Abstract
Replication of cowpea mosaic virus (CPMV) is associated with small membranous vesicles that are induced upon infection. The effect of CPMV replication on the morphology and distribution of the endomembrane system in living plant cells was studied by expressing green fluorescent protein (GFP) targeted to the endoplasmic reticulum (ER) and the Golgi membranes. CPMV infection was found to induce an extensive proliferation of the ER, whereas the distribution and morphology of the Golgi stacks remained unaffected. Immunolocalization experiments using fluorescence confocal microscopy showed that the proliferated ER membranes were closely associated with the electron-dense structures that contain the replicative proteins encoded by RNA1. Replication of CPMV was strongly inhibited by cerulenin, an inhibitor of de novo lipid synthesis, at concentrations where the replication of the two unrelated viruses alfalfa mosaic virus and tobacco mosaic virus was largely unaffected. These results suggest that proliferating ER membranes produce the membranous vesicles formed during CPMV infection and that this process requires continuous lipid biosynthesis.
Many positive-stranded RNA viruses modify intracellular membranes of their host cells to create a membrane compartment in which RNA replication takes place. Modifications include proliferation and reorganization of different membranes, including the early and late endomembrane system (26, 36, 44, 54), the nuclear envelope (11), the peroxisomal membrane (7), the chloroplasts (49), and the mitochondrial membrane (7). Furthermore, the importance of membranes for viral replication is evident from the observation that the activity of most purified viral RNA-dependent RNA polymerases (RdRps) depends on the presence of membranes and/or phospholipids (27, 32, 59). It was proposed that the membranes play both a structural role and a functional role in the replication complex. Although the modification of intracellular membranes seems an essential part of the viral replicative cycle, little is known about the mechanisms by which the virus converts intracellular membranes for its own use.
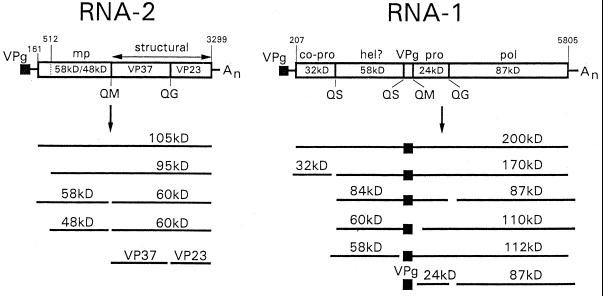
Cowpea mosaic virus (CPMV), a bipartite positive-stranded RNA virus, is the type member of the comoviruses and bears strong resemblance to animal picornaviruses both in gene organization and in amino acid sequence of replicative proteins (1, 15). Both RNA1 and RNA2 express large polyproteins, which are proteolytically cleaved into the different cleavage products by the 24-kDa (24K) protease (Fig. 1). The proteins encoded by RNA1 are necessary and sufficient for replication, whereas RNA2 codes for the capsid proteins and the movement protein.
FIG. 1.
Genetic organization and translational expression of the CPMV genome. Open reading frames in the RNA molecules (open bars) and VPg (black square) are indicated. Nucleotide positions of start and stop codons are indicated. Abbreviations: mp, movement protein; pro, proteinase; co-pro, cofactor for proteinase; hel?, putative helicase; pol, RdRp.
Upon infection of cowpea plants with CPMV, a typical cytopathological structure is formed adjacent to the nucleus, consisting of an amorphous matrix of electron-dense material which is traversed by rays of small membranous vesicles (10). The membranous vesicles are closely associated with CPMV RNA replication, as was revealed by autoradiography in conjunction with electron microscopy on sections of CPMV-infected leaves treated with [3H]uridine (10). Furthermore, the physical association of the viral replicase with membranes became apparent in the purification of the CPMV replication complex, where the viral RdRp activity was found to cofractionate with the crude membrane fraction of infected leaves (14, 60). These experiments moreover revealed that only a small proportion of the viral nonstructural proteins found in CPMV-infected cowpea leaves are active in CPMV replication (14). It was proposed that the electron-dense structures, which were shown to contain the bulk of the nonstructural proteins (57), represent a deposit of inactive replicative proteins which accumulate after having been functional in replication (52).
The origin of the CPMV-induced membranous vesicles, which serve as sites for viral RNA replication, is unknown, but it is tempting to draw an analogy with membrane proliferation in poliovirus-infected cells, as poliovirus and CPMV might use similar strategies for the induction of the membranous vesicles. This suggestion is based on the observation that expression of poliovirus proteins 2BC and 2C in HeLa cells (8) and 2C in yeast cells (2) resulted in an extensive proliferation of membranous vesicles in the cytoplasm, while the 2C homologue of the CPMV 60K protein expressed in insect cells using the baculovirus expression system gave rise to the formation of small vacuoles scattered in the cytoplasm (51).
