Abstract
Human immunodeficiency virus type 1 (HIV-1) Vpr regulates nuclear transport of the viral preintegration complex, G2 cell cycle arrest, and transcriptional transactivation. We asked whether phosphorylation could affect Vpr activity. Vpr was found to be phosphorylated on serine residues in transiently transfected and infected cells. Residues 79, 94, and 96 were all found to be phosphorylated, as assessed by alanine mutations. Mutation of Ser-79 to Ala abrogated effects of Vpr on cell cycle progression, whereas mutation of Ser-94 and Ser-96 had no effect. Simultaneous mutation of all three Vpr serine residues attenuated HIV-1 replication in macrophages, whereas single and double Ser mutations had no effect.
Human immunodeficiency virus 1 (HIV-1) primarily targets CD4 T cells and macrophages (10, 30). Like genomes of other lentiviruses, its genome exhibits greater complexity than those of oncoretroviruses. In addition to the common retroviral structural proteins (Gag, Pol, and Env), HIV-1 contains at least six auxiliary proteins (Vif, Vpr, Tat, Rev, Vpu, and Nef) that are important in viral replication and infection (9, 51). HIV-1 Vpr is a 96-amino-acid protein present at a high copy number in HIV-1 virions (8, 56). Virion-associated Vpr plays an important role in the early stages of HIV-1 infection in terminally differentiated macrophages. Several investigators have shown that Vpr is a nuclear protein (22, 32, 37, 38, 57). Both traditional and nontraditional nuclear localization signals (NLSs) have been identified within Vpr (27, 45). Further studies indicated that the coordination of HIV-1 Vpr with other nucleophilic virion proteins including the Gag matrix (MA) and integrase (IN) proteins account for the infection of nonreplicating cells (5, 16, 17, 22). In addition, de novo and virion-packaged Vpr can enhance virus production by upregulating the HIV long terminal repeat or a wide range of other promoters (1, 23, 44, 52). Another important feature of HIV Vpr is its ability to arrest infected cells in the G2 phase, resulting in increased HIV-1 production (11, 21, 46, 50). Other proposed activities of HIV-1 Vpr include formation of ion channels (43), interference with DNA mutation (18, 28, 40), and induction of micronuclei and aneuploidy (49).
Phosphorylation plays a critical role in the NLS-mediated nuclear transport, cell cycle progression, and gene expression (20, 26, 42, 48). The phosphorylation-regulated NLSs have been shown to control nuclear transport in eukaryotic cells from yeast and plants to higher mammals (26). For example, the archetypal NLS-containing simian virus 40 large T antigen is regulated, by the CcN motif, which comprises the T-antigen NLS (N), a casein kinase II (CKII) phosphorylation site (C) 13 amino acids N terminal to the NLS, modulating the rate of the nuclear import, and a cyclin-dependent kinase site (c) adjacent to the NLS regulating the maximal level of nuclear accumulation. Phosphorylation regulates cell cycle progression and gene expression by changing the nuclear localization of various proteins, as well as their association with transcriptional activation factors. The importance of protein phosphorylation has been shown for other HIV-1 proteins, such as MA, capsid, Rev, Nef, and Vpu (6, 17, 33, 41). In addition, protein kinases have been found in HIV virions, suggesting that protein phosphorylation may be important for regulating virion maturation and supporting early steps of infection (6, 25). This study provides the first evidence that Vpr is phosphorylated and that this modification is important for its biological activity.
MATERIALS AND METHODS
DNA constructs.
The vpr gene of HIV-1 NL4-3 and the SRIG mutant with a deletion of codons 79 to 82 were cloned into pTM-3 or pcDNA1.1 to generate pTM-VPR, pTM-VPR(SRIG), and pcDNA-Vpr as previously described (32, 53, 59). pTM-VPR79A, pTM-VPR94A, pTM-VPR96A, pTM-VPR79S, pTM-VPR94S, and pTM-VPR96S mutants were generated by PCR mutagenesis using primers Vpr79Adn (5′-TGTCGACATGCTAGAATAGGC-3′), Vpr94Aup (5′-CAGGATCCTAGGATCTGNCGGCTCCATT-3′), Vpr96Aup (5′-CAGGATCCTAGNCTCTTGAGGCTCCATT-3′), and Vpr94/96Aup (5′-CAGGATCCTAGNCTCTGNCGGCTCCATT-3′) (2). The PCR products were digested with SalI/BamHI and then cloned into pTM-VPR SalI/BamHI sites. pTM-28S was generated by PCR mutagenesis using primers Vpr79Adn (5′-TGTCGACATGCTAGAATAGGC-3′) and Vpr94/96Aup (5′-CAGGATCCTAGNCTCTGNCGGCTCCATT-3′) and then cloned in pTM-VPR SalI and BamHI sites. pTM-VPR28A was constructed by sequential PCR using the primer pairs Vpr-NC-dn (5′-AATACCATGGAACAAGCCCCAGAAGA-3′)/Vpr28up (5′-TCTAACAGCTTCAGCCTTAAGTTCCTC-3′) and Vpr28dn (5′-GAGGAACTTAAGGCTGAAGCTGTTAGA-3′)/Vpr-down (5′-GATGCTTCCAGGGATCCGTCTAGGATCTACTG-3′) for the first round. Vpr28A (Vpr fragment 28A mutation) was obtained by a second round PCR with Vpr-NC-dn/Vpr-down using the mixture of the first round PCR products as the template (2). The PCR product was digested with NcoI/EcoRI and cloned into pTM-VPR NcoI/EcoRI sites. To obtain pTMVPR4S(−), vpr SalI/BamHI fragment from pTM-VPR28S was cloned into the pTM-VPR28A SalI/BamHI site.
To construct the p125-TTK proviral mutant clone, sequential PCR was used (22, 54). The KKQ-to-TTK mutations at residues 26 to 28 were introduced into HIV-1 p125 MA by first-round PCR with p125-628dn (5′-TCTAGCAGTGGCGCCCGA-3′)/MA-TTKup (5′-AAGGCCAGGGGGAACGACAAAATATAAACTAAAACAT-3′) and MA-TTKdn, (5′-ATGTTTTAGTTTATATTTTGTCGTTCCCCCTGGCCTT-3′)/p24-Bamup (5′-CCGGATCCTACAAAACTCTTGCTTTAT-3′). The second-round PCR using primer pair p125-628dn/p24-BamHIup was carried out to obtain the mutated MA coding sequence. The PCR product was digested with BssHII and SphI to clone into p125 proviral DNA digested with the same restriction enzymes. To disrupt Vpr expression without perturbing the Vif coding sequence, the Vpr start codon, ATG, was mutated to GTG by sequential PCR mutagenesis using primer pairs HXB2-5251dn (5′-TCAGGGAGTCTCCATA-3′)/Vpr(−)up (5′-GCCTTGTTCCACATATCCT-3′) and Vpr(−)dn (5′-AGGATAGGTGGAACAAGCC-3′)/HXB2-5999up (5′-CTTCGTCGCTGTCTCC-3′) followed by PCR with primer pairs HXBII-5251dn/HXBII5999up. The PCR product was digested with PflMI/SalI and cloned into p125 or p125-TTK PflMI/SalI sites to get p125-Vpr(−) or p125-TTK/Vpr(−).
