Abstract
Many plant RNA viruses are associated with one or more subviral RNAs. Two subviral RNAs, satellite RNA C (satC) and defective interfering RNA G (diG) intensify the symptoms of their helper, turnip crinkle virus (TCV). However, when the coat protein (CP) of TCV was replaced with that of the related Cardamine chlorotic fleck virus (CCFV), both subviral RNAs attenuated symptoms of the hybrid virus TCV-CPCCFV. In contrast, when the translation initiation codon of the TCV CP was altered to ACG and reduced levels of CP were synthesized, satC attenuated symptoms while diG neither intensified nor attenuated symptoms. The determinants for this differential symptom modulation were previously localized to the 3′-terminal 100 bases of the subviral RNAs, which contain six positional differences (Q. Kong, J.-W. Oh, C. D. Carpenter, and A. E. Simon, Virology 238:478–485, 1997). In the current study, we have determined that certain sequences within the 3′-terminal stem-loop structures of satC and diG, which also serve as promoters for complementary strand synthesis, are critical for symptom modulation. Furthermore, the ability to attenuate symptoms was correlated with weakened binding of TCV CP to the hairpin structure.
Many plant RNA viruses are associated with one or more nonessential subviral RNAs, including defective interfering RNAs (DI RNAs) and satellite RNAs (satRNAs), which depend on a helper virus for replication, encapsidation, and movement in plants (26). DI RNAs are fairly ubiquitous in animal virus systems and relatively rare among plant viruses (34, 57). On the other hand, satRNAs are almost exclusively associated with plant viruses (33). DI RNAs are generated as a consequence of errors in viral genome replication, and interference with the replication of the helper virus frequently results in substantial symptom attenuation (6, 9, 14, 35). However, some DI RNAs increase the symptom severity of their helper viruses, such as the DI RNA of broad bean mottle virus (32), DI RNA G (diG) of turnip crinkle virus (TCV) (19) and the DI RNA of bovine diarrhea virus (50). Unlike DI RNAs, which are shortened versions of viral genomic RNAs, satRNAs usually share little sequence similarity with their helper virus. As molecular parasites of their helper viruses, satRNAs can have dramatic effects on symptoms, ranging from amelioration to severe exacerbation (33).
There are several mechanisms that satRNAs can use to attenuate symptoms. In many hosts, disease attenuation by cucumber mosaic virus (CMV) satRNAs is accompanied by a reduction in virus accumulation (12, 13). In contrast, CMV satRNA symptom attenuation of the closely related tomato aspermy virus is not always accompanied by a noticeable decrease in the level of viral RNA (25). Symptom modulation by satRNAs is thought to involve a trilateral interaction among the host plant, satRNA, and helper virus. The involvement of the host was demonstrated when particular subspecies of tobacco determined whether the Y-satellite RNA of CMV produced yellow or green mosaic symptoms (22). The involvement of the helper virus in satRNA symptom modulation was demonstrated by Sleat et al. (43), who showed that symptom modulation of CMV satRNAs mapped to RNA2 of CMV. However, it is not known if cis sequences on RNA2 or if one of the encoded products is the determinant for symptom modulation. The smaller satRNAs, including those of CMV and TCV, do not encode any functional open reading frames (ORFs), indicating that the satRNA determinants for symptom modulation must lie within the RNA sequence. Sequences in satRNAs responsible for symptom intensification have been mapped to specific nucleotide residues which, when altered, can affect the host response (10, 21, 28, 30, 37, 41–43, 58). In contrast, sequence determinants important for symptom attenuation are less well defined.
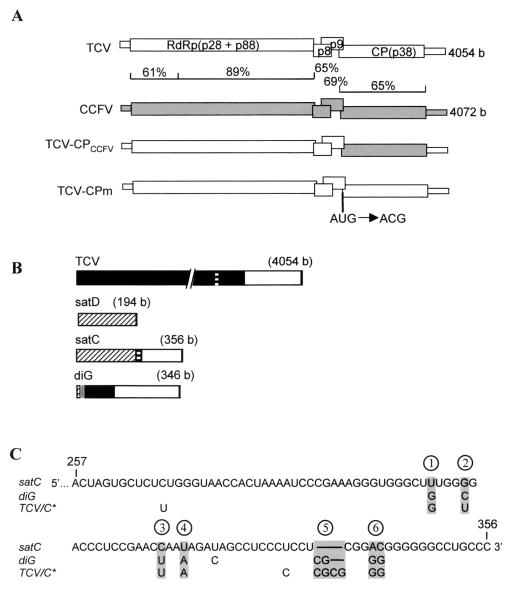
We are studying symptom modulation of TCV subviral RNAs in the host Arabidopsis thaliana. TCV is a single-stranded, positive-sense RNA virus with a 4,054-base genome that serves as the mRNA for the two subunits (p28 and p88) of the viral RNA-dependent RNA polymerase that is required for virus replication (56) (Fig. 1A). Two subgenomic RNAs are synthesized during virus infection (4, 52) that encode the movement proteins p8 and p9 (18) and the coat protein (CP) p38 (Fig. 1A), which is also required for virus movement (8). In addition to the subgenomic RNAs, TCV is associated with several subviral RNAs (Fig. 1B). satD is a typical satRNA that shares little sequence similarity with the helper TCV genomic RNA (Fig. 1B) and has no effect on virus symptoms (20, 36, 38). satC is an atypical satRNA that contains a 5′ domain of 189 bases, similar to the entire sequence of the avirulent satD (88% similarity) and a 3′ domain of 166 bases, similar to two regions of mostly untranslated sequence at the 3′ end of TCV genomic RNA (90% sequence similarity) (Fig. 1B and C). satC is a virulent satRNA, intensifying symptoms in all hosts where TCV produces visible symptoms (20). For example, satC altered the normal slight stunting and slightly mottled and crinkle leaves associated with TCV infection of turnip to severe stunting with dark-green, very stunted and crinkled leaves (19, 37). In most ecotypes of A. thaliana, infection by TCV alone caused moderate stunting symptoms, while inclusion of satC in the inoculum resulted in the death of the plant by about 16 days postinoculation (dpi) (20, 39).
FIG. 1.
Genomic and subviral RNAs used in this study. (A) Genomic RNAs. ORFs and untranslated regions are represented by thick and thin boxes, respectively. The percent sequence similarity between TCV and CCFV genomes is shown. TCV-CPm has a point mutation in the CP initiation codon as indicated, which causes translation initiation at an upstream CUG codon resulting in two additional N-terminal amino acids and a reduction in CP levels to 20% of that of the wt (54). (B) Subviral RNAs associated with TCV. Similar sequences among TCV genomic and subviral RNAs are shaded alike. The sizes of the RNAs are given. (C) Alignment of the 3′-end sequences of satC, diG, and the TCV genomic RNA, which is identical to satC* (TCV/C∗) (16). Only differences among the RNAs are indicated. Lines indicate absence of the bases in satC and diG, compared with TCV or satC∗. The six positional differences between satC and diG are shaded. The last two positions (5 and 6) each have two consecutive nucleotide differences between the two subviral RNAs.
While satC intensified the symptoms of wild-type (wt) TCV, it attenuated the moderate symptoms induced by TCV-CPCCFV (Fig. 1A), a variant in which the TCV CP is replaced with the CP of the related Cardamine chlorotic fleck virus (CCFV) (15). Symptom attenuation was correlated with an 80% reduction in TCV-CPCCFV levels in inoculated leaves and protoplasts (15, 17). satC also attenuated the symptoms of about 70% of plants inoculated with TCV-CPm (Fig. 1A), a variant with a single nucleotide alteration in the translation initiation codon of the CP (17). Lack of TCV-CPm symptoms was correlated with nearly undetectable levels of viral genomic RNA in uninoculated leaves (16, 17). Unlike attenuation in TCV-CPCCFV, satC attenuation of TCV-CPm symptoms did not involve a large reduction in virus accumulation in inoculated leaves or protoplasts but rather was associated with a reduction in virus long-distance movement (17). Plants infected with TCV-CPm accumulated 20% of the wt level of a CP that has two additional amino acids at its N terminus (54). However, symptom attenuation by satC was mainly due to the reduced levels of CP synthesized in TCV-CPm-infected plants, as opposed to the altered N terminus (54).
diG, a second virulent subviral RNA associated with TCV, shares a similar but not identical 3′-terminal segment with satC. It is composed (from 5′ to 3′) of 10 bases from the 5′ end of satD, 12 bases of unknown origin, 99 bases from near the 5′ end of TCV, and then 225 bases of a TCV untranslated 3′-terminal sequence (94% sequence similarity with TCV), including an imperfect repeat of 36 bases (Fig. 1B and C). As with satC, diG intensifies the symptoms of wt TCV and attenuates the symptoms of TCV-CPCCFV. However, unlike satC, diG has no effect on symptoms of TCV-CPm (16).
