Abstract
Background
The left ventricular outflow tract (LVOT) and ascending aorta are spatially complex, with distinct pathologies and embryologic origins. Prior work examined the genetics of thoracic aortic diameter in a single plane.
Objectives
We sought to elucidate the genetic basis for the diameter of the LVOT, aortic root, and ascending aorta.
Methods
Using deep learning, we analyzed 2.3 million cardiac magnetic resonance images from 43,317 UK Biobank participants. We computed the diameters of the LVOT, the aortic root, and at six locations of ascending aorta. For each diameter, we conducted a genome-wide association study and generated a polygenic score. Finally, we investigated associations between these scores and disease incidence.
Results
79 loci were significantly associated with at least one diameter. Of these, 35 were novel, and a majority were associated with one or two diameters. A polygenic score of aortic diameter approximately 13mm from the sinotubular junction most strongly predicted thoracic aortic aneurysm (n=427,016; mean HR=1.42/SD; CI=1.34–1.50; P=6.67×10−21). A polygenic score predicting a smaller aortic root was predictive of aortic stenosis (n=426,502; mean HR=1.08/SD; CI=1.03–1.12; P=5×10−6).
Conclusions
We detected distinct genetic loci underpinning the diameters of the LVOT, aortic root, and at several segments of ascending aorta. We spatially defined a region of aorta whose genetics may be most relevant to predicting thoracic aortic aneurysm. We further described a genetic signature that may predispose to aortic stenosis. Understanding genetic contributions to proximal aortic diameter may enable identification of individuals at risk for aortic disease and facilitate prioritization of therapeutic targets.
Keywords: Aorta, left ventricular outflow tract, machine learning, cardiovascular disease, genetics
Condensed abstract
The left ventricular outflow tract (LVOT) and ascending aorta are spatially complex. We used deep learning to analyze magnetic resonance images from 43,317 individuals. We then computed eight sequential diameters spanning the LVOT, aortic root, and ascending aorta. For each diameter, we conducted genetic studies. 79 loci were associated with at least one diameter; 35 of these were novel. We spatially defined a region whose genetics may be most relevant to predicting thoracic aortic aneurysm. We further described a genetic signature that may predispose to aortic stenosis. This work may help identify individuals at risk for aortic disease.
The ascending thoracic aorta is a developmentally complex organ arising from the septation of the truncus arteriosus and the bulbus cordis in the later stages of cardiogenesis(1). Contributions from two separate germ layers are required for proper aortic morphogenesis, and abnormalities of cardiac ventricular formation or semilunar valve development can influence ascending aortic size and morphology. The phenotypic range in aortic size is wide, from pulmonary stenotic disorders such as tetralogy of Fallot associated with large aortas to congenital aortic valve stenosis and hypoplastic left heart syndrome which can result in an ascending aorta no larger than necessary to transport coronary blood flow(2, 3). Beyond developmental influences, deficits in aortic homeostasis occur over the lifespan under pathogenic processes such as atherosclerosis or hypertension and can result in aortic growth and aneurysm. Ascending thoracic aneurysm develops asymptomatically, but is associated with a risk of aortic dissection, an important cause of sudden cardiac death. Approximately 50% of patients with a type A dissection of the ascending aorta die prior to arrival at a hospital(4). Therefore, understanding the epidemiological and genetic contributions to ascending aortic risk may be important to the development of preventative strategies to avoid sudden cardiac death(5). Clinical studies have noted that aneurysms of the ascending aorta occur in younger patients than descending thoracic or abdominal aortic aneurysms, often associated with pathogenic genetic predisposition(6–8). Genetic variants in several genes have been associated with ascending aortic aneurysms, including highly penetrant Mendelian loci identified in family studies and common variants identified via GWAS(9–12). Prior studies have also described the clinical actionability of genes previously associated with thoracic aortic disease(5). However, the majority of patients with thoracic aortic aneurysm do not carry a variant known (or felt likely) to be pathogenic for the disease.
In prior work, we used deep learning to evaluate the dimensions of the thoracic aorta in 4.6 million cardiac magnetic resonance (CMR) images from the UK Biobank(13). Using short axis images we conducted GWAS in up to 39,688 individuals and identified 82 loci associated with ascending thoracic aortic diameter and 47 loci with descending thoracic aortic diameter.
These results contributed to an understanding of the genetic basis of the diameter of the thoracic aorta. However, the short axis view used in our prior work limited measurement of ascending and descending aorta diameters to a single location. As the proximal aorta is known to be spatially complex, consisting of unique anatomical subregions with distinct embryologic origins, we sought to study the structure in greater detail(14–17). Therefore we undertook a fine-grained evaluation of ascending aortic dimensions using a deep learning architecture to better understand genetic risk and association with ascending aortic diseases.
Methods
Study design
Analysis was approved by the Mass General Brigham institutional review board (2003P001563). The UK Biobank is a population-based cohort of 500,000 individuals aged 40–69(18), to which access was provided under application 7089.
Pathogenic ascending aortic morphology is variable but occurs in reproducible patterns including enlargement of the aortic root, ascending aorta, or combinations of both. Aortic valvular malformations can be associated with smaller or larger aortic diameters. We hypothesized that diameters of the contiguous tract spanning approximately from the aortic annulus to the aortic arch are complex traits with contributions from common genetic variants. Because this segment is composed of structures known to have distinct biological origins and clinical risk factors for disease, we chose to quantify LVOT, aortic root, and ascending aorta separately. We further hypothesized that within the ascending aorta subsegment, serial calibers spanning approximately from the aortic root to the arch would contain distinct genetic underpinnings with varying degrees of correlation to disease. Therefore, we chose to measure up to six diameters of aorta per participant, depending on the length of aorta captured in the image (see Supplemental Methods).
Semantic segmentation with deep learning
First, 250 CMR still-frame images capturing the LVOT, aortic root, and ascending aorta in long axis were randomly selected from the UK Biobank. The images were then manually annotated (MN) under the supervision of a cardiologist (JPP). This annotation is known as semantic segmentation: the task of identifying and labeling all pixels in an image.
We used these annotations to train a deep learning model to perform the same semantic segmentation task using a U-Net architecture(19, 20). As a form of transfer learning, this model’s encoder had been pre-trained on ImageNet: a natural-image classification dataset(21). Therefore, instead of starting with random weights, the model was initialized with weights that are helpful for processing images, reducing the amount of manual annotation and model training necessary to achieve good model performance(19, 22). Using the LVOT as a benchmark, the model achieved 99.8% pixel segmentation accuracy (Jaccard 84.3%) in a held-out validation set comprising 20% of the annotations (See Supplemental Methods).