Proviral constructs p125-TVpr3S(−), p125-TVpr79A, p125-TVpr94G (Vpr94Gdn, 5′-ATGGAGCCGGTAGATCCTAG-3′; Vpr94Gup, 5′-CTAGGATCTACCGGCTCCAT-3′), p125-TVpr96P (Vpr96Pdn, 5′-ATGGAGCCAGTAGACCCTAG-3′; Vpr94Pup, 5′-CTAGGGTCTACTGGCTCCAT-3′), p125-TVpr79S (Vpr94G/96Pdn, 5′-ATGGAGCCGGTAGACCCTAG-3′; Vpr94G/96Pup 5′-CTAGGGTCTACCGGCTCCAT-3′), p125-TVpr94S, p125-TVpr96S were constructed in two steps. First, the mutations were introduced into vpr by PCR (79A and 79S constructs) or sequential PCR mutagenesis using Vpr-NC-dn and HXBII-5999up as first- and second-round PCR primers. The PCR products were digested with SalI/EcoNI (positions 5785 to 5891), mixed with p125 EcoNI/BamHI fragments (positions 5891 to 8474), and cloned into pUC12 SalI/BamHI sites. Second, the SalI/BamHI fragments (positions 5785 to 8474) were cloned into p125-TTK SalI/BamHI (positions 5891 to 8474) sites to obtain the proviral DNAs with different vpr mutations. All proviral constructs were confirmed by restriction mapping with Bsu36I/HincII, and the PCR fragments were sequenced.
To express Vpr in the ecdysone-inducible expression system, pTM-VPR was digested with NcoI and treated with Klenow DNA polymerase followed by BamHI digestion. The Vpr fragment was then cloned into pIND (Invitrogen) HindIII/BamHI sites after blunting the HindIII site with Klenow DNA polymerase. To generate the episomal plasmid with the ecdysone response element, pIND-VPR was digested with BglII/XhoI and cloned into pZY-1 BglII/XhoI sites to get pZY-VPR. (The pZY-1 vector was derived from two plasmids, pIND [Invitrogen] and pDR2 [Clontech]. pDR2 was digested with NruI to purify the DNA fragment with EBNA1/oriP. The pIND vector was digested with HincII, and the fragment with the ecdysone-inducible promoter and G418 resistant gene was purified. After ligation, the structure of pZY-1 was confirmed by restriction enzyme mapping.) The relevant mutation sequences in pUC12 as described earlier were digested with SalI/EcoNI and cloned into pZY-Vpr SalI/BamHI sites after the BamHI site was partially filled in with Klenow DNA polymerase I Exo− in the presence of dGTP, dATP, and TTP. The additional Vpr mutants at residues 78, 79, and 80 were generated by PCR mutagenesis using p125-79S as the template with primers Vpr79D (5′-GGGTGTCGACATGACAGAATAGGCGT-3′), Vpr79F (5′-GGGTGTCGACATTTCAGAATAGGCGT-3′), Vpr79T (5′-GGGTGTCGACATACCAGAATAGGCGT-3′), Vpr79Q (5′-GGGTGTCGACATCAGAGAATAGGCGT-3′), Vpr80K (5′-GGGTGTCGACATAGCAAAATAGGCGT-3′), Vpr80A (5′-GGGTGTCGACATAGCGGAATAGGCGT-3′), and Vpr78Q (5′-GGGTGTCGACAAAGCAGAATAGGCGT-3′). The SalI/EcoNI fragments were cloned into pZY-Vpr SalI/BamHI as described earlier.
PCR.
PCRs were carried out with 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 1.5 mM MgCl2, 10 pmol of each primer, template plasmid DNA, 0.25 mM deoxynucleoside triphosphate, and 2.5 U of Taq polymerase (GIBCO BRL, Gaithersburg, Md.). The reaction was carried out in a total volume of 50 μl. The first cycle, at 94°C for 3 min, 72°C for 2 min, and 55°C for 1.5 min, was followed by 30 cycles of 94°C for 45 s, 72°C for 2 min, and 55°C for 1 min. The last (extension) cycle was at 72°C for 10 min.
Cell lines and DNA transfection.
HEK293, 293T, BSC40, MAGI, and MAGI5 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U of penicillin-streptomycin per ml, plus the selection agent as described elsewhere (59). 293VE (ecdysone-inducible) cell lines were obtained by cotransfecting pVgRXR (Invitrogen), which encodes the ecdysone receptor, and pCMV-EBNA1 (Clontech), which encodes the Epstein-Barr virus nuclear antigen 1, with Lipofectamine into HEK293 cells (59). The transfected cells were split into 96-well plates (50 to 100 cells/well) and cultured for 2 days followed by Zeocin (200 μg/ml) selection for 12 days. The individual colonies were amplified and tested for the gene expression according to the protocol provided by manufacturer (Invitrogen). The cell lines were maintained in DMEM supplemented with 10% FCS, 100 U of penicillin-streptomycin per ml, and 100 μg of Zeocin (Invitrogen) per ml. To induce gene expression, 1 μM muristerone A (Invitrogen) was used.
Immunoprecipitation.
The infection-transfection protocol for the vaccinia virus expression system was as described elsewhere (59). Briefly, BSC40 cells were grown to 90% confluence on 60-mm-diameter plates, infected for 1 h at 37°C with vTF7-3, a vaccinia virus expressing T7 RNA polymerase (15), at a multiplicity of infection of 10, and then transfected with pTM-VPR or relevant Vpr mutant constructs. Four hours after transfection, the cells were labeled for 16 h with 1.5 ml of Met/Cys-free DMEM containing 150 μCi of Tran[35S]label (ICN) or phosphate-free DMEM containing 0.5 mCi of 32Pi (ICN), 10% dialyzed FCS, and antibiotics. HEK293 cells were transfected with pcDNA-VPR and labeled with Tran[35S]label or 32Pi as described earlier. The cells were harvested and lysed into 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer (0.125 M Tris-HCl [pH 6.8], 20% glycerol, 10% [vol/vol] 2-mercaptoethanol, 4% [wt/vol] SDS); with 0.2 mM Na2VO4 added to 32Pi-labeled samples). After boiling for 10 min and a brief microcentrifugation, the sample was diluted 40-fold with radioimmunoprecipitation (RIPA) buffer without SDS (1% [vol/vol] Triton X-100, 0.5% [wt/vol] deoxycholate, and 0.2 mM phenylmethylsulfonyl fluoride in PBS, with 0.2 mM Na2VO4 added to 32Pi-labeled samples). Then 5 μl of anti-Vpr antiserum was added, and the mixture was incubated for 4 h or overnight at 4°C (32). Twenty microliters of immobilized protein A beads (rproteinA; Repligen Co., Boston, Mass.) was added, and the mixture was incubated for 60 min at 4°C with gentle rotation. Immunoprecipitates were collected at 500 × g for 30 s at room temperature and washed three times with RIPA buffer and once with PBS. All buffers contained 0.2 mM Na2VO4 for 32Pi-labeled samples. The beads were resuspended in 30 μl of SDS-PAGE sample buffer treated at 100°C for 10 min before loading onto a 15% polyacrylamide gel. The gels were dried on filter paper and exposed to a XAR-5 film (Kodak) at −80°C or exposed on a Bio-Rad phosphorimager. Due to high background from the 32P-labeled cells, secondary immunoprecipitation was carried out to detect the labeled protein from the cell lysates. After extensive washing of the beads, the antibody-Vpr complex was eluted with 360 μl of glycine buffer (50 mM glycine, 0.1% Triton X-100, 150 mM NaCl [pH 2.8]) and neutralized with 40 μl of 1 M Tris-HCl (pH 9.0). Then 20 μl of protein A beads in 800 μl of RIPA buffer was added, and the mixture was incubated at 4°C for another 30 to 60 min. The beads were washed twice with RIPA buffer and once with PBS before sample loading.