In this report, we have determined that two positions located within the 3′-terminal stem-loop structures of satC and diG are responsible for their different symptom modulation properties. Furthermore, symptom modulation can be directly correlated with affinity of CP binding to the 3′-terminal stem-loop structures. These results provide the first direct link between virus-encoded products and symptom modulation by subviral RNAs.
MATERIALS AND METHODS
Virus strains and plasmid constructions.
Plasmids containing full-length cDNAs of TCV (pT7TCVms) (29), TCV-CPm (pT7TCV-CPm) (17), TCV-CPCCFV (pT7T/C) (15), satC [pT7satC(+)] (44), satC* (pT7satC*) (16), and diG (pT7diG) (19) downstream from a T7 RNA polymerase promoter have been described.
For construction of pT7satC56G, oligonucleotide C56G (oligonucleotides are listed in Table 1) was used with oligonucleotide C5′ in a PCR with pT7satC(+) as a template. The PCR product was digested with SpeI and SmaI, and the smaller fragment was gel purified and ligated to pT7satC(+), which had been previously digested with SpeI and SmaI. Plasmids pT7satC5C* and pT7satC6G were generated in a similar fashion, except that oligonucleotide C56G was replaced with C5C* and C6G, respectively. Plasmids pT7diG56C, pT7diG5C, and pT7diG6C were generated in a similar fashion, except that the template for PCR was replaced with pT7diG, oligonucleotide C5′ was replaced with G5′, and oligonucleotide C56G was replaced with G56C, G5C, and G6C, respectively.
TABLE 1.
Oligonucleotides used in this study
| Application | Oligonucleotide | Position in satC or diG | Sequencea | Polarityb |
|---|---|---|---|---|
| Site-directed mutagenesis | C5′ | 1–19 (satC) | 5′ GGGATAACTAAGGGTTTCT 3′ | + |
| C5C* | 329–356 (satC) | 5′ GGGCAGGCCCCCCCCCCGCGCGAGGAGGGAGG 3′ | − | |
| C6G | 329–356 (satC) | 5′ GGGCAGGCCCCCCCCCCGAGGAGGGAGG 3′ | − | |
| C56G | 329–356 (satC) | 5′ GGGCAGGCCCCCCCCCCGCGAGGAGGGAGG 3′ | − | |
| G5′ | 1–19 (diG) | 5′ GGGATAAAAAGGAGGCTTA 3′ | + | |
| G5C | 313–344 (diG) | 5′ GGGCAGGCCCCCCCCCCG—AGGAGGGAGGCT 3′ | − | |
| G6C | 313–344 (diG) | 5′ GGGCAGGCCCCCCGTCCGCGAGGAGGGAGGCT 3′ | − | |
| G56C | 313–344 (diG) | 5′ GGGCAGGCCCCCCGTCCG—AGGAGGGAGGCT 3′ | − | |
| RT-PCR | DM4 | 206–223 (satC) | 5′ GGGACCAAAAACGGCGGC 3′ | + |
| GM4 | 206–223 (diG) | 5′ TGGCAGCACTGTCTAGCT 3′ | + | |
| T17 adapter | 5′ CCACTCGAGTCGACATCGA(T)17 3′ | |||
| Adapter | 5′ CCACTCGAGTCGACATCGA 3′ | |||
| (dT)18 | 5′ TTTTTTTTTTTTTTTTTT 3′ | |||
| 284 | 284–301 (satC) | 5′ ATCCCGAAAGGGTGGGCT 3′ | − | |
| RNA gel blot analysis | 293 | 274–293 (satC) | 5′ CTTTCGGGATTTTAGTGGTT 3′ | − |
| 260–279 (diG) | ||||
| CP-RNA binding | T7CG3′ | 292–301 (satC) | 5′ GTAATACGACTCACTATAGGGTGGGCT 3′ | + |
| CG3′ | 344–356 (satC) | 5′ GGGCAGGCCCCCC 3′ | − |
Underlined, boldface sequences are inserted or mutated sequences. —, deletion of the nucleotides present in the wt diG sequence in G5C and G56C.
Polarity refers to homology (+) or complementarity (−) with plus strands of subviral RNAs.
Preparation and inoculation of A. thaliana protoplasts.
Protoplasts were prepared from callus cultures derived from A. thaliana ecotype Col-0 embryos as previously described (17). Protoplasts (5 × 106) were inoculated with 20 μg of genomic RNA transcripts synthesized by T7 RNA polymerase in vitro as previously described (17).
RNA gel blot analysis.
Four micrograms of total RNA isolated from protoplasts (39) was denatured by heating in 50 to 70% formamide and then subjected to electrophoresis through nondenaturing 1.5% agarose gels. RNA was then transferred to a NitroPlus membrane (Micron Separations, Inc., Westboro, Mass.) and subjected to hybridization with an oligonucleotide probe specific for TCV and its associated subviral RNAs (Table 1) or a probe specific for plant ribosomal RNAs (39) as previously described (52, 53).
Plant growth and inoculations.
Plants were grown in growth chambers at 20°C as described by Li and Simon (20). Seedlings of A. thaliana ecotypes Col-0 and Di-0 at the six- to eight-leaf stage were subjected to mechanical inoculation of the oldest leaf pair with 4 μl of inoculation buffer containing 0.15 mg of the helper virus RNA transcripts per ml, as described previously (17). For experiments examining the effects of wt and mutant subviral RNAs on symptom modulation, full-length transcripts (0.015 mg/ml) synthesized from the cloned cDNA of the respective subviral RNA were included in the 4-μl inoculum.
3′-end sequence analysis by reverse transcriptase (RT) PCR.
Five micrograms of total RNA isolated from Col-0 plants infected with TCV and a respective subviral RNA were denatured in 50 to 70% formamide and separated on a 1.5% nondenaturing agarose gel, and the RNA species corresponding to the subviral RNA was purified. Poly(A) tails were added to the 3′ ends with poly(A) polymerase (Amersham) as previously described (2). cDNAs were synthesized with a primer containing a 19-base sequence joined to 17 thymidylate residues (Table 1) as previously described (2). PCR was carried out with a primer specific for either satC (DM4) or diG (GM4) (Table 1) and either the 19-base sequence described above or oligonucleotide (dT)18 (Table 1). PCR products were cloned into the SmaI site of pUC19 and sequenced with oligonucleotide 284(−), which is specific for both satC and diG (Table 1).
Virus purification.
Approximately 10 g of TCV-or CCFV-infected A. thaliana Col-0 plants harvested at 14 dpi was mixed with 30 ml of 0.2 M sodium acetate (pH 5.2) and 30 μl of β-mercaptoethanol and ground in a mortar on ice. The supernatant was collected by being filtered through two layers of cheesecloth and incubated on ice for 30 min followed by centrifugation at 8,000 rpm in an SS34 rotor (Sorvall) to pellet cell debris. To precipitate virus particles, 10 ml of fresh 40% polyethylene glycol (molecular weight, 6,000; Sigma) dissolved in 1 M NaCl was added per 40 ml of supernatant, and the mixture was incubated on ice for 30 min, followed by centrifugation at 8,000 rpm in the rotor described above. The pellet was resuspended in 10 to 20 ml of 0.05 M sodium acetate (pH 5.5), incubated on ice for 1 h, and then subjected to centrifugation at 8,000 rpm in the rotor described above. The supernatant was then subjected to further centrifugation at 37,000 rpm for 90 min at 4°C in an SW41 rotor (Beckman). The pellet was resuspended in 0.01 M sodium phosphate buffer (pH 7.0) containing 2.5% glycerol and then clarified by a brief centrifugation. The dissolved virions were subjected to chromatography on a DEAE-agarose gel A (Bio-Rad) column (1.5-cm diameter by 10-cm height) preequilibrated with the same buffer. After the column was washed with 6 column volumes of buffer, elution was carried out with an NaCl gradient (0 to 1 M of NaCl in the same buffer) over 5 h with a flow rate of 0.35 ml/min. Fractions (1.5 ml each) were collected, and virion concentrations were determined by measuring the absorbance at 260 nm. About 2 μg of virions was subjected to electrophoresis on a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel, and the fractions containing CP free of contaminating host proteins were collected. The virions in the fractions were then reprecipitated by mixing with a one-quarter volume of 40% polyethylene glycol in 1 M NaCl, as described above.
