs/BM-iMOS/SP-MOs versus SP-MODCs. For this analysis, we used the JI as indicator of similarity between the sets of genes of different MO populations. This parameter is expressed in percentage that is given by the Venn diagrams intersection, shared Venn diagrams Union and the extent of shared genes between the comparisons (JI values close to number 1 or 100% correspond to similarities between comparisons). The yellow and red dots in the diagram of Fig. 2C illustrates the expected quantity of shared genes between the DEGs: higher and lower in proportion, respectively. Considering the expected proportions of shared genes between the tested DEGs, we observed JI for the highest expected proportion of 0.53 (53%) and the lowest expected proportion of 0.10 (10%) when testing hypothesis 2 (Table 1), suggesting that this is the most likely correct 1 and differentiation of MOs into MODCs occurs predominantly in the SP.
TABLE 1.
Jaccard index to test what of the 2 hypotheses about MODC differentiation route during PbA infection is more probable to happen
| Pairwise contrasts | JI | Expected | Observed |
|---|---|---|---|
| Hypothesis 1 | |||
| [BM-MOxBM-iMO] × [BM-MOxSP-MODC] | 0.16 | Highest proportion | Lowest proportion |
| [BM-MOxBM-iMO] × [BM-iMOxSP-MODC] | 0.18 | Low proportion | Low proportion |
| [BM-MOxSP-MODC] × [BM-iMOxSP-MODC] | 0.48 | Low proportion | Highest proportion |
| Hypothesis 2 | |||
| [BM-MOxSP-MODC] × [SP-MOxSP-MODC] | 0.53 | Highest proportion | Highest proportion |
| [BM-MOxSP-MO] × [BM-MOxSP-MODC] | 0.15 | Low proportion | Low proportion |
| [BM-MOxSP-MO] × [SP-MOxSP-MODC] | 0.10 | Lowest proportion | Lowest proportion |
Jaccard Index (JI) express the similarity and diversity of sample sets, which is calculated as intersection DEGs/union DEGs. JI = 1 is the highest similarity between contrasts. Expected highest and lowest proportion means closer of 1 and 0, respectively. A concordance between what is observed in relation to what is expected indicates which hypothesis is true.
3.3 |. Identification of DEMS, DEGs, and DEMs-Target/DEGs pairs in each cell population
Analysis with Benjamini–Hochberg false discovery rate (FDR) adjusted P ≤ 0.05 and FC cutoff of 1.5 revealed the highest number of both DEGs and DEMs in the contrast SP-MODC versus SP-MO. The total number of DEGs was 2,704 DEGs (1,648 up-regulated and 1,056 down-regulated) and 70 DEMs (40 up-regulated and 30 down-regulated). The comparison between BM-iMOs versus BM-MOs showed a total of 1,686 DEGs (930 up-regulated and 756 down-regulated), and 36 DEMs (20 up-regulated and 16 down-regulated). Supplemental Table 1 lists all DEGs and DEMs described above. Next, we used a miRNA target prediction tool from ingenuity pathway analysis software (IPA) to screen putative targets of DEMs within the list of DEGs for each cell population. This analysis pairs the DEMs with their putative target DEGs considering an inverse expressing pairing relationship between miRNA and its target gene (if the miRNA is up-regulated, its gene target must be down-regulated and vice versa). We only considered those target DEGs that have been highly predicted as targets (from TargetScan database) and/or experimentally validated as targets (from miRecords, TarBase, and direct acquisition from the literature by Ingenuity knowledge Base-IKB). From the list of 70 DEMs identified on SP-MODCs we found targeting information of 42 of them. We thus obtained a list of 496 target DEGs of the 42 DEMs in SP-MODCs. For the BM-iMO cells, from the list of 36 DEMs after filtering we obtained a list of 18 DEMs targeting a total number of 122 DEGs (Supplemental Table 2). The top 2 miRNAs with the highest number of targets DEGs in each cell group were: mmu-miR-16–5p and mmu-miR-29b for BM-iMO targeting 32 and 21 DEGs, respectively. For the MODCs mmu-miR-16–5p and mmu-miR-491 were among the most represented miRNAs, with 64 and 51 target DEGs, respectively.
