Abstract
The increasing prevalence of human immunodeficiency virus type 1 (HIV-1) subtype C infection worldwide calls for efforts to develop a relevant animal model for evaluating strategies against the transmission of the virus. A chimeric simian/human immunodeficiency virus (SHIV), SHIVCHN19, was generated with a primary, non-syncytium-inducing HIV-1 subtype C envelope from a Chinese strain in the background of SHIV33. Unlike R5-tropic SHIV162, SHIVCHN19 was not found to replicate in rhesus CD4+ T lymphocytes. SHIVCHN19 does, however, replicate in CD4+ T lymphocytes of pig-tailed macaques (Macaca nemestrina). The observed replication competence of SHIVCHN19 requires the full tat/rev genes and partial gp41 region derived from SHIV33. To evaluate in vivo infectivity, SHIVCHN19 was intravenously inoculated, at first, into two pig-tailed and two rhesus macaques. Although all four animals became infected, the virus replicated preferentially in pig-tailed macaques with an earlier plasma viral peak and a faster seroconversion. To determine whether in vivo adaptation would enhance the infectivity of SHIVCHN19, passages were carried out serially in three groups of two pig-tailed macaques each, via intravenous blood-bone marrow transfusion. The passages greatly enhanced the infectivity of the virus as shown by the increasingly elevated viral loads during acute infection in animals with each passage. Moreover, the doubling time of plasma virus during acute infection became much shorter in passage 4 (P4) animals (0.2 day) in comparison to P1 animals (1 to 2 days). P2 to P4 animals all became seropositive around 2 to 3 weeks postinoculation and had a decline in CD4/CD8 T-cell ratio during the early phase of infection. In P4 animals, a profound depletion of CD4 T cells in the lamina propria of the jejunum was observed. Persistent plasma viremia has been found in most of the infected animals with sustained viral loads ranging from 103 to 105 per ml up to 6 months postinfection. Serial passages did not change the viral phenotype as confirmed by the persistence of the R5 tropism of SHIVCHN19 isolated from P4 animals. In addition, the infectivity of SHIVCHN19 in rhesus peripheral blood mononuclear cells was also increased after in vivo passages. Our data indicate that SHIVCHN19 has adapted well to grow in macaque cells. This established R5-tropic SHIVCHN19/macaque model would be very useful for HIV-1 subtype C vaccine and pathogenesis studies.
Subtype C viruses have become the most prevalent human immunodeficiency virus type 1 (HIV-1) genotype globally (49). UNAIDS has estimated that there are now eight million subtype C infections worldwide, mainly in sub-Saharan Africa and Asia. In these respective geographic areas, subtype C is more common than any other subtype, and it now accounts for about 40% of all new HIV-1 infections in the world. In one recent study in two cities in southern China, 22 of 23 infected patients were found to carry subtype C viruses (Z. Chen, Y. Cao, L. Zhang, and D. Ho, unpublished data). Despite mounting efforts, it remains unclear why this subtype has gained dominance so quickly and whether means can be developed to slow down its spread. To address these questions effectively, a relevant animal model to study HIV-1 subtype C would be very useful.
One of the current animal models for AIDS research consists of Asian macaques experimentally infected with simian immunodeficiency virus (SIV) (13, 14). Indeed, several molecular clones of SIV are pathogenic in vivo, causing a fatal AIDS-like disease in macaques (25, 26). For this reason, the model has been widely used to evaluate various vaccine strategies and to study AIDS pathogenesis (4, 12, 14, 19, 29, 36, 39). Nevertheless, because the env genes of SIV and HIV-1 show significant sequence diversity (28), the SIV/macaque model is of limited utility for in vivo analyses of the phenotypic and immunological properties of HIV-1 envelope. Some groups have attempted to adapt HIV-1 in macaques (2, 3, 6, 16). These efforts, however, were largely unsuccessful. The value of the macaque model has increased since the development of a chimeric simian/human immunodeficiency virus (SHIV) (31, 34, 44).
Traditionally, SHIV is a chimeric lentivirus that uses pathogenic SIVmac239 as a genetic background, except that its tat, rev, and env genes are replaced by the corresponding regions of HIV-1 (23, 32, 34, 44). Since SHIV retains the ability to infect macaques, it provides a unique in vivo model for studying the pathogenic properties of HIV-1 envelope and for examining the efficacy of HIV-1 vaccines based on envelope glycoproteins. Several SHIV strains have been constructed, and their pathogenicity in nonhuman primates has been evaluated. Most current SHIV constructs utilize envelope genes derived from HIV-1 subtype B strains, either from lab-adapted, syncytium-inducing (SI), T-tropic viruses (HIV-1HXB2 and HIV-1NL43) or from primary, non-syncytium-inducing (NSI), M-tropic (HIV-1162), SI T-tropic (HIV-133), and dual-tropic (HIV89.6 and HIV-1DH12) isolates (23, 32, 34, 44). Since these chimeras retain biological properties of corresponding parental HIV-1 env, they have been used to reveal envelope-determined differences in the replication capacity of the SHIVs in vivo and in the induction of various virus-specific immune responses. These SHIV/macaque models have allowed researchers to explore the significance of HIV-1 env variation, as well as to evaluate vaccines based on HIV-1 Env antigens. In addition to SHIVs based on subtype B, one has been successfully developed for subtype E (27). However, there has been no SHIV for subtype C.
In this study, approaches similar to those used for constructing subtype B and E SHIVs were adopted to make a subtype C envelope-based SHIV. We focused on primary, NSI HIV-1 subtype C viruses, as they have been demonstrated to use CCR5 for entry (52). This subtype was selected because of its emerging dominance in the epidemic, and the particular NSI, R5-tropic phenotype was selected because it represents the dominant type of HIV-1 strains transmitted sexually (54, 55). Moreover, it has been demonstrated recently that R5-tropic viruses cause distinct pathogenic effects in comparison to X4-tropic ones (20, 50). Here, we report that a replication-competent SHIVCHN19 was generated by using HIV-1 subtype C envelope in the background of SHIV33. SHIVCHN19 was found to be different from SHIV162 in that the new virus did not infect rhesus peripheral blood mononuclear cells (PBMC) despite CD8+ T-cell depletion. The virus was, however, replication competent in CD4+ T lymphocytes of pig-tailed macaques. To test its in vivo growth capacity, SHIVCHN19 was inoculated into two pig-tailed and two rhesus macaques. We found that SHIVCHN19 replicated preferentially in pig-tailed macaques. To determine whether in vivo adaptation would enhance the infectivity of SHIVCHN19, serial passages were carried out in three groups of two pig-tailed macaques each, via intravenous blood-bone marrow transfusion. In comparison to two passage 1 (P1) pig-tailed macaques, the passages were successful as shown by (i) the increasingly elevated levels of plasma viremia in animals from later passages, (ii) the shortened doubling time of plasma virus during acute infection with each passage, (iii) faster seroconversion in P2 to P4 animals, (iv) higher levels of sustained viral load in animals from later passages, (v) the enhanced viral infectivity in rhesus PBMC, and (vi) profound CD4+ T-cell depletion in the jejunal lamina propria of P4 animals. Importantly, the serial passages did not change the viral phenotype as determined by the persistence of the R5 tropism of SHIVCHN19 isolated from the two P4 animals. Our data indicate the establishment of the first R5-tropic SHIV/macaque model for HIV-1 subtype C vaccine and pathogenesis studies.
MATERIALS AND METHODS
Amplification of HIV-1 tat/rev/env using a nested PCR method.