Coat protein purification.
Virions (5 mg/ml) were dissociated by incubation on ice for 1.5 h in 0.1 M Tris-HCl (pH 8.5), 5 mM EDTA, and 1.0 M NaCl with a protease inhibitor mixture (0.5 μM phenylmethylsulfonyl fluoride [PMSF], 50 μM tosylphenylchloroketone [TPCK], and 10 μg of α2-macroglobulin per mg of virus). Fifty milliliters of a 50% solution of polyethyleneimine (Sigma) was dispersed in 350 ml of deionized water, adjusted to pH 8.5, and diluted to a final concentration of 5% (wt/vol) (11). The polyethyleneimine solution was added by rapid mixing to dissociated virus to a final concentration of 0.5 mg/ml, and incubation on ice was continued for 15 min. RNA was then removed by centrifugation for 10 min at 15,000 × g. The resulting supernatant containing TCV or CCFV CP was applied to a 1.5- by 30-cm column of Sephacryl S200HR equilibrated with a solution of 0.1 M Tris-HCl (pH 8.5), 0.5 M NaCl, and 5 mM EDTA. Fractions (1.5 ml) were collected and analyzed by SDS-polyacrylamide gel electrophoresis, and those with intact CP free of RNA were pooled and precipitated with an equal volume of saturated (NH4)2SO4 by incubation on ice for 15 min, followed by centrifugation at 4°C in a microcentrifuge at maximum speed for 30 min. The pellet was resuspended in 50% saturated (NH4)2SO4 and subjected to additional centrifugation to collect the precipitate. The final pellet was resuspended in 250 μl of 0.01 M Tris-HCl (pH 8.0) and 25 mM NaCl and then dialyzed for 16 h at 4°C against two changes of the same buffer. CP concentrations were determined by the Bradford assay (1).
Preparation of radiolabeled and unlabeled RNAs for CP-RNA binding.
DNA templates used to generate RNA transcripts of C3′ and C56G3′ were amplified from cDNA clones pT7satC(+) (44) and pT7satC56G (this study), respectively, by PCR with oligonucleotide primers T7CG3′ and CG3′ (Table 1). The DNA template used to generate RNA transcripts of SK70 was pBluescript SK(+) (Stratagene) previously digested with SmaI.
For preparation of radiolabeled RNAs, in vitro transcription reactions were performed with a final volume of 20 μl containing 40 mM Tris-HCl (pH 8.0); 6 mM MgCl2; 10 mM NaCl; 2 mM spermidine; 10 mM dithiothreitol; 20 U of RNasin (Promega); 0.5 mM each of ATP, GTP, and CTP; 19 μM UTP; 50 μCi of [α-32P]UTP (3,000 Ci/mmol; Amersham); 40 U of T7 RNA polymerase (New England Biolabs); and either 2.5 μg of plasmid DNA or 0.625 μg of PCR-generated template. Reactions were incubated for 1.5 to 2 h at 37°C. Transcription products were extracted once with phenol-chloroform and precipitated with ammonium acetate-isopropanol in the presence of 5 μg of yeast tRNA. The pellets were resuspended in a small volume of deionized water and applied to a 8% polyacrylamide gel containing 7 M urea. The gel was covered with Saran Wrap and subjected to brief autoradiography. Gel slices containing radiolabeled RNAs were excised and soaked overnight with constant shaking in a buffer containing 25 mM Tris-HCl (pH 7.5), 400 mM NaCl, and 0.1% SDS. Supernatants were extracted with phenol-chloroform once, followed by ammonium acetate-isopropanol precipitation.
For preparation of unlabeled competitor RNAs, in vitro transcription reactions were performed as described above except that the 50 μCi of [α-32P]UTP was replaced with 481 μM UTP. Transcripts were treated with DNase I (Gibco BRL) to remove template DNA, followed by phenol-chloroform extraction and ammonium acetate-isopropanol precipitation in the presence of 5 μg of yeast tRNA.
CP-RNA binding assays.
32P-labeled RNAs were incubated in 0.2 M Tris-HCl (pH 8.0), 2 mM MgCl2, and 160 mM KCl for 10 min at 65°C and cooled to room temperature over 15 min. The binding of CP to RNA was performed as described previously (40) with the following modifications: the reaction volume was increased to 20 μl with half of the reaction mixture applied to the gel; 5% polyacrylamide was used for electrophoresis at 4°C. For quantitative analysis, several independent experiments were performed, autoradiograms were scanned by a laser densitometer, and results quantified by Molecular Analysis software (Bio-Rad).
For competition assays, binding reactions were initiated by the addition of a saturating amount of CP to mixtures containing 32P-labeled C3′ RNA (100 pM) and increasing concentrations (0 to 1.62 μM) of unlabeled competitor RNAs (C3′ and C56G3′ RNA). The incubation mixtures were fractionated by polyacrylamide gel electrophoresis and analyzed by autoradiography.
RESULTS
Differential modulation of TCV-CPm symptoms by satC and diG maps to the 3′-terminal hairpins that are also the promoters for minus-strand synthesis.
The symptom intensification property of satC was mapped previously to the TCV-similar region of the satRNA, most of which is also shared with diG (37). To determine if this region was also responsible for the differential symptom modulation properties of satC and diG when associated with TCV-CPm, satC*, a hybrid RNA composed of the 5′ 256 bases of satC joined to the 3′ 103 bases of TCV, was previously constructed (TCV and diG share very similar 3′ ends) (Fig. 1B and C). satC* behaved like diG by intensifying the symptoms of wt TCV, attenuating the symptoms of TCV-CPCCFV, and not affecting the symptoms of TCV-CPm (16). These results suggested that the 3′-terminal 100 bases of satC and the corresponding regions in satC* and diG, which contain six positional differences (Fig. 1C), were responsible for differential symptom modulation.
To further define the sequences in the 3′-terminal 100 bases of satC and diG responsible for symptom modulation differences, mutations were introduced into each of the six positional differences to convert the nucleotides in one subviral RNA to the other. Corresponding alterations at positions 1 through 4 had no effect on the symptom modulation properties of satC or diG when coinoculated with TCV, TCV-CPm, or TCV-CPCCFV onto A. thaliana ecotype Col-0 (TCV-susceptible) and Di-0 (TCV-resistant) plants (data not shown; while Di-0 plants are resistant to TCV, they are fully susceptible to TCV-CPm and TCV-CPCCFV [15, 29], since high levels of TCV CP are necessary to induce resistance [54]). The variances at positions 1 through 4 also had no effect on accumulation of satC or diG in protoplasts coinoculated with TCV (data not shown).
Additional satC and diG mutants were constructed by converting one or both of the variances at positions 5 and 6 into the corresponding nucleotides from satC, diG, or satC*. Reciprocal exchanges of position 5 and/or 6 between satC and diG generated mutants satC6G (satC with position 6 of diG) and satC56G (satC with positions 5 and 6 of diG; other mutants are similarly named), as well as diG mutants diG5C, diG6C, and diG56C. Conversion of position 5 of satC to that of satC* generated mutant satC5C*. When TCV was the helper virus, all mutant subviral RNAs mimicked their parental wt subviral RNAs in intensifying the symptoms of Col-0 (Fig. 2A and data not shown). To determine if the alterations were maintained during subviral RNA accumulation in plants, subviral RNAs accumulating in uninoculated leaves at 14 dpi were cloned by RT-PCR as described in Materials and Methods. All satC56G and satC5C* mutants that were sequenced maintained their original mutations (Table 2). diG6C and diG56C were mainly stable, with five of six clones retaining the original alterations. satC6G and diG5C were unstable with only 1 of 18 and 5 of 12 clones remaining unchanged, respectively (Table 2). These results indicate that the subviral RNAs were most stable when both positions 5 and 6 were either satC- or diG like. In addition, satC tolerated the 4-base addition from satC* at position 5, and diG tolerated the two base changes at position 6. Because of the instability of satC6G and diG5C, these mutants were eliminated from further analysis.