Several lines of evidence imply that the poliovirus-induced membranous vesicles are derived from the secretory pathway. First, electron microscopy analysis of poliovirus-infected cells showed morphological changes of the endoplasmic reticulum (ER) and a disappearance of structures with the typical morphology of Golgi stacks (42). Second, the poliovirus-induced vesicles contained molecular markers occurring throughout the secretory pathway (ER, trans-Golgi stacks, trans-Golgi network, and lysosomes) (3, 44). Finally, poliovirus infection blocked protein secretion (13), and brefeldin A, a specific inhibitor of the formation of coat protein-coated vesicles, appeared to be a potent inhibitor of poliovirus RNA replication (12, 20, 29). This suggests that the membranous vesicles arise by a mechanism resembling vesicle-mediated intracellular transport. Additionally, it seems that the vesicle formation in poliovirus-infected cells involves de novo membrane synthesis rather than a modification of preexisting membranes, as it was found that cerulenin, a fungal antibiotic which prevents de novo phospholipid biosynthesis and exerts its action by specifically inhibiting β-ketoacyl-acyl carrier protein synthase (31), proved to be a strong inhibitor of poliovirus RNA replication (18). Furthermore, poliovirus infection stimulated the biosynthesis of phosphatidylcholine (53).
To study the effect of CPMV replication on the morphology and distribution of membranes of the secretory pathway, we have expressed the green fluorescent protein (GFP) targeted to the ER and the Golgi and used confocal microscopy to visualize the organelles in living plant cells infected by CPMV. Furthermore, we have tested the action of cerulenin on virus replication.
MATERIALS AND METHODS
Construction of plasmids.
pMON-ERD2-smYFP contains a plant optimized mutant of GFP fused to the coding sequence of the Arabidopsis ERD2 homologue (25) in pMON999. The plant expression vector pMON999 contains a multiple cloning site between the double cauliflower mosaic virus (CaMV) 35S promoter and a terminator sequence of the nopaline synthase gene (Tnos) (52). pMON-smYFP1 (generously provided by G. van der Krogt, Wageningen University, Wageningen, The Netherlands) contains an EcoRI site immediately downstream of the ATG start codon of the coding sequence of the soluble modified yellow fluorescent protein (smYFP), constructed by PCR overlap extension introducing the S65T, V68L, S72A, and T203Y mutations in smGFP (9), which improve the fluorescence intensity of the GFP, using standard fluorescein isothiocyanate (FITC) settings. The coding sequence of the Arabidopsis ERD2 homologue (25) was PCR amplified from an Arabidopsis two-hybrid cDNA library (Clontech) using the following primers, which created additional restriction sites (bold; the start codon is underlined): GGTCTAGATCAACCATGAATATCTTTAGATTTG and GGGAATTCAGCCGGAAGCTTAAGTTTGGTG. This PCR fragment was digested with XbaI and EcoRI and cloned in pMON-smYFP1 digested with the same enzymes. The resulting clone was designated pMON-ERD2-smYFP.
pMON-STtmd-eYFP contains the first 53 amino acids of the rat sialyltransferase (ST) gene containing the transmembrane domain fused to a mutant of GFP. pMON-eYFP (generously provided by G. van der Krogt) contains an NcoI site overlapping the start codon of the coding sequence of enhanced YFP obtained by PCR using yellow cameleon-2 (30) (generously provided by R. Y. Tsien, University of California, San Diego) as template. The complete cDNA of the rat ST was released from pSMH4 (generously provided by S. Munro, Cambridge University, Cambridge, England) using HindIII and XbaI and subcloned in pBSK(−), creating a ClaI recognition site directly upstream of the 5′ nontranslated region of ST. Subsequent digestion with ClaI and NcoI created a fragment containing the 5′ nontranslated region and the coding region for the first 53 amino acids of ST, and this fragment was cloned in pMON-eYFP digested with the same restriction enzymes. The resulting clone was designated pMON-STtmd-eYFP.
pUC-mGFP5-ER contains the plant optimized GFP5 (47) with an N-terminal Arabidopsis thaliana basic chitinase signal sequence and a C-terminal HDEL ER retention signal (19). This region was released from pBIN m-GFP5-ER (generously provided by J. Haseloff, Cambridge University) and cloned as an XbaI-SstI fragment in the smBFP vector (9) (obtained from the Arabidopsis Biological Research Center at The Ohio State University), replacing the GFP present in the original construct. The resulting clone contains mGFP5-ER between a CaMV 35S promoter and a Tnos in the high-copy-number plasmid pUC118 and is referred to as pUC-mGFP5-ER.
Fluorescence microscopy.
A Zeiss LSM 510 confocal microscope was used to obtain images. Standard filters for FITC and rhodamine were used to detect GFP5/YFP and Cy3 in fixed protoplasts or GFP5/YFP and the chlorophyll in living protoplasts (FITC: excitation wavelength 488 nm, emission band pass filter 505 to 550 nm; rhodamine: excitation wavelength 543 nm, emission long pass filter 560 nm).
Transfection of cowpea protoplasts and infection of plants.
Cowpea (Vigna unguiculata L.) mesophyll protoplasts were prepared and transfected by polyethylene glycol-mediated transformation as described previously (52). Five-week-old Nicotiana benthamiana plants carrying the mGFP5-ER transgene (41) (generously provided by D. Baulcombe, John Innes Centre, Norwich, England) were dusted with carborundum and inoculated with a homogenate of CPMV-infected leaves.