3.4 |. Target DEGs set enrichment analysis reveals potential pathways regulated by miRNAs during MODC differentiation in PbA-infected mice
In Figs. 3A–C, Venn diagrams show the number of common and unique DEGs, DEMs, and target DEGs between BM-iMOs and MODCs. Supplemental Fig. 3 depict these molecules lists. To explore the gene pathways that were enriched in BM-iMOs and SP-MODCs during PbA infection, we carried a functional and canonical pathway analysis using the IPA software. A heatmap representing the enriched pathways shows that MODCs and iMOs exhibit multiple pathways in common; however, some pathways, such as, inflammatory responses and hematologic system development and function were highly activated in MODCs (bigger boxes with high z-score seen in orange) and not in iMOs (Fig. 3D). Canonical pathway analysis of the DEGs was performed for each cell group. In Fig. 3E, we show 12 canonical pathways among the most enriched pathways found for each cell type contrast displaying their predicted activation/inhibition status based on their z-score (activated processes are orange, whereas inhibited processes are blue). The stacked bar charts show the percentage of target DEMs down-regulated (in green) and up-regulated (red) in each 1 of the enriched pathways for each cell population (Fig. 3F). For both MO subsets, we observed overrepresentation of canonical pathways related to immune processes. For the SP-MODCs, IFN signaling, neuroinflammation, TREM1, and NF-kB signaling and, most importantly, DC maturation pathway were predicted to be upregulated (orange), whereas PPAR and chemokine signaling were predicted to be down-regulated (blue). Furthermore, in BM-iMOs, IFN signaling, IRF and PPARα pathways were predicted to be activated, whereas innate and acquired immune response, TREM1 and NF-kB signaling were down-regulated.
FIGURE 3. Differentially expressed genes and miRNAs and functional pathways enriched during MODC differentiation.
(A) Venn diagram showing the number of differentially expressed genes and (B) miRNAs in SP-MODCs and BM-iMOs and intersected genes. (C) Venn diagram with the number of targets DEGs (high predicted and experimentally validated targets) in iMOs and MODCs as well as those that are shared between these 2 cell populations. (D) Enriched ingenuity pathway analysis (IPA) categories of DEGs in BM-iMOs and SP-MODCs. Diseases and biologic function pathways colored by z-score that measure the activation state of these processes (blue: inhibited and orange: activated); sized by the number of genes: the bigger the box the more genes of the provided list it contains. On the scale blue means lower activation levels and orange higher activation levels. (E) Ingenuity pathway analysis shown for differentially expressed genes (DEGs) and (F) microRNAs (DEMs). Pathway analysis for the comparison SP-MODC versus SP-MO or BM-iMO versus BM-MO. For DEGs, we shown canonical pathways most significantly enriched colored by their activation (z-score) the activated pathways (high z-score) are in orange and inhibited (low z-score) are in blue. To DEMs target analysis, the numerical value in the top of each pathway bar represents the total number of genes in that canonical pathway. The stacked bar charts display the percentage of target DEGs that are positively (red), negatively (green), no change (black), and no overlap with IPA database (gray). In all analysis, the Benjamini–Hochberg false discovery rate (FDR) was used with adjusted P ≤ 0.05 and a fold change (FC) cutoff ≥ 1.5
3.5 |. Upstream regulator analysis and DEM/DEG networks revealed IFNγ as a key molecule during PbA infection
The upstream regulator analysis tool from IPA was used to identify potential upstream regulators by analyzing linkage to DEM targets through coordinated expression. The analysis indicated that IFNγ is the top upstream expression regulator, followed by other cytokines like IL-4, TNFα, CSF2, IL-6, IL-1β, IL-3, IL-2, IL-5, and CD40LG predicted to be activated (orange) or inhibited (blue), based on their z-score value (Fig. 4A). In order to investigate the possible role of miRNAs in regulating key DEGs in MODC activation and differentiation, we built a DEM-DEG network for the MODC population, with the top 2 miRNAs found to potentially regulate the highest number of targets DEGs (mmu-miR-16–5p and mmu-miR-491). The built network and prediction analysis revealed the potential role/connection of these 2 miRNAs and their target DEGs in regulating key biologic and pathophysiologic process in the malaria context: DC maturation and neuroinflammation, both predicted to be activated (orange) (Fig. 4B). The network shows the potential targets for mmu-miR-16–5p: Protein kinase Akt-3 (AKT3), amyloid precursor protein (APP), CD40 receptor (CD40), C-X-C motif chemokine ligand 10 (CXCL10), major histocompatibility complex, class II (HLA-DQA1), IFN gamma receptor 2 (IFNGR2), IL-1 receptor-associated kinase 2 (IRAK2), mitogen-activated protein kinase 9 (MAPK9), prostaglandin endoperoxide synthase 2 (PTGS2), and TLR1, all connected to neuroinflammation pathway. For the mmu-miR-491–5p the targets DEGs present in the network were: TLR9, innate immune signal transduction adaptor (MYD88), intercellular adhesion molecule 1 (ICAM1), major histocompatibility complex (HLA-DQA1), class II, DQ beta 1 and DM alpha (HLA-DQB1, HLA-DMA), and fascin actin-bundling protein 1 (FSCN1), connected to DC maturation. Importantly, IFNγ was shown to be a node interconnecting a great number of DEMs and DEGs, suggesting that this cytokine is a central molecule in the process of MODC differentiation and neuroinflammation during PbA infection.