FIG. 2.
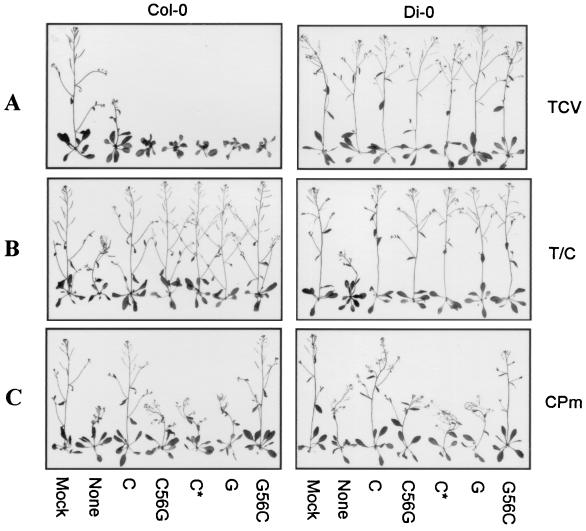
Symptom modulation by the TCV subviral RNAs coinoculated with the helper TCV (A), TCV-CPCCFV (B), and TCV-CPm (C). Seedlings of A. thaliana ecotypes Col-0 and Di-0 at the six- to eight-leaf stage were inoculated with buffer alone (Mock), helper virus genomic RNA without any subviral RNA (None), or with satC (C), satC56G (C56G), satC∗ (C∗), diG (G), and diG56C (G56C), as indicated below the plants. Representative plants were photographed at 17 dpi. The helper virus used to inoculate the plant is shown on the right. Di-0 plants are resistant to infection by TCV. T/C, TCV-CPCCFV; CPm, TCV-CPm.
TABLE 2.
Stability of the mutations in the 3′ ends of satC and diG
| Mutants | Analyzed clones | No. of clones with original mutations | No. of clones with alterationsa |
|---|---|---|---|
| satC5C* | 9 | 9 | 0 |
| satC6G | 18 | 1 | 16(C); 1(C56G) |
| satC56G | 8 | 8 | 0 |
| diG5C | 12 | 5 | 7(G56C) |
| diG6C | 6 | 5 | 1(G56C) |
| diG56C | 6 | 5 | 1(G6C) |
The nature of the altered clone is indicated in parentheses. C, wt satC.
All subviral RNA variants attenuated the symptoms of TCV-CPCCFV, as did the parental satC and diG (Fig. 2B). When TCV-CPm was the helper virus, satC attenuated the symptoms of TCV-CPm, while satC56G exhibited the symptom modulation ability of diG by not affecting symptoms (Fig. 2C). This result indicates that conversion of the nucleotides in positions 5 and 6 from satC to diG in an otherwise wt satC molecule was sufficient to convert satC to the phenotype of diG. diG56C, which has the reciprocal exchange in diG, gained the ability to attenuate the symptoms of TCV-CPm (Fig. 2C). These results indicate that positions 5 and 6 were responsible for the differential modulation of TCV-CPm symptoms by the subviral RNAs. satC5C* and diG6C, however, gave inconsistent results: in Col-0, 50% of plants inoculated with TCV-CPm and satC5C* had attenuated symptoms, with the remaining plants exhibiting only TCV-CPm symptoms. On the other hand, diG6C acquired the phenotype of satC. Using ecotype Di-0, 50% of the plants inoculated with TCV-CPm and diG6C had attenuated symptoms, while satC5C* exhibited diG-like symptom modulation properties (data not shown). These latter results are possibly due to structural considerations, described in the Discussion.
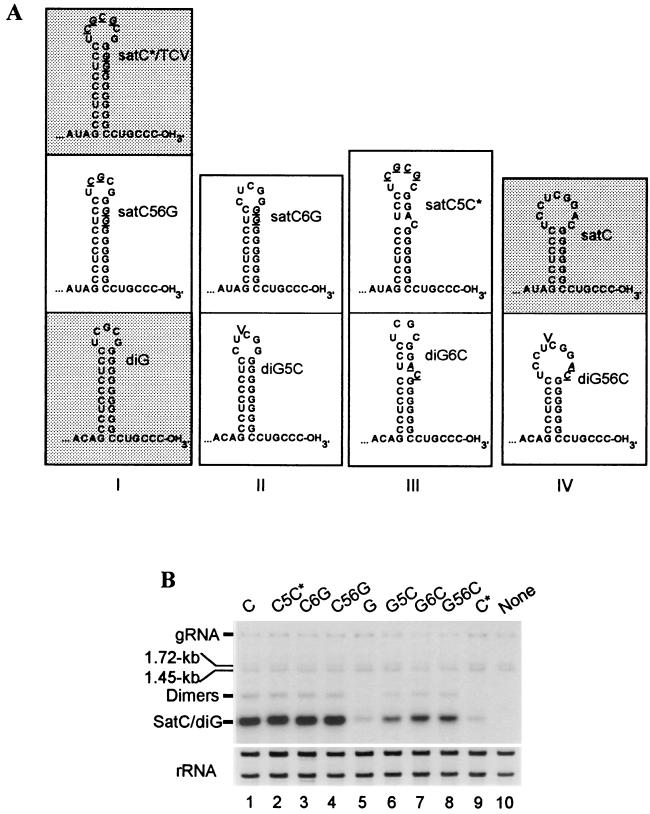
The location of positions 5 and 6 coincides with a 3′-terminal hairpin that comprises a major portion of the promoter for minus-strand satC synthesis (3, 45, 46). The nucleotide differences in this region between satC and diG affect the size of the hairpin (Fig. 3A), according to computer-generated predictions (59). In wt satC, the lack of 2 nucleotides (nt) in position 5 and the presence of AC as opposed to GG residues in position 6 result in a hairpin with a 7-bp stem and a nine-base loop. diG, on the other hand, contains a 10-bp stem and a five-base loop, similar to the hairpin of TCV (or satC*), which is composed of a 10-bp stem and a seven-base loop (Fig. 3A). Since the mutations altered the structure of the hairpin promoter, the effect of the alterations on subviral RNA accumulation in protoplasts was assessed (Fig. 3B). At 40 h postinoculation, all satC derivatives accumulated to similar levels (Fig. 3B, lanes 2 to 4). In contrast, alterations in diG to the corresponding satC nucleotides increased the accumulation of diG, although the levels reached (lanes 6 to 8) were still lower than those of satC and satC-derived mutants (lanes 1 to 4). These results indicate that the introduced mutations did not negatively affect the accumulation of the subviral RNAs in protoplasts.
FIG. 3.
3′-terminal stem-loop structures and replication of satC, diG, and mutant subviral RNAs. (A) 3′-end stem-loop structures of wt and mutant subviral RNAs as predicted by the computer structure program MFOLD (Genetics Computer Group, University of Wisconsin, Madison). Similar 3′-terminal stem-loop structures are assigned to the same class (class I through IV). Subviral RNAs that have been previously studied for symptom modulation (16) are shaded. Mutations in each mutant subviral RNA are shown in italics and underlined. V, location of deleted nucleotides in diG5C and diG56C that are present in wt diG. (B) Accumulation of wt and mutant subviral RNAs in protoplasts. A. thaliana protoplasts (5 × 106) were inoculated with 20 μg of wt TCV with (lanes 1 to 9) or without (lane 10) the addition of 2 μg of wt or mutant subviral RNAs, as shown above each lane. Total RNA extracted at 40 h postinoculation was subjected to RNA gel blot analysis with a probe specific for TCV and the subviral RNAs (Table 1) or rRNA (39). Species corresponding to TCV genomic RNA (gRNA), the two subgenomic RNAs (1.72 and 1.45 kb), and the subviral RNAs and their dimer forms are indicated.