To label the HIV-1-infected cells and virions, 5 × 106 Jurkat cells were infected for 7 days with 500 ng of viruses derived from p120 and then labeled for 16 h with 1.5 ml of Met/Cys-free RPMI 1640 containing Tran[35S]label (50 μCi/ml) or phosphate-free RPMI 1640 containing 32Pi (0.5 mCi/ml), 10% dialyzed FCS, and antibiotics. The culture supernatants were removed and centrifuged at 2,500 rpm for 15 min in a Beckman GS-6 rotor to remove cellular debris. Particles were concentrated by sedimentation through a 20% sucrose cushion prepared in PBS at 25,000 rpm for 90 min at 4°C in an SW28.1 rotor. The pellets were resuspended with RIPA buffer (0.1% SDS, 1% [vol/vol] Triton X-100, 0.5% [wt/vol] deoxycholate, and 0.2 mM phenylmethylsulfonyl fluoride in PBS, with 0.2 mM Na2VO4 added to 32Pi-labeled samples) plus 5 μl of anti-Vpr polyclonal antibodies.
To examine phosphorylation of S79 in the Vpr position 78, 79, and 80 mutants, 293VE cells were transfected with different Vpr mutant plasmids and then selected with G418 sulfate (750 μg/ml; GIBCO BRL) for 5 days. Then 5 × 105 G418-resistant cells were plated into six-well plates, induced for Vpr expression for 24 h with 1 μM of muristerone A, and then transfected with p125-Vpr(−) proviral DNA. The cells were continued in culture for 24 h in the presence of the inducer and labeled with 32Pi or Tran[35S]label overnight. The supernatant containing HIV-1 virions was harvested to test for Vpr expression and phosphorylation as described earlier.
Phosphoamino acid analysis.
To determine the phosphorylated residue(s), the 32P-labeled Vpr was immunoprecipitated from pcDNA-VPR-transfected HEK293 cells, separated by SDS-PAGE, and transferred to an Immobilon-P membrane (Millipore). The Vpr bands were excised and hydrolyzed in 6 N HCl at 110°C for 2 h. The hydrolysate was dried and dissolved in 10 μl of H2O containing 0.5 μg each of phosphoserine, phosphothreonine, and phosphotyrosine and then spotted onto a cellulose plate to perform phosphoamino acid analysis (PAA) (2). The first-dimension electrophoresis was run in pH 1.9 buffer (0.58 M formic acid, 1.36 M acetic acid). The second-dimension electrophoresis was carried out in pH 3.5 buffer (0.87 M acetic acid, 0.5% pyridine, 0.5 mM EDTA). The three phosphoamino acid locations were shown by staining the plate with 0.25% ninhydrin, while the 32P-phosphoamino acids were shown by exposure of the plate to a phosphorimager screen (Bio-Rad).
Infection of human PBMCs and macrophages.
All viral stocks were prepared by transfecting 293T cells overnight using Lipofectamine (GIBCO BRL). The next day, fresh medium was added and cultured for 3 days. The supernatants were harvested and spun (1,000 × g) for 10 min to discard cell debris. Viral titers were determined by a p24-specific enzyme-linked immunosorbent assay (ELISA) kit (Coulter, Miami, Fla.) or MAGI5 infection and then aliquoted into a small volume and stored at −80°C for later analysis as described elsewhere (24).
Human peripheral blood mononuclear cells (PBMCs) from HIV-1-negative blood donors were isolated by Ficoll-Hypaque (Sigma, St. Louis, Mo.) gradient centrifugation and stimulated with phytohemagglutinin (PHA; 10 μg/ml; Difco, Detroit, Mich.) for 3 days. Activated PBMCs were maintained in RPMI 1640 medium (GIBCO-BRL) supplemented with 10% FCS, 4 mM l-glutamine penicillin (100 U/ml), streptomycin (100 μg/ml), and interleukin-2 (50 U/ml). Human monocyte-derived macrophages (MDMs) from healthy donors were cultured in Iscove's modified Dulbecco's medium supplemented with 10% human serum (Biocell, Rancho Dominguez, Calif.), 4 mM l-glutamine (BioWhittaker, Walkersville, Md.), antibiotics, and recombinant murine colony-stimulating factor (500 U/ml) (24).
PHA-stimulated PBMCs were seeded in 24-well plates at 105 cells/well on the day of infection. Viral supernatants normalized with MAGI5 titration (about 5 ng of p24 equivalent was added to each well and adsorbed for 4 h. The next day, viral inocula were removed and replaced with PBMC medium. Cultured supernatants were collected every 2 to 3 days, and the level of HIV-1 p24 was determined by the HIV-1 p24 ELISA kit. Monocytes were plated at a density of 2 × 106 to 3 × 106 cells/ml in RPMI 1640 medium containing 1% human AB serum (heat inactivated) for 1 h, then washed three times, and maintained in macrophage medium. The monocytes were allowed to differentiate in vitro for 7 to 14 days and fed with fresh medium every 3 days before infection. Infection of MDMs was carried out in the same way as infection of PBMCs.
RESULTS
HIV-1 Vpr is phosphorylated in vivo.
We hypothesized that the different Vpr functions (nuclear transport of the preintegration complex, G2 cell cycle arrest, transactivation, etc.) may be performed by differently modified versions of HIV-1 Vpr. Computer analysis of HIV-1 NL4-3 Vpr using the ScanProsite program (ExPASy molecular biology server) revealed a CKII phosphorylation domain, TYGD, at positions 49 to 52 of Vpr (the positions are according to HIV-1 NL4-3 Vpr) (Fig. 1). This suggested that threonine 49 (T49) may be phosphorylated in vivo. Further analysis indicated that the potential phosphorylation sites (47Y, 49T, 50Y, 53T, and 79S) are highly conserved among almost all of the HIV and simian immunodeficiency virus (SIV) isolates. 94S and 96S are found in HIV-1 isolates but not in HIV-2 and SIV Vpr. Other sites (15Y, 19T, 28S, and 84T) are much less conserved between different isolates (Fig. 1). It is interesting that the highly conserved residues are mostly located in the C-terminal half of Vpr while the relative poorly conserved sites are located in the N-terminal half of the protein.
FIG. 1.
Alignment of HIV-1, HIV-2, and SIV Vpr genes. The sequences are as downloaded from the online server site (hiv-web.lan1.gov), with modifications. All potential phosphorylated amino acid residues are underlined and printed in bold. The putative CKII domain, TYGD, is double underlined. LR indicates a leucine-rich region. HIV-1-A, -B, -C, -D, -F, -H, -O, and -U are the consensus sequences from each HIV-1 subtype. HIV-2-A and -B are the consensus sequences from HIV-2 subtypes. SIV-CPZ, SIV-SD, and SIV-STM represent the consensus sequence for SIV subtypes. NL4-3 is the Vpr sequence from HIV-1 NL4-3, and HIV-2(ROD) shows the Vpr sequence from HIV-2 ROD. −, sequence is the same as shown for HIV-1 NL4-3 or HIV-2 (ROD); ., a gap for that position; ?, no residue homologous to the consensus sequence.