FIGURE 4. Upstream regulator and DEM/DEG network of MODC differentiation during PbA infection.
(A) Upstream regulator analysis of the DEM targets in BM-iMOs and SP-MODCs. Orange or blue color represent activated or inhibited, respectively, according to z-score prediction statistical calculation of the upstream regulator activation state in both cell populations. (B) DEM-DEG networks for SP-MODC transcriptome and microRNA data using IPA software. Built network contain the top 2 miRNAs found to potentially regulate the highest number of targets DEGs (mmu-miR-16–5p and mmu-miR-491) in the context of DC maturation and neuroinflammation, both predicted to be activated (orange). Relationship between molecules is represented as a line (direct or indirect relationship). The molecules colors in graduation of red and green represent if their fold change is up or down regulated, respectively
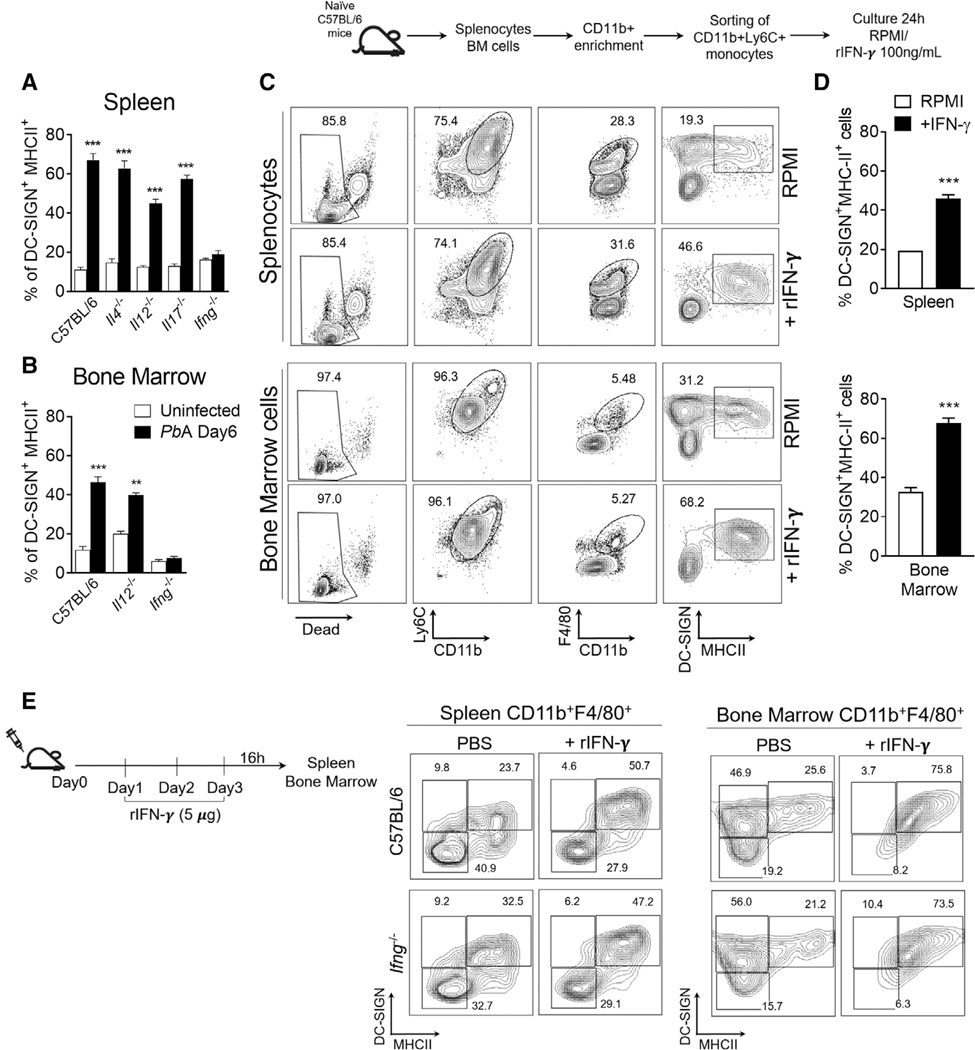
3.6 |. IFNγ is a key factor for the differentiation of MODCs during PbA infection
To test the hypothesis that IFNγ is the key factor on MODC differentiation, we infected knockout mice for either IL-4, IL-12, IL-17 and IFNγ genes and at 6 days post-infection, SP and BM were used to determine the frequency of MODCs. Notably, only IFNγ−/− mice fail to differentiate splenic MODCs (Figs. 5A and B). IFNγ deficiency leaded to an accumulation of splenic iMOs that did not differentiated into MODCs (Supplemental Fig. 3A). We also show that CD4+ and CD8+ T cells are the main source of IFNγ in the SP.11,18,29 The IFNγ producing CD4+ and CD8+ T cells were also found in a low number in the BM of infected mice. NK cells were present in both compartments but represented a smaller fraction of the IFNγ producing cells (Supplemental Figs. 3B and C). During PbA infection CD4+ and CD8+ T cells represents approximately 18 and 8% of total splenic cells, respectively; and comprise less than 4% of total BM cells (data not shown), indicating that the production of IFNγ in the SP is far more abundant. Next, we tested if IFNγ would be sufficient to induce MOs to differentiate into MODCs in vitro and in vivo. Culturing SP-MOs or BM-MOs (CD11b+Ly6C+) isolated from naïve mice in the presence of rIFNγ induced the expression of DC-SIGN and MHCII indicating a differentiation towards MODCs (Fig. 5C and D). In addition, naïve WT (Fig. 5E—top panel) or IFNγ−/− (Fig. 5E—bottom panel) mice treated with rIFNγ presented an increase of CD11b+F4/80+DC-SIGNhiMHCIIhi cells both in the SP and BM. In summary, we observed that IFNγ is a central DEM–DEG network hub, which indicate the importance of this molecule in the biologic process of MODC differentiation during PbA infection.