TCV CP preferentially binds to the 3′-terminal hairpin of diG.
The ability of TCV subviral RNAs to reduce virus systemic movement and thereby attenuate symptoms depends on the presence of reduced levels of the TCV CP (54). Reduction in TCV CP levels was achieved by either altering the initiation codon of the CP ORF (16, 17), mutating the subgenomic RNA promoter for the CP mRNA (54), or substituting the CP of the related virus CCFV for the TCV CP (15). The involvement of the TCV CP in satC- but not diG-mediated symptom attenuation led to the suggestion that the CP might bind differentially to the 3′ regions of satC and diG (54). The model suggested that lower affinity of binding to satC might allow, in the presence of reduced amounts of TCV CP, the binding of a factor required for TCV movement.
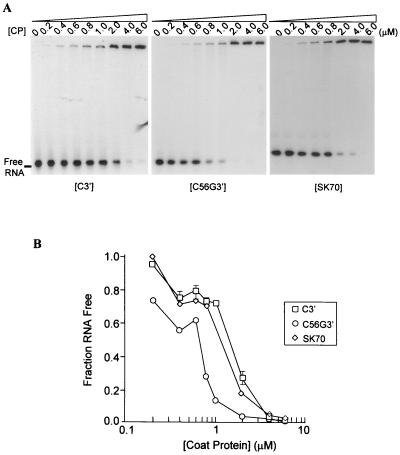
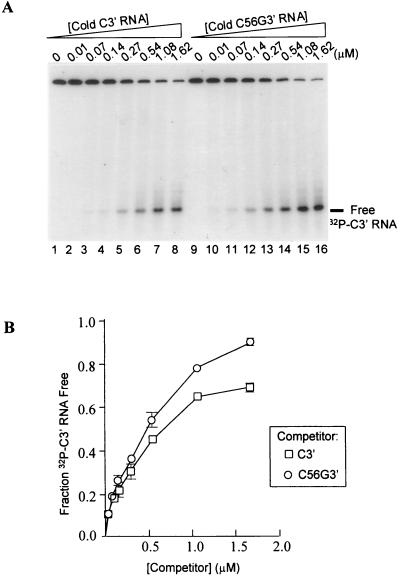
To determine if there is differential CP affinity for the 3′ ends of satC and satC56G (which is phenotypically like diG) and whether any differences can be correlated with symptom modulation, gel retardation experiments were performed with a 64-nt fragment from the 3′-terminal region of satC (C3′) and a 66-nt fragment from the 3′-terminal region of satC56G (C56G3′) (Fig. 4). The TCV CP bound C3′ with an affinity similar to that of a nonviral control RNA of 70 nt (SK70), indicating that the CP binds nonspecifically to the 3′ end of satC. The affinity of the CP for the diG-like 3′ end of C56G3′ was consistently twofold higher than the nonspecific binding to C3′. The higher affinity of the TCV CP for the diG-like 3′ end was confirmed by competition experiments, where increasing concentrations of unlabeled competitor RNAs were added to a fixed amount of 32P-labeled C3′ RNA in the presence of saturating levels of CP. In repeated experiments, C56G3′ RNA was a better competitor for CP binding than C3′ (Fig. 5).
FIG. 4.
Gel retardation analysis of TCV CP binding to RNA fragments. (A) TCV CP binding to the C3′ (64 nt), C56G3′ (66 nt) and SK70 (70 nt) RNAs. 32P-labeled RNAs (100 pM) were incubated in the presence of a series of increasing concentrations of CP, indicated above each gel. Incubation mixtures were subjected to electrophoresis on a 5% polyacrylamide gel that was fixed and dried prior to autoradiography. (B) Quantification of the results presented in panel A and two additional independent experiments (data not shown). Autoradiograms were scanned by densitometry, and the fractions of unbound RNA remaining in the presence of different CP concentrations were determined and plotted against CP concentrations. Each point represents the average of three experiments. Standard deviation bars that are not within the limits of the symbols are shown.
FIG. 5.
Competition for CP binding between 3′-terminal RNA fragments of satC and satC56G. (A) Autoradiogram of a representative competition binding assay between the 32P-labeled C3′ RNA and increasing concentrations (indicated above each lane) of unlabeled C3′ (lanes 1 to 8) and C56G3′ (lanes 9 to 16) RNAs. (B) Quantification of the results from two independent experiments. Autoradiograms were scanned by densitometry, and the fractions of unbound 32P-labeled C3′ RNA in the presence of different competitor concentrations were determined and plotted against the competitor concentrations. Each point represents the average of two experiments. Standard deviation bars that are not within the limits of the symbols are shown.
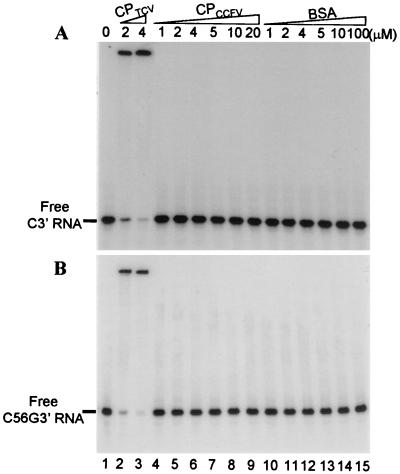
Since the symptoms of TCV-CPCCFV were attenuated by all wt and mutant subviral RNAs, the binding of the CCFV CP to C3′ and C56G3′ RNAs was tested (Fig. 6A and B). No binding of CCFV CP to radiolabeled C3′ or C56G3′ RNA was observed in the presence of up to 20 μM CP (Fig. 6, lanes 4 to 9). Altogether, these results indicate a correlation between CP binding affinity for the 3′-terminal stem-loops of satC and diG and symptom modulation, with reduced or absence of binding corresponding to symptom attenuation by the subviral RNAs.
FIG. 6.
The TCV CP and CCFV CP differentially bind to C3′ (A) and C56G3′ (B) RNAs. 32P-labeled C3′ or C56G3′ RNA (100 pM) in lanes 1 was incubated with increasing concentrations (indicated above each lane) of TCV CP (lanes 2 and 3), CCFV CP (lanes 4 to 9), and bovine serum albumin (lanes 10 to 15). Incubation mixtures were subjected to electrophoresis on a 5% polyacrylamide gel that was fixed and dried prior to autoradiography.
DISCUSSION
3′-terminal stem-loops of satC and diG are involved in symptom modulation.
The differential symptom modulation properties of satC and diG, when coinoculated with TCV-CPm, have allowed for the determination of the region of the subviral RNAs that is involved in symptom modulation. When satC contained positions 5 and 6 of diG, the satRNA acquired the modulation properties of diG. Likewise, diG with positions 5 and 6 of satC gained the ability to attenuate the symptoms of TCV-CPm. Symptom attenuation properties were not correlated with replication competence in protoplasts. While diG and satC* (nonattenuating) replicated more poorly than satC (attenuating), nonattenuating satC56G accumulated to levels equal to those of attenuating satC (Fig. 3B).
Alterations at positions 5 and 6 affected replication by being located within a stable 3′-terminal stem-loop structure of satC, which together with a six-base single-stranded tail comprises the promoter for minus-strand synthesis (Fig. 3A) (3, 45, 46). The 3′ ends of diG and TCV genomic RNA (as well as all other sequenced viral genomic RNAs of the genus Carmoviridae) can also fold into hairpins that likely perform similar functions (45). Bases in position 5 and 6 of satC are located in the loop of the 3′-terminal hairpin. In contrast, while bases in position 5 are within the diG loop, the GG residues in position 6 of diG contribute to forming a longer stem.
The satC and diG mutants with alterations at positions 5 and/or 6 can be divided into four classes according to the composition of their 3′-terminal stem-loops (Fig. 3A). When satC contains positions 5 and 6 from diG (satC56G), the hairpin has the 10-bp stem and five-base loop of diG (class I). satC6G and diG5C have hairpins with 9-bp stems and five-base loops (class II). The hairpins of satC5C* and diG6C have a 10-bp stem that is interrupted with a single-base asymmetrical bulge and a six- or four-base loop, respectively (class III). diG56C contains the shorter hairpin (7-bp stem and nine-base loop) of satC (class IV).