The specimens for this experiment were PBMC derived from individuals naturally infected with HIV-1 in Yunnan, China. These primary strains were subsequently classified as subtype C viruses based on sequencing analysis of env gp120 and the p17 region of gag (Z. Chen, Y. Cao, L. Zhang, and D. Ho, unpublished data). High-molecular-weight genomic DNA from uncultured PBMC was extracted by using a DNA/RNA extraction kit (United States Biochemical Corp., Cleveland, Ohio). HIV-1 tat/rev/vpu/env fragments were amplified from genomic DNA by using EXPAND high-fidelity DNA polymerase according to the manufacturer's specifications (Boehringer Mannheim, Indianapolis, Ind.) in a 100-μl total volume containing 0.5 to 1 μg of DNA; 10 mM Tris-HCl (pH 8.5); 50 mM KCl; 1.5 mM MgCl2; 0.1% Triton X-100; 200 μM each dATP, dGTP, dCTP, and dTTP; 20 pmol of each primer; and 2.5 U of EXPAND high-fidelity DNA polymerase. The first-round PCR consisted of an initial cycle of 95°C for 2 min, followed by 30 cycles of 95°C for 20 s, 45°C for 45 s, and 72°C for 5 min, using outer primer pair YX1 (5′-CAG AAT TGG GTG CCA GCA TAG C-3′) and YX2 (5′-AAT TAA CCC TTC CAG TCC CCC C-3′). The second-round PCR, using inner primer pair VprEnd (5′-TGCC GAATTC GC ATG CTA TAG A TAG AGA AGA GCA AG-3′) and EnvEnd (5′-TGTT CTCGAG TT TAT TGC AAA GCT GCT TCA AAG-3′), consisted of another 30 cycles of 94°C for 20 s, 56°C for 45 s, and 72°C for 3.5 min. In the primer sequences, the introduced stop codons are underlined and the restriction enzyme sites are italicized. For both rounds of PCR, amplification was completed by incubation of the PCR mixture at 72°C for 9 min. Products from the second-round amplifications were run on a 1% agarose gel, and positive bands were visualized by staining with ethidium bromide.
Genetic analysis and phenotypic characterization of the amplified HIV-1 genes.
Full-length tat/rev/vpu/env amplicons were cloned into the expression vector pcDNA-I/Amp (Invitrogen, Carlsbad, Calif.). The fragments were subsequently sequenced by using an automated DNA sequencer (ABI Prism 377). To rule out possible intersubtype recombination, each full-length env gene was subjected to phylogenetic analysis by a neighbor-joining method and a recombination identification program (HIV sequence database). Ultimately, nonrecombinant amplicons with full-length HIV-1 subtype C envelope were selected for phenotypic characterization.
To test the expression and phenotypic characteristics of env genes, 293T cells were transiently transfected with pcDNA-I/Amp-based vectors encoding different HIV-1 Envs. After 24 h, the transfected cells (2 × 104) were mixed in 24-well dishes preseeded with an equal number of GHOS.CD4 cells expressing either CCR5 or CXCR4 or other coreceptors, including CCR1, CCR2b, CCR3, CCR4, BOB, and Bonzo, respectively (10). After 12 h of cocultivation, the cultures were photographed by phase-contrast microscopy to detect syncytium formation. Functional env genes that were able to mediate the maximum R5-CD4-cell fusion were subsequently chosen for SHIV construction.
Construction and characterization of SHIV subtype C strains.
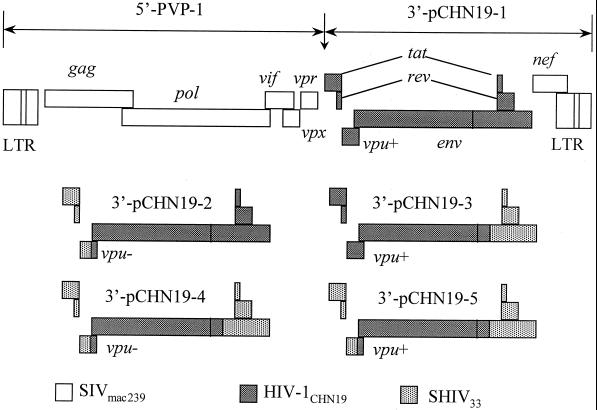
The strategy for the construction of subtype C SHIV was similar to those utilized in generating subtype B SHIVs (34). Since the 5′-SIVmac239 genome in plasmid PVP-1 was not changed, the reconstruction was concentrated on the 3′-SHIV33 genome. The genetic organization of each 3′-SHIV construct is depicted in Fig. 1. As 3′-pCHN19-1 did not result in replication-competent virus, the 5′-tat/rev and partial vpu were replaced with the corresponding regions of 3′-SHIV33 by an overlapping PCR without disturbing the subtype C env gene from initiation codon ATG. The 3′-tat/rev of 3′-SHIVCHN19 was replaced by the counterparts of 3′-SHIV33 by using restriction enzyme AvrII, which cuts at a site in the gp41 region shared by both SHIV33 and HIV-1CHN19. Each construct was analyzed by restriction enzyme cutting and was further confirmed by DNA sequencing. In order to enhance the viral replication in vivo, the mutant vpu gene of SHIV33 was opened by a PCR mutagenesis method to ensure vpu expression.
FIG. 1.
Schematic representation of subtype C SHIV construction. The viral genome is constructed in two plasmids, 5′-PVP-1 and 3′-pCHN19. Since the structure of plasmid PVP-1 was unchanged, the reconstruction was limited to the 3′-SHIV33 genome. The 5′-tat/rev and partial vpu were replaced with the corresponding regions of 3′-SHIV33 by an overlapping PCR without disrupting the subtype C env gene. The 3′-tat/rev of 3′-pCHN19 was replaced by the counterpart of 3′-SHIV33 by using restriction enzyme AvrII, which cuts at a site in the gp41 region shared by SHIV33 and HIV-1CHN19. In order to enhance the viral replication in vivo, the mutant vpu gene of SHIV33 was opened by a PCR mutagenesis method to ensure vpu expression (vpu+).
To generate replication-competent SHIV strains, linear 5′-PVP-1 and 3′-chimeric constructs were cotransfected into 293T cells. After 48 h, 5 × 106 phytohemagglutinin (PHA)-activated human PBMC were infected for about 12 h with the viral supernatants from transfected cells. Meanwhile, the rest of the transfected 293T cells were subjected to the cell fusion assay to test the expression and correct phenotype of Env as described above. The growth of each SHIV was assessed by monitoring the production of p27 antigen production in the PMBC culture by using a commercial enzyme-linked immunosorbent assay kit (Cellular Products, Inc., Buffalo, N.Y.). The SI capacity of the isolate was examined by cocultivation with MT-2 cells. The coreceptor usage of the chimeric viruses was established by infection of various GHOS.CD4+ cell lines engineered to selectively express one of a panel of coreceptors as described above. In addition, to test the infectivity of newly generated SHIV strains in monkey cells, 2 × 106 macaque CD4+ T lymphocytes (CD8+ T lymphocytes were depleted) or PBMC were infected with a virus input equal to 100 50% tissue culture infective doses (TCID50). The replication of SHIV in macaque cells was also monitored by using the p27 assay.
Inoculation of Asian macaques with SHIVCHN19.
The animals used in this study included eight juvenile pig-tailed macaques and two juvenile rhesus macaques. The animal protocol was reviewed and approved by the Institutional Animal Care and Use Committee at the Tulane Regional Primate Research Center, where the experimental animals were housed. The viral titer of SHIVCHN19 was determined by infecting human PBMC. A dose equal to 3 × 104 TCID50 of cell-free viral supernatant and 2 × 108 autologous monkey PBMC, which were preinfected with 3,000 TCID50 of SHIVCHN19 for 12 h, was infused intravenously back into each of two pig-tailed macaques and two rhesus macaques. For serial passages, 10 ml of whole blood and 5 ml of bone marrow collected from each of the P1 pig-tailed macaques at 2 weeks postinoculation (p.i.) were transfused intravenously into two naïve pig-tailed macaques, which served as P2 animals. Subsequent passages for P3 and P4 animals were carried out in the same manner as for P2 animals.