FIGURE 5. Role of IFNγ on the differentiation of MODCs during PbA infection.
Spleens and BMs were harvested 6 days after PbA infection. (A and B) Splenocytes or BM cells were obtained from C57BL/6, IL-4−/−, IL-12−/−, IL-17−/−, and IFNγ−/− mice. Total cells were first gated for CD11b+F4/80+ cells and then for DC-SIGN+MHCII+ cells. Bar graphs correspond to frequency of DC-SIGN+MHCII+ cells within total monocytes CD11b+F4/80+. The data shown are representative of 2 independent experiments. Results are expressed as average ± S.E.M. Two-way ANOVA analysis of variance comparing splenic versus BM cells in infected C57BL/6 mice. ***P < 0.0001; **P < 0.001. (C) Splenocytes and BM cells were collected from naïve C57BL/6 mice for sorting of monocytes CD11b+Ly6C+. Isolated cells were cultured with IFNγ (100 ng/ml) for 24 h and then analyzed for MODC differentiation by flow cytometry. Contour plot shows live monocytes (CD11b+ Ly6C+F4/80+) being evaluated for the expression of DC-SIGN and MHCII (MODC phenotype). (D) Bar graphs correspond to frequency of DC-SIGN+MHCII+ cells within total monocytes CD11b+F4/80+ in cultures treated with IFNγ or not (RPMI). (E) C57BL/6 mice were administered recombinant IFNγ or PBS once per day for 3 days. Spleen and BM were collected 18 h after the final injection. Contour plot show the frequency of DC-SIGN+MHCII+ cells within total monocytes CD11b+F4/80+and histogram show the expression of CD11c on this cell. Statistical analysis was performed by 2-tailed nonparametric unpaired t-test at 95% CI. The data shown are representative of 3 independent experiments. ***P < 0.0001
3.7 |. Differentiation of splenic MO into MODCs during PbA infection
According to our hypothesis, MODCs differentiate from resident SP-MOs. By using an adoptive cell transfer experiments, we evaluate the ability of SP-MOs to differentiate into MODCs in vivo during PbA infection (Figs. 6A–C). MOs (F4/80+CD11b+DC-SIGN−MHCII−) were enriched from SPs of uninfected CD45.2 C57BL/6 mice and 2 × 106 cells were transferred to either uninfected or PbA-infected CD45.1 congenic mouse. Two days later, the frequencies of donor CD45.2+ MODCs were evaluated in the SP and BM. In infected mice, like host cells (CD45.1+), most splenic CD45.2+ MOs differentiated into splenic MODCs (F4/80+CD11b+DC-SIGNhiMHCIIhi). Likewise, these MOs migrate to the BM, where they also express DC-SIGN and MHCII. Thus, we favor the hypothesis that SP-MOs are main precursor cells or, at least, enough to generate the pathogenic MODCs that migrate and promote ECM in the PbA model.
FIGURE 6. Differentiation of splenic MO into MODCs during PbA infection.
