Abstract
Attachment of measles virus (MV) to its cellular receptor is mediated by the viral envelope glycoprotein hemagglutinin (H). H exists at the viral surface as a disulfide-linked dimer which may associate into a tetramer. We aimed to define regions of H essential for its homo-oligomerization. To delineate these more precisely, we have generated a series of H ectodomain truncation mutants and studied their abilities to form both homotypic complexes and heterotypic complexes with full-length H. We define a “minimal unit” which is sufficient for MV H dimerization as that encompassing residues 1 to 151. This unit forms both homodimers and heterodimers with full-length H protein, although neither is transported to the cell surface even in the presence of other MV proteins. We show that cysteine residues at positions 139 and 154 are both critical in mediating covalent dimerization, not only of the truncated H mutants but also of full-length MV H protein. Even those cysteine mutants unable to form covalent intermolecular interactions are biologically active, mediating the formation of syncytia, albeit at a reduced rate. We demonstrate that this impaired capacity to mediate cell-to-cell fusion is based mainly on a reduced transport rate of the mutant molecules to the cell surface, indicating a role for covalent intermolecular interactions in efficient transport of MV H dimers to the cell surface.
Measles virus (MV) belongs to the Paramyxoviridae, a family of enveloped, negative-stranded RNA viruses. The surface of the virion is composed of two envelope glycoproteins, hemagglutinin (H) and fusion protein (F), which together mediate virus-cell attachment and entry (31). The H protein is responsible for binding to the cell surface receptor (21, 31), which for the most extensively studied strain of MV, the vaccine strain MV Edmonston (MVEdm), is the regulator of complement activation CD46 (6, 20). H is also involved in supporting the ability of F to mediate virus-cell fusion subsequent to CD46 binding (15).
The H protein is a type II transmembrane glycoprotein, believed to be functional at the virion surface as a disulfide-linked dimer which may associate into a tetramer (16, 18, 24). H protein dimerization and acquisition of conformation-dependent antigenic epitopes are both thought to occur in the endoplasmic reticulum (ER) prior to transport to the Golgi complex (13). The mature H protein comprises a short cytoplasmic tail of 34 amino acids preceding a single hydrophobic transmembrane region and a large C-terminal ectodomain (1, 4, 9). Although lacking neuraminidase activity, MV H protein may show some structural similarity with the hemagglutinin-neuraminidase (HN) proteins of all other members of the Paramyxoviridae (16).
The regions of MV H protein essential for its homo-oligomerization have yet to be mapped. Residues 2 to 14 of the cytoplasmic tail cannot be required for MV H oligomerization, since viruses with this deletion are capable of both dimerizing and mediating cell-cell fusion (3). Residues 35 to 58 of H are buried in the lipid bilayer, forming the transmembrane domain (1). It has been proposed that amino acids 59 to 181 of the H protein form a slender stalk encompassing a highly protease-sensitive region at residues 135 to 181, which may be exposed to the outside, forming the “hinge” of the molecule (16, 24). Furthermore, study of soluble forms of H generated by protease digestion from infected cells suggests that the region between amino acids 135 and 173 may be involved in oligomerization and that cysteine 139 may be critical in this interaction (24). Beyond amino acid 181 lies the globular head of the molecule comprising the CD46 binding site and proposed neuraminidase-like domain.
Taken together, these data and structural predictions led us to propose that the stalk region of the ectodomain of MV H protein may be essential for its efficient dimerization. To test this hypothesis we generated a series of progressive H ectodomain truncation mutants and studied their abilities to form both homotypic complexes and heterotypic complexes with full-length H. All of these mutants include the membrane-proximal region of the ectodomain defined as the fusion-promoting region of paramyxovirus H or HN proteins (5, 26–28, 30). Although incapable of mediating fusion support, the truncated H proteins have given insights into the mechanism underlying MV H dimerization. We define a “minimal unit” which is sufficient for MV H dimerization as that encompassing residues 1 to 151. Furthermore, we show that cysteines at positions 139 and 154 are responsible for mediating dimerization in the context of both truncated and full-length H proteins. This covalent dimerization is demonstrated to be a prerequisite for efficient transport to the cell surface and hence for efficient function.
MATERIALS AND METHODS
Cell culture and transfection.
Vero (African green monkey kidney) cells were maintained in Dulbecco's modified Eagle's medium containing 5% fetal bovine serum, penicillin, and streptomycin at 37°C and 5% CO2. The cells were transiently transfected by Lipofection (Superfect; Qiagen) and analyzed 18 to 24 h posttransfection. Unless otherwise stated, cotransfection of different plasmids was performed using equimolar quantities of DNA corresponding to each construct.
Plasmid construction and site-specific mutagenesis.
Parental plasmids for mutagenesis and all experiments were MVEdm H and F proteins subcloned into the pCG vector (2). In all cases, site-directed mutagenesis was performed using the quick change system (Stratagene) according to the manufacturer's instructions, and the integrity of mutant constructs was confirmed by DNA sequencing and Western analysis. Table 1 shows the plus strand of the mutagenesis primers used in this study. To generate truncated H constructs, designated H stems 1 to 7 we introduced a six-histidine epitope followed by two stop codons. The six-histidine epitope was designed as a tool to detect H stem proteins at the cell surface. For amino-terminal Flag-tagging of H we inserted the sequence encoding the Flag epitope (DYKDDDDK) downstream of the ATG start codon of H. Cysteine-to-serine exchanges (H C139S and H C154S) were introduced by converting the codon TGT to TCT at the indicated positions.
TABLE 1.