SHIVCHN19 isolation.
The isolation of SHIV strains from infected macaques was performed according to a standard protocol (8, 34). Briefly, viruses were isolated from infected macaques by cocultivating 106 of their PBMC with 2 × 106 donor PBMC, which were isolated from naïve pig-tailed macaques. The donor PBMC were isolated 3 days in advance by Ficoll-Hypaque density gradient centrifugation and were stimulated with 6 μg of PHA (Sigma) per ml in RPMI 1640 medium supplemented with 10% fetal calf serum and 1% penicillin-streptomycin. In some cases, the donor cells were washed with phosphate-buffered saline containing 2% fetal calf serum, and CD8+ T cells were depleted by binding to anti-CD8 monoclonal antibody-coated magnetic beads according to manufacturer's instructions (Dynal, Oslo, Norway). The growth kinetics of SHIVCHN19 were monitored by measuring p27 production in the culture supernatants.
Western blot analysis.
To monitor the seroconversion in SHIVCHN19-infected animals, a homemade Western blot assay was used. Briefly, SHIVCHN19 was propagated in large quantities by infecting CEMx174.Hu-CCR5 cells in vitro. The production of SHIVCHN19 was measured by the p27 assay. At the peak level of p27 production, viral supernatants were harvested, and virons were subsequently concentrated and purified by a sucrose gradient ultracentrifugation method (9). The virions were suspended in a lysis buffer, and the viral proteins were subsequently heat denatured and loaded into a sodium dodecyl sulfate–12% polyacrylamide gel. The gel transfer and detection were otherwise standard as described elsewhere (9). The plasma samples from SHIV-infected animals were heat inactivated and diluted 1:100 before use. The results were further confirmed by a commercially available HIV blot 2.2 kit (Genelabs Diagnostics, Redwood City, Calif.).
Quantitation of SHIV RNA load and proviral load.
The level of plasma viremia was measured by an in-house real-time PCR method (21, 30). Briefly, to measure the viral load of infected monkeys, plasma was separated from whole blood collected in EDTA-containing tubes. Plasma samples were initially spun at 5,000 rpm in a Sorvall Microspin 24S centrifuge for 10 min to remove residual cells. Plasma (500 μl) was ultracentrifuged at 17,000 rpm in a Heraeus Sepatech 17RS centrifuge for 60 min at 4°C. Supernatant was removed to leave ∼140 μl of plasma and a viral pellet. Viral RNA was extracted by using the QIAamp viral RNA kit (QIAGEN, Inc., Valencia, Calif.). Reverse transcription (RT) of viral RNA was then performed in 96-well plates. Each 30-μl reaction mixture contained 10 μl of viral RNA; 1× Taqman Buffer A (Perkin-Elmer, Norwalk, Conn.); 5 mM MgCl2; 2.5 μM random hexamers (Perkin-Elmer); 0.5 mM each dATP, dCTP, dGTP, and dTTP; 20 U of RNasin (Promega, Madison, Wis.); and 20 U of Moloney murine leukemia virus reverse transcriptase (Superscript; Gibco BRL, Gaithersburg, Md.). One round of RT (25°C for 15 min, 42°C for 40 min, and 75°C for 5 min) was performed. To quantify the copy number of viral RNA, a molecular beacon was used in combination with real-time PCR. This method of detection using molecular beacons and a fluorescence detector system has been previously described (21, 30). PCR was carried out in an ABI 7700 PRISM spectrofluorometric thermal cycler (Applied Biosystems, Inc.) that monitored changes in the fluorescence spectrum of each reaction tube, while simultaneously carrying out programmed temperature cycles.
For the quantification of proviral load in infected animals, the same real-time PCR assay was used as described above. In this case, since the RT step was not required, genomic DNA extracted from monkey PBMC served directly as the PCR template. In order to accurately determine the number of input cells, the copy number of chemokine receptor CCR5 was also measured by using a similar assay (53). As each cell contains two copies of CCR5, the copy number of proviral genomes can be normalized by the cell number.
FACS analysis.
To quantify the absolute number of peripheral CD4+ and CD8+ lymphocytes as well as CD4/CD8 ratios in SHIV-infected animals, TruCount lymphocyte phenotyping of each cell population was performed on all blood samples collected according to protocol. Briefly, 50 μl of whole blood (prior to plasma separation and well mixed) was transferred to a TruCount tube (Becton Dickinson, San Jose, Calif.). Then, 10 μl of leu-3a-PE anti-CD4 (Becton Dickinson), 10 μl of leu-2a-PerCP anti-CD8 (Becton Dickinson), and 10 μl of fluorescein isothiocyanate anti-CD3ɛ (Pharmingen, San Diego, Calif.) antibodies were added, and the mixture was incubated at room temperature in the dark for 20 min. Then, 450 μl of 1× fluorescence-activated cell sorter (FACS) lysing solution (Becton Dickinson) was added, and another incubation at room temperature for 15 min was carried out to ensure the total lysis of red blood cells. The sample was then subjected to FACS analysis by using a FACS Calibur flow cytometer with Cellquest software (Becton Dickinson). Absolute CD4+ and CD8+ T-lymphocyte counts in the blood sample were calculated according to manufacturer's instructions. To analyze T-lymphocyte populations from gut tissues, the specimens were collected and processed immediately after jejunal biopsy based on a previously published method (50, 51).
RESULTS
Sequence analysis of HIV-1 subtype C tat/rev/vpu/env fragments.
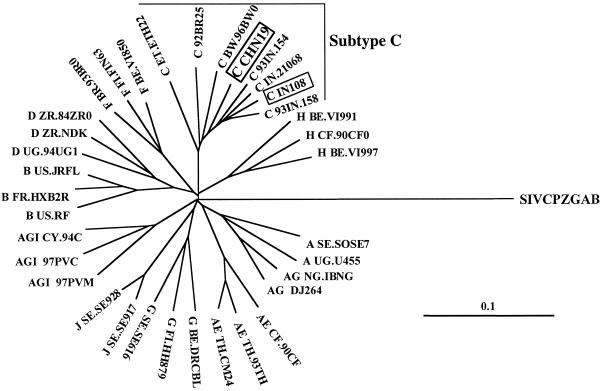
Before SHIV construction, the full-length tat/rev/vpu/env fragments of HIV-1CHN19 were cloned into plasmid vector pcDNA I/Amp and were subsequently sequenced. The sequence results revealed open reading frames for tat/rev/vpu/env genes of HIV-1CHN19. No mutations were found to alter the open reading frames which might cause frameshifts or premature stops. In comparison to vpu+ SHIV33, HIV-1CHN19 had 29 amino acid differences in Tat, 17 differences in Rev, and 13 differences in Vpu. To avoid recombinant env in SHIV constructs, the gene was further analyzed phylogenetically as described previously (9). The env gene from HIV-1CHN19 clustered tightly within the subtype C subfamily by a neighbor-joining method (Fig. 2). That it is not a recombinant env was further confirmed by a recombination identification program (HIV sequence database). Notably, viruses found in China and India cluster closely on the phylogenetic tree without intermingling with sequences obtained from other parts of the world. In contrast, sequences from other countries, including Africa and Brazil, are quite divergent from each other. This analysis indicates that the subtype C viruses in China and India are closely related. In addition, HIV-1CHN19 is still tightly related to the most recent and dominant epidemic subtype C strains in Southern China (Z. Chen, Y. Cao, L. Zhang, and D. Ho, unpublished data). The limited genetic distance of HIV-1CHN19 env from other Chinese C strains makes it a representative virus of the subtype C strains that are now prevalent in Southern China.