Phenotype extraction
Having identified which pixels of the given images represented structures of interest, we then sought to extract diameters of the LVOT, aortic root, and serial diameters of the ascending aorta. Using classical image processing algorithms, we first computed the centerline of the contiguous structure comprising the left ventricle, LVOT, aortic root, and ascending aorta(23, 24) We then drew lines orthogonal to the midline at LVOT, aortic root, and up to six points of the aorta. Serial measurements of the aorta were spaced relative to each participant’s height, approximately 1cm apart for a participant of average height. Finally, we took note of the intersection points of these orthogonal lines and the boundaries of their respective structures of interest. Each normal yielded two intersection points; the Euclidean distance between these two points was taken to represent a diameter of interest. For brevity, we refer to these eight extracted diameters as follows, in order of most proximal to most distal: LVOT, Aortic root, and Aorta 0 through 5. Measurements of aorta were serially defined in order of distance from the sinotubular junction. For most participants, Aorta 0–2 represent proximal aorta; Aorta 3–4 represent mid-ascending aorta, and Aorta 5 represents the proximal aortic arch. (See Central Illustration, Supplemental Table 5, Supplemental Methods for illustration of diameter locations). Additional aortic measurements were precluded by the fact that more distal aorta was not reliably captured in-frame. We excluded biologically implausible measurements and out-of-plane images, and we selected images taken near end-diastole.
Central Illustration: Overview of study steps.
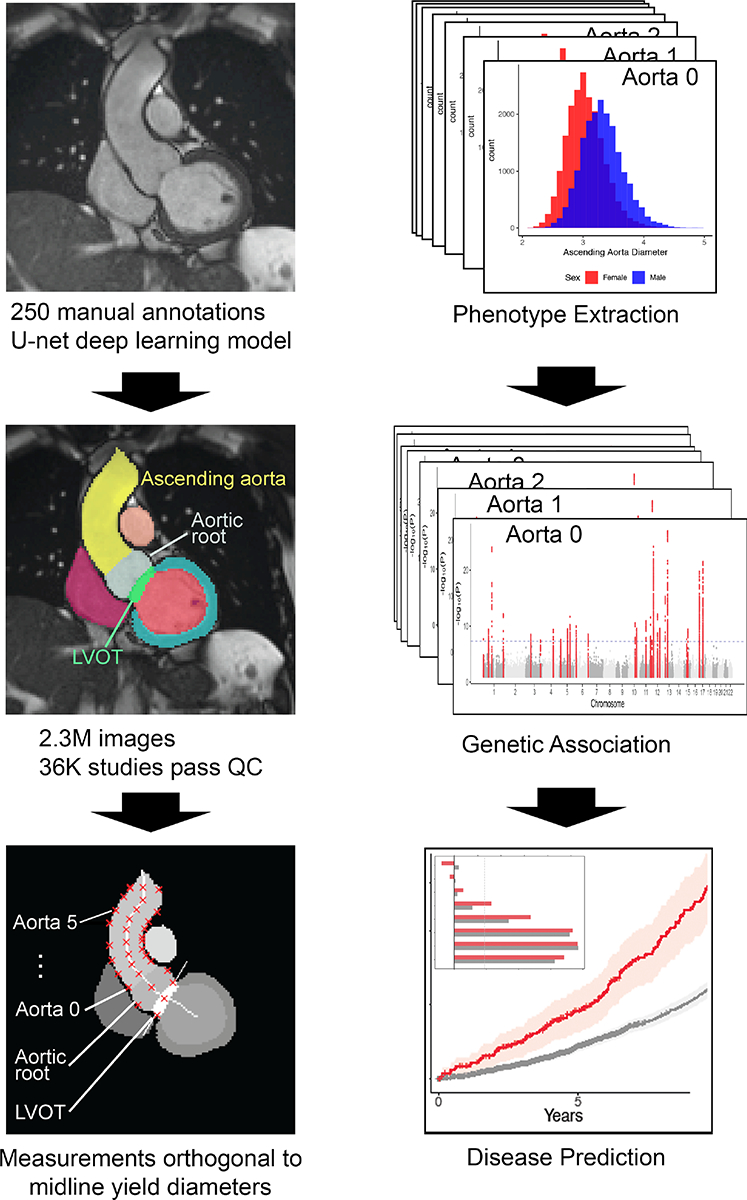
Flow diagram of major study steps, visualized by representative examples. Left column, from top: CMR image; deep learning output; output for the diameter-extracting algorithm. Right column, from top: phenotype histograms; output of multiple GWAS; disease prediction models (overlaid: prediction strength comparison).
After quality control, phenotypes were available for up to 33,870 participants. Because the length of aorta captured in the MRI frame is variable, fewer phenotypes were available for more distal measurements of the ascending aorta (Supplemental Table 3). We inspected the phenotypes’ distributions and found them biologically plausible (Supplemental Figure 2).
We sought to validate our diameters by investigating their correlations to prevalent disease (Supplemental Table 6). Only 12 participants had both phenotypic data available and diagnoses corresponding to thoracic aortic aneurysm (99 for aortic stenosis). Nonetheless, all scaled diameters were positively correlated with presence of thoracic aortic aneurysm, and negatively correlated with the presence of aortic stenosis, in univariate models (see Supplemental Methods).
Genotyping, genome-wide association studies, and rare variant analysis
Genotype information was imputed into the Haplotype Reference Consortium panel and the UK10K+1000 Genomes panel(25). We calculated heritability using BOLT-REML and genome-wide association using REGENIE, in both cases using as covariates age, the first five principal components of ancestry, the genotyping array, and the MRI scanner’s identifier(26, 27). The GWAS assessed 11.2 million genotyped and imputed SNPs with minor allele frequency (MAF)>0.005 (Supplemental Table 4). We used a commonly-used genome-wide significance threshold (p<5×10−8). Lead SNPs were tested for deviation from Hardy-Weinberg equilibrium at p<1×10−6. We considered SNPs novel if they were both 500kb away from and not in linkage disequilibrium with a previously described association. Rare variant contributions were analyzed using over 200,000 UK Biobank exomes to detect and assess the impact of loss-of-function variants(28–30) (See Supplemental Methods).
Polygenic scores and correlation with disease
We used ICD-10 and OPCS-4 codes to define diseases of interest, including thoracic aortic aneurysm and aortic stenosis. For each described phenotype, we used the independently significant SNPs of the respective GWAS to construct a polygenic score(31). Participants whose data were used for the GWAS were excluded. Finally, we analyzed the relationship between polygenic scores and incident disease using both unadjusted survival analyses and Cox proportional hazards models that were adjusted for clinical factors. These covaiariates included sex, genotypic array, the first five principal components of ancestry, and the cubic natural splines of BMI, age (and its interaction with age), blood pressure, height, and weight. We used accelerated failure time models to confirm the results of Cox proportional hazards models.