Enriched splenic F4/80+CD11b+DC-SIGN−MHCII− cells from uninfected CD45.2+ donor mice were adoptively transferred i.v. into CD45.1+ uninfected or PbA-infected mice. Dot plot of enriched monocytes show that ∼99% of the cells were undifferentiated monocytes DC-SIGN−MHCII− (MO). After 48 h, spleens and BM of recipient mice were obtained and frequency of CD45.2+ donor-derived MODCs were compared with recipient-derived CD45.1+ MODCs. Bar graphs show frequency of MODC in F4/80+CD11b+ populations from CD45.2+ or CD45.1+ cells of 2 pooled experiments that yielded similar results
4 |. DISCUSSION
The parasite–host interaction is complex and should be tightly regulated from the immunologic point of view: parasites evade the immune response to avoid elimination from the host; whereas antiparasite immune responses try to limit the invasion and parasite proliferation without eliciting significant collateral damage. Among the innate immune cells, MOs and DCs play an important role in host resistance to Plasmodium infection and pathogenesis of malaria.30–32 The activation of these innate immune cells and consequent systemic inflammation lead to the initial signs and symptoms of malaria. Activation of MOs, cytokinemia, cytoadherence of iRBCs, and anemia associated with the severe and lethal forms of the disease, including cerebral malaria. While the extensive heterogeneity of these cells is evident, the functional significance of their differentiation during infection remains elusive.
The aim of this study was to integrate both miRNome and transcriptome data from MO subpopulations to broaden our understanding and gain insight in how miRNAs translation during differentiation of the plastic MO populations during PbA infection. Splenic MODCs differentiated during PbA infection are characterized by the phenotype CD11b+F4/80+DC-SIGNhiMHCIIhi, expression of different levels of Ly6C and DC maturation and activation markers, such as, CD11c, CD80, CD40. In addition, splenic MODCs express the receptor CCR5, essential for migration to the CNS 11 and the chemokines CXCL9 and 10, involved in the recruitment of activated T lymphocytes expressing CXCR3 and development of ECM.13 On the other hand, iMOs are CD11b+F4/80+DC-SIGNintMHCII−CD80− and display low phagocytic activity compared to MODCs.11,32 Here, we have also identified another cell population MODC-like, which share some markers with MODCs, such as DC-SIGN and MHCII, but lack markers of mature DCs, i.e. CD40, CD86, CCR5, CXCL9, or CXCL10. In addition, splenic MODCs, but not BM MODC-like, produce high levels of cytokines and express iNOS in response to LPS, indicating that BM MODC-like cells are not mature or functional MODCs.
Moreover, PbA infection induces expansion of newly differentiated iMOs in the BM. Importantly, the egress from BM and the emergence of iMOs in nonlymphoid as well as peripheral lymphoid organs was previously shown to be CCR2 dependent.28 However, the generation of splenic MODCs is only partially compromised, whereas the migration of MODCs to the CNS and the development of ECM are not affected in PbA-infected CCR2−/− mice.11 In addition, the high frequency of iMOs (CD11b+F4/80+CD11c−DC-SIGNintMHCII−) and not MODCs (CD11b+F4/80+CD11c+ DC-SIGNhiMHCIIhi) in the BM from PbA-infected mice further suggest that MODCs are differentiating elsewhere.
MicroRNAs have been found to be fine tuners of different biologic processes by post-transcriptionally controlling gene expression. They can indirectly control the immune system homeostasis by targeting cytokines, enzymes, transcription factors, and other regulatory molecules and their dysregulated expression is associated with different pathologies, including infectious diseases and immune disorders.
Here, we identified DEMs and DEGs profiles to determine the potential site(s), regulatory molecules and processes related to SP-MODCs differentiation during PbA infection. The transcriptome profiling showed a great number of DEMs and DEGs. The unsupervised analyses (PCA, hierarchical clustering and Jaccard index) of both transcriptomes’ profiles were able to segregate the different MO subsets. The Jaccard index (JI) also called the Jaccard similarity coefficient is a measure of similarity used to compare members from 2 sets showing which ones are shared and which are distinct. The JI has been previously used as a measure of similarity between 2 sets of data in a great number of studies, for example, tumorigenesis,33,34 systematic phenotyping of human cells in different diseases 35 and re-annotation of Drosophila genomes.36 The analysis supported the hypothesis that the SP-MOs are likely to be the main progenitors for SP-MODCs and differentiation of MOs into MODCs occurs predominantly in the SP. This hypothesis is corroborated by earlier studies showing that MODC differentiation may occur in the site of infection,26,37 and is further supported by our data showing that MODCs still emerge in the SP of PbA-infected CCR2−/− mice that have an impaired egress of iMOs from the BM.11 We also used the adoptive cell transfer experiments, and the results showed that SP-MOs differentiate into SP-MODCs, validating this hypotheses.