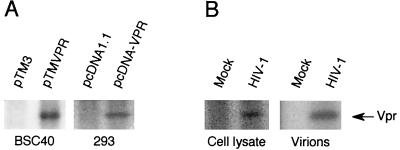
To test whether HIV-1 Vpr protein is phosphorylated, Vpr was expressed from pTM-VPR in BSC40 cells using the vaccinia virus expression system or pcDNA-VPR in HEK293 cells, and labeled with 32Pi (0.5 mCi/ml) for 6 h. The cells were harvested, lysed, and immunoprecipitated with anti-Vpr antibodies. Vpr was separated by SDS-PAGE, and the phosphorylated Vpr was detected by exposure of the dried gel to X-ray film. A 32P band was evident in the cells expressing Vpr but not in the negative controls (Fig. 2A). To confirm that Vpr is also phosphorylated in the HIV-1-infected cells, Jurkat cells were infected with HIV-1 p120 for 7 days (54). Both the infected and noninfected Jurkat cells were labeled with 32Pi, and the cellular and virion-incorporated forms of Vpr were analyzed. In both cases, a 32P-labeled Vpr band was detected in the HIV-1-infected Jurkat cells, indicating that Vpr is phosphorylated either in transiently expressed cells or in HIV-1-infected cells (Fig. 2B).
FIG. 2.
Phosphorylation of Vpr in BSC40, HEK293, and HIV-1-infected Jurkat cells. (A). HIV-1 Vpr was expressed from pTM-VPR in BSC40 cells by using the vaccinia virus expression system or pcDNA-VPR in HEK293 cells and labeled with 0.5 mCi of [32P]H3PO4 per ml for 6 h. The cells were harvested, lysed, and immunoprecipitated with anti-Vpr antibodies. Vpr was separated by SDS-PAGE, and the phosphorylated Vpr was detected by exposure of the dried gel to X-ray film. (B) Both HIV-1 p120-infected (7 days postinfection) and noninfected Jurkat cells were labeled with 0.5 mCi of [32P]H3PO4 per ml for 6 h. The cells were harvested, lysed, and immunoprecipitated with anti-Vpr antibodies. The supernatants were passed through 20% sucrose cushions and then immunoprecipitated with anti-Vpr antibodies. The labeled Vpr was detected by exposure of the dried gel to X-ray film and indicated by an arrow.
Serine residue(s) phosphorylation.
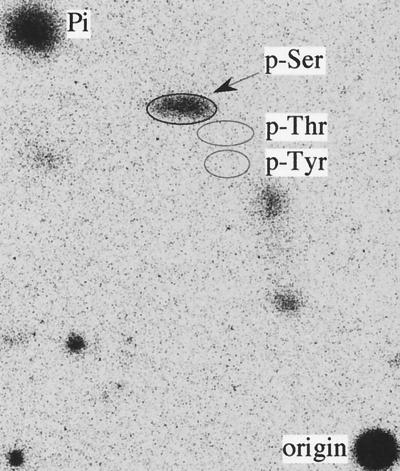
To determine the phosphorylated residue(s), the 32P-labeled Vpr was immunoprecipitated from pcDNA-VPR-transfected HEK293 cells, separated by SDS-PAGE, and transferred to an Immobilon-P membrane to perform PAA. Only [32P]phosphoserine was detected; no 32P signal was detected at the position of phosphotyrosine or phosphothreonine (Fig. 3). The same result was obtained from the Vpr expressed with the vaccinia virus expression system (data not shown). In both cases, no signal was detected in the three phosphoamino acid positions in the negative controls. Therefore, the serine residue(s) is phosphorylated in HIV-1 Vpr, while the putative CKII phosphorylation site, T49, is not modified.
FIG. 3.
PAA of Vpr. The 32P-labeled Vpr was immunoprecipitated from pcDNA-VPR-transfected HEK293 cells, separated by SDS-PAGE, and transferred to an Immobilon-P membrane. The Vpr band was excised and hydrolyzed. The hydrolysate was dried, dissolved in 10 μl of H2O containing 0.5 μg each of phosphoserine (p-Ser), phosphothreonine (p-Thr), and phosphotyrosine (p-Tyr), and then spotted onto a cellulose plate to perform PAA. The three phosphoamino acid locations are shown by ovals; the 32P-phosphoamino acids were revealed by exposure of the plate to a phosphorimager screen.
Mapping the phosphorylation site(s).
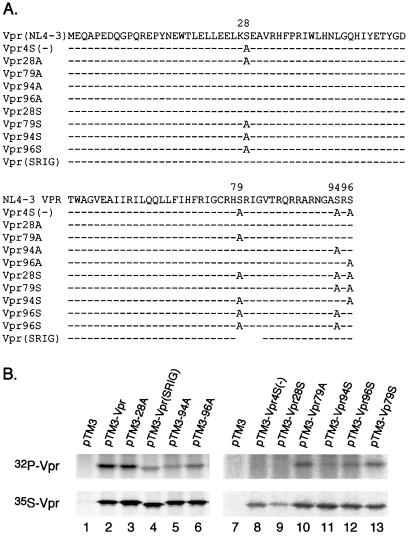
To map the phosphorylation site(s), all four serine residues at positions 28, 79, 94, and 96 in HIV-1 Vpr were mutated to alanines to construct Vpr4S(−) (Fig. 4A). Vpr4S(−) was expressed in BSC40 cells with the vaccinia virus expression system and labeled with Tran[35S]label or 32Pi. No Vpr phosphorylation was detected from 32Pi-labeled pTM-VPR4S(−)-transfected cells although Vpr was expressed well in the Tran[35S]label cells (Fig. 4B, lane 8). This result further confirmed that only serine residues are phosphorylated. However, none of the single mutants (Vpr28A, Vpr79A, Vpr94A, and Vpr96A) or the SRIG deletion mutant could completely abolish the phosphorylation, indicating that multiple sites are phosphorylated (Fig. 4B, lanes 3 to 6 and 10). To further define the phosphorylation sites, the triple serine mutants with only one serine remaining, Vpr28S, Vpr79S, Vpr94S, and Vpr96S, were expressed and labeled. Besides Vpr28S, the other three mutants could be phosphorylated, but the signal was lower than that of wild-type Vpr (Fig. 4B, lanes 9, 11, 12, and 13). The 35S-labeling results indicated that all of the mutant Vpr proteins could be expressed well. These results indicated that S79, S94, and S96 are phosphorylation sites but S28 is not.
FIG. 4.
Mapping the Vpr phosphorylation sites. (A) Schematic representations of HIV-1 NL4-3 Vpr and various mutations. Serines at positions 28, 79, 94, and 96 were mutated to alanines. Dashes indicate amino acid residues that are the same as in Vpr(NL4-3); Vpr(SRIG) is a SRIG deletion mutant. (B) Wild-type Vpr and relevant mutants were expressed in BSC40 cells using the vaccinia virus expression system. The cells were labeled with 35S or 32P overnight, and labeled Vpr was detected with anti-Vpr antibodies.
Phosphorylation and infection of nonreplicating cells.