Summary of key statistical methods
As detailed above, pixel accuracy and Jaccard agreement were used to gauge performance of the deep learning output. Linear models were used to assess correlation between phenotypes and disease prevalence, using significance threshold p<0.05. During genomic studies, deviation from Hardy-Weinberg equilibrium was tested using significance threshold p<1×10−6. Genome-wide association was tested using significance threshold p<5×10−8. Correlation between polygenic scores and disease incidence was tested using unadjusted survival analysis and Cox proportional hazards models that adjusted for cofactors as above, using significance threshold p<0.05.
Results
GWAS and rare variant association tests of LVOT, aortic root, and ascending aorta diameters
We sought to define the genetic basis for variation of size of the LVOT, aortic root, and proximal ascending aorta. In total, 33,870 participants had data that passed quality control and contributed to genetic analysis of at least one phenotype (Table 1).
Table 1:
GWAS participant characteristics
| Female (N=18403) |
Male (N=15467) |
All (N=33870) |
|
|---|---|---|---|
| Age at time of MRI | 63.9 (7.5) | 64.9 (7.8) | 64.3 (7.7) |
| BMI (kg/m^2) | 25.7 (4.4) | 26.7 (3.7) | 26.2 (4.1) |
| Height (cm) | 163 (6) | 176 (7) | 169 (9) |
| Weight (kg) | 68.1 (12.2) | 82.7 (12.6) | 74.8 (14.4) |
| Systolic Blood Pressure (mmHg) | 136 (19) | 142 (17) | 139 (19) |
| Diastolic Blood Pressure (mmHg) | 76.9 (9.9) | 80.9 (9.7) | 78.7 (10.0) |
| LVOT diameter (mm) | 23.4 (2.6) | 26.4 (3.1) | 24.8 (3.2) |
| Aortic root diameter (mm) | 28.3 (3.6) | 32.4 (4.0) | 30.2 (4.3) |
| Ascending aorta diameter 0 (mm) | 24.7 (3.4) | 27.5 (3.8) | 26.0 (3.8) |
| Ascending aorta diameter 1 (mm) | 26.6 (3.2) | 29.1 (3.5) | 27.7 (3.5) |
| Ascending aorta diameter 2 (mm) | 27.9 (3.3) | 30.4 (3.5) | 29.0 (3.6) |
| Ascending aorta diameter 3 (mm) | 29.0 (3.8) | 31.5 (4.1) | 30.1 (4.1) |
| Ascending aorta diameter 4 (mm) | 28.7 (4.2) | 30.9 (4.2) | 29.7 (4.3) |
| Ascending aorta diameter 5 (mm) | 27.8 (3.9) | 29.9 (4.0) | 28.8 (4.1) |
Demographic information is shown for UK Biobank participants with genetic and cardiac MRI data that passed quality control as detailed in the sample flow diagram in Supplementary Figure 1. For count data, values shown are N (%). For quantitative data, values shown are mean (SD).
We confirmed that all phenotypes were heritable traits, with SNP heritability estimates ranging from 22% to 49% (Supplemental Table 9). We further verified that genetic correlation between the inverse normal of extracted traits was generally greater for traits in closer anatomic proximity, and that genetic correlation between these values and the inverse normal of height was modest (R range=0.22–0.38, Supplemental Figure 3).
We identified 66 independent loci associated with one or more diameters of the ascending aorta. Of these, 15 were novel as defined by lack of detection in the only other study (to our knowledge) that investigated the genetics of ascending aorta diameter at a comparable anatomic location (see Supplemental Methods)(13). In the aortic root, we identified 28 independent genome-wide significant loci. Of these, 17 were novel as defined by lack of detection in the only other study that investigated the genetics of aortic root diameter at a comparable anatomic location. In the LVOT, we identified 6 independent genome-wide significant loci(32). (Figure 1, Table 2). To our knowledge, no previous GWAS has investigated LVOT diameter. No lead SNPs deviated from Hardy-Weinberg equilibrium (HWE) at the commonly used threshold p<1×10−6.
Figure 1:
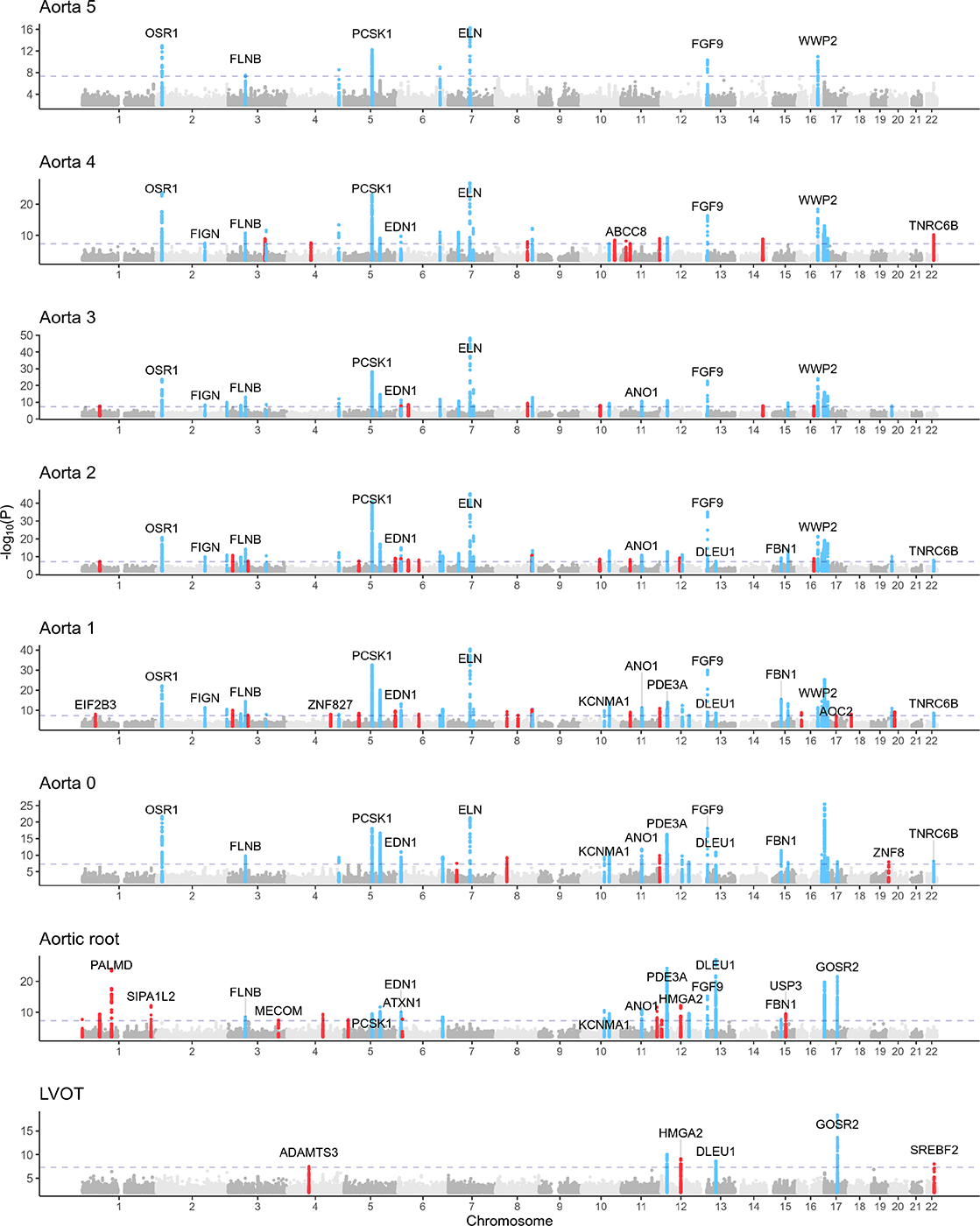
Genetic associations with LVOT, aortic root, and ascending aorta diameters
Loci with P<5×10−8 are shown in blue (if also associated at genome-wide significance with two or more anatomically contiguous traits) or red (if associated at genome-wide significance with up to one anatomically contiguous trait. A selected subset of nearest genes of loci with P < 5×10−8 are overlaid.