Immunofluorescent analysis of transfected protoplasts.
Immunofluorescent detection of accumulation of viral proteins to determine the number of infected cells using the inhibitor cerulenin was performed as described previously (52). For the double-labeling experiments, a different method of fixation was used to retain the GFP fluorescence as follows.
At 42 h posttransfection, the protoplasts were harvested for immunofluorescent staining. The protoplasts were allowed to settle on poly-l-lysine-coated coverslips, and 1 volume of fixing solution (4% paraformaldehyde, 0.1% glutaraldehyde, 0.25 M mannitol, 50 mM sodium phosphate) was added to the protoplast suspension. After incubation for 15 min, the liquid was removed, replaced with fixing solution, and allowed to incubate for another 30 min. The cells were washed three times with phosphate-buffered saline (PBS) and permeabilized with a 0.5% Triton X-100 solution in PBS for 10 min. Nonspecific antibody binding was reduced by incubation for 10 min in blocking solution (1% bovine serum albumin and 0.8% gelatin from cold water fish skin in PBS). Subsequently, the protoplasts were incubated for 1 h with dilutions of the primary anti-48K (58), anti-VPg (14), or anti-110K (51) antibody in blocking solution. After three washes with PBS, the protoplasts were incubated with goat anti-rabbit antibodies conjugated to Cy3 (Sigma) for another hour. After two washes with PBS, the coverslips were mounted on microscope slides using Citifluor.
Electron microscopy analysis of N. benthamiana mesophyll cells infected with CPMV.
For electron microscopy, samples were cut from infected leaf tissue and fixed in glutaraldehyde-paraformaldehyde, followed by postfixation with osmium tetroxide and uranyl acetate, dehydration with ethanol, and embedding in LR White as described by van Lent et al. (55). For immunogold staining, thin sections were treated with saturated sodium metaperiodate for 1 h at room temperature, washed with distilled water, and subsequently labeled and stained as described by van Lent et al. (55), using commercially available polyclonal antibodies to GFP (Molecular Probes).
RESULTS
Morphological changes of the ER but not the Golgi apparatus upon CPMV infection.
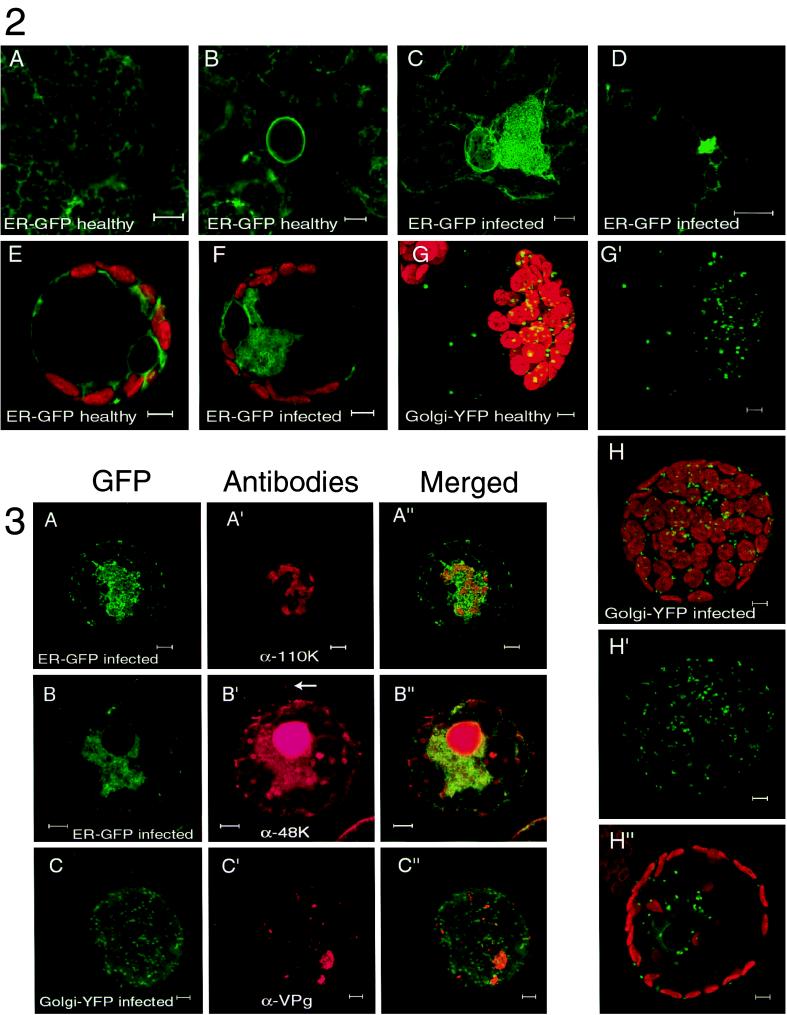
Transgenic N. benthamiana plants, expressing GFP targeted to the lumen of the ER (19), were mechanically inoculated with CPMV. Three days postinfection (p.i.), the leaves were examined by confocal fluorescence microscopy and compared to mock-inoculated leaves. As expected, in epidermal cells of mock-inoculated leaves, green fluorescence was detected in the typical stationary cortical ER network (Fig. A), the nuclear envelope (Fig. B), and the ER tubules traversing the cytoplasmic threads (data not shown). In contrast, in the epidermis of CPMV-infected leaves, we found clusters of cells which contained, in addition to the ER structures as occurred in the mock-inoculated cells, a large GFP-containing structure often located near the nucleus. High-resolution imaging showed that this structure was connected to the cortical ER network and consisted of a ball of tubular ER (Fig. C). Once formed, this structure remained present in the infected cell for the 2-week duration of the experiment. To obtain insight into morphological changes of the ER preceding the formation of this large fluorescent structure, a cluster of infected cells 4 days p.i. was examined. From the center to the periphery, such a cluster of infected cells represents an approximate time course of infection because CPMV infection starts at a single epidermal cell and subsequently spreads to neighboring cells within 2 days p.i. (56; unpublished results). Newly infected cells at the periphery of the cluster showed several small, highly mobile bodies of fluorescence, which were connected to the cortical ER network (Fig. D shows a close-up). In these cells, the large stationary fluorescent body present in cells infected for a longer period, more inward to the cluster of infected cells, was absent. It should be noted that the cortical ER network in both newly infected cells (Fig. D) and cells infected for a longer period (Fig. C) remained intact despite the formation of these highly fluorescent bodies, suggesting that they are formed by a process of proliferation of preexisting ER membranes rather than aggregation of these membranes.