Primers used for site-directed mutagenesis.
| Construct | Primer sequence (plus strand)a |
|---|---|
| H stem 1 | 5-CCACTCTTCAAAATCATCGGTCATCACCACCATCACCATTAATGACAGAGATTCACTGACCTAGTG-3 |
| H stem 2 | 5-CCTTAATCCGGATAGGGAGTACCATCACCACCATCACCATTAATGAATCAACCCGCCAGAGAGAATC-3 |
| H stem 3 | 5-CCAGAGAGAATCAAATTGGATTATCATCACCACCATCACCATTAATGAGCTGAAGAGCTCATGAATGC-3 |
| H stem 4 | 5-GTGAACTCAACTCTACTGCATCACCACCATCACCATTAATGACTAGCTGTCTCAAAGGG-3 |
| H stem 5 | 5-GTCACTATGACATCCCAGCATCACCACCATCACCATTAATGAGTGGAAAAGCCTAATCTG-3 |
| H stem 6 | 5-CCTGGGTCCCCTTATCAACGCATCACCACCATCACCATTAATGATACCTCTCATCTCACAGAGGTG-3 |
| H stem 7 | 5-GGATACCGAGATTCAAGGTTAGTCATCACCACCATCACCATTAATGAAAGGAAGCAGGCGAAGACTGC-3 |
| HFlagb | 5-GGGTGCAAGATCATCGATAATGGACTACAAGGATGACGATGACAAGTCACCACAACGAGACCGGATAAATGC-3 |
| H C139S | 5-CGACTTCAGAGATCTCACTTGGTCTATCAACCCGCCAGAGAGAATC-3 |
| H C154S | 5-GAATCAAATTGGATTATGATCAATACTCTGCAGATGTGGCTGCTGAAGAG-3 |
For H stem 1 to 7, boldface type highlights the two stop codons; for HFlag, boldface type highlights the Flag tag epitope, for H C139S and H C154S, boldface type highlights cysteine-to-serine substitutions.
HFlag, full-length Flag-tagged H.
Western analysis and deglycosylation experiments.
For Western analysis of MV proteins, Vero cells were transfected with plasmids encoding F and either full-length or mutated H protein as indicated and incubated in 3.5-cm-diameter tissue culture wells at 37°C. Subsequent to cell lysis for 5 min at 4°C in lysis buffer (50 mM Tris, pH 8.0; 62.5 mM EDTA; 0.4% deoxycholate; 1% Igepal [Sigma]), protease inhibitors (Complete mix [Boehringer] and 1 mM phenylmethylsulfonyl fluoride [PMSF]) were added and the supernatant was clarified by centrifugation at 5,000 × g for 10 min at 4°C. The resulting postnuclear supernatant was mixed with an equal volume of urea buffer (200 mM Tris, pH 6.8; 8 M urea; 5% sodium dodecyl sulfate [SDS]; 0.1 mM EDTA; 0.03% bromphenol blue) containing 1.5% dithiothreitol (DTT) and incubated for 25 min at 50°C in a thermomixer. Samples were fractionated on SDS-polyacrylamide gels as indicated, blotted to polyvinyl difluoride membranes (Millipore) and subjected to enhanced chemiluminescence detection (Amersham Pharmacia Biotech).
For endoglycosidase H (endo H) treatment, samples were mixed subsequent to lysis with denaturing buffer (final concentration, 0.5% SDS, 1% β-mercaptoethanol) and incubated at 55°C for 25 min. Deglycosylation buffer (final concentration, 50 mM sodium citrate, pH 5.5) and 0.5 U of endo H were added prior to incubation at 37°C for 12 h. Reactions were stopped by addition of equal volumes urea buffer containing 1.5% DTT, samples were fractionated by polyacrylamide gel electrophoresis, blotted to polyvinylidene difluoride membranes, and analyzed by enhanced chemiluminescence detection as described above.
Syncytium formation.
Vero cells were cotransfected in duplicate with 1.5 μg of plasmid DNA encoding H stem constructs, MVEdm H (HEdm), or empty plasmids for control and 1.5 μg of plasmid DNA encoding FEdm. Cells were incubated at 32°C to prevent cells from reaching a 100% confluency prior to appearance of syncytia. The amount of syncytia in representative fields (approximately 20% of a 3.5-cm-diameter tissue culture well) was determined at the indicated times.
Metabolic labeling and immunoprecipitation.
Vero cells transfected as described in six-well tissue culture plates were incubated for 30 min in labeling medium lacking cysteine, methionine, and ammonium sulfate and then metabolically labeled by incubation in labeling medium containing [35S]methionine (Amersham Pharmacia Biotech) at a final concentration of 100 μCi/ml for 45 min at 37°C. Subsequently, labeling medium was replaced by chase medium containing 5% fetal calf serum and the cells were incubated at 37°C for various periods as indicated.
For direct immunoprecipitation cells were lysed in radioimmunoprecipitation assay buffer (10 mM Tris, pH 7.4; 1% deoxycholate; 1% Triton X-100; 0.1% SDS; 150 mM sodium chloride; protease inhibitors [Complete mix; Boehringer Mannheim]; 1 mM PMSF) for 15 min at 4°C, and lysates were subjected to centrifugation for 30 min at 20,000 × g. For coimmunoprecipitation experiments cells were lysed in coimmunoprecipitation buffer (50 mM HEPES, pH 7.3; 100 mM sodium chloride; 10 mM n-dodecyl β–d-maltoside; protease inhibitors [Complete mix; Boehringer Mannheim]; 1 mM PMSF).
Proteins were precipitated from cell lysates with agarose-conjugated anti-Flag antibodies (M2; Sigma) or with antibodies directed against various cytosolic or extracellular epitopes of MV F and H proteins as indicated. Lysates were incubated with antibodies for 1 to 2 h at 4°C in lysis buffer. If not coupled to agarose, immune complexes were absorbed subsequently to protein G-Sepharose beads for 1 h at 4°C. Precipitates were washed four times in lysis buffer prior to resuspension in urea buffer containing 1.5% DTT for 25 min at 50°C and fractionation on polyacrylamide gels as indicated. For nonreducing gel electrophoresis precipitates were incubated in urea buffer lacking DTT. Dried gels were exposed to highly sensitive films (Kodak Biomax) or quantified using a Storm imaging system (Molecular Dynamics).
Surface immunoprecipitation.
For selective immunoprecipitation of plasma membrane-localized MV glycoproteins, Vero cells were transfected in six-well tissue culture plates and metabolically labeled as described above. Subsequent to a 4-h chase period at 37°C, cells were detached from plates and washed three times in ice-cold phosphate-buffered saline containing 0.05% sodium azide. Intact cells were incubated with antibodies directed against extracellular epitopes of MV H for 1 h at 4°C, washed five times with ice-cold phosphate-buffered saline (pH 7.2), and lysed by incubation in coimmunoprecipitation buffer containing protease inhibitors (Complete mix [Boehringer Mannheim] and 1 mM PMSF) for 15 min at 4°C. Debris was removed by centrifugation at 20000 × g for 30 min at 4°C, and the supernatant was incubated with protein G-Sepharose beads for 1 h at 4°C. Surface precipitates were washed as described above, and the supernatant of the first washing cycle containing the intracellular fraction of the respective MV glycoprotein was subjected to a second precipitation using agarose-conjugated anti-Flag antibodies. All precipitates were finally resuspended in urea buffer containing 1.5% DTT for 25 min at 50°C and subjected to gel electrophoresis and autoradiography as described above.