When the symptom modulation properties of the wt and mutant subviral RNAs are viewed in terms of their 3′-terminal hairpins, a correlation was found between class I and class IV hairpins and the ability to attenuate the symptoms of TCV-CPm. satC containing a hairpin similar to diG or TCV (satC56G and satC*) lost the ability to attenuate the symptoms of TCV-CPm, while diG with the 3′-terminal hairpin of satC (diG56C) gained the satC ability to attenuate TCV-CPm symptoms (Fig. 2C).
The mutants with asymmetrical bulges in their 3′-terminal hairpins (class III) gave mixed results in their ability to attenuate TCV-CPm symptoms. The inconsistent results obtained may be due to the mixed presence of satC- and diG-like 3′-terminal stem-loop structures due to the location of the asymmetrical bulge, which may destabilize the upper stem region. Thus, the subviral RNAs may spend a portion of time with a satC-like 7-bp stem and the remaining portion with a longer diG-like 10-bp stem. It is possible that early events in the infection process dictate whether TCV is able to initiate a systemic infection and that these events are influenced by a relatively small number of subviral RNA and/or genomic RNA interactions.
The alterations present in class II mutants satC6G and diG5C were not stable in plants (Table 2). Based on analysis of the computer-predicted structures (Fig. 3A), it is not clear why this instability exists. Given the apparent stability of the hairpins pictured, it is unlikely that the mutations altered the structure of full-length satC or diG such that the 3′-terminal hairpins no longer formed. Both mutants contained the five-base loop sequence CUCGG. Evidence from in vivo genetic selection studies of the 3′-terminal hairpin suggests that the sequence of the loop may play a role in the fitness of the hairpin to promote minus-strand synthesis (3). However, satC6G accumulated in protoplasts to levels equal to those of satC, and diG5C accumulated better in protoplasts than did diG. While it is possible that the instability of the class II mutants in plants was also manifested during 40 h of replication in protoplasts, additional as-yet-unknown parameters regarding the viability of subviral RNAs with class II hairpins cannot be ruled out.
The involvement of a 3′-terminal hairpin structure of a satRNA in symptom modulation has been found for several other virus systems. For example, structural studies of a necrogenic strain of CMV satRNA identified one helix and two tetraloop regions at 3′ ends of the plus-strand RNAs correlating with necrogenicity (31). A more recent report (48) showed that tomato necrosis, but not chlorosis, is induced by high levels of minus-strand necrogenic D4 satRNA expressed via a potato virus X vector in the absence of natural helper CMV. The necrogenic determinant was identified as an octanucleotide loop and adjacent base-paired stem of a thermodynamically stable hairpin in the 5′ end of the minus strand (corresponding to the 3′ end of the plus strand) of all necrogenic satRNAs (48). Studies of peanut stunt virus satRNA-mediated symptom modulation in tobacco also revealed that a hairpin located in the 3′ end of the satRNA is a determinant for symptom attenuation (27, 28). Attenuation and/or nonattenuation of a particular peanut stunt virus satRNA was related to the stabilization or destabilization of the stem region of the hairpin. It was suggested that the hairpin forms a tertiary structure with a distantly located bulged loop which then interacts with putative host and/or viral components, leading to the suppression of viral symptoms (28). However, there is yet no evidence for host involvement (5, 23).
Mechanisms of symptom modulation by TCV-associated subviral RNAs.
The level of TCV CP synthesized by the helper virus determines whether the subviral RNAs intensify, attenuate, or have no effect on symptoms associated with TCV (15, 54). Our current finding that the structure of the 3′-terminal hairpin is important for symptom modulation suggested an interaction between the CP and the hairpin structure. Analysis of TCV CP binding affinity to the 3′-terminal stem-loop structures of satC and satC56G in vitro (Fig. 4 through 6) revealed differential binding that correlated with the ability to attenuate symptoms. The affinity of TCV CP for the 3′-terminal stem-loop of satC56G was twofold greater than for that of the similar region of satC or a nonviral RNA fragment. This result suggests that in the presence of reduced levels of CP, the hairpin of satC might not be bound as extensively to CP, leaving it free to bind to an additional viral or host factor and leading to a reduction in systemic movement of TCV genomic RNA and symptom attenuation, as previously proposed (54). The inability of up to 20 μM of CCFV CP to bind to the 3′ end of either satC or satC56C supports the model, since both subviral RNAs are able to restrict the systemic movement of TCV-CPCCFV.
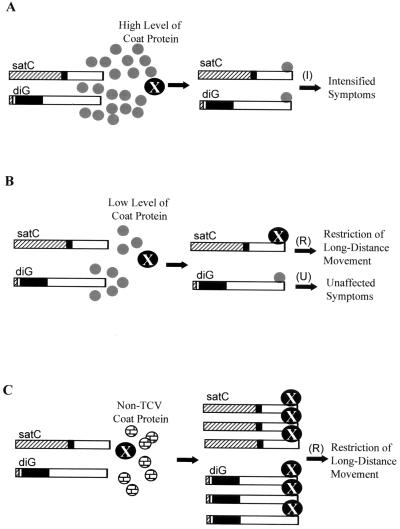
As depicted in Fig. 7, the model suggests that the 3′-terminal stem-loop structures of both satC and diG may be targeted by either the viral CP or a putative host factor (X) that is involved in virus long-distance movement. The presence of high levels of viral CP (as in TCV-infected plants) may exclude the binding of X to the 3′ ends of these two subviral RNAs. Therefore, the putative X is available to assist systemic movement of viral RNA through the plant (Fig. 7A). In contrast, in the presence of reduced levels of CP (as in TCV-CPm-infected plants), X outcompetes the CP for binding to the 3′-terminal stem-loop structure of satC, and sequestration of X by satC leads to restriction of virus long-distance movement and results in symptom attenuation (Fig. 7B). On the other hand, the higher affinity of the stem-loop structure of diG for the TCV CP excludes the binding of X, leading to systemic infection by TCV-CPm (Fig. 7B). Why the symptoms are not then intensified by diG is not known. When TCV-CPCCFV is the helper virus, X is able to bind, and movement is restricted (Fig. 7C) because the CCFV CP, which shares only 65% identity with TCV CP (29), does not bind to the 3′-terminal stem-loop structures of satC and diG (Fig. 6).
FIG. 7.
Putative model for symptom modulation by TCV subviral RNAs in Arabidopsis. In this model, the TCV CP (indicated by small gray circles) binds directly to the 3′ ends of satC and diG and competes for binding with a putative host factor, X (indicated by large black circles), that is involved in virus long-distance movement. I, symptom intensification; R, resistance (i.e., symptom attenuation); U, unaffected symptoms. (A) When high levels of CP are present, CP outcompetes X for binding to the 3′-terminal stem-loops of both satC and diG, leaving X available for virus long-distance movement (i.e., systemic infection). (B) When low levels of CP are present, X outcompetes the CP for binding to the 3′-terminal stem-loop of satC but not that of diG, and sequestration of X by satC restricts virus movement and results in symptom attenuation. In contrast, CP outcompetes X for binding to the 3′-terminal stem-loop of diG, due to the higher affinity of the diG-like hairpin for binding to CP. Why symptoms are not intensified under these conditions is not known. (C) X outcompetes non-TCV CP such as CCFV CP (indicated by small brick-patterned circles) for binding to the 3′ ends of both satC and diG, and sequestration of X by the subviral RNAs restricts virus movement and results in symptom attenuation. Note that satC and diG are more abundant in the absence of TCV CP than in the presence of TCV CP as previously found (16).
There are several other possible ways that access to the free 3′ ends of the subviral RNAs could restrict virus movement. First, while the packaging signals on the subviral RNAs have not been mapped, it is possible that free subviral RNA 3′ ends could interfere with the packaging of the genomic RNA. However, this mechanism is unlikely, since few, if any, virions are present in TCV-CPm-infected cells (17, 54), and thus TCV must be able to spread systemically without a packaging requirement (although CP is required for TCV movement) (8).
Alternatively, since virions are not detected in TCV-CPm-infected protoplasts (17, 54), some other type of viral RNA-CP complex could be engaged in virus movement. Symptom attenuation by satC of TCV variants producing reduced levels of CP could therefore result from satC competing with the viral genomic RNA for limited amounts of CP, thus reducing or eliminating genomic RNA-CP complexes required for systemic movement of the virus.