To confirm that CPMV-induced ER proliferation also takes place in cells from the natural host plant V. unguiculata L., cowpea mesophyl protoplasts were isolated and transfected with pUC-mGFP5-ER alone or together with CPMV RNA. Typically 40% of the protoplasts showed GFP fluorescence whereas over 80% of the protoplasts were infected, as tested 2 days p.i. by immunofluorescence with antibodies directed against the RNA1-encoded 110K protein. The organization of the ER in uninfected living cowpea protoplasts was examined 42 h posttransfection and differed somewhat from the epidermal cells in the transgenic N. benthamiana plants. Again the cortical ER network and nuclear envelope were readily visible, but the majority of fluorescence was seen in disordered ER tubules in the cytoplasm surrounding the nucleus and the chloroplasts (Fig. E). In the CPMV-infected protoplasts, additionally a large body of fluorescence often near the nucleus was observed in the majority of the pUC-mGFP5-ER-transfected cells (Fig. F). Observation of the infected protoplasts at an earlier time point (18 h p.i.) did not reveal changes of the ER structure in comparison to uninfected cells, although the disorderly nature of the ER in the cytoplasm could have obscured minor changes in ER structure. Next we tested whether RNA1 replication alone, without expression of the viral capsid proteins and movement proteins encoded by RNA2, was able to induce the observed changes in ER structure. For this purpose, RNA transcripts of an infectious cDNA clone of RNA1 were cotransfected with pUC-mGFP5-ER and at 2 days p.i. were observed with a fluorescence microscope. Again the large regions of proliferated ER membranes were apparent in protoplasts infected with RNA1 alone (data not shown).
Since in plant cells the Golgi stacks are closely associated with the ER (4), it was of interest to study the distribution of the individual Golgi stacks and the possible proliferation of Golgi-derived membranes in CPMV-infected cells. For this purpose, two different Golgi markers were used in vivo: an A. thaliana homologue of the yeast HDEL receptor fused to GFP, which accumulates mainly in the Golgi stacks and partly in the ER (pMON-ERD2-smYFP) (4), and the N-terminal transmembrane domain of the rat ST fused to GFP, which accumulates exclusively in the Golgi stacks (pMON-STtmd-eYFP) (4). In uninfected cowpea protoplasts, the numerous Golgi stacks as visualized by both ERD2-smYFP and STtmd-eYFP were scattered uniformly throughout the cytoplasm, mainly surrounding the chloroplasts and in cytoplasmic threads (ERD2-smYFP [Fig. G and G′]; STtmd-eYFP [data not shown]). Using the faint background staining of the ER by ERD2-smYFP, we verified that the Golgi stacks were associated with ER tubules as was reported previously (4) (data not shown). In CPMV-infected protoplasts, the amount and distribution of the Golgi stacks did not differ from uninfected cells (ERD2-smYFP [Fig. H to H"]; STtmd-eYFP [data not shown]) despite the presence of the large region of proliferated ER observed with ERD2-smYFP (Fig. H").
The data indicate that CPMV infection causes a strong proliferation of ER membranes starting at the cortical ER network, ultimately leading to a large region of densely packed ER membranes often near the nucleus. The changes in ER morphology can be induced by infection with RNA1 alone, suggesting a role for these structures in viral RNA replication. The distribution and morphology of the Golgi stacks remain unaffected in CPMV-infected cells.
RNA1 proteins involved in replication colocalize with the ER but not the Golgi stacks.