Canonical pathway and integration of DEGs and DEMs analysis showed an enrichment in pathways related to immune response. Importantly, the DC maturation signaling pathway was exclusively enriched on SP-MODCs and other canonical pathways shared between the 2 cell groups were: IFN, TLR, activation of IRF, PPAR activation, NF-κb, and neuroinflammation signaling pathways. Computational and integration analysis revealed the number of DEGs, DEMs, and Target DEGs in common and unique for each cell population. We observed that a great number of DEGs are potential DEMs targets in both cell populations with a higher number in SP-MODCs.
Our analysis revealed 2 top miRNAs (both down-regulated in SP-MODC), mmu-miR-16–5p and mmu-miR-491, as potentially regulators of the majority of target DEGs on SP-MODCs. The built network and prediction analysis showed that mmu-miR-16–5p have 10 putative target DEGs (AKT3, APP, CD40, CXCL10, HLA-DQA1, IFNGR2, IRAK2, MAPK9, PTGS2, and TLR1) all node molecules are up-regulated and indirectly connected to activation of neuroinflammation signaling. The DEM mmu-miR-491–5p showed 6 target DEGs molecules (TLR9, MYD88, ICAM1, HLA-DQB1, HLA-DMA, and FSCN1), and indirectly connected to DC maturation. Importantly, the network also showed IFNγ as a node interconnecting a great number of DEMs and DEGs in the network. An upstream regulator analysis based on the DEGs profiles also indicated that IFNγ is the top 1 potential upstream key cytokine modulating the observed transcriptional changes. These both findings indicate that this cytokine is a central molecule in the process of MODC differentiation and neuroinflammation during PbA infection. The mouse miRNA-16–5p (mmu-miR-16–5p) (ID: MIMAT0000527) is down-regulated in SP-MODCs. This miRNA is part of the mir-15 gene family, which include 6 highly conserved miRNAs (miR15a/15b/16/195/497), which are clustered on 3 separate chromosomes and their mature miRNAs have the same seed sequence.
The precursors of mmu-miR-16–5p are mmu-mir-16–1 and mmu-mir-16–2 located in chromosome 14 and 3, respectively.38 It was first described by Calin et al.39 as deleted or down-regulated in chronic lymphocytic leukemia. The entire family appears to function as cell proliferation in lymphoid and nonlymphoid cells. It was also found down-regulated in plasma of P. vivax infected patients and may be potential biomarkers for malaria infection.40 Our network analysis showed that miR-16–5p and miR-491–5p have potential targets related to neuroinflammation and DC maturation: MiRNA-491–5p has 2 targets, TLR9 and MYD88, crucial molecules for the development of protective immunity to malaria.41 Among the miR-16–5p target genes are CD40, CXCL10, IFNGR2, and PTGS2, all previously described as implicated in the pathogenesis of ECM.42,43
One of the limitations of the analysis performed here is that is based in a broad systematic analysis providing an overview of miRNA–mRNA networks, and correlation is not proof of causality. So, we decided to experimentally validated IFNγ regulatory function in MODC differentiation by treating MOs with IFNγ either in vitro or in vivo and we observed that the treatment alone was enough to induce MODCs differentiation. Similar effect was observed in the SP and BM, indicating BM-MOs can complete their differentiation to MODCs, if provided sufficient levels of IFNγ. During ECM and MA-ARDS, CD4+, and CD8+ T cells are the main source of IFNγ in the SP and lungs.11,12,46 Since the T lymphocytes are in high frequency in the SP, but not in the BM, we believe that in IFNγ-treated mice, but not in PbA-infected mice, this cytokine reaches high enough levels to induce BM-MO differentiation into BM-MODC.20,47 Moreover, a recent study demonstrates that IFNGR2 expression on brain epithelial cells are also important for the development of ECM.45 These data are intriguing, taking that migration of MO-DCs expressing high levels of IFNGR2 is required for the development of ECM and impairment of MODC differentiation is observed in IFNγR1−/− mice that are resistant to ECM.11,44 These findings suggest that IFNγR1 may be more relevant for MOs differentiation and migration of MODCs, to the CNS during PbA infection.
In conclusion, we have shown that IFNγ is the key factor on MODC differentiation. The built network implies IFNγ as a central molecule (node molecule) interconnecting a great number of DEMs and DEGs being an upstream regulator. We also provide experimental evidences that IFNγ is a main factor orchestrating the differentiation of MODCs during PbA infection. Our data also support that MODCs differentiate primarily from SP-MOs, contrasting with a more conventional route of differentiation from BM-iMOs and recruited to peripheral lymphoid organs and non-lymphoid tissue. Finally, by using a combination of functional in vivo and in vitro assays with an in silico mRNA and miRNA network predictions, our data favor the hypothesis that microRNAs are important regulators of genes relevant to malaria pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
The author Patricia A Assis developed the present work at University of Massachusetts Medical School, USA and Fundação Oswaldo Cruz - Minas, Brazil. We thank Dr. Andrew D. Luster from Massachusetts General Hospital (MGH) and Dr. John E. Harris from Umass Medical School (UMMS) for providing mouse lines used in this study. This work was supported by the US NIH (R01NS098747, R01AI079293, U19AI089681, and R21AI131632), a Brazilian National Institute of Science and Technology for Vaccines (INCT/CNPq), Fundação de Pesquisa do Estado de Minas Gerais (465293/2014-0), and Fundação de Amparo de Pesquisa do Estado de São Paulo (2016/23618-8).