Preparation of MV stocks.
Vero cells (80% confluent in 10-cm-diameter tissue culture dishes) were infected at a multiplicity of infection (MOI) of 0.1 PFU/cell with MVEdm and incubated at 37°C until approximately 90% of cells were found in syncytia. Cells were resuspended in 3 ml of low-serum medium (Opti-MEM; Gibco) and scraped into 15-ml centrifugation tubes, and particles were released by three repeated freeze-thaw cycles in liquid nitrogen and at 37°C. Stock titers were determined by 50% tissue culture infective dose titration, and stocks were stored at −70°C.
Purification and analysis of MV particles.
Vero cells in 10-cm-diameter tissue culture dishes were infected at a multiplicity of infection of 1.0 PFU/cell with MVEdm and incubated at 37°C. Four hours postinfection (p.i.) cells were transfected with 15 μg of plasmid DNA encoding full-length or truncated MV H carrying an amino-terminal Flag epitope. Seven hours p.i. labeling medium containing [35S]methionine at a final concentration of 75 μCi/ml was added, and cells were incubated at 37°C. Forty-five hours p.i. medium was harvested, cell debris was removed by low-speed centrifugation (20,000 × g, 20 min, 4°C), and virus particles were concentrated at the interphase of a two step 20% and 60% sucrose gradient in TNE buffer (10 mM Tris, pH 7.8; 100 mM sodium chloride; 1 mM EDTA) by centrifugation at 100,000 × g, 4°C for 90 min. The virus-containing fraction was diluted with TNE buffer to less than 30% sucrose, and particles were pelleted at 100,000 × g at 4°C for 90 min. The pellet was resuspended in radioimmunoprecipitation assay buffer and subjected to immunoprecipitation, gel fractionation, and autoradiography as described above.
RESULTS
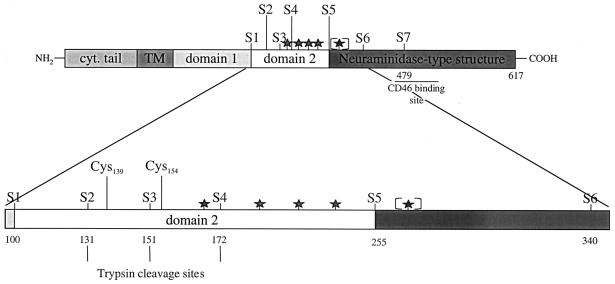
To identify a minimal region of the MV H ectodomain required for stable H intermolecular interaction, we generated progressive deletion mutants of the HEdm. Schematic diagrams of the full-length HEdm protein and the seven ectodomain mutants designated H stem 1 to 7, indicated as S1 to S7, are shown in Fig. 1. H stem 7 lies immediately N-terminal to the region of H identified as the CD46 binding site (11, 17, 22). H stems 2, 3, and 4 were designed on the basis of natural trypsin cleavage sites in H (24), and H stems 1, 5, and 6 were based on the structural predictions of Langedijk and colleagues (16), such that H stem 1 comprises the first proposed extracellular folding domain, H stem 5 encompasses both first and second such domains, and H stem 6 comprises all of the protein N-terminal to the proposed neuraminidase-like β-sheet region.
FIG. 1.
Schematic diagram of the predicted domain structure of the HEdm protein and the seven ectodomain H stem mutants (S1 to S7) generated in this study. Trypsin cleavage sites (vertical lines), N-linked glycosylation motifs (stars), and the postulated CD46 binding site (horizontal line) are indicated. The fifth potential glycosylation site (in brackets) is not recognized by the ER glycosylation machinery (12). cyt., cytoplasmic.
Expression, stability, and intracellular transport of H stem mutants.
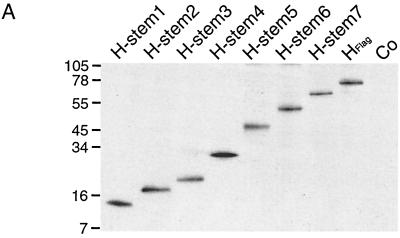
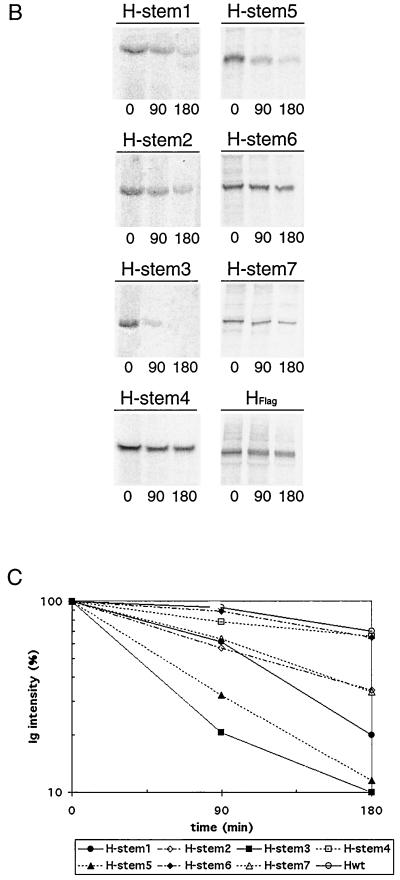
Vero cells were transiently transfected with all H stem constructs, and Western blot analysis confirmed their expression with the expected molecular weight (Fig. 2A). Stability of the seven H stem proteins was further characterized by pulse-chase analysis (Fig. 2B). H stems 4 and 6 showed a degree of stability similar to that reported for HEdm (Fig. 2C) (half-life [t1/2] ≈ 3 h). H stems 1, 2, and 7 displayed an intermediate stability (t1/2 = 106, 122, and 122 min respectively), while H stems 3 and 5 were the least stable (t1/2 = 40 and 55 min, respectively), suggesting their rapid degradation, most probably initiated by the ER quality control system (23).