CPMV RNA replication occurs on clusters of smooth membranous vesicles located near the large electron-dense structures that contain the bulk of the replicative RNA1-encoded proteins (10). To determine whether the observed bodies of proliferated ER membranes colocalize with this cytopathological structure, CPMV-infected cowpea protoplasts transfected with pUC-mGFP5-ER were fixed and immunostained with antibodies raised against the RNA1-encoded proteins VPg and 110K. In the infected cells, intermediate cleavage products accumulate; of these, the anti-VPg antibody recognizes the 170K, 112K, 84K, and 60K proteins and the anti-110K antibody recognizes the 170K, 112K, 87K, and 84K proteins (Fig. 1). Furthermore, an antibody was used against the RNA2-encoded 48K movement protein to determine possible colocalization with the proliferated ER membranes.
GFP retained its green fluorescence during the immunostaining procedure, and the antibodies raised against the viral proteins were stained with red fluorescence by treatment with goat anti-rabbit-Cy3 as secondary antibody. Confocal microscopy showed that in the majority of cells the replicative proteins were localized in one or several large fluorescent bodies per cell (Fig. A′ and C′) probably corresponding to the matrix of electron-dense structures observed by electron microscopy. The proliferated ER membranes stained by mGFP5-ER were always found to surround and traverse these bodies (anti-110K [Fig. A to A"]; anti-VPg [data not shown]). A small proportion of the proteins recognized by anti-48K colocalized with the proliferated ER membranes, but the majority was localized in the nucleus and in the typical tubular structures formed by the movement protein (Fig. B to B"). On the other hand, no colocalization was found for the replicative proteins visualized with anti-VPg and the Golgi markers STtmd-eYFP and ERD2-smYFP (STtmd-eYFP [Fig. C to C"]; ERD2-smYFP [data not shown]).
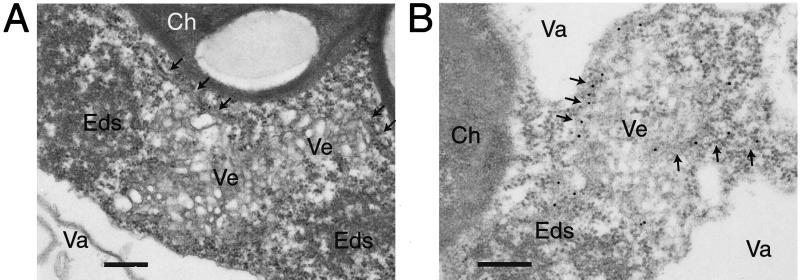
To further analyze the presence of proliferated ER in the cytopathological structures, leaf tissues of CPMV-infected N. benthamiana plants transgenic with ER-GFP were prepared for electron microscopy. ER tubules could be distinguished based on their morphology and were shown to be present near the electron-dense structures and the small membranous vesicles (Fig. 4A). Immunolabeling with anti-GFP confirmed that ER-GFP was present in ER tubules near these structures (Fig. 4B). No specific labeling of ER-GFP was found in the small membranous vesicles.
FIG. 4.
Electron microscopy of cytopathological structures in CPMV-infected N. benthamiana mesophyll cells carrying the mGFP5-ER transgene. (A) ER tubules (arrows) located near electron-dense structures (Eds) and small membranous vesicles (Ve). (B) Immunolabeling with anti-GFP shows labeling of the ER tubules (arrows) and not the vesicles. Ch, chloroplasts; Va, vacuole. Bars = 300 nm.
These data show that the cytopathological structures are enriched with proliferated ER membranes but not with Golgi membranes, suggesting that the ER produces the small membranous vesicles which are the sites of viral RNA replication.
Replication of CPMV requires continuous lipid biosynthesis.
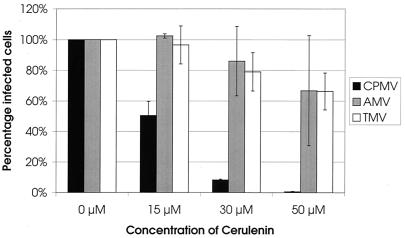
To examine whether the proliferation of ER membranes is essential for viral RNA replication, the effect of cerulenin, an inhibitor of de novo lipid synthesis (35, 45), on the replication of CPMV was tested. For this purpose, cowpea protoplasts were infected with virus and divided in four portions, and cerulenin was added in different concentrations to the incubation medium. Two days p.i., the protoplasts were fixed and stained with antibodies raised against the viral proteins and the percentage of fluorescent cells was calculated. Because viral proteins accumulate to detectable levels only when replication of the virus takes place, the percentage of fluorescent cells is considered to correspond to the percentage of infected cells (52). Figure 5 summarizes the results of two independent experiments. Infection of the sample in which no cerulenin was added was normalized to 100%. The presence of 15 μM cerulenin in the incubation medium markedly decreased the infection rate to 50%, while concentrations of 30 and 50 μM further decreased the infection rate to 10 and 0%, respectively. These results show that CPMV replication is strongly inhibited by cerulenin. To exclude the possibility that the observed inhibitory effect is due to a reduced viability of the protoplasts and not the inhibition of de novo lipid synthesis per se, the effect of cerulenin on the replication of the unrelated viruses tobacco mosaic virus (TMV) and alfalfa mosaic virus (AMV) was tested using identical concentrations of cerulenin. As shown in Fig. 5, cerulenin slightly decreased the infection rates of AMV and TMV to 65% at the highest concentration of 50 μM. The requirement of continuous lipid biosynthesis for CPMV replication suggests that the formation of new membranes plays an essential role in the viral replicative cycle.