Table 2:
Loci not previously identified in aortic root or aortic diameter GWAS
| Trait | SNP | CHR | BP | Effect/Non-effect allele | Closest gene | P |
|---|---|---|---|---|---|---|
| Aorta 4 | rs34266187 | 3 | 123975301 | G/A | KALRN | 1.25E-09 |
| Aorta 2 | rs540082300 | 5 | 51196504 | C/CT | ISL1 | 2.83E-08 |
| Aorta 2 | 5:173276788_AT_A | 5 | 173276788 | AT/A | CPEB4 | 8.21E-10 |
| Aorta 2 | rs12195791 | 6 | 72205132 | G/A | OGFRL1 | 7.64E-09 |
| Aorta 2 | rs2137537 | 12 | 71113087 | T/C | PTPRR | 8.19E-12 |
| Aorta 1 | rs747347287 | 1 | 45418228 | AT/A | EIF2B3 | 7.73E-09 |
| Aorta 1 | rs1979974 | 4 | 146800815 | A/G | ZNF827 | 9.39E-09 |
| Aorta 1 | rs13158444 | 5 | 51201361 | T/C | ISL1 | 2.95E-09 |
| Aorta 1 | rs62376928 | 5 | 173288146 | T/C | CPEB4 | 2.67E-10 |
| Aorta 1 | rs143917622 | 6 | 72205158 | A/AT | OGFRL1 | 7.80E-09 |
| Aorta 1 | rs112621658 | 8 | 38774696 | C/A | PLEKHA2 | 4.70E-10 |
| Aorta 1 | rs40430 | 10 | 79179345 | A/T | KCNMA1 | 1.88E-10 |
| Aorta 1 | rs10743356 | 12 | 20235039 | A/G | PDE3A | 2.92E-11 |
| Aorta 1 | rs7304603 | 12 | 71114400 | T/C | PTPRR | 3.78E-13 |
| Aorta 1 | 16:14505488_CT_C | 16 | 14505488 | CT/C | PARN | 1.36E-09 |
| Aorta 1 | rs35296742 | 17 | 40999303 | G/A | AOC2 | 4.41E-08 |
| Aorta 0 | rs917275 | 7 | 28658522 | A/G | CREB5 | 2.91E-08 |
| Aorta 0 | rs112621658 | 8 | 38774696 | C/A | PLEKHA2 | 5.52E-10 |
| Aorta 0 | rs40430 | 10 | 79179345 | A/T | KCNMA1 | 4.34E-10 |
| Aorta 0 | rs10841441 | 12 | 20210632 | C/T | PDE3A | 4.54E-17 |
| Aorta 0 | rs2137537 | 12 | 71113087 | T/C | PTPRR | 1.57E-10 |
| Aorta 0 | rs78033733 | 17 | 45290078 | T/G | MYL4 | 8.66E-09 |
| Aorta 0 | rs57785785 | 19 | 58815158 | T/A | ZNF8 | 1.08E-08 |
| Aortic root | rs76947392 | 1 | 1256608 | G/A | CPSF3L | 2.19E-08 |
| Aortic root | rs1285677 | 1 | 232712355 | A/C | SIPA1L2 | 7.20E-13 |
| Aortic root | rs2686630 | 3 | 58091861 | G/C | FLNB | 3.20E-09 |
| Aortic root | rs73030346 | 3 | 169317856 | C/T | MECOM | 3.54E-08 |
| Aortic root | rs13134800 | 4 | 120900282 | T/C | MAD2L1 | 4.61E-10 |
| Aortic root | 5:15005465_CTCTT_C | 5 | 15005465 | CTCTT/C | ANKH | 2.68E-08 |
| Aortic root | rs4264961 | 5 | 95617018 | C/T | PCSK1 | 3.15E-10 |
| Aortic root | rs1630736 | 6 | 12295987 | C/T | EDN1 | 9.69E-11 |
| Aortic root | rs80036911 | 6 | 16472975 | C/T | ATXN1 | 1.63E-08 |
| Aortic root | rs7754762 | 6 | 152311537 | T/A | ESR1 | 3.06E-09 |
| Aortic root | rs28735 | 10 | 79178044 | G/C | KCNMA1 | 2.80E-11 |
| Aortic root | rs1977289 | 10 | 96301907 | T/C | HELLS | 2.63E-10 |
| Aortic root | rs12280388 | 11 | 121670712 | T/C | SORL1 | 2.48E-12 |
| Aortic root | rs4296081 | 12 | 22005781 | A/G | ABCC9 | 1.94E-10 |
| Aortic root | rs903175 | 12 | 94158719 | G/A | CRADD | 2.73E-10 |
| Aortic root | rs12866004 | 13 | 22865917 | T/A | FGF9 | 5.20E-16 |
| Aortic root | rs17352842 | 15 | 48694211 | C/T | FBN1 | 1.93E-08 |
| LVOT | rs35631249 | 4 | 73443865 | C/A | ADAMTS3 | 3.28E-08 |
| LVOT | rs7139226 | 12 | 20180749 | C/T | AEBP2 | 9.36E-11 |
| LVOT | rs10400419 | 12 | 66389968 | T/C | HMGA2 | 7.77E-10 |
| LVOT | rs2740516 | 13 | 50761619 | G/A | DLEU1 | 2.27E-09 |
| LVOT | rs17677363 | 17 | 45036112 | A/T | GOSR2 | 4.11E-19 |
| LVOT | rs62240962 | 22 | 42259524 | C/T | SREBF2 | 9.75E-09 |
Novel lead SNPs from GWASs for the eight traits of interest. SNP = the rsID of the variant, where available; for variants that are not in dbSNP, this column displays [chromosome] : [genomic position] _ [effect allele _ non-effect allele]. CHR = chromosome. BP = genomic position, keyed to GRCh37.