FIG. 2.
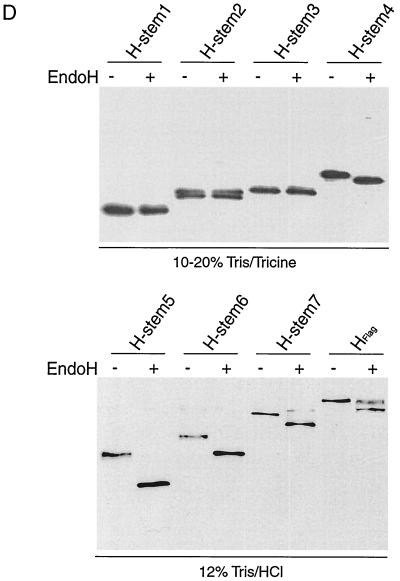
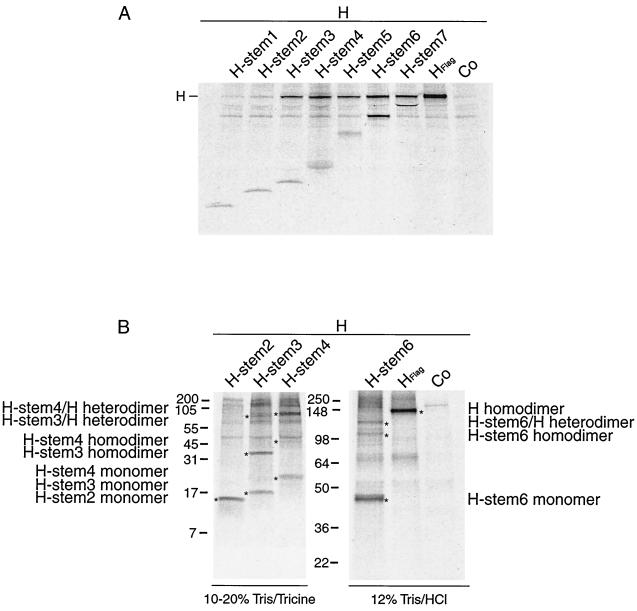
Truncated H stem proteins are expressed with the expected molecular weight but reveal different stability and no carbohydrate chain trimming. (A) Vero cells were transfected with Flag-tagged H stem constructs as indicated or were mock-transfected for control (Co). Western analysis of truncated MV HEdm proteins using anti-Flag antibodies was carried out. Polypeptides were separated on 10 to 20% Tris-Tricine gels. Numbers to the left of the gel correspond to the molecular weight (in thousands). (B) Two H stem proteins demonstrate full-length HEdm-like stability. Vero cells transfected with H stem constructs were radiolabeled with [35S]methionine for 45 min and then incubated in chase medium for the indicated times. MV H was immunoprecipitated from the cell lysates and separated on 10 to 20% Tris-Tricine (H stem 1, 2, 3, and 4) or on 12% Tris-HCl (H stem 5, 6, 7, and HFlag) gels. (C) Quantification of the radioactivity of the precipitated polypeptides was achieved using a Storm phosphoimager. (D) Western analysis of H stem constructs subsequent to incubation with (+) or without (−) endo H for 12 h. Samples were analyzed on 10 to 20% Tris-Tricine or on 12% Tris-HCl gels as indicated.
Endo H treatment (Fig. 2D) revealed proper glycosylation of all H stems carrying N-linked glycosylation sites, namely, H stems 4 to 7. Thus, truncation of H does not interfere with its recognition by the ER glycosylation machinery. Furthermore, complete sensitivity of all H stems to endo H treatment demonstrated a lack of Golgi-type carbohydrate chain trimming, suggesting that the H stem proteins were not exported from the ER. In contrast, HEdm displayed approximately 50% resistance to endo H, similar to that previously reported (3, 4), demonstrating that the HEdm protein was transported.
Amino acids 1 to 151 define a minimal region of MV H required for intermolecular interaction.
Despite their lack of transport to the cell surface, the ability of each H stem mutant to interact with MV HEdm could be studied since H dimerization occurs rapidly after integration of the nascent molecules into the ER membrane (4). Flag-tagged versions of each H stem mutant were coexpressed with nontagged HEdm in Vero cells. We have previously verified that the addition of Flag tag to the N terminus of HEdm does not affect its expression, transport, or activity (data not shown). Cells were labeled with [35S]methionine, and then subjected to coimmunoprecipitation using an anti-Flag antibody and analysis by SDS-polyacrylamide gel electrophoresis (Fig. 3A). Differences in the intensities of immunoprecipitated H stem bands reflect different numbers of methionine residues in the stem proteins, rather than differences in expression levels. While HEdm could barely be coimmunoprecipitated with H stem mutants 1 and 2, H stems 3 to 7 were all capable of efficient interaction with HEdm. Thus, a minimal region of MV H required for its stable interaction with other H monomers can be defined as H stem 3, or amino acids 1 to 151.
FIG. 3.
Identification of a minimal H stem unit required for dimerization with full-length HEdm. (A) H stems 3 to 7 are capable of interacting with HEdm. Vero cells doubly transfected with Flag-tagged H stem constructs and with full-length HEdm as indicated were radiolabeled and then incubated in chase medium for 45 min prior to lysis with coimmunoprecipitation buffer. Samples were fractionated on 10 to 20% Tris-Tricine gels. Full-length HEdm was coimmunoprecipitated with H stems using anti-Flag antibodies. Co, mock-transfected control. (B) H stem 3, 4, and 6 proteins are capable of efficient oligomerization. Vero cells were double transfected with Flag-tagged H stem constructs and full-length HEdm as indicated, radiolabeled, and chased for 1 h at 37°C. Complexes were coimmunoprecipitated using anti Flag antibodies and fractionated on 10 to 20% Tris-Tricine or 12% Tris-HCl gels under nonreducing conditions. Labeled (∗) bands correspond to the indicated complexes.