Abbreviations:
- BM
bone marrow
- CMP
common myeloid progenitor
- DC
dendritic cell
- DEGs
differentially expressed genes
- DEMs
differentially expressed miRNAs
- ECM
experimental cerebral malaria
- iMO
inflammatory monocyte
- iRBCs
infected red blood cells
- JI
Jaccard Index
- MA-ARDS
malaria-associated acute respiratory distress syndrome
- MO
monocyte
- MODC
monocyte-derived dendritic cell
- PbA
Plasmodium berghei ANKA
- SP
spleen
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327(5966):656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Borgne M, Etchart N, Goubier A, et al. Dendritic cells rapidly recruited into epithelial tissues via CCR6/CCL20 are responsible for CD8+ T cell crosspriming in vivo. Immunity. 2006;24(2):191–201. [DOI] [PubMed] [Google Scholar]
- 3.Davies LC, Taylor PR. Tissue-resident macrophages: then and now. Immunology. 2015;144(4): 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheong C, Matos I, Choi JH, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143(3):416–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai SM, Sheng J, Gupta P, et al. Organ-specific fate, recruitment, and refilling dynamics of tissue-resident macrophages during blood-stage malaria. Cell Rep. 2018;25(11):3099–3109. [DOI] [PubMed] [Google Scholar]
- 6.Nakano H, Lin KL, Yanagita M, et al. Blood-derived inflammatory dendritic cells in lymph nodes stimulate acute T helper type 1 immune responses. Nat Immunol. 2009;10(4):394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobbs KR, Embury P, Vulule J, et al. Monocyte dysregulation and systemic inflammation during pediatric falciparum malaria. JCI Insight. 2017;2(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonelli LR, Leoratti FM, Costa PA, et al. The CD14+CD16+ inflammatory monocyte subset displays increased mitochondrial activity and effector function during acute Plasmodium vivax malaria. PLoS Pathog. 2014;10(9):e1004393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schumak B, Klocke K, Kuepper JM, et al. Specific depletion of Ly6C(hi) inflammatory monocytes prevents immunopathology in experimental cerebral malaria. PLoS One. 2015;10(4):e0124080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pai S, Qin J, Cavanagh L. Real-time imaging reveals the dynamics of leukocyte behaviour during experimental cerebral malaria pathogenesis. PLoS Pathog. 2014;10(7):e1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirako IC, Ataide MA, Faustino L, et al. Splenic differentiation and emergence of CCR5(+)CXCL9(+)CXCL10(+) monocyte-derived dendritic cells in the brain during cerebral malaria. Nat Commun. 2016;7:13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galvao-Filho B, de Castro JT, Figueiredo MM, Rosmaninho CG, Antonelli LRDV, Gazzinelli RT. The emergence of pathogenic TNF/iNOS producing dendritic cells (Tip-DCs) in a malaria model of acute respiratory distress syndrome (ARDS) is dependent on CCR4. Mucosal Immunol. 2019;12(2):312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campanella GS, Tager AM, El Khoury JK, et al. Chemokine receptor CXCR3 and its ligands CXCL9 and CXCL10 are required for the development of murine cerebral malaria. Proc Natl Acad Sci USA. 2008;105(12):4814–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE. Pairing beyond the seed supports MicroRNA targeting specificity. Mol Cell. 2016;64(2):320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehl T, Backes C, Kern F, et al. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget. 2017;8(63):107167–107175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groom JR, Richmond J, Murooka TT, et al. CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity. 2012;37(6):1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin BS, Ishizaka ST, Lamphier M, et al. Therapeutical targeting of nucleic acid-sensing Toll-like receptors prevents experimental cerebral malaria. Proc Natl Acad Sci USA. 2011;108(9):3689–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cossarizza A, Chang HD, Radbruch A, et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur J Immunol. 2019;49(10):1457–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Askenase MH, Han SJ, Byrd AL, et al. Bone-marrow-resident NK cells prime monocytes for regulatory function during infection. Immunity. 2015;42(6):1130–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 25.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43(W1):W566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19(1):59–70. [DOI] [PubMed] [Google Scholar]