FIG. 2.
Phylogenetic analysis of full-length env genes of HIV-1CHN19 and HIV-1IN108. The reference sequences were obtained from the HIV sequence database in GenBank (28). The tree was constructed by using full-length env nucleotide sequences by a neighbor-joining method as described previously (9). The branches are additive, and they represent genetic distances between viruses. The solid scale bar indicates a genetic distance of 10%. The accession number of the HIV-1CHN19 env gene is AF268277.
Phenotypic characterization of HIV-1 subtype C env.
The tat/rev/vpu/env genes were directly amplified from PBMC of a Chinese patient (CHN19) without any in vitro manipulation. As the pcDNA I/Amp vector in which the tat/rev/vpu/env genes of HIV-1CHN19 were cloned is an expression vector in mammalian cells, it enabled us to assess Env-mediated coreceptor usage by a syncytium formation assay. Syncytium formation was observed after 293T cells transfected with env expression vectors were mixed with GHOS.CD4.Hu-CCR5 cells (data not shown). In contrast, no syncytium formation was found using GHOST.CD4 cells expressing a panel of coreceptors, including CXCR4, CCR1, CCR2b, CCR3, CCR4, BOB, and Bonzo. These data indicate that the functional env genes for SHIV construction have an R5 tropism phenotype. This phenotype was also confirmed by testing the env genes in a viral entry assay using pseudotyped viruses as described before (11). The observed pattern of CCR5 usage by HIV-1CHN19 env reinforces the conclusion that the parent HIV-1CHN19 must be a primary NSI strain.
Construction of replication-competent SHIVCHN19.
To generate a replication-competent virus, linear 5′-PVP-1 was cotransfected with various 3′-chimeric constructs into 293T cells (Fig. 1). Interestingly, p27 production, ranging from 10 to 40 ng/ml, was readily detected in the culture supernatants 48 h posttransfection. Nevertheless, viral supernatants made from 3′ constructs pCHN19-1, pCHN19-2, and pCHN19-3 did not result in productive infection in PHA-stimulated human PBMC even after CD8+ T lymphocytes were depleted. In contrast, viruses made of constructs pCHN19-4 and pCHN19-5 readily infect human PBMC. The experimental results were consistent when 3′ constructs were made of subtype C HIV-193IN108 genes in parallel (data not shown). Therefore, our findings with the subtype C SHIV construction do not reveal a HIV-1CHN19 strain-specific phenomenon. The SHIVCHN19 described in this paper was made from pCHN19-5, which is vpu+ (Fig. 3A).
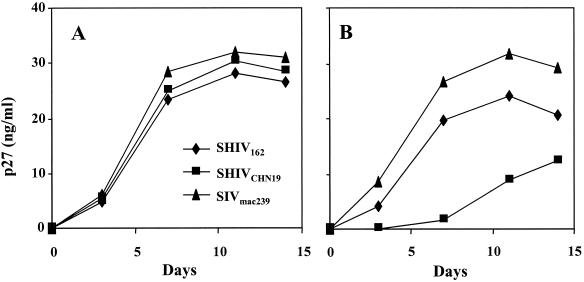
FIG. 3.
The replication kinetics of SHIVCHN19 in human PBMC (A) and in CD4+ T lymphocytes derived from a pig-tailed macaque (B). Human PBMC or macaque CD4+ T lymphocytes (2 × 106) were infected with a virus input of 100 TCID50. The replication of SHIV and SIV was monitored by using the p27 assay (Cellular Products). The y axis represents the level of p27 production in the culture supernatants.
To test the infectivity of SHIVCHN19 in macaque PBMC, SHIVCHN19 was used to infect PBMC of multiple monkeys. SHIVCHN19 replicated in CD4+ T lymphocytes of pig-tailed macaque origin but with delayed viral peaks in comparison to control viruses SHIV162 and SIVmac239 (Fig. 3B). Replication, however, was not observed in PBMC of multiple rhesus macaques even when 1,000 TCID50 of SHIVCHN19 was used for infection (data not shown). Since the proviral load was detected in genomic DNA extracted from infected rhesus CD4+ T lymphocytes by a nested PCR assay (data not shown), the blockage of SHIVCHN19 is likely to occur after RT or integration of the viral genome. In contrast, both SHIV162 and SIVmac239 replicated well in rhesus cells. To determine if the blockage is a strain-specific phenomenon, we further tested R5-tropic SHIV93IN108, a strain carrying a subtype C envelope derived from HIV-193IN108 (Fig. 2). This test yielded similar results: SHIV93IN108 infected pig-tailed CD4+ T lymphocytes but not the cells of rhesus macaque origin. Therefore, the blockage might be directly related to HIV-1 subtype C envelopes.
SHIVCHN19 replicates preferentially in pig-tailed macaques.
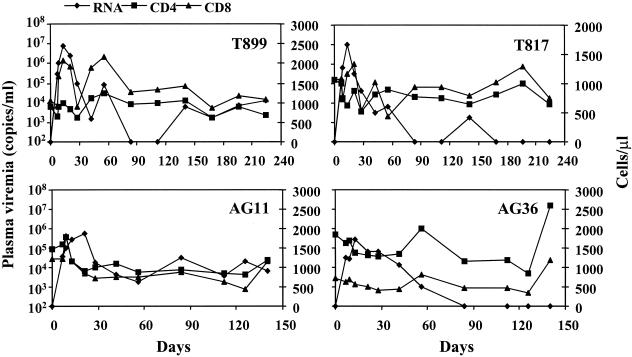
Speculating that the poor replication of SHIVCHN19 in rhesus PBMC might not necessarily reflect in vivo susceptibility, we sought to infect two rhesus monkeys and two pig-tailed monkeys with equal amounts of virus via intravenous infusion. Longitudinal specimens of peripheral blood from each of the four animals were collected and analyzed for plasma viremia and for changes in CD4/CD8 counts. Eventually, all four animals became infected as shown by the occurrence of viremia within 1 week p.i. (Fig. 4). Productive viral replication was found in four macaques with 105 to 107 copies of viral RNA per ml of plasma during the acute phase of infection. Viral load, however, was about 10-fold higher in the plasma of two pig-tailed macaques (∼107 copies/ml) than in that of the rhesus macaques (∼105 to 106 copies/ml) during the period of viral peak, indicating more productive SHIVCHN19 replication in the former. This result was consistent with the finding that PBMC from pig-tailed macaques were more susceptible to SHIVCHN19 infection in vitro as described above. Moreover, although rhesus peripheral CD4+ T lymphocytes were not supportive of SHIVCHN19 replication, the productive infection of rhesus macaques by SHIVCHN19 suggests that the distribution of cells susceptible to viral infection might exist outside peripheral blood in these animals. Viremia in four P1 animals began to decline about 2 to 3 weeks p.i. The changes in the absolute numbers of CD4+ and CD8+ T lymphocytes varied among the animals (Fig. 4). An initial expansion of both CD4+ and CD8+ T lymphocytes was observed in three of four animals (AG11, T899, and T817) while viral loads peaked. The seemingly parallel changes of CD4+ and CD8+ T-cell numbers made the CD4/CD8 ratio relatively stable in T817 but slightly decreased in T899 during the early phase of infection (Fig. 5). Nevertheless, no significant CD4+ T-cell loss was found in the peripheral blood of these animals over time up to 6 months postinfection. In addition, all four animals seroconverted p.i. Anti-Gag, anti-gp120 and anti-gp41 antibodies were detected in two pig-tailed macaques about 3 weeks p.i. (data not shown), whereas similar levels of antibodies were not found in the rhesus macaques until 6 weeks later. The higher viral load and earlier seroconversion suggested that SHIVCHN19 replicated preferentially in pig-tailed macaques. Nonetheless, a low level of viremia lasted for about 2 months (more than 4 months in AG11), and proviral DNA was still detected 6 months p.i. in all four animals. The plasma viremia rebounded in T899 4 months p.i. and remained at a relatively higher level (104).