We assessed whether the interactions observed between H stem proteins and HEdm were based on dimerization. H stems 2, 3, 4, and 6 were chosen for this analysis, since 2 does not interact with HEdm, 3 is the minimal unit for interaction, and 4 and 6 are the most stably expressed H stems. Flag-tagged H stems 2, 3, 4, and 6 were coexpressed in Vero cells with nontagged HEdm. Subsequent to labeling with [35S]methionine and coimmunoprecipitation with anti-Flag antibody, proteins were resolved under nonreducing conditions. For H stem 2, no higher-order complexes were observed, whereas for H stems 3, 4, and 6, both homodimers and heterodimers formed with HEdm were observed (Fig. 3B). The identity of the complexes was inferred from their estimated molecular weight and by comparison to the same samples resolved under reducing conditions (data not shown). Thus, dimerization accounts for the interaction between H stems and HEdm.
Influence of MV proteins on transport and incorporation into viral particles of H stem mutants.
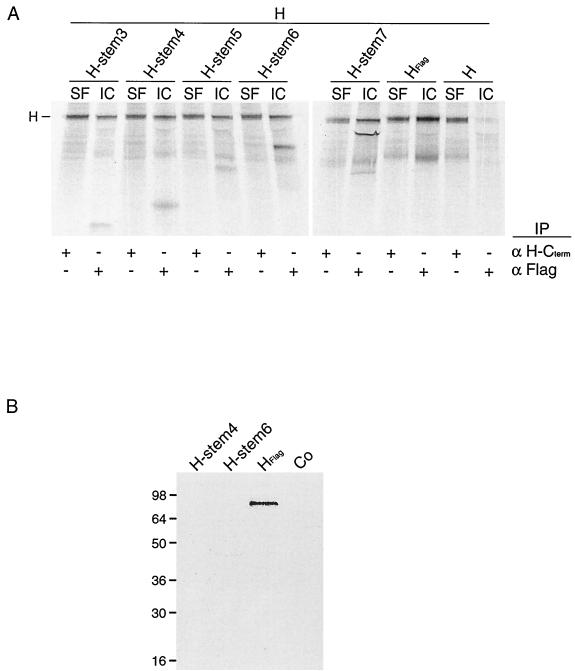
Considering the ability of H stems 3 to 7 to interact with HEdm, we investigated whether the presence of full-length, transport-competent HEdm could partially restore transport of these H stem proteins to the cell surface. Therefore, Flag-tagged H stems 3 to 7 were coexpressed with nontagged HEdm in Vero cells, and subsequent to [35S]methionine labeling, a surface coimmunoprecipitation was carried out. Whole cells were first incubated with an antibody directed against the extracellular C terminus of H and then were washed, lysed, and coimmunoprecipitated to detect surface HEdm and interacting H stems before a second coimmunoprecipitation step using anti-Flag antibody was performed to detect intracellular H stems and bound HEdm (Fig. 4A). None of the H stem mutants could be found at the cell surface in a complex with HEdm protein, but in contrast, intracellular nontagged HEdm could be coprecipitated with each of the H stem mutants 3 to 7, confirming our previous findings. Notably, none of the H stems seems to act as a strong dominant-negative inhibitor of transport of HEdm to the cell surface. This suggests a reversible interaction of the H stems with HEdm in the early secretory system.
FIG. 4.
Presence of full-length HEdm or other viral proteins does not restore transport of H stem constructs to the cell surface. (A) Vero cells were double transfected with Flag-tagged H stem constructs and HEdm, radiolabeled, chased for 4 h at 37°C, and incubated with antibodies directed against an epitope located at the extracellular carboxy-terminus of HEdm. Subsequent to lysis, immune complexes representing cell surface (SF)-localized MV glycoproteins were precipitated, and supernatants were subjected to a second precipitation using anti-Flag antibodies to isolate intracellular (IC) antigenic material. All samples were fractionated on 10 to 20% Tris-Tricine gels. (B) H stem proteins are not incorporated into viral particles. Vero cells were infected with MVEdm, transfected with Flag-tagged H stem constructs 4 or 6 or full-length Flag-tagged H 4 h p.i., radiolabeled 7 hours p.i., and incubated at 37°C. Viral particles were purified 45 h p.i., subjected to immunoprecipitation using anti-Flag antibodies, and analyzed on 12% Tris-HCl gels. Co, mock-transfected control.
While coexpression of HEdm was not sufficient to restore transport of the H stem proteins to the plasma membrane, it was conceivable that by interaction with other viral components at least the most stable H stem proteins, 4 and 6, may be transported to the cell surface and incorporated into viral particles. Therefore, Flag-tagged H stems 4 and 6 and Flag-tagged HEdm were transiently expressed in Vero cells which were labeled with [35S]methionine and infected with MVEdm. Viral particles were purified from the extracellular medium, lysed, and subjected to immunoprecipitation using anti-Flag antibodies. While transiently expressed Flag-tagged HEdm could be detected, indicating its uptake into viral particles, neither H stem 4 nor 6 was found in virions (Fig. 4B).
Cysteines 139 and 154 are responsible for covalent interaction of MV H monomers.
The region between H stems 2 and 3 which defines the minimal region for interaction between H monomers includes one cysteine residue at position 139. We proposed that this cysteine and the cysteine at position 154, which lies in the region between stems 3 and 4, may be responsible for intermolecular disulfide bonding between H monomers.
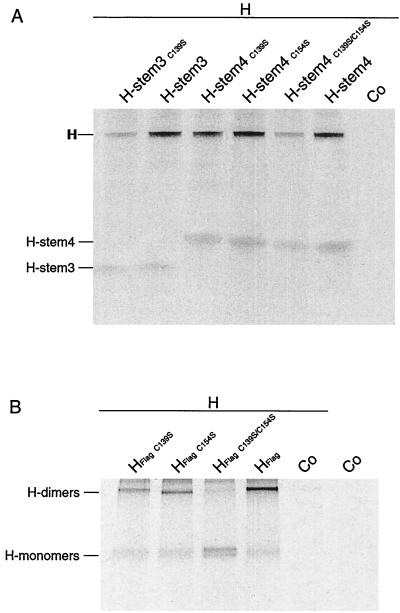
To address this question, we mutated cysteines 139 and 154 in Flag-tagged H stems 3 and 4 singly and in combination. Cysteines were replaced by serine, since this substitution results in minimal secondary effects on protein structure: the mutation consisting of a sulfur-to-oxygen substitution (10, 29). Mutant constructs H stem 3 C139S, H stem 4 C139S, H stem 4 C154S, and H stem 4 C139S-C154S were transfected in Vero cells, and Western blot analysis confirmed all proteins were expressed at the expected molecular weight (data not shown). By coimmunoprecipitation, both H stem 3 C139S and H stem 4 C139S-C154S demonstrated drastically reduced abilities to interact with HEdm compared with the parental H stem 3 and H stem 4 proteins (Fig. 5A). The singly mutated H stem 4 C139S and H stem 4 C154S, however, showed no reduction in their interaction with HEdm, suggesting that both cysteines contribute equally to MV H intermolecular interaction and that lack of one can be compensated for by presence of the other.