- 27.Jakubzick C, Tacke F, Ginhoux F, et al. Blood monocyte subsets differentially give rise to CD103+ and CD103-pulmonary dendritic cell populations. J Immunol. 2008;180(5):3019–3027. [DOI] [PubMed] [Google Scholar]
- 28.Sponaas AM, Freitas do Rosario AP, Voisine C, et al. Migrating monocytes recruited to the spleen play an important role in control of blood stage malaria. Blood. 2009;114(27):5522–5531. [DOI] [PubMed] [Google Scholar]
- 29.Hirako IC, Assis PA, Hojo-Souza NS, et al. Daily rhythms of TNFalpha expression and food intake regulate synchrony of plasmodium stages with the host circadian cycle. Cell Host Microbe. 2018;23(6):796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amorim KN, Chagas DC, Sulczewski SB, Boscardin F. Dendritic cells and their multiple roles during malaria infection. J Immunol Res. 2016;2016:2926436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chua CL, Brown G, Hamilton JA, Rogerson S, Boeuf P. Monocytes and macrophages in malaria: protection or pathology?. Trends Parasitol. 2013;29(1):26–34. [DOI] [PubMed] [Google Scholar]
- 32.Hirako IC, Assis PA, Galvão-Filho B, Luster AD, Antonelli LRV, Gazzinelli RT. Monocyte-derived dendritic cells in malaria. Curr Opin Microbiol. 2019;52:139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aoto Y, Okumura K, Hachiya T, et al. Time-series analysis of tumorigenesis in a murine skin carcinogenesis model. Sci Rep. 2018;8(1):12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hjaltelin JX, Izarzugaza JMG, Jensen LJ, Russo F, Westergaard D, Brunak S. Identification of hyper-rewired genomic stress non-oncogene addiction genes across 15 cancer types. NPJ Syst Biol Appl. 2019;5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gapp BV, Konopka T, Penz T, et al. Parallel reverse genetic screening in mutant human cells using transcriptomics. Mol Syst Biol. 2016;12(8):879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, Jaime M, Polihronakis M, et al. Re-annotation of eight Drosophila genomes. Life Sci Alliance. 2018;1(6):e201800156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leon B, Lopez-Bravo M, Ardavin C. Monocyte-derived dendritic cells formed at the infection site control the induction of protective T helper 1 responses against Leishmania. Immunity. 2007;26(4):519–531. [DOI] [PubMed] [Google Scholar]
- 38.Porrello ER, Johnson BA, Aurora AB, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. 2011;109(6): 670–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24): 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chamnanchanunt S, Kuroki C, Desakorn V, et al. Downregulation of plasma miR-451 and miR-16 in Plasmodium vivax infection. Exp Parasitol. 2015;155:19–25. [DOI] [PubMed] [Google Scholar]
- 41.Franklin BS, Rodrigues SO, Antonelli LR, et al. MyD88-dependent activation of dendritic cells and CD4(+) T lymphocytes mediates symptoms, but is not required for the immunological control of parasites during rodent malaria. Microbes Infect. 2007;9(7):881–890. [DOI] [PubMed] [Google Scholar]
- 42.Piguet PF, Kan CD, Vesin C, Rochat A, Donati Y, Barazzone C, et al. Role of CD40-CVD40L in mouse severe malaria. Am J Pathol. 2001;159(2):733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anyona SB, Hengartner NW, Raballah E, et al. Cyclooxygenase-2 haplotypes influence the longitudinal risk of malaria and severe malarial anemia in Kenyan children from a holoendemic transmission region. J Hum Genet. 2020;65(2):99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amani V, Vigário AM, Belnoue E, et al. Involvement of IFN-gamma receptor-medicated signaling in pathology and anti-malarial immunity induced by Plasmodium berghei infection. Eur J Immunol. 2000;30(6):1646–1655. [DOI] [PubMed] [Google Scholar]
- 45.Villegas-Mendez A, Strangward P, Shaw TN, et al. Gamma interferon mediates experimental cerebral malaria by signaling within both the hematopoietic and nonhematopoietic compartments. Infect Immun. 2017;85(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belnoue E, Potter SM, Rosa DS, et al. Control of pathogenic CD8+ T cell migration to the brain by IFN-gamma during experimental cerebral malaria. Parasite Immunol. 2008;30(10):544–553. [DOI] [PubMed] [Google Scholar]
- 47.Belyaev NN, Biró J, Langhorne J, Potocnik AJ. Extramedullary myelopoiesis in malaria depends on mobilization of myeloid-restricted progenitors by IFN-gamma induced chemokines. PLoS Pathog. 2013;9(6):e1003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.