FIG. 4.
The plasma viremia and the absolute CD4+ and CD8+ T-cell counts in four P1 animals p.i. Animals T899 and T817 (upper panel) are two P1 pig-tailed macaques, whereas AG11 and AG36 (lower panel) are two P1 rhesus macaques. The primary y axis represents viral RNA copies per milliliter of plasma. The secondary y axis indicates the absolute number of CD4+ or CD8+ T cells per microliter of whole blood.
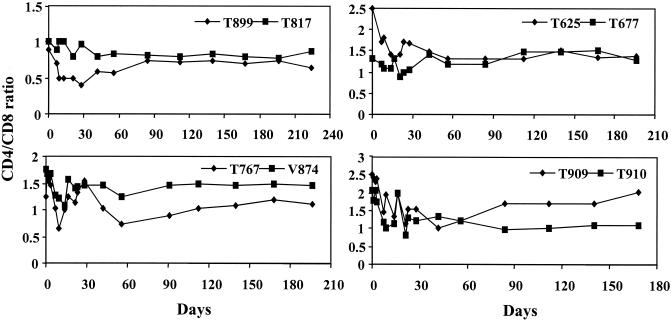
FIG. 5.
The changes in CD4/CD8 ratios over time in P1 to P4 pig-tailed macaques postinfection. Animals from the same passage are plotted in the same panel, which include two P1 (T899 and T817, upper left), two P2 (T625 and T677, upper right), two P3 (T767 and V874, lower left), and two P4 (T909 and T910, lower right) macaques.
Serial passages of SHIVCHN19 in macaques.
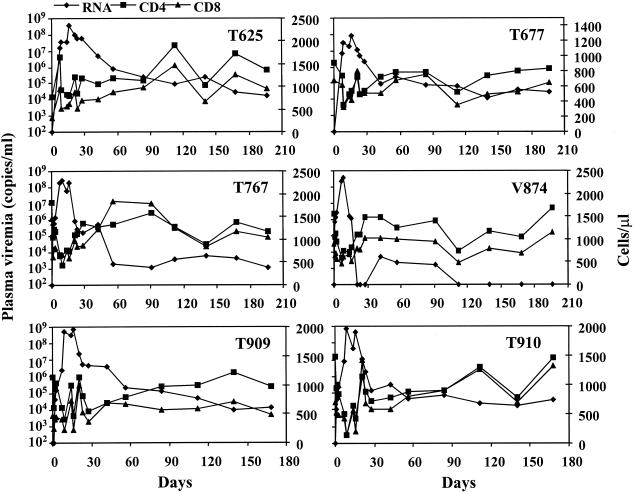
To enhance the infectivity of SHIVCHN19 in macaques, whole blood and bone marrow from P1 pig-tailed macaques (T899 and T817) were mixed and infused into naïve pig-tailed macaques. Such in vivo passaging was carried out successively twice more as described elsewhere (7, 20, 43), using two animals with each passage. The inclusion of bone marrow transfusion was based on an earlier finding that some virulent viral strains might reside there (22). Both P2 animals (T625 and T677) developed productive infection characterized by a burst of viral replication during the initial 1 to 3 week period following the blood-bone marrow transfusion (Fig. 6). In comparison to P1 macaques, T625 and T677 had increased peak viral loads of ∼108 copies of viral RNA per ml of plasma. Further successive in vivo passages of SHIVCHN19 significantly enhanced viral infectivity, as shown by viral peaks between 108 and 109 copies of viral RNA per ml of plasma in both P3 (T767 and V874) and P4 (T909 and T910) macaques (Fig. 6). Likewise, the doubling times for plasma virus shortened to about 0.2 to 0.5 days, which are substantially faster than those in P1 animals (1 to 2 days), as calculated by means previously described (46). Two P2 animals sustained plasma viremia between 104 and 105 viral RNA copies per ml of plasma for over 6 months. Three of the four P3 and P4 macaques (T767, T909, and T910) had similar levels of persistent viremia. The viral RNA load in one P3 macaque (V874) became undetectable 3 weeks p.i. and rebounded 2 weeks later to a level between 103 and 104 copies per ml of plasma for about 2 months, then diminished again.
FIG. 6.
The plasma viremia and the absolute CD4+ and CD8+ T-cell counts in P2 to P4 pig-tailed macaques p.i. Animals include two P2 (T625 and T677, upper panel), two P3 (T767 and V874, middle panel), and two P4 (T909 and T910, lower panel) macaques. The primary y axis represents viral RNA copies per milliliter of plasma. The secondary y axis indicates the absolute number of CD4+ or CD8+ T cells per microliter of whole blood.
Most P2 to P4 monkeys had a parallel drop in the absolute numbers of CD4+ and CD8+ T lymphocytes while viral load reached its peak (Fig. 6). These cell populations, however, returned to near baseline values at 4 to 6 weeks p.i. As with P1 animals, there was a trend of a gradual drop in the CD4/CD8 ratio during the early phase of infection (Fig. 5). This drop was seen to bounce back to near the normal level as the viral load reached a steady state. All P2 to P4 animals became seropositive 2 to 3 weeks posttransfusion (data not shown). The data indicate that the higher levels of viremia in P2 to P4 animals result from the adaptation of SHIVCHN19 in pig-tailed macaques.
Unchanged phenotype and enhanced infectivity of SHIVCHN19 after serial passages.
To further determine phenotypic changes in SHIVCHN19 after in vivo passages, viruses were isolated from two P4 animals by cocultivation with PBMC from naïve pig-tailed macaques. To determine the viral coreceptor usage, the isolates were used to infect MAGI and MAGI-Rh-CCR5 cell lines, both of which have high levels of endogenous CXCR4 expression. We found that isolates from T909 and T910 infected MAGI-Rh-CCR5 cells but not MAGI cells (Table 1). Furthermore, the exclusive R5 usage of the viruses was demonstrated with GHOS.CD4 cells expressing various specific coreceptors, including CXCR4, CCR1, CCR2b, CCR3, CCR4, CCR5, BOB, and Bonzo. The P4 viral isolates were able to infect those cells expressing CCR5 but not ones expressing other coreceptors (Table 1), a pattern similar to that of the parental SHIVCHN19. These results suggest that, despite in vivo adaptation, SHIVCHN19 has maintained CCR5 specificity with no evidence of altered or expanded coreceptor utilization.
TABLE 1.
Comparison of coreceptor usage of four SHIV strains
| Strain | Usage of coreceptor:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GHOS.Hu-CD4
|
Hela.Hu-CD4
|
|||||||||
| CCR5 | CXCR4 | CCR1 | CCR2b | CCR3 | CCR4 | BOB | Bonzo | MAGI | MAGI.CCR5 | |
| SHIVCHN19 | + | − | − | − | − | − | − | − | − | + |
| SHIVCHN19Pa | + | − | − | − | − | − | − | − | − | + |
| SHIV162 | + | − | − | − | − | − | − | − | − | + |
| SHIV33 | − | + | − | − | − | − | − | − | + | + |
Isolated from P4 macaque T910.