FIG. 5.
Cysteine residues 139 and 154 are equally sufficient for hetero-oligomerization of H-stem constructs and of HEdm. Vero cells doubly transfected with Flag-tagged H stem constructs and with full-length HEdm as indicated were radiolabeled and then incubated in chase medium for 45 min prior to lysis with coimmunoprecipitation buffer. (A) Coimmunoprecipitation of full-length HEdm with H stem constructs carrying cysteine to serine exchanges as indicated using anti Flag antibodies. (B) Exchange of both cysteine residues 139 and 154 prevents oligomerization of full-length HEdm. Immunoprecipitation of Flag-tagged full-length HEdm carrying exchanges as indicated using anti-Flag antibodies was carried out. Samples were analyzed on a 10% Tris-HCl gel under nonreducing conditions. Co, mock-transfected control.
To evaluate the relevance of these data obtained with the truncated H stem proteins for oligomerization of full-length H protein, we generated Flag-tagged HEdm carrying the C139S and C154S exchanges singly and in combination. The ability of the mutated full-length H to oligomerize was analyzed by immunoprecipitation and fractionation under nonreducing conditions. Both singly mutated HEdm C139S and HEdm C154S were shown to dimerize (Fig. 5B), consistent with our previous observations. However, the doubly mutated HEdm C139S-C154S protein was incapable of dimerization, confirming that the minimal requirement for H oligomerization identified using the truncated H stem constructs is relevant in the context of full-length protein.
Covalent interaction between HEdm monomers is required for efficient transport but not for functionality.
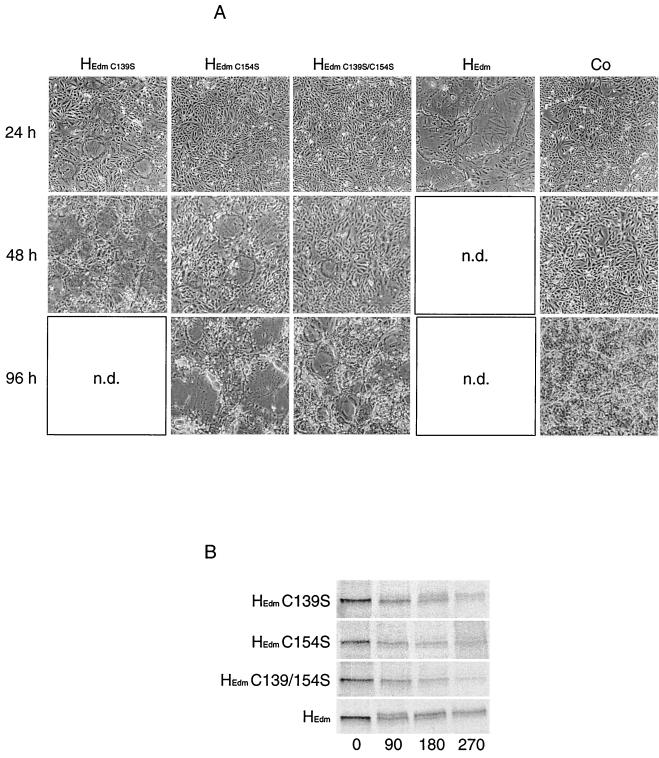
To assess the consequences for biological activity of these cysteine mutations, the mutant HEdm proteins were coexpressed in Vero cells with MV F protein, and the presence of syncytia was scored every 24 h for 8 days. Strikingly, even the HEdm C139S-C154S double mutant, although incapable of covalent interaction, mediated cell-to-cell fusion (Fig. 6A). Consistent with previous reports (4), coexpression of HEdm with FEdm in Vero cells resulted in the formation of large syncytia, comprising 90% of all cell nuclei 24 h posttransfection. After cotransfection with FEdm, 90% of cells were found in syncytia 2 days posttransfection for the singly mutated HEdm C139S, 7 days for the HEdm C154S mutant, and 8 days for the doubly mutated HEdm C139S-C154S. These data suggest that the C154S mutation has a greater limiting effect on H functionality than the C139S exchange.
FIG. 6.
HEdm mutants carrying single or combined cysteine-to-serine exchanges at positions 139 and 154 are biologically active. Vero cells were double transfected with HEdm species as indicated and FEdm. (A) Syncytium formation was analyzed by light microscopy after incubation for 24, 48, and 96 h at 32°C. n.d., not done. (B) Pulse-chase analysis reveals a reduced ER-to-Golgi transport rate of HEdm constructs carrying cysteine-to-serine exchanges. Cells were radiolabeled with [35S]methionine for 45 min and then incubated in chase medium for indicated times. MV H species were immunoprecipitated from the cell lysates and separated on 7.5% Tris-HCl gels.
To investigate whether the basis for this phenotype lies in reduced transport of the mutated molecules to the cell surface, we performed a pulse-chase analysis to determine the kinetics of carbohydrate chain trimming, indicating ER-to-Golgi transport (Fig. 6B). Following the fate of HEdm, 50% conversion to Golgi-type glycosylation was detectable after approximately 100 min of chase time, whereas HEdm C139S reached 50% Golgi-type glycosylation only after 200 min. Even after 270 min of chase time, a barely detectable fraction of HEdm C154S and HEdm C139S-C154S was converted. These data indicate that the reduced biological activity of HEdm carrying the cysteine mutations is attributable rather to a reduced rate of transport than to a lack of complex formation.