FIG. 5.
Cerulenin inhibits CPMV but not TMV or AMV replication in cowpea protoplasts. Infected protoplasts were divided in four equal portions and incubated for 48 h in the presence of 0, 15, 30, or 50 μM cerulenin. Subsequently the protoplasts were processed for indirect immunofluorescence using antisera against CPMV 110K, TMV coat protein, and AMV coat protein, and the percentage of infected cells was determined. For each virus two independent experiments were performed; error bars indicate the standard deviation.
DISCUSSION
In this study, we have shown, by confocal fluorescence microscopy using living tissue expressing GFP targeted to the ER and the Golgi, that CPMV infection induces a massive proliferation of ER resulting in the formation of a cytopathological structure highly enriched in ER membranes but not Golgi stacks. The Golgi stacks were moving in close association with the cortical ER network, which remained intact during the whole infection process. Remarkably, they were excluded from the region of proliferating ER membranes, indicating that this region is a distinct subdomain of the ER disturbed in its normal function of protein trafficking through the secretory pathway. The rapid formation of such a subregion of the ER specialized in viral replication exemplifies the remarkable versatility and adaptability of this organelle. In plants, as many as 16 ER domains can be distinguished based on morphological features and their known or postulated functions (48).
The proliferated ER membranes were present in the cytopathological structure, as shown with immunolabeling experiments using confocal microscopy and electron microscopy. The enrichment of ER membranes in this region suggests that they produce small membranous vesicles, which are the sites of viral replication (10). Since in plant cells the ER is the site where the synthesis of the majority of the phospholipids takes place (34), CPMV might stimulate this synthesis, leading to ER proliferation and vesiculation. ER membranes are implicated in viral replication due to their intimate association with replicative proteins for many positive-stranded RNA viruses, including tobamoviruses (28), bromoviruses (38, 39), flaviviruses (17), potyviruses (43), and nidoviruses (36). The Golgi membranes did not proliferate, and the amount and distribution of Golgi stacks did not differ from those of uninfected cells, suggesting at most a minor role for this organelle in CPMV-induced vesiculation. In contrast, poliovirus infection causes a complete disassembly of the Golgi stacks (42), and isolated membranous vesicles involved in replication were shown to contain molecular markers from throughout the secretory pathway, including the ER, trans-Golgi stacks, trans-Golgi network, and lysosomes (44). The clusters of vesicles in the cytopathological structures in CPMV-infected tobacco leaf cells transgenic with mGFP5-ER did not show this marker in electron microscopy. Also, the poliovirus-induced vesicles proven to originate from the ER (3) contained a relatively low amount of the luminal ER marker protein disulfate isomerase (44). This indicates that during the generation of these vesicles, luminal ER proteins are excluded.
Investigation of a cluster of infected cells on transgenic N. benthamiana leaves expressing GFP targeted to the ER allowed us to observe different stages of the infection cycle. At the border of infection sites, several fluorescent bodies at the cortical ER were found to be the first signs of proliferation of ER membranes. These bodies then aggregate into one large fluorescent body, usually near the nucleus. Also, for poliovirus it has been described that RNA replication starts at small clusters of membranous vesicles distributed through most of the cytoplasm and that viral RNA associated with these vesicles later in infection migrate to the center of the cell (6).
The unique opportunity to observe virus-mediated changes of ER in planta using mGFP5-ER has been exploited for several viruses, including potato virus X (PVX), tobacco etch virus (TEV) and TMV (5, 37, 43). The morphological changes of the ER induced by infection with these viruses all involved the formation of large fluorescent structures as seen with CPMV, but comparison brings to light some remarkable differences. For TMV and TEV infection, it was reported that the formation of the large fluorescent structures coincides with the disappearance of the typical cortical ER network (37, 43), suggesting that preexisting ER membranes aggregate to form the structure. In contrast, during PVX (5) and CPMV infection, the typical cortical ER network remains present, indicating that the fluorescent bodies are formed by proliferation of membranes. Our results showing that CPMV, but not TMV, replication requires formation of new membranes, as tested with the inhibitor cerulenin, are consistent with this observation. Furthermore, TMV-induced changes of the ER network are transient and seem to coincide with the synthesis and subsequent degradation of the movement protein (28, 37), while the CPMV-induced changes are permanent and independent of expression of the movement protein of CPMV. The latter was shown in cowpea cells where infection with RNA1 alone showed ER proliferation. Previous experiments using electron microscopy showed that in RNA1-infected protoplasts, small membranous vesicles were formed (40).