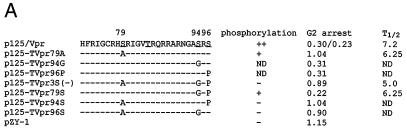
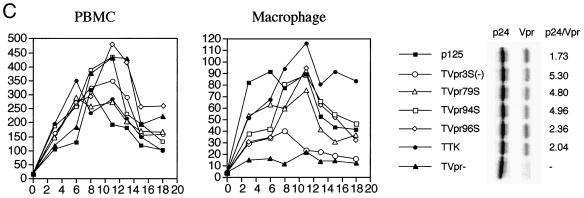
Since phosphorylated Vpr was detected in the virion, it could play a role in the infection of macrophages. To determine the role of phosphorylation of Vpr on infection of nonreplicating cells, positions S79, S94, and S96 were changed to A, G, and P, respectively, to avoid any amino acid changes in the overlapping tat gene (Fig. 5A). The mutants were cloned into a macrophagetropic proviral plasmid, p125-TTK, a chimeric virus with HIV-1 NL4-3 Vpr and ADA envelope V3 sequence with KKQ-to-TTK mutations at the putative MA NLS domain (22). Viruses stocks from HIV-1 p125 containing MA-NLS and Vpr mutations (Fig. 5B) were prepared and titered with MAGI5 cells (24). Equal numbers of blue plaque units of viruses were used to infect PBMCs or terminally differentiated MDMs. The infection was analyzed by the p24 antigen assay. All of the mutant viruses could infect PBMCs at similar levels (Fig. 5C). However, differences were observed in macrophage infection. The Vpr-ablated virus, p125-TVpr(−), exhibited a much lower level of infection than p125-TTK but was still detectable. The Vpr phosphorylation site mutant viruses p125-TVpr79S, -TVpr94S, and -TVpr96S infected macrophages similarly to wild-type viruses (Fig. 5C). However, the triple Vpr mutant virus, p125-TVpr3S(−), infected macrophages at a lower level, similar to that of p125-TVpr(−) virus (Fig. 5C).
FIG. 5.
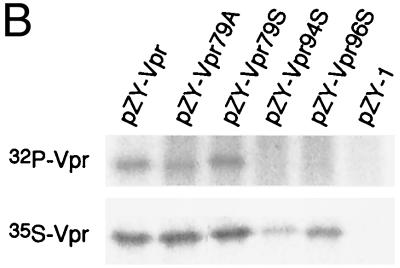
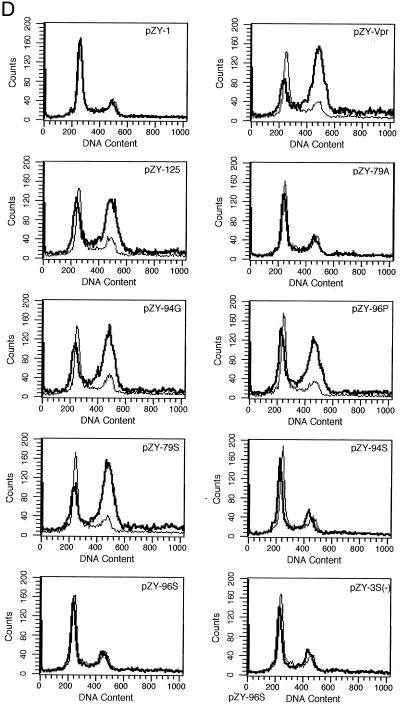
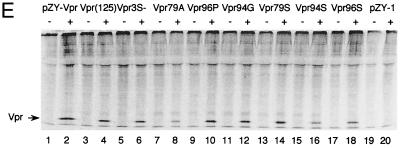
Phosphorylation and Vpr biological functions. (A) Mutant Vpr sequences in the p125-TTK proviral construct. Numbers above the p125 sequence show serine positions; each serine is underlined. +, Vpr is phosphorylated; −, Vpr is not phosphorylated; ND, not done. The data shown in the G2 arrest column represent the ratio of G1/(G2/M) with inducer to G1/(G2/M) without inducer. The data are the averages from two independent experiments. Vpr protein stability, shown in the T1/2 (half-life, in hours) column, is the average of two independent experiments. (B) Both 32P- and 35S-labeled forms of Vpr were detected as described in Materials and Methods and indicated by arrows on the left. (C) PBMC and macrophage infections by the mutant p125 viruses shown in panel A. The p24 volume is from the average of two different clones of the same mutant. The ratio of p24 to Vpr from the immunoprecipitation was determined by phosphorimager using the Multi-Analyst 1.0.1 program (Bio-Rad) and normalized to the amount of Met/Cys in p24 (11 of total Met/Cys) and Vpr (2 of total Met/Cys) by dividing the p24 counts by 6.5. The data represent the average of two independent experiments. (D) FACscan analysis profiles from one of the two independent experiments. The thick and thin lines represent the presence and absence of the inducer, muristerone A, respectively. The average G2 cell cycle arrest result is also shown in panel A. (E) Vpr and mutant proteins were detected by immunoprecipitation of the 35S-labeled cells in the absence (−) or presence (+) of muristerone A.
To determine the amount of Vpr packaged into virions, the proviral constructs were transfected into 293T cells and labeled with Tran[35S]label for 24 h in the presence of inducer. The supernatants were harvested and passed through a 20% sucrose cushion to pellet the virions. Virion-associated p24 and Vpr were immunoprecipitated and quantitated by a phosphorimager (Fig. 5C). p125-TTK and p125-TVpr96S packaged Vpr as well as p125, while p125-TVpr79S, -TVpr94S, and -TVpr3S(−) only packaged about on third as much Vpr as p125 or p125-TTK.
Position 79 phosphorylation is important for Vpr G2 cell cycle arrest.
To test whether phosphorylation is important for Vpr-mediated G2 cell cycle arrest, Vpr79A, -94G, -96P, -79S, -94S, -96S, and -3S(−) (Fig. 5A) were separately cloned into pZY-1 and then transfected into 293VE cells. After a 5-day selection with G418, the cells were split and continued in culture for 36 h in the presence or absence of the inducer, 1 μM muristerone A. Cell cycle arrest activity was analyzed by propidium iodide staining and FACScan analysis (59) (Fig. 5D). In the absence of muristerone A, no effect on the cell cycle was observed. However, when wild-type Vpr was expressed from one of two different wild-type constructs, pZY-Vpr or pZY-125 (pZY-125 has additional 170 bp downstream of Vpr from the HIV-1 provirus), the cells were arrested in the G2 phase (Fig. 5D). The three single mutants Vpr79A, -94G, and -96P had different effects on the cell cycle. Vpr79A was unable to arrest the cell cycle, but Vpr94G and Vpr96P could arrest cells as well as the wild-type Vpr. These results suggested that 79S is critical for the G2 cell cycle arrest (Fig. 5D). Vpr79S could arrest the cell cycle as well as wild-type Vpr, but neither Vpr94S nor Vpr96S could arrest the cell cycle, further suggesting that 79S, but not 94S and 96S, is critical for cell cycle arrest. As expected, Vpr3S(−), with mutations at all three phosphorylation sites, had no significant effect on the cell cycle.
To exclude the possibility that the differences in protein expression levels could account for these findings, the transfected cells were labeled with Tran[35S]label and Vpr proteins were detected by immunoprecipitation. In the absence of inducer, we detected no Vpr protein; in the presence of the inducer, similar levels of Vpr proteins were detected (Fig. 5E). To further confirm that differences in protein stability do not account for the effects on G2 cell cycle arrest of pZY-79A, -79S, and -3S(−), the resistant cells were labeled overnight with Tran[35S]label and chased in the absence of radioisotope. The cells were harvested at different time points, and immunoprecipitations were carried out. The proportion of Vpr protein or mutant Vpr protein at different points was determined by a phosphorimager (Fig. 5A). Wild-type Vpr, 79A, and 79S were similarly stable, while Vpr3S(−) was slightly less stable.