DISCUSSION
Using a series of progressive ectodomain truncation mutants, we have characterized a minimal region of the HEdm protein which is sufficient for its intermolecular interaction and oligomerization. This unit encompasses 151 amino acids, including the cytoplasmic tail, transmembrane domain, and the membrane-proximal 93 amino acids of the ectodomain. We have further delineated the requirements for interaction, demonstrating that cysteine 139 is critical in mediating a stable intermolecular interaction of this minimal unit with full-length H protein. In the absence of cysteine 139, the inability of this minimal unit to efficiently dimerize can be overcome by extending the unit to include a cysteine at position 154. Significantly, we have confirmed that both cysteines are critical in mediating efficient dimerization not only of the minimal interacting unit, but also the full-length MV H protein.
One concern of our approach must be whether the H stems are conformationally acceptable and whether their lack of cell surface transport may be due to a global loss of conformation. The ability of H stems 3, 4, and 6 to form dimers with themselves, and more significantly with full-length HEdm, suggests that their lack of transport is not due to their recognition as unassembled and therefore improperly folded protein complex subunits by the ER quality control system (23), since they are clearly recognizable as part of H. It may be conceivable that the truncated H stems expose hydrophobic regions which would normally be buried in the full-length molecule and that recognition of these domains by molecular chaperones results in ER retention (7). Furthermore, the conclusions we have reached concerning disulfide bond formation using the H stem proteins have proven transferable to full-length HEdm. This validates them as relevant in the context of full-length MV H protein.
Although our data define a minimum requirement for stable H intermolecular interaction, we expect that other regions of H also contribute to the interaction. Firstly, our findings suggest that the transmembrane region of MV H protein may contribute to its oligomerization. The H stem 3 C139S mutant and the H stem 4 C139S-C154S double mutant display some residual ability to form both homotypic dimers and heterotypic dimers with HEdm. This strongly indicates that even in the absence of cysteine residues, the minimal unit is capable of mediating some intermolecular interaction. A previous report demonstrating the ability of H deleted in part of its cytoplasmic tail to oligomerize and mediate fusion help (3) suggests that the residual binding capacity does not reside in the N-terminal half of the H tail but rather in the transmembrane and membrane-proximal regions.
Additionally, our data suggest that regions of H C-terminal to those studied here may also contribute to H oligomerization, since the efficiency with which H stems 3, 4, and 6 form either homotypic or heterotypic dimers is less than that observed for HEdm homodimer formation. A mutational analysis in which each of the 13 cysteine residues in the MV H protein was individually replaced with serine was interpreted as suggesting that cysteines 287, 300, 381, 394, 494, 579, and 583 may be important for dimerization (14). However, the H proteins mutated in these cysteine residues lost all ability to bind conformation-dependent antibodies, indicating that a loss in overall conformational structure, rather than loss of a specific intermolecular interaction, may account for their inability to dimerize. This work (14) also reported that mutating either cysteine 139 or cysteine 154 had no adverse effect on H dimerization, leading the authors to propose that neither residue is involved in H intermolecular interaction. However, our data show that these two cysteines are sufficient for H oligomerization in a reciprocal manner, such that the lack of one can be compensated for by the presence of the other.
Although it is incapable of covalent intermolecular interaction, we show that full-length MV H protein carrying mutated cysteines 139 and 154 maintains some functionality. Thus, noncovalent interactions between MV H monomers are sufficient to confer some biological activity on the molecules. Interestingly, certain strains of the related paramyxovirus Newcastle disease virus (NDV) have been isolated which lack covalent dimerization of the HN protein (25). In other NDV strains, however, a cysteine residue at position 123 of the HN protein has been identified as mediating intermolecular disulfide bond formation (19). Thus, intermolecular disulfide bonding is not essential for the functionality of NDV HN protein or for virus viability. Given the reduction in kinetics of syncytium formation of our mutated MV H protein, we suggest that in contrast to NDV HN, covalent intermolecular interactions are required for efficient MV H protein function. The sequence of the first two proposed extracellular folding domains of MV H cannot be readily aligned with that of the HN of other paramyxoviruses (16); thus, making direct comparisons of the structure of these proteins difficult. However, that MV has conserved through evolution two closely related cysteine residues which are both required for its covalent dimerization, whereas only some NDV HN proteins covalently dimerize, further underlines the critical role of these residues in the virus life cycle.
Our data demonstrate that covalent dimerization of MV H is required for efficient transport to the cell surface rather than for functionality per se, since ER-to-Golgi transport of the mutant H protein lacking the critical cysteine residues 139 and 154 is delayed and since complete fusion can be reached after an increased incubation time. Considering the t1/2 of MV H at the plasma membrane of approximately 10 h (8), the steady-state levels of our mutant H proteins at the cell surface must be minimal. We therefore postulate that only a few MV glycoprotein complexes rather than a large network are required to be present at the plasma membrane in order to support syncytium formation.
ACKNOWLEDGMENTS
We thank Stephen J. Russell for critical reading of the manuscript.
This work was supported by a grant from the Siebens Foundation to R.C. and a career development award from the Deutsche Forschungsgemeinschaft to R.K.P. (grant PL293/1-1).