A third possibility explaining how the subviral RNAs restrict TCV movement is that a free 3′-terminal hairpin of the subviral RNA activates a homology-based posttranscriptional gene-silencing mechanism, a mechanism which involves a sequence-specific degradation process that affects all highly homologous transcripts (7, 24, 55). Cosuppression was suggested as one of the mechanisms of transgenic resistance, mediated by a mild variant satRNA of groundnut rosette virus (49). However, satC only moderately affects the level of viral RNA in either inoculated protoplasts (54) or inoculated leaves (17). In addition, although diG has greater sequence similarity (94%) with the TCV genome in the 3′-terminal region than satC (88%) (Fig. 1C), it does not attenuate the symptoms of TCV-CPm (Fig. 2C) (16).
In summary, symptom attenuation by TCV subviral RNAs is correlated with weakened and/or absent binding of the helper virus CP to the 3′-terminal stem-loop structure of the subviral RNAs. This study provides the first direct evidence linking subviral RNA symptom modulation with determinants from the helper virus. Studies of how interaction between the virus CP and the 3′-terminal stem-loop of the subviral RNA interferes with virus movement and whether a host factor(s) is involved are currently underway.
ACKNOWLEDGMENTS
This work was supported by National Science Foundation grants MCB-9630191 and MCB-9728277 to A.E.S.
We thank Rashie Athuko, a former undergraduate student in the lab, for his assistance in purification of CCFV coat protein.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter C D, Simon A E. In vivo restoration of biologically active 3′ ends of virus-associated RNAs by nonhomologous RNA recombination and replacement of a terminal motif. J Virol. 1996;70:478–486. doi: 10.1128/jvi.70.1.478-486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter C D, Simon A E. Analysis of sequences and predicted structures required for viral satellite RNA accumulation by in vivo genetic selection. Nucleic Acids Res. 1998;26:2426–2432. doi: 10.1093/nar/26.10.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrington J C, Morris T J, Stockley P G, Harrison S C. Structure and assembly of turnip crinkle virus. IV. Analysis of the coat protein gene and implications of the subunit primary structure. J Mol Biol. 1987;194:265–276. doi: 10.1016/0022-2836(87)90374-3. [DOI] [PubMed] [Google Scholar]
- 5.Ferreiro C, Ostrowka K, Lopez-Moya J J, Diaz-Ruiz J R. Nucleotide sequence and symptom modulating analysis of a peanut stunt virus-associated satellite RNA from Poland: high level of sequence identities with the American PSV satellites. Eur J Plant Pathol. 1996;102:779–786. [Google Scholar]
- 6.Finnen R L, Rochon D M. Sequence and structure of defective interfering RNAs associated with cucumber necrosis virus infections. J Gen Virol. 1993;74:1715–1720. doi: 10.1099/0022-1317-74-8-1715. [DOI] [PubMed] [Google Scholar]
- 7.Grant S R. Dissecting the mechanisms of posttranscriptional gene silencing: divide and conquer. Cell. 1999;96:303–306. doi: 10.1016/s0092-8674(00)80541-3. [DOI] [PubMed] [Google Scholar]
- 8.Hacker D L, Petty I T D, Wei N, Morris T J. Turnip crinkle virus genes required for RNA replication and virus movement. Virology. 1992;186:1–8. doi: 10.1016/0042-6822(92)90055-t. [DOI] [PubMed] [Google Scholar]
- 9.Hillman B I, Carrington J C, Morris T J. A defective interfering RNA that contains a mosaic of a plant virus genome. Cell. 1987;51:427–433. doi: 10.1016/0092-8674(87)90638-6. [DOI] [PubMed] [Google Scholar]
- 10.Jaegle M, Devic M, Longstaff M, Baulcombe D. Cucumber mosaic virus satellite RNA (Y strain): analysis of sequences which affect yellow mosaic symptoms on tobacco. J Gen Virol. 1990;71:1905–1912. doi: 10.1099/0022-1317-71-9-1905. [DOI] [PubMed] [Google Scholar]
- 11.Jendrisak J. The use of polyethyleneimine in protein purification. In: Burgess R R, editor. Protein purification: micro to macro. A. R. New York, N.Y: Liss, Inc.; 1987. [Google Scholar]
- 12.Kaper J M, Tousignant M E. Cucumber mosaic virus-associated RNA 5. I. Role of host plant and helper strain in determining amount of associated RNA5 with virions. Virology. 1977;80:186–195. doi: 10.1016/0042-6822(77)90391-9. [DOI] [PubMed] [Google Scholar]
- 13.Kaper J M, Collmer C W. Modulation of viral plant diseases by secondary RNA agents. In: Domingo E, Holland J J, Ahlquist P, editors. RNA genetics. 3. Variability of RNA genomes. Boca Raton, Fla: CRC Press; 1988. pp. 171–194. [Google Scholar]
- 14.Kollar A, Dalmay T, Burgyan J. Defective interfering RNA-mediated resistance against cymbidium ringspot tombusvirus in transgenic plants. Virology. 1993;193:313–318. doi: 10.1006/viro.1993.1127. [DOI] [PubMed] [Google Scholar]
- 15.Kong Q, Oh J-W, Simon A E. Symptom attenuation by a normally virulent satellite RNA of turnip crinkle virus is associated with the coat protein open reading frame. Plant Cell. 1995;7:1625–1634. doi: 10.1105/tpc.7.10.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong Q, Oh J-W, Carpenter C D, Simon A E. The coat protein of turnip crinkle virus is involved in subviral RNA-mediated symptom modulation and accumulation. Virology. 1997;238:478–485. doi: 10.1006/viro.1997.8853. [DOI] [PubMed] [Google Scholar]
- 17.Kong Q, Wang J, Simon A E. Satellite RNA-mediated resistance to turnip crinkle virus in Arabidopsis involves a reduction in virus movement. Plant Cell. 1997;9:2051–2063. doi: 10.1105/tpc.9.11.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W-Z, Qu F, Morris T J. Cell-to-cell movement of turnip crinkle virus is controlled by two small open reading frames that function in trans. Virology. 1998;244:405–416. doi: 10.1006/viro.1998.9125. [DOI] [PubMed] [Google Scholar]
- 19.Li X H, Heaton L, Morris T J, Simon A E. Defective interfering RNAs of turnip crinkle virus intensify viral symptoms and are generated de novo. Proc Natl Acad Sci USA. 1989;86:9173–9177. doi: 10.1073/pnas.86.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X H, Simon A E. Symptom intensification on cruciferous hosts by the virulent satellite RNA of turnip crinkle virus. Phytopathology. 1990;80:238–242. [Google Scholar]
- 21.Masuta C, Takanami Y. Determination of sequence and structural requirements for pathogenicity of a cucumber mosaic virus satellite RNA (Y-satRNA) Plant Cell. 1989;1:1165–1173. doi: 10.1105/tpc.1.12.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuta C, Suzuki M, Kuwata S, Takanami Y, Koiwai A. Yellow mosaic symptoms induced by Y satellite RNA of cucumber mosaic virus are regulated by a single incompletely dominant gene in wild Nicotiana species. Phytopathology. 1993;83:411–413. [Google Scholar]
- 23.Militao V, Moreno I, Rodriguez-Cerezo E, Garcia-Arenal F. Differential interactions among isolates of peanut stunt cucumovirus and its satellite RNA. J Gen Virol. 1998;79:177–184. doi: 10.1099/0022-1317-79-1-177. [DOI] [PubMed] [Google Scholar]
- 24.Montgomery M K, Fire A. Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet. 1998;14:255–258. doi: 10.1016/s0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- 25.Moriones E, Diaz I, Rodriguez-Cerezo E, Fraile A, Garcia-Arenal F. Differential interactions among strains of tomato aspermy virus and satellite RNAs of cucumber mosaic virus. Virology. 1992;186:475–480. doi: 10.1016/0042-6822(92)90012-e. [DOI] [PubMed] [Google Scholar]
- 26.Murphey F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy: sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. [Google Scholar]