In total, 79 independent loci displayed genome-wide significant association with one or more of the eight extracted diameters. Of these, a majority (41) were significantly associated with only one or two diameters, and only 26 were significantly associated with four or more diameters. Hierarchical clustering of absolute effect sizes for significant SNPs visualizes the observation that the SNPs significantly associated with a given diameter were largely distinct from those significantly associated with other diameters (Supplemental Figure 4). A subset of these SNPs are significantly associated with more than one diameter (46/157); these SNPs demonstrate the shared genetic basis across the LVOT and ascending aorta (Figure 2A). To further visualize the spatial distribution of genetic determinants, we binned the genes closest to the loci significantly associated with proximal diameters (LVOT, Aortic root), middle diameters (Aorta 0–2), and distal diameters (Aorta 3–5). We discovered a set of loci independently associated with all three sets (proximal, middle, and distal) as well as associations shared between adjacent pairs of sets (proximal and middle, middle and distal). Notably, no loci were significantly associated with the proximal and distal sets without concurrent association with the middle set (Figure 2B).
Figure 2:
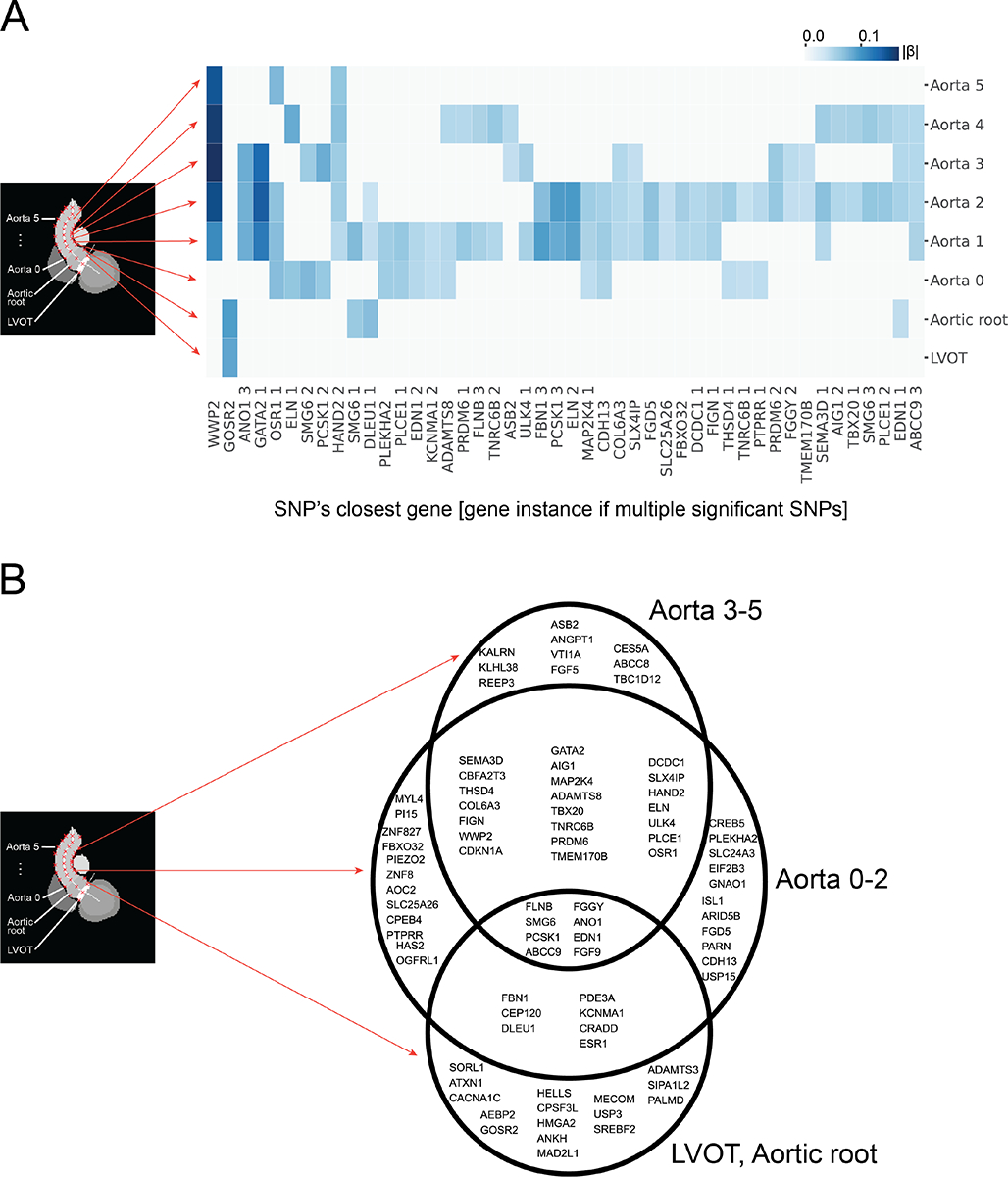
Shared genetic determinants across the LVOT and ascending aorta
A: Rows denote associated GWAS as shown with arrows. Columns denote SNPs associated with two or more traits at p<5×10−8; labels display closest genes. Where multiple SNPs share a closest gene, a number displays the gene’s instance in the combined significant GWAS results. See Supplemental Table 8 for corresponding SNP names. Color indicates absolute effect size (|β|) per standard error of a significantly associated SNP. To illustrate anatomic relationship of significant SNPs, column position is hierarchically clustered to group associations of similar significance (p). For this illustrative purpose, SNPs not meeting genome-wide significance are assigned p=0. B: Venn diagram of genes nearest to loci found to be associated at p<5×10−8 with one or more traits. Traits are binned into groups based on proximity to the heart as labeled. Loci are defined using a 500 kilobase radius (see Supplemental Methods: Genotyping and genome-wide association studies).
To investigate contributions from rare genetic variants to trait variation, we conducted rare variant association tests in 18,461 UK Biobank participants with both LVOT view imaging and exome sequencing data. No gene achieved Bonferroni significance in an exome-wide analysis.
Genetic prediction of computed diameters are associated with thoracic aortic aneurysm
We next sought to investigate the relationship between common-variant genetics underlying the diameters of the LVOT, aortic root, and ascending aortic tract and the incidence of aortic disease. Excluding participants whose data was used for the GWAS, we computed a polygenic risk score using the GWAS of each described phenotype using the autosomal, independently significant SNPs of the respective GWAS.