REFERENCES
- 1.Alkhatib G, Briedis D J. The predicted primary structure of the measles virus hemagglutinin. Virology. 1986;150:479–490. doi: 10.1016/0042-6822(86)90312-0. [DOI] [PubMed] [Google Scholar]
- 2.Cathomen T, Buchholz C J, Spielhofer P, Cattaneo R. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology. 1995;214:628–632. doi: 10.1006/viro.1995.0075. [DOI] [PubMed] [Google Scholar]
- 3.Cathomen T, Naim H Y, Cattaneo R. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J Virol. 1998;72:1224–1234. doi: 10.1128/jvi.72.2.1224-1234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattaneo R, Rose J K. Cell fusion by the envelope glycoproteins of persistent measles viruses which caused lethal human brain disease. J Virol. 1993;67:1493–1502. doi: 10.1128/jvi.67.3.1493-1502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng R, Wang Z, Mirza A M, Iorio R M. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology. 1995;209:457–469. doi: 10.1006/viro.1995.1278. [DOI] [PubMed] [Google Scholar]
- 6.Dorig R E, Marcil A, Chopra A, Richardson C D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 7.Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 8.Fujinami R S, Sissons J G, Oldstone M B. Immune reactive measles virus polypeptides on the cell's surface: turnover and relationship of the glycoproteins to each other and to HLA determinants. J Immunol. 1981;127:935–940. [PubMed] [Google Scholar]
- 9.Gerald C, Buckland R, Barker R, Freeman G, Wild T F. Measles virus haemagglutinin gene: cloning, complete nucleotide sequence analysis and expression in COS cells. J Gen Virol. 1986;67:2695–2703. doi: 10.1099/0022-1317-67-12-2695. [DOI] [PubMed] [Google Scholar]
- 10.Giese N A, Robbins K C, Aaronson S A. The role of individual cysteine residues in the structure and function of the v-sis gene product. Science. 1987;236:1315–1318. doi: 10.1126/science.3035718. [DOI] [PubMed] [Google Scholar]
- 11.Hsu E C, Sarangi F, Iorio C, Sidhu M S, Udem S A, Dillehay D L, Xu W, Rota P A, Bellini W J, Richardson C D. A single amino acid change in the hemagglutinin protein of measles virus determines its ability to bind CD46 and reveals another receptor on marmoset B cells. J Virol. 1998;72:2905–2916. doi: 10.1128/jvi.72.4.2905-2916.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu A, Cattaneo R, Schwartz S, Norrby E. Role of N-linked oligosaccharide chains in the processing and antigenicity of measles virus haemagglutinin protein. J Gen Virol. 1994;75:1043–1052. doi: 10.1099/0022-1317-75-5-1043. [DOI] [PubMed] [Google Scholar]
- 13.Hu A, Kovamees J, Norrby E. Intracellular processing and antigenic maturation of measles virus hemagglutinin protein. Arch Virol. 1994;136:239–253. doi: 10.1007/BF01321055. [DOI] [PubMed] [Google Scholar]
- 14.Hu A, Norrby E. Role of individual cysteine residues in the processing and antigenicity of the measles virus haemagglutinin protein. J Gen Virol. 1994;75:2173–2181. doi: 10.1099/0022-1317-75-9-2173. [DOI] [PubMed] [Google Scholar]
- 15.Lamb R A. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 16.Langedijk J P, Daus F J, van Oirschot J T. Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J Virol. 1997;71:6155–6167. doi: 10.1128/jvi.71.8.6155-6167.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecouturier V, Fayolle J, Caballero M, Carabana J, Celma M L, Fernandez-Munoz R, Wild T F, Buckland R. Identification of two amino acids in the hemagglutinin glycoprotein of measles virus (MV) that govern hemadsorption, HeLa cell fusion, and CD46 downregulation: phenotypic markers that differentiate vaccine and wild-type MV strains. J Virol. 1996;70:4200–4204. doi: 10.1128/jvi.70.7.4200-4204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malvoisin E, Wild T F. Measles virus glycoproteins: studies on the structure and interaction of the haemagglutinin and fusion proteins. J Gen Virol. 1993;74:2365–2372. doi: 10.1099/0022-1317-74-11-2365. [DOI] [PubMed] [Google Scholar]
- 19.McGinnes L W, Morrison T G. The role of the individual cysteine residues in the formation of the mature, antigenic HN protein of Newcastle disease virus. Virology. 1994;200:470–483. doi: 10.1006/viro.1994.1210. [DOI] [PubMed] [Google Scholar]
- 20.Naniche D, Varior-Krishnan G, Cervoni F, Wild T F, Rossi B, Rabourdin-Combe C, Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nussbaum O, Broder C C, Moss B, Stern L B, Rozenblatt S, Berger E A. Functional and structural interactions between measles virus hemagglutinin and CD46. J Virol. 1995;69:3341–3349. doi: 10.1128/jvi.69.6.3341-3349.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson J B, Scheiflinger F, Manchester M, Yilma T, Oldstone M B. Structural and functional studies of the measles virus hemagglutinin: identification of a novel site required for CD46 interaction. Virology. 1999;256:142–151. doi: 10.1006/viro.1999.9644. [DOI] [PubMed] [Google Scholar]
- 23.Plemper R K, Wolf D H. Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem Sci. 1999;24:266–270. doi: 10.1016/s0968-0004(99)01420-6. [DOI] [PubMed] [Google Scholar]
- 24.Sato T A, Enami M, Kohama T. Isolation of the measles virus hemagglutinin protein in a soluble form by protease digestion. J Virol. 1995;69:513–516. doi: 10.1128/jvi.69.1.513-516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheehan J P, Iorio R M, Syddall R J, Glickman R L, Bratt M A. Reducing agent-sensitive dimerization of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus correlates with the presence of cysteine at residue 123. Virology. 1987;161:603–606. doi: 10.1016/0042-6822(87)90158-9. [DOI] [PubMed] [Google Scholar]
- 26.Tanabayashi K, Compans R W. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J Virol. 1996;70:6112–6118. doi: 10.1128/jvi.70.9.6112-6118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsurudome M, Ito M, Nishio M, Kawano M, Okamoto K, Kusagawa S, Komada H, Ito Y. Identification of regions on the fusion protein of human parainfluenza virus type 2 which are required for haemagglutinin-neuraminidase proteins to promote cell fusion. J Gen Virol. 1998;79:279–289. doi: 10.1099/0022-1317-79-2-279. [DOI] [PubMed] [Google Scholar]
- 28.Tsurudome M, Kawano M, Yuasa T, Tabata N, Nishio M, Komada H, Ito Y. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology. 1995;213:190–203. doi: 10.1006/viro.1995.1559. [DOI] [PubMed] [Google Scholar]
- 29.Wang A, Lu S D, Mark D F. Site-specific mutagenesis of the human interleukin-2 gene: structure-function analysis of the cysteine residues. Science. 1984;224:1431–1433. doi: 10.1126/science.6427925. [DOI] [PubMed] [Google Scholar]
- 30.Wild T F, Fayolle J, Beauverger P, Buckland R. Measles virus fusion: role of the cysteine-rich region of the fusion glycoprotein. J Virol. 1994;68:7546–7548. doi: 10.1128/jvi.68.11.7546-7548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wild T F, Malvoisin E, Buckland R. Measles virus: both the haemagglutinin and fusion glycoproteins are required for fusion. J Gen Virol. 1991;72:439–442. doi: 10.1099/0022-1317-72-2-439. [DOI] [PubMed] [Google Scholar]