- 27.Naidu R A, Collins G B, Ghabrial S A. Symptom-modulating properties of peanut stunt virus satellite RNA sequence variants. Mol Plant-Microbe Interact. 1991;4:268–275. doi: 10.1094/mpmi-4-268. [DOI] [PubMed] [Google Scholar]
- 28.Naidu R A, Collins G B, Ghabrial S A. Peanut stunt virus satellite RNA: analysis of sequences that affect symptom attenuation in tobacco. Virology. 1992;189:668–677. doi: 10.1016/0042-6822(92)90590-l. [DOI] [PubMed] [Google Scholar]
- 29.Oh J-W, Kong Q, Song C, Carpenter C D, Simon A E. Open reading frame of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol Plant-Microbe Interact. 1995;8:979–987. doi: 10.1094/mpmi-8-0979. [DOI] [PubMed] [Google Scholar]
- 30.Palukaitis P. Pathogenicity regulation by satellite RNAs of cucumber mosaic virus: minor nucleotide sequence changes alter host response. Mol Plant-Microbe Interact. 1988;1:175–181. doi: 10.1094/mpmi-1-175. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Alvarado G, Roossinck M J. Structural analysis of a necrogenic strain of cucumber mosaic cucumovirus satellite RNA in planta. Virology. 1997;236:155–166. doi: 10.1006/viro.1997.8731. [DOI] [PubMed] [Google Scholar]
- 32.Romero J, Huang Q, Pogany J, Bujarski J J. Characterization of defective interfering RNA components that increase symptom severity of broad bean mottle virus infections. Virology. 1993;194:576–584. doi: 10.1006/viro.1993.1297. [DOI] [PubMed] [Google Scholar]
- 33.Roossinck M J, Sleat D E, Palukaitis P. Satellite RNAs of plant viruses: structures and biological effects. Microbiol Rev. 1992;56:265–279. doi: 10.1128/mr.56.2.265-279.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roux L, Simon A E, Holland J J. Effects of defective interfering RNAs on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res. 1991;40:181–211. doi: 10.1016/S0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russo M, Burgyan J, Martelli P G. The molecular biology of Tombusviridae. Adv Virus Res. 1994;44:382–424. doi: 10.1016/s0065-3527(08)60334-6. [DOI] [PubMed] [Google Scholar]
- 36.Simon A E, Howell S H. The virulent satellite RNA of turnip crinkle virus has a major domain homologous to the 3′ end of the helper virus genome. EMBO J. 1986;5:3423–3428. doi: 10.1002/j.1460-2075.1986.tb04664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon A E, Engel H, Johnson R P, Howell S H. Identification of regions affecting virulence, RNA processing and infectivity in the virulent satellite of turnip crinkle virus. EMBO J. 1988;7:2645–2651. doi: 10.1002/j.1460-2075.1988.tb03117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon A E, Engel H, Howell S H. Turnip crinkle virus satellite domains involved in virulence and processing. In: Staskowitz B, Ahlquist P, Yoder O, editors. Molecular Biology of Plant-Pathogen Interactions. New York, N.Y: Alan R. Liss, Inc.; 1989. pp. 217–227. [Google Scholar]
- 39.Simon A E, Li X H, Lew J E, Stange R, Zhang C, Polacco M, Carpenter C D. Susceptibility and resistance of Arabidopsis thaliana to turnip crinkle virus. Mol Plant-Microbe Interact. 1992;5:496–503. [Google Scholar]
- 40.Skuzeski J M, Morris T J. Quantitative analysis of the binding of turnip crinkle virus coat protein to RNA fails to demonstrate binding specificity but reveals a highly cooperative assembly interaction. Virology. 1995;210:82–90. doi: 10.1006/viro.1995.1319. [DOI] [PubMed] [Google Scholar]
- 41.Sleat D E, Palukaitis P. Site-directed mutagenesis of a plant viral satellite RNA changes its phenotype from ameliorative to necrogenic. Proc Natl Acad Sci USA. 1990;87:2946–2950. doi: 10.1073/pnas.87.8.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sleat D E, Palukaitis P. Induction of tobacco chlorosis by certain cucumber mosaic virus satellite RNAs is specific to subgroup II helper strain. Virology. 1990;176:292–295. doi: 10.1016/0042-6822(90)90256-q. [DOI] [PubMed] [Google Scholar]
- 43.Sleat D E, Zhang L, Palukaitis P. Mapping determinants within cucumber mosaic virus and its satellite RNA for the induction of necrosis in tomato plants. Mol Plant-Microbe Interact. 1994;7:189–195. doi: 10.1094/mpmi-7-0189. [DOI] [PubMed] [Google Scholar]
- 44.Song C, Simon A E. RNA-dependent RNA polymerase from plants infected with turnip crinkle virus can transcribe (+)- and (−)-strands of virus-associated RNAs. Proc Natl Acad Sci USA. 1994;91:8792–8796. doi: 10.1073/pnas.91.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song C, Simon A E. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J Mol Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- 46.Stupina V, Simon A E. Analysis in vivo of turnip crinkle virus satellite RNA C variants with mutations in the 3′-terminal minus-strand promoter. Virology. 1997;238:470–477. doi: 10.1006/viro.1997.8850. [DOI] [PubMed] [Google Scholar]
- 47.Taliansky M E, Robinson D J. Trans-acting untranslated elements of groundnut rosette virus satellite RNA are involved in symptom production. J Gen Virol. 1997;78:1277–1285. doi: 10.1099/0022-1317-78-6-1277. [DOI] [PubMed] [Google Scholar]
- 48.Taliansky M E, Ryabov E V, Robinson D J, Palukaitis P. Tomato cell death mediated by complementary plant viral satellite RNA sequences. Mol Plant-Microbe Interact. 1998;11:1214–1222. [Google Scholar]
- 49.Taliansky M E, Ryabov E V, Robinson D J. Two distinct mechanisms of transgenic resistance mediated by groundnut rosette virus satellite RNA sequences. Mol Plant-Microbe Interact. 1998;11:367–374. [Google Scholar]
- 50.Tautz N, Thiel H J, Dubovi E J, Meyers G. Pathogenesis of mucosal disease: a cytopathogenic pestivirus generated by an internal deletion. J Virol. 1994;68:3289–3297. doi: 10.1128/jvi.68.5.3289-3297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor J M. Human hepatitis delta virus: an agent with similarities to certain satellite RNAs of plants. Curr Top Microbiol Immunol. 1999;239:108–122. doi: 10.1007/978-3-662-09796-0_6. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Simon A E. Analysis of the two subgenomic RNA promoters for turnip crinkle virus in vivo and in vitro. Virology. 1997;232:174–186. doi: 10.1006/viro.1997.8550. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Carpenter C D, Simon A E. Minimal sequence and structural requirements of a subgenomic RNA promoter for turnip crinkle virus. Virology. 1999;253:327–336. doi: 10.1006/viro.1998.9538. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Simon A E. Symptom attenuation by a satellite RNA in vivo is dependent on reduced levels of virus coat protein. Virology. 1999;259:234–245. doi: 10.1006/viro.1999.9781. [DOI] [PubMed] [Google Scholar]
- 55.Wassenegger M, Pelissier T. A model for RNA-mediated gene silencing in higher plants. Plant Mol Biol. 1998;37:349–362. doi: 10.1023/a:1005946720438. [DOI] [PubMed] [Google Scholar]
- 56.White K A, Skuzeski J M, Li W-Z, Morris T J. Immunodetection, expression strategy and complementation of turnip crinkle virus p28 and p88 replication components. Virology. 1995;211:525–534. doi: 10.1006/viro.1995.1434. [DOI] [PubMed] [Google Scholar]
- 57.White K A, Morris T J. Defective and defective interfering RNAs of monopartite plus-strand RNA plant viruses. Curr Top Microbiol Immunol. 1999;239:1–18. doi: 10.1007/978-3-662-09796-0_1. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L, Kim C H, Palukaitis P. The chlorosis-induction domain of the satellite RNA of cucumber mosaic virus: identifying sequences that affect accumulation and the degree of chlorosis. Mol Plant-Microbe Interact. 1994;7:208–213. doi: 10.1094/mpmi-7-0208. [DOI] [PubMed] [Google Scholar]
- 59.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]