Although the movement protein plays no role in the induction of the modified ER membranes, a small proportion of the proteins recognized by anti-48K colocalized with this structure in CPMV-infected protoplasts. It should be noted that anti-48K recognizes both the 48K movement protein and the co-C-terminal 58K protein that has been implicated in replication of RNA2 (50). Possibly the observed colocalization with proliferated ER membranes reflects primarily 58K proteins present in replication complexes at the site of viral replication. Previous experiments, however, indicate that the bulk of the 58K proteins accumulates in the nucleus (23). Alternatively, the colocalization reflects 48K and 58K proteins, which are recently synthesized and not yet distributed to the tubules and the nucleus, as it is likely that the site of replication corresponds to the site of translation of viral proteins (50). Like the 58K protein, the N-terminal 2A protein of grapevine fanleaf virus, another member of the family Comoviridae, is required for RNA2 replication (16). Moreover, it was shown that a 2A-GFP fusion protein, transiently expressed from a plant expression vector, partly colocalized with the sites of viral replication. This observation led the authors to suggest that this domain within the polyprotein is responsible for targeting RNA2 to the replication site (16).
The observation of this radical virus-induced proliferation of ER membranes, which the virus uses for its replication, raises the question of how the virus accomplishes the disturbance of a normal function of healthy cells. The CPMV-induced ER proliferation resembles the drastic changes in ER morphology which occur after overexpression of certain endogenous ER-resident membrane proteins in yeast and in animal cells (e.g., 3-hydroxymethyl-3-methylglutaryl coenzyme A reductase [22, 24], cytochrome P-450 [33], and malfolded cytochrome P-450 [21]). It was shown in these cases that overcrowding of the ER membrane and/or improper folding of the overexpressed integral ER membrane proteins affected the triggering of the so-called unfolded protein response (UPR). This is a mechanism to relieve ER stress by both the up-regulation of ER-resident chaperone proteins like the heavy-chain binding protein (BiP), protein disulfate isomerase, and KAR2 and an increase in phospholipid synthesis (for a review, see reference 46). We speculate that CPMV infection triggers the UPR, which leads to the observed CPMV-induced proliferation of ER membranes and rationalizes the requirement of CPMV replication for de novo phospholipid biosynthesis. The viral protein responsible for triggering the UPR might be the RNA1-encoded 60K protein because it was shown that this protein expressed in insect cells using the baculovirus expression system induces the formation of small vesicles (51).
FIG. 2-3.
Confocal fluorescence micrographs of healthy (A, B, E, G, and G′) and CPMV-infected (C, D, F, H, H′, and H") plant cells expressing GFP or YFP targeted to the ER (A to F) or the Golgi (G to H"). The confocal images were collected with focal depth of 1 μm, using standard FITC filter settings to detect GFP or YFP (pseudo-colored green) and standard rhodamine filter settings to detect autofluorescence of the chlorophyll (pseudo-colored red). (C, G, G′, H, and H′) Projections of serial optical sections; (A, B, D, E, F, and H") projections of single optical sections. (A to D) N. benthamiana mGFP5-ER epidermal cells. (A) Reticulate pattern of cortical ER network. (B) Fluorescent halo of mGFP5-ER in nuclear envelope. (C) Large body of proliferated ER adjacent to nucleus in CPMV-infected cell. (D) Small cortical body of proliferated ER early in infection. (E to H") Cowpea mesophyll protoplasts. (E) Disorganized ER tubules in cytoplasm surrounding chloroplasts and nucleus in uninfected cells. (F) Large body of proliferated ER in CPMV-infected cell. (G and G′) Golgi stacks in uninfected cell scattered through cytoplasm visualized by ERD2-smYFP fluorescence, shown in combination with autofluorescence of chloroplasts (G) or alone (G′). (H to H") Similar distribution in CPMV-infected cells using ERD2-smYFP shown in combination with autofluorescence of chloroplasts (H) or alone (H′). As illustrated in a single optical section, ERD2-smYFP faintly stains the ER, showing the nuclear envelope and the CPMV-induced large body of proliferated ER (H"). Bars = 5 μm. Immunofluorescence double labeling showing the intracellular distribution of mGFP5-ER targeted to the ER (A and B) and STtmd-eYFP targeted to the Golgi (C) and viral proteins in CPMV-infected cowpea protoplasts. Cells were fixed 48 h p.i. and processed for indirect immunofluorescence using rabbit antibodies against the viral proteins followed by anti-rabbit antibodies conjugated to Cy3. GFP or YFP retained its fluorescence throughout the procedure. Rows show GFP or YFP (left), viral protein (middle), and their superposition (right) of a representative cell. The antibodies used were raised against the replicative proteins 110K (A′) and VPg (C′) and the 48K movement protein (B′). The arrow indicates the tubular structure formed by the 48K movement protein. (C to C") Projections of serial optical sections; (A to B") projections of single optical sections. Bars = 5 μm.
ACKNOWLEDGMENTS
We are grateful to David Baulcombe, Jim Haseloff, Gerard van der Krogt, Sean Munro, and Roger Tsien for providing the biological material indicated in Materials and Methods. Ton Bisseling is acknowledged for critically reading the manuscript.
This work was supported by The Netherlands Foundation of Chemical Research with financial aid from The Netherlands Organisation for Scientific Research.