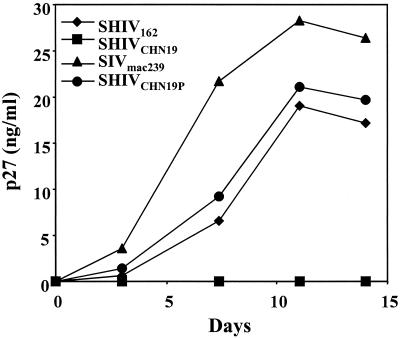
Given that the parental SHIVCHN19 could not infect rhesus PBMC, it was interesting and important to determine whether in vivo passages would change its viral replication capability in rhesus cells. Indeed, a SHIV/rhesus macaque model would be of greater practical use to researchers, since so many more reagents are relevant only to this species. The viral isolate from T910 was tested in parallel with three control viruses, SHIVCHN19, SHIV162, and SIVmac239. Interestingly, with an equal amount of virus input, the parental SHIVCHN19 still did not infect rhesus PBMCs, whereas replication of the T910 isolate occurred with kinetics slightly higher than that of SHIV162 (Fig. 7). Comparable results were obtained by using PBMC derived from pig-tailed macaques (data not shown). Therefore, our in vitro data provided further evidence that the infectivity of SHIVCHN19 in macaque cells has been dramatically enhanced after serial passages of the virus in pig-tailed macaques.
FIG. 7.
Comparison of the replication kinetics of the viral isolate (SHIVCHN19P) from P4 animal T910 to parental SHIVCHN19 in rhesus PBMC. Rhesus PBMC (2 × 106) were infected with a virus input of 100 TCID50. The y axis represents the level of p27 production in the culture supernatant.
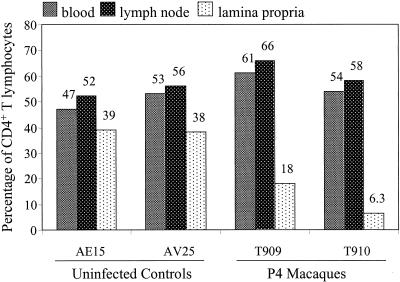
Since CD4+ T-cell depletion in the lamina propria of the gut has been described with SHIV162, we looked for a similar effect in our P4 animals compared to two normal controls. As shown in Fig. 8, no impact was found on the CD4+ lymphocyte population in blood or lymph nodes. However, SHIVCHN19 infection induced profound CD4+ T-cell depletion in the lamina propria of the jejunum. Thus, this feature represents one pathological consequence of SHIVCHN19 infection in vivo.
FIG. 8.
Comparison of percentage of CD4+ cells among total CD3+ T lymphocytes from different compartments. Four animals were tested, including two P4 (T909 and T910) and two naïve (AE15 and AV25) pig-tailed macaques. T lymphocytes derived from three compartments, including blood, colonic lymph node, and jejunal lamina propria, were assayed. The y axis represents the percentage of CD4+ T lymphocytes. The number above each vertical bar indicates the average percentage of two separate stainings of the same specimen. Specimens from the two P4 animals were collected 2 weeks p.i.
DISCUSSION
Subtype C has become the dominant HIV-1 genotype in the world. After being initially identified in Africa, HIV-1 subtype C was subsequently introduced into Asia, including India, China, and many other countries (11, 18, 35, 38, 42, 45, 48, 49). The rapid spread of this particular subtype in these heavily populated regions has resulted in over five million infections in Asia alone. Combination antiretroviral therapy has greatly reduced transmission and death rates in developed countries, but this approach is far too expensive for less-developed nations to afford. Apart from educational campaigns, basic scientific research offers the most hope for finding solutions. Towards that end, it is crucial that we develop an animal model to determine alternative strategies in preventing HIV-1 transmission, particularly against the dominant genotype. In the present study, a relevant SHIVCHN19/macaque model has been developed by using an env gene of a HIV-1 subtype C strain derived from an epidemic area in Yunnan, China. Since the majority of new infections in the study area were caused by HIV-1 subtype C strains and the viruses are genetically closely related, the animal model generated will directly benefit studies to test various strategies (e.g., vaccines, topical microbicides, and entry inhibitors) to prevent the viral spread at an early stage of the epidemic. It is known that most subtype C HIV-1 strains are NSI and R5 tropic (1, 5, 17, 33, 37, 40, 41, 52). The latter property confers, perhaps, an advantage in transmission of the virus through the mucosal route (54, 55). Therefore, we chose a virus with such a phenotype for our SHIVCHN19 construction so that future vaccine and transmission studies would be more relevant to the natural situation of HIV-1.
Using a traditional method, we sought to create a SHIV by replacing tat/rev/env genes of SIVmac239 with the corresponding regions of HIV-1 subtype C strains. Although this method has been successfully used to establish multiple SHIV/macaque models by using the genes of various HIV-1 subtype B strains (31, 34), replication-competent subtype C SHIV strains were not generated by the same technique. The basis for this observation remains to be determined. Possible explanations may be related to the functional failure of tat or rev of subtype C strains on SIV-responsive elements or to the less-efficient interaction between subtype C gp41 and SIV matrix protein during viral packaging (15). We tried to swap 5′ or 3′ halves of tat/rev-encoding regions to minimize the loss of subtype C env genes, but still no replication-competent subtype C SHIV emerged. However, replication-competent subtype C SHIV was obtained only when the full tat/rev, partial vpu, and gp41 C-terminal regions were derived from SHIV33 subtype B. The resulting SHIVCHN19 is, therefore, a chimeric virus with genes originated from HIV-1CHN19, SHIV33, and SIVmac239. This finding is consistent with the strategy that was used to generate a subtype E SHIV (27). Because SHIVCHN19 includes the entire gp120 and partial gp41 of HIV-1CHN19, which comprises the determinant regions for viral entry and phenotype, the new SHIV will be very useful for studies involving primary HIV-1 subtype C envelopes.
The envelope of HIV-1 subtype C determines the replicative capacity of SHIVCHN19 in macaque cells. Unlike R5-tropic SHIV162, which grows readily in rhesus PBMC (20, 34), productive replication was never achieved when SHIVCHN19 was added to CD4+ T lymphocytes derived from multiple rhesus macaques. Nevertheless, besides growing in human PBMC, SHIVCHN19 could infect CD4+ T lymphocytes of pig-tailed macaques. These in vitro results are in agreement with the finding that SHIVCHN19 replicates preferentially in pig-tailed macaques. Since SHIVCHN19 differs from SHIV162 only in env, the failure of SHIVCHN19 to grow in rhesus cells is undoubtedly envelope mediated.
The infectivity of SHIVCHN19 was enhanced dramatically in pig-tailed macaques after serial passages. Some previous studies tried to adapt HIV-1 directly into pig-tailed macaques. But viral replication diminished promptly and markedly, even after serial passages in pig-tailed macaques (3). In contrast, SHIVCHN19 adapted nicely to pig-tailed macaques, as demonstrated by several findings. First, the quantity of viral production increased greatly with successive passages. In comparison to P1 animals, the peak viral loads during acute infection increased more than 1 log unit in P2 and 2 log units in P3 and P4 animals. Moreover, five of six P2 to P4 animals maintained relatively higher levels of viremia over time. Second, the speed of viral replication was also greatly enhanced, as evidenced by the shortened viral doubling time from P1 (1 to 2 days) to P4 (0.2 to 0.5 day) animals. Third, a variant of SHIVCHN19, isolated from animal T910, was able to grow rapidly in vitro in both rhesus and pig-tailed macaque PBMC, even without the depletion of CD8+ T cells, unlike the parental SHIVCHN19. Therefore, these findings provide direct evidence that the infectivity of SHIVCHN19 has been substantially enhanced by serial adaptation in macaques. The precise molecular determinants for the adaptation, however, remain to be determined.