To confirm that Vpr could be phosphorylated in the same pattern in 293VE as in BSC40 cells, 32Pi labeling was also performed. The single mutation Vpr79A could be phosphorylated, indicating that 94S and/or 96S are phosphorylated (Fig. 5B, lane 2). Furthermore Vpr79S could be phosphorylated, confirming that 79S is a phosphorylation site. However, no phosphorylation signal could be detected from the double mutations Vpr94S and Vpr96S. Since, Vpr79A could be phosphorylated, it is possible that the 94G or 96P mutation disrupted the phosphorylation motifs at the C terminus of Vpr.
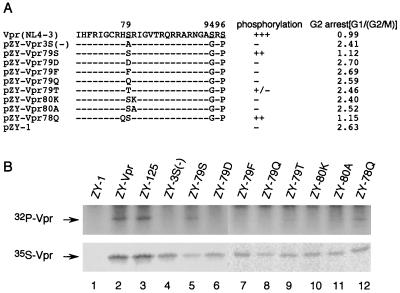
Since our results indicated that 79S is critical for the Vpr G2 cell cycle arrest function, it is possible that either the serine residue is required at this position or phosphorylation is important for the Vpr cell cycle arrest. To study the correlation between position 79 phosphorylation and Vpr G2 cell cycle arrest function, additional Vpr mutants at positions 78, 79, and 80 were generated and cloned into pZY-1 (Fig. 6A). Since phosphorylation of serines at positions 94 and 96 has no effect on Vpr G2 cell cycle arrest, to simplify the analysis, positions 94S and 96S were mutated to G and P, respectively for all mutants. The plasmids were transfected into 293VE cells, and the resistant cells were tested for G2 cell arrest as described earlier. As shown earlier, Vpr and Vpr79S could arrest cells in the G2/M phase (Fig. 6A). All mutants at positions 79 or 80 disrupted the ability of Vpr to arrest the cell cycle in the G2 phase. However, the Vpr78Q mutation still had full G2 cell cycle arrest activity (Fig. 6A). To analyze whether or not serine 79 was phosphorylated in these Vpr mutants, the stable transfected cells were transfected with p125-Vpr(−) constructs in the presence of the inducer to test virion incorporation of 32P-labeled Vpr (Fig. 6B). Mutants pZY-Vpr79S and pZY-Vpr78Q exhibited a phosphorylation signal that was strong but weaker than that of wild-type Vpr (Fig. 6B, lanes 5 and 12). Mutant 79T exhibited much less phosphorylation (lane 9), while all other mutants were not phosphorylated. These results further indicated that there is a good correlation between G2 cell cycle arrest and phosphorylation.
FIG. 6.
Mutational analysis of the role of residues 78, 79, and 80 on position 79 phosphorylation and G2 cell cycle arrest. (A) Schematic diagram of different Vpr mutants at positions 78, 79, and 80. +, Vpr is phosphorylated; −, Vpr is not phosphorylated. Average ratios of G1 to G2/M are listed under the G2 arrest column. The data are from an average of two individual constructs of the same mutants. (B) Both 32P- and 35S-labeled forms of Vpr were detected as described in Materials and Methods and are indicated with arrows.
DISCUSSION
In this study, we have shown that Vpr is a phosphorylated protein. The mutations pTM-VPR4S(−) and pZY-Vpr3S(−) totally abolished Vpr phosphorylation, suggesting that only the serine residues are phosphorylated. Since we did not detect any threonine phosphorylation, the predicted CKII domains (TYGD) in HIV-1 Vpr may not be phosphorylated. However, we cannot rule out the possibility that CKII may phosphorylate Vpr only under some circumstances. In this study, we did not detect tyrosine phosphorylation, and no tyrosine phosphorylation domain has been predicted in HIV-1 Vpr.
The triple mutant pTM-Vpr79S and the double mutant pZY-Vpr79S are phosphorylated, indicating that 79S is one of several phosphorylation sites. The finding that both pTM-VPR79A and pZY-VPR79A could be phosphorylated implied that at least one site at positions 94S or 96S was phosphorylated, although neither pZY-VPr94S or pZY-Vpr96S could be phosphorylated. However, pTM-Vpr94S and pTM-Vpr96S were phosphorylated, suggesting that both 94S and 96S in the HIV-1 Vpr are phosphorylated. These differing observations could be due to the fact that in the pTM-constructs, both sites were substituted with alanines, while in the pZY- constructs, the sites were changed to glycine and proline, respectively. Since S94 and S96 are separated by only a single residue, different amino acid residues at position 94 could affect 96S phosphorylation or vice versa. It is unlikely that a new serine phosphorylation site was created by mutation of its adjacent site to alanine. Therefore, our results indicate that serines at positions 79, 94, and 96 in the HIV-1 Vpr are phosphorylated.
Phosphorylation may change the protein charge and result in a difference in migration on SDS-PAGE in some but not all cases (13). We did not observe any migration differences in 35S- and 32P-labeled Vpr proteins (data not shown). It is unlikely that Vpr is fully phosphorylated, because Vpr4S(−) and Vpr3S(−) had the same electrophoretic mobilities as the other Vpr mutants as well as wild-type Vpr in the 35S-labeled analysis.
Phosphorylation is one the most important posttranslational modifications for proteins. It plays an important role in protein stability, protein-protein association, protein localization, and transcriptional regulation (26). Kewalramani et al. have shown that HIV-2 Vpr protein has less effect on cell cycle arrest than HIV-1 Vpr due to a lower level of stability (29). Sequence analysis shows that the C-terminal 20 amino acid residues represent the most diverse domains comparing HIV-1 and HIV-2 Vpr, and both 94S and 96S absent from HIV-2 Vpr (Fig. 1). Moreover, truncation of Vpr at position 76 or deletion of the last three amino acids, residues 94 to 96, result in unstable proteins, suggesting that the C terminus of Vpr is important for protein stability (39). However, no difference in protein stability was observed between Vpr79A and Vpr79S, suggesting that the 94G/96P replacement may not cause Vpr instability. Since both Vpr79A and Vpr79S are slightly less stable than wild-type Vpr, the cumulative effect resulted in Vpr3S(−) being somewhat more unstable than any of the other mutants. However, it is still unclear whether Vpr phosphorylation alone regulates HIV-1 Vpr turnover.
Vpr is located in the nucleus and perinuclear membrane (22, 32, 37, 38, 57). Both the N- and C-terminal domains of HIV-1 Vpr are important for nuclear localization (27). The basic domain in the C terminus of Vpr has a putative traditional NLS (31). Furthermore, the C terminus of Vpr has a similar structure as a phosphorylation-regulated protein, the CcN motif (26). Upstream of the putative NLS is 79S, and downstream is 94/96S. Phosphorylation of 79S, 94S, and 96S may regulate this putative NLS function and further control HIV-1 Vpr localization. This may account for the impaired replication of p125-TVpr3S(−) in macrophages. However, protein instability and reduction of Vpr3S(−) packaged into virions may also contribute to the impaired replication of p125TVpr3S(−) in macrophages.