Using Cox proportional hazards models adjusted for clinical risk factors, we analyzed the relationship between the resultant polygenic scores for individuals and incident thoracic aortic aneurysm, dissection, or rupture. We found that, for all eight diameters, polygenic scores corresponding to larger diameters positively correlated with incidence of disease at a significance level of P<0.05. We observed the strongest effect size and significance for the polygenic score of Aorta 1 (ascending aortic diameter approximately 13 mm distal to the sinotubular junction; n=427,016; n(incident disease)=743, representing 0.17% of n; mean HR=1.42 per standard deviation; CI=1.34–1.50, P=6.67×10−21). We further observed a gradient of signal in which both effect size and significance of the remaining seven scores was lesser in sequence with the associated diameter’s distance from Aorta 1 (Figure 3A). Finally, we observed that participants above the 90th percentile for a polygenic score of larger Aorta 1 diameter displayed a significantly increased incidence of thoracic aortic aneurysm relative to remaining participants in an unadjusted survival analysis (Figure 3A). Proportional hazards violations were observed during modeling, so the results were confirmed with accelerated failure time models, which showed similar results (Supplementary Table 10, Supplemental Methods).
Figure 3:
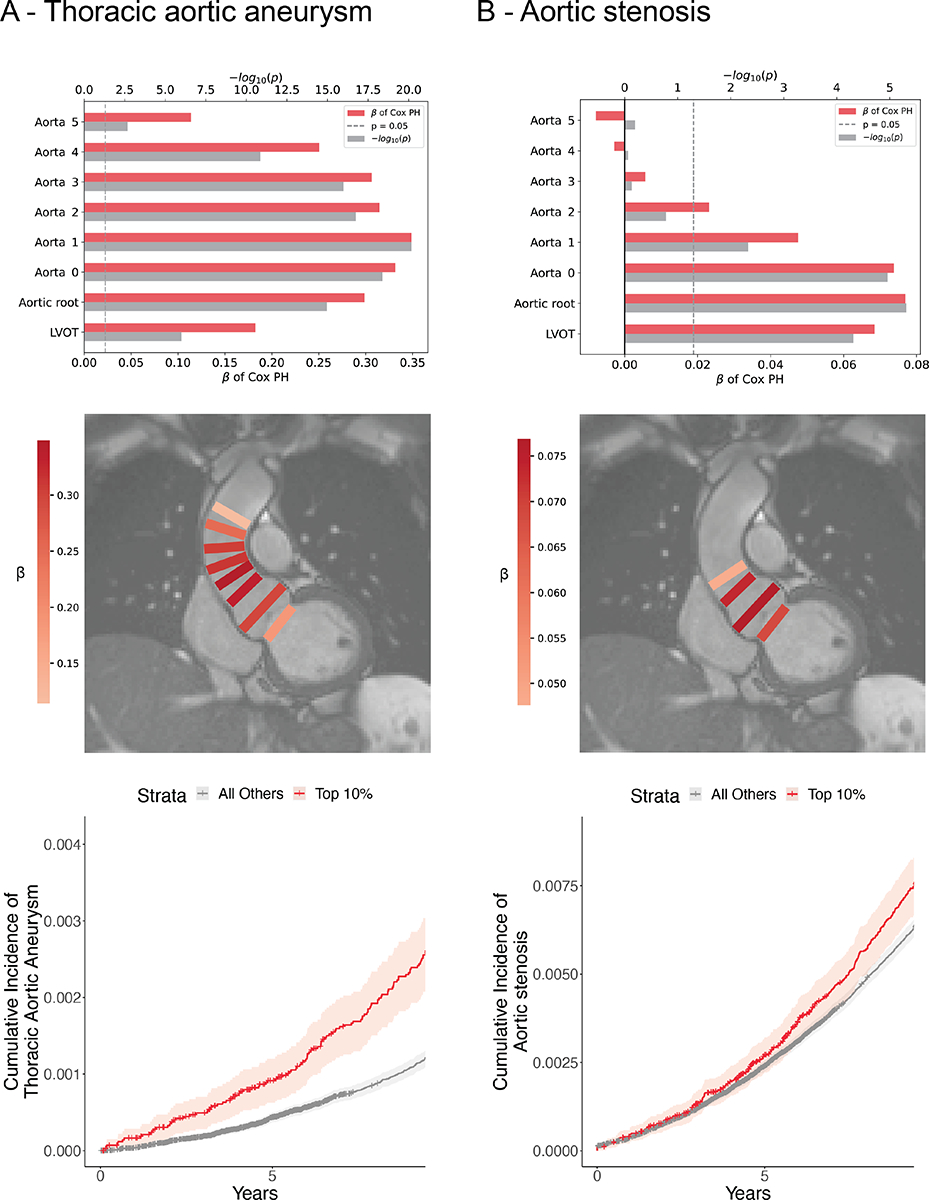
Disease prediction
A: Top: Cox proportional hazards models predicting incidence of thoracic aortic aneurysm using polygenic scores of larger diameters and covariates. Vertical position denotes trait used to create score. Horizontal bars denote effect size (β) and significance. Middle: Overlay of model performance. Rectangle color represents relative effect size (β); location represents trait used to create polygenic score. Bottom: Kaplan-Meier plot: cumulative incidence of thoracic aortic aneurysm for strata of a polygenic score derived from Aorta 1. B: Top: Cox proportional hazards models predicting incidence of aortic stenosis using polygenic scores of smaller diameters and covariates. Vertical position denotes trait used to create score. Horizontal bars denote effect size (β) and significance. Middle: Overlay of model performance. Rectangle color represents relative effect size (β); location represents trait used to create polygenic score. Bottom: Kaplan-Meier plot: cumulative incidence of aortic stenosis for strata of a polygenic score derived from Aortic root.
Genetic prediction of aortic root diameter is associated with aortic stenosis
We used a similar approach leveraging adjusted Cox proportional hazards models to investigate the relationship between the eight derived polygenic scores and incidence of aortic stenosis. In contrast to thoracic aortic aneurysm, which was positively associated with polygenic scores of larger diameters, we found that the scores corresponding to smaller values of the four most proximal diameters—LVOT, Aortic root, Aorta 0, and Aorta 1—significantly predicted incidence of aortic stenosis. We observed the strongest effect size and significance for the polygenic score of aortic root diameter, (n=426,502; n(incident disease) = 3604, representing 0.85% of n; mean HR=1.08 per standard deviation; CI=1.03–1.12; P=5×10−6). Unlike for thoracic aortic aneurysm, where both proximal and distal polygenic scores were associated with disease, we observed that only aortic root, LVOT, and proximal aortic scores were associated with aortic stenosis. Participants above the 90th percentile of polygenic score derived from smaller aortic root diameter displayed a significantly increased incidence of aortic stenosis relative to remaining participants in an unadjusted survival analysis (Figure 3B). Similar to the models for thoracic aortic aneurysm, proportional hazards violations prompted confirmatory analysis with accelerated failure time models, which showed similar results (Supplementary Table 10, Supplemental Methods).
Discussion
We used deep learning to assess the size of the LVOT, aortic root, and ascending aorta using MRI data in a large population-based biobank. We identified 6 novel loci in the LVOT, 17 novel loci in the aortic root, and 23 novel loci in the ascending aorta, and assessed their association with thoracic aortic aneurysm and aortic stenosis. These findings permit several conclusions.