Despite robust replication of SHIVCHN19 through in vivo passages, no significant CD4 lymphopenia was found in the blood of infected animals. One of the major concerns about SHIV/macaque models is the pathogenicity of the virus in vivo. Given that the absolute number of CD4+ T cells in blood did not drop significantly in our infected macaques, future studies will need to examine tissue compartments in detail to address the pathogenic effects mediated by our subtype C SHIV. Previous studies have demonstrated that, unlike pathogenic X4-tropic and X4R5-tropic SHIV strains which induce severe CD4+ T-cell loss in the periphery (20, 24, 47), pathogenic R5-tropic SIV or SHIV strains cause similar effects preferentially in gut-associated lymph tissues (20, 50, 51). A preliminary analysis has shown similar CD4+ T-cell loss in two P4 macaques (Fig. 8).
In conclusion, an R5-tropic subtype C SHIVCHN19/macaque model has been established. This model will provide a unique opportunity for studies related to HIV-1 subtype C vaccinology and pathogenesis in the future.
ACKNOWLEDGMENTS
We thank C. Cheng-Mayer and P. Luciw for helpful discussions, T. Zhu for providing patient DNA samples, S. Lewin and L. Zhang for developing SIV viral load assays, Y. Guo for DNA sequencing, and D. Gurner for editorial input.
This work was supported by a gift from Donald Pels. Additional funds were provided by National Institutes of Health grants (1F32AI10256, AI 40387, AI 42848 [CFAR], and AI 43042), the Irene Diamond Fund, the Bristol-Myers Squibb Foundation, and the Rockefeller Foundation via the Population Council. Z. Chen was also supported by the Charles H. Revson/Norman and Rosita Winston Foundation Fellowship.
REFERENCES
- 1.Abebe A, Demissie D, Goudsmit J, Brouwer M, Kuiken C L, Pollakis G, Schuitemaker H, Fontanet A L, Rinke de Wit T F. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. AIDS. 1999;13:1305–1311. doi: 10.1097/00002030-199907300-00006. [DOI] [PubMed] [Google Scholar]
- 2.Agy M B, Frumkin L R, Corey L, Coombs R W, Wolinsky S M, Koehler J, Morton W R, Katze M G. Infection of Macaca nemestrina by human immunodeficiency virus type-1. Science. 1992;257:103–106. doi: 10.1126/science.1621083. [DOI] [PubMed] [Google Scholar]
- 3.Agy M B, Schmidt A, Florey M J, Kennedy B J, Schaefer G, Katze M G, Corey L, Morton W R, Bosch M L. Serial in vivo passage of HIV-1 infection in Macaca nemestrina. Virology. 1997;238:336–343. doi: 10.1006/viro.1997.8832. [DOI] [PubMed] [Google Scholar]
- 4.Almond N M, Heeney J L. AIDS vaccine development in primate models. AIDS. 1998;12(Suppl. A):S133–S140. [PubMed] [Google Scholar]
- 5.Bjorndal A, Sonnerborg A, Tscherning C, Albert J, Fenyo E M. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res Hum Retrovir. 1999;15:647–653. doi: 10.1089/088922299310944. [DOI] [PubMed] [Google Scholar]
- 6.Bosch M L, Schmidt A, Agy M B, Kimball L E, Morton W R. Infection of Macaca nemestrina neonates with HIV-1 via different routes of inoculation. AIDS. 1997;11:1555–1563. doi: 10.1097/00002030-199713000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Cayabyab M, Karlsson G B, Etemad-Moghadam B A, Hofmann W, Steenbeke T, Halloran M, Fanton J W, Axthelm M K, Letvin N L, Sodroski J G. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2) J Virol. 1999;73:976–984. doi: 10.1128/jvi.73.2.976-984.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Telfer P, Reed P, Zhang L, Getti A, Ho D D, Marx P A. Isolation and characterization of the first simian immunodeficiency virus from a feral sooty mangabey (Cercocebus atys) in West Africa. J Med Primatol. 1995;24:108–115. doi: 10.1111/j.1600-0684.1995.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Telfier P, Gettie A, Reed P, Zhang L, Ho D D, Marx P A. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Csillag C. HIV-1 subtype C in Brazil. Lancet. 1994;344:1354. [PubMed] [Google Scholar]
- 12.Daniel M D, Desrosiers R C. Use of simian immunodeficiency virus for evaluation of AIDS vaccine strategies. AIDS. 1989;3(Suppl. 1):S131–S133. doi: 10.1097/00002030-198901001-00019. [DOI] [PubMed] [Google Scholar]
- 13.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 14.Desrosiers R C, Letvin N L. Animal models for acquired immunodeficiency syndrome. Rev Infect Dis. 1987;9:438–446. doi: 10.1093/clinids/9.3.438. [DOI] [PubMed] [Google Scholar]
- 15.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frumkin L R, Patterson B K, Leverenz J B, Agy M B, Wolinsky S M, Morton W R, Corey L. Infection of Macaca nemestrina brain with human immunodeficiency virus type 1. J Gen Virol. 1995;76:2467–2476. doi: 10.1099/0022-1317-76-10-2467. [DOI] [PubMed] [Google Scholar]
- 17.Gao F, Robertson D L, Carruthers C D, Morrison S G, Jian B, Chen Y, Barre-Sinoussi F, Girard M, Srinivasan A, Abimiku A G, Shaw G M, Sharp P M, Hahn B H. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao F, Yue L, Craig S, Thornton C L, Robertson D L, McCutchan F E, Bradac J A, Sharp P M, Hahn B H. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. WHO Network for HIV Isolation and Characterization. AIDS Res Hum Retrovir. 1994;10:1359–1368. doi: 10.1089/aid.1994.10.1359. [DOI] [PubMed] [Google Scholar]
- 19.Gardner M B. Simian and feline immunodeficiency viruses: animal lentivirus models for evaluation of AIDS vaccines and antiviral agents. Antivir Res. 1991;15:267–286. doi: 10.1016/0166-3542(91)90009-g. [DOI] [PubMed] [Google Scholar]
- 20.Harouse J M, Gettie A, Tan R C, Blanchard J, Cheng-Mayer C. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science. 1999;284:816–819. doi: 10.1126/science.284.5415.816. [DOI] [PubMed] [Google Scholar]
- 21.Jin X, Bauer D E, Tuttleton S E, Lewin S, Gettie A, Blanchard J, Irwin C E, Safrit J T, Mittler J, Weinberger L, Kostrikis L G, Zhang L, Perelson A S, Ho D D. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joag S V, Li Z, Foresman L, Pinson D M, Raghavan R, Zhuge W, Adany I, Wang C, Jia F, Sheffer D, Ranchalis J, Watson A, Narayan O. Characterization of the pathogenic KU-SHIV model of acquired immunodeficiency syndrome in macaques. AIDS Res Hum Retrovir. 1997;13:635–645. doi: 10.1089/aid.1997.13.635. [DOI] [PubMed] [Google Scholar]
- 23.Joag S V, Li Z, Foresman L, Stephens E B, Zhao L J, Adany I, Pinson D M, McClure H M, Narayan O. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J Virol. 1996;70:3189–3197. doi: 10.1128/jvi.70.5.3189-3197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joag S V, Li Z, Wang C, Jia F, Foresman L, Adany I, Pinson D M, Stephens E B, Narayan O. Chimeric SHIV that causes CD4+ T cell loss and AIDS in rhesus macaques. J Med Primatol. 1998;27:59–64. doi: 10.1111/j.1600-0684.1998.tb00227.x. [DOI] [PubMed] [Google Scholar]