Phosphorylation regulates the cell cycle progression. Vpr G2 cell cycle arrest function appears to be regulated through phosphorylation of S79. This is in agreement with previous studies that showed that the C terminus of HIV-1 Vpr is important for G2 cell cycle arrest (11, 37, 59). Furthermore, the H(S/F)RIG domains has been identified to be important for G2 cell cycle arrest (3, 34, 35). Deletion of the SRIG (79 to 82) sequence or mutation of all arginines in the C-terminal portion of Vpr impaired the Vpr G2 arrest activity (59). S79I and R80A mutations were also defective in G2 cell cycle arrest activity (14). In addition, the HSRIG sequence is highly conserved in all HIV-1, HIV-2, and SIV isolates (Fig. 1).
The loss of Vpr G2 cell cycle arrest in our mutants could be due to mutation of the phosphorylation site, S79, or effects on phosphorylation by mutations in the adjacent sequences. Both Vpr80A and Vpr80K mutations impair G2 cell cycle arrest and phosphorylation, indicating that position 80 belongs to the 79S phosphorylation motif. It is also suggested that the phosphorylation motif is very restricted since similar amino acid residues at position 80, lysine or alanine substitutions abolished phosphorylation and G2 cell cycle arrest activities. The glutamine (Q)-to-histamine (H) substitution at position 78 did not affect Vpr function, suggesting that position 78 is less restrictive on the G2 arrest domain than 79S and 80R. It is interesting that the threonine substitution Vpr79T exhibits only a very low level of phosphorylation, although both serine and threonine may be phosphorylated by the same serine/threonine kinase. Aspartate (D) is structurally similar to phosphoserine and has been used to replace serines in the phosphorylated proteins to mimic phosphoserine function (12). However, Vpr79D could not arrest cells in the G2 phase.
In both yeast and mammalian cells, the HFRIGHSRIG motif in Vpr is critical for G2 cell cycle arrest (3, 34, 35). In the yeast model, Vpr79A arrested the cell cycle in the G2 phase like the wild-type Vpr (7). However, our results indicated that Vpr79A abolished the G2 cell cycle arrest function in mammalian cells. Several cellular factors have been identified to interact with HIV-1 Vpr, such as Vpr-interacting protein, uracil DNA glycosylase, human Vpr-interacting protein, and the human homologue of the yeast repair protein RAD23 (4, 19, 36, 47, 55, 58). These factors may be important for Vpr G2 cell cycle arrest or other Vpr functions. Studies of the interaction of these proteins in mammalian cells with the phosphorylated Vpr may provide insights into their role in cell cycle regulation. It is also not known whether one of these factors has the kinase activity for Vpr. The identification of the Vpr kinase will be important for defining the mechanisms of Vpr arrest of cells in the G2 phase.
ACKNOWLEDGMENTS
We thank Robert Horton, Nancy Vander Heyden, and Mark Dalton for critical technique support.
This work is supported by PHS grants, and Yi Zhou is supported by NIH training grant CA09547-12.
REFERENCES
- 1.Agostini I, Navarro J M, Rey F, Bouhamdan M, Spire B, Vigne R, Sire J. The human immunodeficiency virus type 1 Vpr transactivator: cooperation with promoter-bound activator domains and binding to TFIIB. J Mol Biol. 1996;261:599–606. doi: 10.1006/jmbi.1996.0485. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1989. [Google Scholar]
- 3.Berglez J M, Castelli L A, Sankovich S A, Smith S C, Curtain C C, Macreadie I G. Residues within the HFRIGC sequence of HIV-1 vpr involved in growth arrest activities. Biochem Biophys Res Commun. 1999;264:287–290. doi: 10.1006/bbrc.1999.1511. [DOI] [PubMed] [Google Scholar]
- 4.Bouhamdan M, Benichou S, Rey F, Navarro J M, Agostini I, Spire B, Camonis J, Slupphaug G, Vigne R, Benarous R, Sire J. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camaur D, Gallay P, Swingler S, Trono D. Human immunodeficiency virus matrix tyrosine phosphorylation: characterization of the kinase and its substrate requirements. J Virol. 1997;71:6834–6841. doi: 10.1128/jvi.71.9.6834-6841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M, Elder R T, Yu M, O'Gorman M G, Selig L, Benarous R, Yamamoto A, Zhao Y. Mutational analysis of Vpr-induced G2 arrest, nuclear localization, and cell death in fission yeast. J Virol. 1999;73:3236–3245. doi: 10.1128/jvi.73.4.3236-3245.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen E A, Dehni G, Sodroski J G, Haseltine W A. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J Virol. 1990;64:3097–3099. doi: 10.1128/jvi.64.6.3097-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen B R, Green W C. Functions of the auxiliary gene products of the human immunodeficiency virus type-1. Virology. 1990;178:1–5. doi: 10.1016/0042-6822(90)90373-y. [DOI] [PubMed] [Google Scholar]
- 10.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 11.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N R. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumaz N, Meek D W. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002–7010. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eissenberg J C, Ge Y W, Hartnett T. Increased phosphorylation of HP1, a heterochromatin-associated protein of Drosophila, is correlated with heterochromatin assembly. J Biol Chem. 1994;269:21315–21321. [PubMed] [Google Scholar]
- 14.Felzien L K, Woffendin C, Hottiger M O, Subbramanian R A, Cohen E A, Nabel G J. HIV transcriptional activation by the accessory protein, VPR, is mediated by the p300 co-activator. Proc Natl Acad Sci USA. 1998;95:5281–5286. doi: 10.1073/pnas.95.9.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 18.Goh W C, Rogel M E, Kinsey C M, Michael S F, Fultz P N, Nowak M A, Hahn B H, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 19.Gragerov A, Kino T, Ilyina-Gragerova G, Chrousos G P, Pavlakis G N. HHR23A, the human homologue of the yeast repair protein RAD23, interacts specifically with Vpr protein and prevents cell cycle arrest but not the transcriptional effects of Vpr. Virology. 1998;245:323–330. doi: 10.1006/viro.1998.9138. [DOI] [PubMed] [Google Scholar]
- 20.Graves J D, Krebs E G. Protein phosphorylation and signal transduction. Pharmacol Ther. 1999;82:111–121. doi: 10.1016/s0163-7258(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 21.Gummuluru S, Emerman M. Cell cycle- and Vpr-mediated regulation of human immunodeficiency virus type 1 expression in primary and transformed T-cell lines. J Virol. 1999;73:5422–5430. doi: 10.1128/jvi.73.7.5422-5430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hrimech M, Yao X J, Bachand F, Rougeau N, Cohen E A. Human immunodeficiency virus type 1 (HIV-1) Vpr functions as an immediate-early protein during HIV-1 infection. J Virol. 1999;73:4101–4109. doi: 10.1128/jvi.73.5.4101-4109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hung C S, Pontow S, Ratner L. Relationship between productive HIV-1 infection of macrophages and CCR5 utilization. Virology. 1999;264:278–288. doi: 10.1006/viro.1999.0013. [DOI] [PubMed] [Google Scholar]