First, serial diameters of the LVOT-ascending aorta region demonstrated distinct genetic underpinnings detectable even over short intervals of 10 millimeters or less. Where available, corroboration with prior knowledge suggests that these diverse genetic signatures are meaningful. For example, we found that a majority of loci significantly associated with ascending aortic diameter reproduce the results of our prior work investigating ascending aorta diameter via short axis imaging(13). We also observed genetic associations with aortic root diameter in agreement with prior echocardiography-driven work. For example, SNPs near GOSR2 have previously been implicated in variations in aortic valve area, aortic root diameter, and other cardiac dimensions(32–35). Further, PALMD has been implicated as a susceptibility gene for calcific aortic stenosis(32, 36, 37). This agreement lends credence to interesting novel loci, such as aortic root diameter’s association with SNP rs80036911 on MECOM, a gene previously associated with left ventricular trabeculation(34). Developing a better understanding of the distinct drivers along the course of the LVOT-ascending aorta tract may ultimately enable more targeted therapies for subtypes of aortic pathology, such as root-predominant and root-sparing aortic aneurysm.
Second, we demonstrated spatial localization of a region of ascending aorta whose underlying genetics may be most relevant to incidence of aneurysm. Prediction was optimized with a polygenic score from Aorta 1 (a diameter about 13mm distal to the sinotubular junction) and signal sequentially diminished with subsequent diameters both proximally and distally. Loci found to be uniquely associated with this diameter include rs35296742 on AOC2, an amine oxidase, and rs1979974 on ZNF827 which encodes a zinc finger protein observed to be involved in telomere synthesis and cell viability(38, 39). Beyond individual targets, localizing the aorta diameter most salient for incidence of pathology may guide future efforts to develop imaging protocols to screen asymptomatic individuals at risk of thoracic aneurysm.
Third, we demonstrated that a polygenic score associated with a smaller aortic root diameter predicted incidence of aortic stenosis. Weaker predictive signals were seen for scores created for diameters of LVOT and proximal ascending aorta diameters, but we found no predictive signal for scores of more distal aorta diameters. Similar to the prediction of aneurysm, this finding implicates loci uniquely associated with aortic root diameter, or those uniquely associated with proximal diameters, as appealing targets for screening and therapeutics for aortic stenosis. One locus (lead SNP rs139939693, intronic to USP3) presents an interesting target for downstream research. USP3, encoding ubiquitin carboxyl-terminal hydrolase 13, has been previously implicated in atrial fibrillation, ECG morphology, and aortic root size (via echocardiography)(32, 40, 41).
Our work was subject to a number of limitations. All data were derived from deep learning models that contain imprecision that could be reduced with further training data. Like any deep learning model, these models can produce non-physiologic measurements when presented with images containing features not seen in the training data. An advantage of the semantic segmentation approach is that outliers can be visually inspected and the model re-trained as needed. The diameter measurements are one-dimensional estimates of three-dimensional structures and therefore cannot capture complete information about tract size and morphology. With our quality control measures, we have attempted to ensure that the images selected for phenotype extraction display the LVOT and aorta near their maximal diameters, i.e. that the tract is not foreshortened. However, variations in anatomy, distance between diameters of interest, and the complex motion of the heart introduce imprecision. Because the utilized CMR view did not capture a standardized length of aorta, samples are smaller for distal traits (Supplemental Table 3); though we performed steps to mitigate this (e.g. including only diameters available for a majority of the population), it is possible that comparisons of regional genetics and associations with disease are partially biased by differential power. The deep learning models have not been tested outside of the specific devices and imaging protocols used by the UK Biobank and may not generalize to other data sets without additional fine-tuning. The study population is largely of European ancestry, similar to the remainder of UK Biobank, thus necessitating validation in more diverse study populations before findings are further assessed for clinical value; failure to do so may result in misdiagnosis or mistreatment of aortic disease in non-European populations. The individuals who underwent MRI in the UK Biobank tend to be healthier than the remainder of the UK Biobank population, which itself is healthier than the general population. Because we have used hospital-based diagnosis codes and procedural codes to identify individuals with disease our disease definitions are susceptible to misclassification. Finally, because echocardiography is the clinical standard for diagnosis of pathology of or near the aortic valve, it is possible that comparisons of polygenic scores’ ability to predict disease are biased in favor of scores derived from areas visible via echocardiography.
In summary, we used machine learning to obtain serial measurements of LVOT, aortic root, and ascending aorta diameter in a large population-based biobank. We found that the genetics underlying these measurements may be clinically relevant for the prediction of thoracic aortic aneurysm and aortic stenosis. For both aneurysm and stenosis, future work, including validation in more ancestrally diverse study populations, is warranted to determine whether a model incorporating a polygenic score and clinical risk factors might identify high-risk, asymptomatic individuals who would benefit from screening via thoracic imaging.
Supplementary Material
Sources of Funding:
This work was supported by the Fondation Leducq (14CVD01), and by grants from the National Institutes of Health to Dr. Pirruccello (K08HL159346), Dr. Ellinor (1RO1HL092577, K24HL105780) and Dr. Ho (R01HL134893, R01HL140224, K24HL153669). This work was also supported by a Sarnoff Cardiovascular Research Foundation Scholar Award to Dr. Pirruccello. Dr. Nauffal is supported by NIH grant 5T32HL007604-35. Dr. Lubitz is supported by NIH grant R01HL139731 and American Heart Association 18SFRN34250007. This work was supported by a grant from the American Heart Association Strategically Focused Research Networks to Dr. Ellinor. Dr. Lindsay is supported by the Fredman Fellowship for Aortic Disease and the Toomey Fund for Aortic Dissection Research. This work was funded by a collaboration between the Broad Institute and IBM Research.
Disclosure Statement:
Dr. Pirruccello has served as a consultant for Maze Therapeutics. Dr. Lubitz receives sponsored research support from Bristol Myers Squibb / Pfizer, Bayer AG, Boehringer Ingelheim, and Fitbit, and has consulted for Bristol Myers Squibb / Pfizer and Bayer AG, and participates in a research collaboration with IBM. Dr. Ng is employed by IBM Research. Dr. Batra receives sponsored research support from Bayer AG and IBM, and has consulted for Novartis and Prometheus Biosciences. Dr. Ho has received research grant support from Bayer AG focused on machine-learning and cardiovascular disease, and research supplies from EcoNugenics. Dr. Philippakis is supported by a grant from Bayer AG to the Broad Institute focused on machine learning for clinical trial design. Dr. Ellinor has received sponsored research support from Bayer AG and from IBM Research. Dr. Ellinor has also served on advisory boards or consulted for Bayer AG, MyoKardia and Novartis. Remaining authors report no disclosures.