- 25.Johnson P R, Goldstein S, London W T, Fomsgaard A, Hirsch V M. Molecular clones of SIVsm and SIVagm: experimental infection of macaques and African green monkeys. J Med Primatol. 1990;19:279–286. [PubMed] [Google Scholar]
- 26.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 27.Klinger J M, Himathongkham S, Legg H, Luciw P A, Barnett S W. Infection of baboons with a simian immunodeficiency virus/HIV-1 chimeric virus constructed with an HIV-1 Thai subtype E envelope. AIDS. 1998;12:849–857. doi: 10.1097/00002030-199808000-00006. [DOI] [PubMed] [Google Scholar]
- 28.Korber B, Foley B, Leitner T, McCutchan F, Hahn B, Mellors J W, Myers G, Kuiken C. Human retroviruses and AIDS 1997. Theoretical Biology and Biophysics Group T-10. Los Alamos, N.M: Los Alamos National Laboratory; 1997. [Google Scholar]
- 29.Letvin N L. Nonhuman primate models for HIV vaccine development. Immunodefic Rev. 1992;3:247–260. [PubMed] [Google Scholar]
- 30.Lewin S R, Vesanen M, Kostrikis L, Hurley A, Duran M, Zhang L, Ho D D, Markowitz M. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J Virol. 1999;73:6099–6103. doi: 10.1128/jvi.73.7.6099-6103.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Lord C I, Haseltine W, Letvin N L, Sodroski J. Infection of cynomolgus monkeys with a chimeric HIV-1/SIVmac virus that expresses the HIV-1 envelope glycoproteins. J Acquir Immune Defic Syndr. 1992;5:639–646. [PubMed] [Google Scholar]
- 32.Li J T, Halloran M, Lord C I, Watson A, Ranchalis J, Fung M, Letvin N L, Sodroski J G. Persistent infection of macaques with simian-human immunodeficiency viruses. J Virol. 1995;69:7061–7067. doi: 10.1128/jvi.69.11.7061-7067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luciw P A, Pratt-Lowe E, Shaw K E, Levy J A, Cheng-Mayer C. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV) Proc Natl Acad Sci USA. 1995;92:7490–7494. doi: 10.1073/pnas.92.16.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo C C, Tian C, Hu D J, Kai M, Dondero T, Zheng X. HIV-1 subtype C in China. Lancet. 1995;345:1051–1052. doi: 10.1016/s0140-6736(95)90792-0. [DOI] [PubMed] [Google Scholar]
- 36.Miller C, Gardner M B. AIDS and mucosal immunity: usefulness of the SIV macaque model of genital mucosal transmission. J Acquir Immune Defic Syndr. 1991;4:1169–1172. [PubMed] [Google Scholar]
- 37.Mochizuki N, Otsuka N, Matsuo K, Shiino T, Kojima A, Kurata T, Sakai K, Yamamoto N, Isomura S, Dhole T N, Takebe Y, Matsuda M, Tatsumi M. An infectious DNA clone of HIV type 1 subtype C. AIDS Res Hum Retrovir. 1999;15:1321–1324. doi: 10.1089/088922299310223. [DOI] [PubMed] [Google Scholar]
- 38.Novitsky V A, Montano M A, McLane M F, Renjifo B, Vannberg F, Foley B T, Ndung'u T P, Rahman M, Makhema M J, Marlink R, Essex M. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J Virol. 1999;73:4427–4432. doi: 10.1128/jvi.73.5.4427-4432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oxford J S, Frezza P, Race E. Challenges and strategies for AIDS vaccine development. Vaccine. 1993;11:612–614. doi: 10.1016/0264-410x(93)90303-f. [DOI] [PubMed] [Google Scholar]
- 40.Peeters M, Vincent R, Perret J L, Lasky M, Patrel D, Liegeois F, Courgnaud V, Seng R, Matton T, Molinier S, Delaporte E. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:115–121. doi: 10.1097/00042560-199902010-00002. [DOI] [PubMed] [Google Scholar]
- 41.Ping L-H, Nelson J A E, Hoffman I F, Schock J, Lamers S L, Goodman M, Vernazza P, Kazembe P, Maida M, Zimba D, Goodenow M M, Eron J J, Jr, Fiscus S A, Cohen M S, Swanstrom R. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J Virol. 1999;73:6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin G, Shao Y, Liu G. Subtype and sequence analysis of the C2-V3 region of gp120 genes among HIV-1 strains in Sichuan province. Chung-Hua Liu Hsing Ping Hsueh Tsa Chih. 1998;19:39–42. . (In Chinese.) [PubMed] [Google Scholar]
- 43.Reimann K A, Li J T, Veazey R, Halloran M, Park I W, Karlsson G B, Sodroski J, Letvin N L. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shibata R, Maldarelli F, Siemon C, Matano T, Parta M, Miller G, Fredrickson T, Martin M A. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J Infect Dis. 1997;176:362–373. doi: 10.1086/514053. [DOI] [PubMed] [Google Scholar]
- 45.Sonnerborg A, Sherefa K. HIV-1 subtype C epidemic in Ethiopia. AIDS. 1997;11:1897. doi: 10.1097/00002030-199715000-00024. [DOI] [PubMed] [Google Scholar]
- 46.Stafford M A, Corey L, Cao Y, Daar E S, Ho D D, Perelson A S. Modeling plasma virus concentration during primary HIV infection. J Theor Biol. 2000;203:285–301. doi: 10.1006/jtbi.2000.1076. [DOI] [PubMed] [Google Scholar]
- 47.Steger K K, Dykhuizen M, Mitchen J L, Hinds P W, Preuninger B L, Wallace M, Thomson J, Montefiori D C, Lu Y, Pauza C D. CD4+-T-cell and CD20+-B-cell changes predict rapid disease progression after simian-human immunodeficiency virus infection in macaques. J Virol. 1998;72:1600–1605. doi: 10.1128/jvi.72.2.1600-1605.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tien P C, Chiu T, Latif A, Ray S, Batra M, Contag C H, Zejena L, Mbizvo M, Delwart E L, Mullins J I, Katzenstein D A. Primary subtype C HIV-1 infection in Harare, Zimbabwe. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:147–153. doi: 10.1097/00042560-199902010-00006. [DOI] [PubMed] [Google Scholar]
- 49.UNAIDS. The global source of HIV/AIDS information. AIDS epidemic update. Geneva, Switzerland: UNAIDS; 1996–1999. [Google Scholar]
- 50.Veazey R S, DeMaria M, Chalifoux L V, Shvetz D E, Pauley D R, Knight H L, Rosenzweig M, Johnson R P, Desrosiers R C, Lackner A A. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 51.Veazey R S, Rosenzweig M, Shvetz D E, Pauley D R, DeMaria M, Chalifoux L V, Johnson R P, Lackner A A. Characterization of gut-associated lymphoid tissue (GALT) of normal rhesus macaques. Clin Immunol Immunopathol. 1997;82:230–242. doi: 10.1006/clin.1996.4318. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, He T, Huang Y, Chen Z, Guo Y, Wu S, Kunstman K J, Brown R C, Phair J P, Neumann A U, Ho D D, Wolinsky S M. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72:9307–9312. doi: 10.1128/jvi.72.11.9307-9312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Lewin S R, Markowitz M, Lin H H, Skulsky E, Karanicolas R, He Y, Jin X, Tuttleton S, Vesanen M, Spiegel H, Kost R, van Lunzen J, Stellbrink H J, Wolinsky S, Borkowsky W, Palumbo P, Kostrikis L G, Ho D D. Measuring recent thymic emigrants in blood of normal and HIV-1-infected individuals before and after effective therapy. J Exp Med. 1999;190:725–732. doi: 10.1084/jem.190.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Brown A J, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]