- 25.Jacque J M, Mann A, Enslen H, Sharova N, Brichacek B, Davis R J, Stevenson M. Modulation of HIV-1 infectivity by MAPK, a virion-associated kinase. EMBO J. 1998;17:2607–2618. doi: 10.1093/emboj/17.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jans D A, Hubner S. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol Rev. 1996;76:651–685. doi: 10.1152/physrev.1996.76.3.651. [DOI] [PubMed] [Google Scholar]
- 27.Jenkins Y, McEntee M, Weis K, Greene W C. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J Cell Biol. 1998;143:875–885. doi: 10.1083/jcb.143.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jowett J B, Xie Y M, Chen I S. The presence of human immunodeficiency virus type 1 Vpr correlates with a decrease in the frequency of mutations in a plasmid shuttle vector. J Virol. 1999;73:7132–7137. doi: 10.1128/jvi.73.9.7132-7137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kewalramani V N, Park C S, Gallombardo P A, Emerman M. Protein stability influences human immunodeficiency virus type 2 Vpr virion incorporation and cell cycle effect. Virology. 1996;218:326–334. doi: 10.1006/viro.1996.0201. [DOI] [PubMed] [Google Scholar]
- 30.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 31.Lu Y L, Bennett R P, Wills J W, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo T, Downing J R, Garcia J V. Induction of phosphorylation of human immunodeficiency virus type 1 Nef and enhancement of CD4 downregulation by phorbol myristate acetate. J Virol. 1997;71:2535–2539. doi: 10.1128/jvi.71.3.2535-2539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macreadie I G, Arunagiri C K, Hewish D R, White J F, Azad A A. Extracellular addition of a domain of HIV-1 Vpr containing the amino acid sequence motif H(S/F)RIG causes cell membrane permeabilization and death. Mol Microbiol. 1996;19:1185–1192. doi: 10.1111/j.1365-2958.1996.tb02464.x. [DOI] [PubMed] [Google Scholar]
- 35.Macreadie I G, Castelli L A, Hewish D R, Kirkpatrick A, Ward A C, Azad A A. A domain of human immunodeficiency virus type 1 Vpr containing repeated H(S/F)RIG amino acid motifs causes cell growth arrest and structural defects. Proc Natl Acad Sci USA. 1995;92:2770–2774. doi: 10.1073/pnas.92.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Kao G D, Muschel R J, Weiner D B. HIV-1 Vpr interacts with a human 34-kDa mov34 homologue, a cellular factor linked to the G2/M phase transition of the mammalian cell cycle. Proc Natl Acad Sci USA. 1998;95:3419–3424. doi: 10.1073/pnas.95.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Weiner D B. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahalingam S, Collman R G, Patel M, Monken C E, Srinivasan A. Functional analysis of HIV-1 Vpr: identification of determinants essential for subcellular localization. Virology. 1995;212:331–339. doi: 10.1006/viro.1995.1490. [DOI] [PubMed] [Google Scholar]
- 39.Mahalingam S, Collman R G, Patel M, Monken C E, Srinivasan A. Role of the conserved dipeptide Gly75 and Cys76 on HIV-1 Vpr function. Virology. 1995;210:495–500. doi: 10.1006/viro.1995.1368. [DOI] [PubMed] [Google Scholar]
- 40.Mansky L M. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology. 1996;222:391–400. doi: 10.1006/viro.1996.0436. [DOI] [PubMed] [Google Scholar]
- 41.Paul M, Jabbar M A. Phosphorylation of both phosphoacceptor sites in the HIV-1 Vpu cytoplasmic domain is essential for Vpu-mediated ER degradation of CD4. Virology. 1997;232:207–216. doi: 10.1006/viro.1997.8541. [DOI] [PubMed] [Google Scholar]
- 42.Peterson R T, Schreiber S L. Kinase phosphorylation: keeping it all in the family. Curr Biol. 1999;9:R521–R524. doi: 10.1016/s0960-9822(99)80326-1. [DOI] [PubMed] [Google Scholar]
- 43.Piller S C, Ewart G D, Premkumar A, Cox G B, Gage P W. Vpr protein of human immunodeficiency virus type 1 forms cation-selective channels in planar lipid bilayers. Proc Natl Acad Sci USA. 1996;93:111–115. doi: 10.1073/pnas.93.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poon B, Grovit-Ferbas K, Stewart S A, Chen I S Y. Cell cycle arrest by Vpr in HIV-1 virions and insensitivity to antiretroviral agents. Science. 1998;281:266–269. doi: 10.1126/science.281.5374.266. [DOI] [PubMed] [Google Scholar]
- 45.Popov S, Rexach M, Zybarth G, Reiling N, Lee M A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Re F, Braaten D, Franke E K, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selig L, Benichou S, Rogel M E, Wu L I, Vodicka M A, Sire J, Benarous R, Emerman M. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J Virol. 1997;71:4842–4846. doi: 10.1128/jvi.71.6.4842-4846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shackney S E, Shankey T V. Cell cycle models for molecular biology and molecular oncology: exploring new dimensions. Cytometry. 1999;35:97–116. doi: 10.1002/(sici)1097-0320(19990201)35:2<97::aid-cyto1>3.3.co;2-x. [DOI] [PubMed] [Google Scholar]
- 49.Shimura M, Onozuka Y, Yamaguchi T, Hatake K, Takaku F, Ishizaka Y. Micronuclei formation with chromosome breaks and gene amplification caused by Vpr, an accessory gene of human immunodeficiency virus. Cancer Res. 1999;59:2259–2264. [PubMed] [Google Scholar]
- 50.Stivahtis G L, Soares M A, Vodicka M A, Hahn B H, Emerman M. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J Virol. 1997;71:4331–4338. doi: 10.1128/jvi.71.6.4331-4338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subbramanian R A, Cohen E A. Molecular biology of the human immunodeficiency virus accessory proteins. J Virol. 1994;68:6831–6835. doi: 10.1128/jvi.68.11.6831-6835.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subbramanian R A, Kessous-Elbaz A, Lodge R, Forget J, Yao X J, Bergeron D, Cohen E A. Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J Exp Med. 1998;187:1103–1111. doi: 10.1084/jem.187.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J J, Lu Y, Ratner L. Particle assembly and Vpr expression in human immunodeficiency virus type 1-infected cells demonstrated by immunoelectron microscopy. J Gen Virol. 1994;75:2607–2614. doi: 10.1099/0022-1317-75-10-2607. [DOI] [PubMed] [Google Scholar]
- 54.Westervelt P, Gendelman H E, Ratner L. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc Natl Acad Sci USA. 1991;88:3097–3101. doi: 10.1073/pnas.88.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Withers-Ward E S, Jowett J B, Stewart S A, Xie Y M, Garfinkel A, Shibagaki Y, Chow S A, Shah N, Hanaoka F, Sawitz D G, Armstrong R W, Souza L M, Chen I S. Human immunodeficiency virus type 1 Vpr interacts with HHR23A, a cellular protein implicated in nucleotide excision DNA repair. J Virol. 1997;71:9732–9742. doi: 10.1128/jvi.71.12.9732-9742.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong-Staal F, Chanda P K, Ghrayeb J. Human immunodeficiency virus type III. The eighth gene. AIDS Res Hum Retroviruses. 1987;3:33–39. doi: 10.1089/aid.1987.3.33. [DOI] [PubMed] [Google Scholar]
- 57.Yao X J, Subbramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao L J, Mukherjee S, Narayan O. Biochemical mechanism of HIV-1 Vpr function. Specific interaction with a cellular protein. J Biol Chem. 1994;269:15577–15582. [PubMed] [Google Scholar]
- 59.Zhou Y, Lu Y, Ratner L. Arginine residues in the C-terminus of HIV-1 Vpr are important for nuclear localization and cell cycle arrest. Virology. 1998;242:414–424. doi: 10.1006/viro.1998.9028. [DOI] [PubMed] [Google Scholar]