Abbreviations
- LVOT
left ventricular outflow tract
- CMR
cardiac magnetic resonance
- GWAS
genome-wide association study/studies
- MAF
minor allele frequency
- HWE
Hardy-Weinberg Equilibrium
- SNP
single nucleotide polymorphism
- ICD-10
International Classification of Diseases, Tenth Revision
- OPCS-4
Office of Population Censuses and Surveys Classification of Interventions and Procedures version 4
- BMI
body mass index
References
- 1.Orts-Llorca F, Puerta Fonolla J, Sobrado J. The formation, septation and fate of the truncus arteriosus in man. J. Anat 1982;134:41–56. [PMC free article] [PubMed] [Google Scholar]
- 2.Grotenhuis HB, Dallaire F, Verpalen IM, van den Akker MJE, Mertens L, Friedberg MK. Aortic Root Dilatation and Aortic-Related Complications in Children After Tetralogy of Fallot Repair. Circ. Cardiovasc. Imaging 2018;11:e007611. [DOI] [PubMed] [Google Scholar]
- 3.Axt-Fliedner R, Kreiselmaier P, Schwarze A, Krapp M, Gembruch U. Development of hypoplastic left heart syndrome after diagnosis of aortic stenosis in the first trimester by early echocardiography. Ultrasound Obstet. Gynecol 2006;28:106–109. [DOI] [PubMed] [Google Scholar]
- 4.Howard DPJ, Banerjee A, Fairhead JF, et al. Population-based study of incidence and outcome of acute aortic dissection and premorbid risk factor control: 10-year results from the Oxford Vascular Study. Circulation 2013;127:2031–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renard M, Francis C, Ghosh R, et al. Clinical Validity of Genes for Heritable Thoracic Aortic Aneurysm and Dissection. J. Am. Coll. Cardiol 2018;72:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fann JI. Descending thoracic and thoracoabdominal aortic aneurysms. Coron. Artery Dis 2002;13:93–102. [DOI] [PubMed] [Google Scholar]
- 7.Guo D-C, Papke CL, He R, Milewicz DM. Pathogenesis of thoracic and abdominal aortic aneurysms. Ann. N. Y. Acad. Sci 2006;1085:339–352. [DOI] [PubMed] [Google Scholar]
- 8.Vapnik JS, Kim JB, Isselbacher EM, et al. Characteristics and Outcomes of Ascending Versus Descending Thoracic Aortic Aneurysms. Am. J. Cardiol 2016;117:1683–1690. [DOI] [PubMed] [Google Scholar]
- 9.Jondeau G, Boileau C. Familial thoracic aortic aneurysms. Curr. Opin. Cardiol 2014;29:492–498. [DOI] [PubMed] [Google Scholar]
- 10.Verstraeten A, Luyckx I, Loeys B. Aetiology and management of hereditary aortopathy. Nat. Rev. Cardiol 2017;14:197–208. [DOI] [PubMed] [Google Scholar]
- 11.Pinard A, Jones GT, Milewicz DM. Genetics of thoracic and abdominal aortic diseases. Circ. Res 2019;124:588–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashvetiya T, Fan SX, Chen Y-J, et al. Identification of novel genetic susceptibility loci for thoracic and abdominal aortic aneurysms via genome-wide association study using the UK Biobank Cohort. PLoS One 2021;16:e0247287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pirruccello JP, Chaffin MD, Chou EL, et al. Deep learning enables genetic analysis of the human thoracic aorta. Nature Genetics 2022;54:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murillo H, Lane MJ, Punn R, Fleischmann D, Restrepo CS. Imaging of the aorta: embryology and anatomy. Semin. Ultrasound CT MR 2012;33:169–190. [DOI] [PubMed] [Google Scholar]
- 15.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol 2005;287:134–145. [DOI] [PubMed] [Google Scholar]
- 16.Waldo KL, Hutson MR, Ward CC, et al. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev. Biol 2005;281:78–90. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development 2000;127:1607–1616. [DOI] [PubMed] [Google Scholar]
- 18.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard J, Gugger S. Fastai: A layered API for deep learning. Information 2020;11:108. [Google Scholar]
- 20.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention Springer, 2015:234–241. [Google Scholar]
- 21.Russakovsky O, Deng J, Su H, et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis 2015;115:211–252. [Google Scholar]
- 22.Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. Imagenet: A large-scale hierarchical image database. In: 2009. IEEE conference on computer vision and pattern recognition Ieee, 2009:248–255. [Google Scholar]
- 23.Zhang TY, Suen CY. A fast parallel algorithm for thinning digital patterns. Commun. ACM 1984;27:236–239. [Google Scholar]
- 24.van der Walt S, Schönberger JL, Nunez-Iglesias J, et al. scikit-image: image processing in Python. PeerJ 2014;2:e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eveborn GW, Schirmer H, Heggelund G, Lunde P, Rasmussen K. The evolving epidemiology of valvular aortic stenosis. the Tromsø study. Heart 2013;99:396–400. [DOI] [PubMed] [Google Scholar]
- 27.Mbatchou J, Barnard L, Backman J, et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet 2021;53:1097–1103. [DOI] [PubMed] [Google Scholar]
- 28.Van Hout CV, Tachmazidou I, Backman JD, et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature 2020;586:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun 2019;10:1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wild PS, Felix JF, Schillert A, et al. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J. Clin. Invest 2017;127:1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Córdova-Palomera A, Tcheandjieu C, Fries JA, et al. Cardiac Imaging of Aortic Valve Area From 34 287 UK Biobank Participants Reveals Novel Genetic Associations and Shared Genetic Comorbidity With Multiple Disease Phenotypes. Circ Genom Precis Med 2020;13:e003014. [DOI] [PubMed] [Google Scholar]
- 34.Meyer HV, Dawes TJW, Serrani M, et al. Genetic and functional insights into the fractal structure of the heart. Nature 2020;584:589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu M, Tcheandjieu C, Georges A, et al. Computational estimates of annular diameter reveal genetic determinants of mitral valve function and disease. JCI Insight 2022;7. Available at: 10.1172/jci.insight.146580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thériault S, Gaudreault N, Lamontagne M, et al. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat. Commun 2018;9:988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Gaudreault N, Arsenault BJ, Mathieu P, Bossé Y, Thériault S. Phenome-wide analyses establish a specific association between aortic valve PALMD expression and calcific aortic valve stenosis. Commun Biol 2020;3:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaitaniemi S, Elovaara H, Grön K, et al. The unique substrate specificity of human AOC2, a semicarbazide-sensitive amine oxidase. Cell. Mol. Life Sci 2009;66:2743–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conomos D, Reddel RR, Pickett HA. NuRD–ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol 2014;21:760–770. [DOI] [PubMed] [Google Scholar]
- 40.Roselli C, Chaffin MD, Weng L-C, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet 2018;50:1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verweij N, Benjamins J-W, Morley MP, et al. The Genetic Makeup of the Electrocardiogram. Cell Syst 2020;11:229–238.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