REFERENCES
- 1.Argos P, Kamer G, Nicklin M J, Wimmer E. Similarity in gene organization and homology between proteins of animal picornaviruses and a plant comovirus suggest common ancestry of these virus families. Nucleic Acids Res. 1984;12:7251–7267. doi: 10.1093/nar/12.18.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barco A, Carrasco L. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3349–3364. doi: 10.1002/j.1460-2075.1995.tb07341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bienz K, Egger D, Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987;160:220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- 4.Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;15:441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- 5.Boevink P, Santa Cruz S, Hawes C, Harris N, Oparka K J. Virus-mediated delivery of the green fluorescent protein to the endoplasmic reticulum of plant cells. Plant J. 1996;10:935–941. [Google Scholar]
- 6.Bolten R, Egger D, Gosert R, Schaub G, Landmann L, Bienz K. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent in situ hybridization. J Virol. 1998;72:8578–8585. doi: 10.1128/jvi.72.11.8578-8585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgyan J, Rubino L, Russo M. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J Gen Virol. 1996;77:1967–1974. doi: 10.1099/0022-1317-77-8-1967. [DOI] [PubMed] [Google Scholar]
- 8.Cho M W, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 9.Davis S J, Vierstra R D. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- 10.De Zoeten G A, Assink A M, van Kammen A. Association of cowpea mosaic virus-induced double-stranded RNA with a cytopathological structure in infected cells. Virology. 1974;59:341–355. doi: 10.1016/0042-6822(74)90449-8. [DOI] [PubMed] [Google Scholar]
- 11.De Zoeten G A, Gaard G, Diez F B. Nuclear vesiculation associated with pea enation mosaic virus-infected plant tissue. Virology. 1972;48:638–647. doi: 10.1016/0042-6822(72)90148-1. [DOI] [PubMed] [Google Scholar]
- 12.Doedens J, Maynell L A, Klymkowsky M W, Kirkegaard K. Secretory pathway function, but not cytoskeletal integrity, is required in poliovirus infection. Arch Virol Suppl. 1994;9:159–172. doi: 10.1007/978-3-7091-9326-6_16. [DOI] [PubMed] [Google Scholar]
- 13.Doedens J R, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggen R, Kaan A, Goldbach R, van Kammen A. Cowpea mosaic virus RNA replication in crude membrane fractions from infected cowpea and Chenopodium amaranticolor. J Gen Virol. 1988;69:2711–2720. [Google Scholar]
- 15.Franssen H, Leunissen J, Goldbach R, Lomonossoff G, Zimmern D. Homologous sequences in non-structural proteins from cowpea mosaic virus and picornaviruses. EMBO J. 1984;3:855–861. doi: 10.1002/j.1460-2075.1984.tb01896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaire F, Schmitt C, Stussi-Garaud C, Pinck L, Ritzenthaler C. Protein 2A of grapevine fanleaf nepovirus is implicated in RNA2 replication and colocalizes to the replication site. Virology. 1999;264:25–36. doi: 10.1006/viro.1999.9927. [DOI] [PubMed] [Google Scholar]
- 17.Grief C, Galler R, Cortes L M, Barth O M. Intracellular localisation of dengue-2 RNA in mosquito cell culture using electron microscopic in situ hybridisation. Arch Virol. 1997;142:2347–2357. doi: 10.1007/s007050050247. [DOI] [PubMed] [Google Scholar]
- 18.Guinea R, Carrasco L. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 1990;9:2011–2016. doi: 10.1002/j.1460-2075.1990.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haseloff J, Siemering K R, Prasher D C, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irurzun A, Perez L, Carrasco L. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology. 1992;191:166–175. doi: 10.1016/0042-6822(92)90178-r. [DOI] [PubMed] [Google Scholar]
- 21.Ishihara N, Yamashina S, Sakaguchi M, Mihara K, Omura T. Malfolded cytochrome P-450(M1) localized in unusual membrane structures of the endoplasmic reticulum in cultured animal cells. J Biochem (Tokyo) 1995;118:397–404. doi: 10.1093/oxfordjournals.jbchem.a124920. [DOI] [PubMed] [Google Scholar]
- 22.Jingami H, Brown M S, Goldstein J L, Anderson R G, Luskey K L. Partial deletion of membrane-bound domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase eliminates sterol-enhanced degradation and prevents formation of crystalloid endoplasmic reticulum. J Cell Biol. 1987;104:1693–1704. doi: 10.1083/jcb.104.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasteel D, Wellink J, Verver J, van Lent J, Goldbach R, van Kammen A. The involvement of cowpea mosaic virus M RNA-encoded proteins in tubule formation. J Gen Virol. 1993;74:1721–1724. doi: 10.1099/0022-1317-74-8-1721. [DOI] [PubMed] [Google Scholar]
- 24.Kochevar D T, Anderson R G. Purified crystalloid endoplasmic reticulum from UT-1 cells contains multiple proteins in addition to 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Biol Chem. 1987;262:10321–10326. [PubMed] [Google Scholar]
- 25.Lee H I, Gal S, Newman T C, Raikhel N V. The Arabidopsis endoplasmic reticulum retention receptor functions in yeast. Proc Natl Acad Sci USA. 1993;90:11433–11437. doi: 10.1073/pnas.90.23.11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackenzie J M, Jones M K, Westaway E G. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J Virol. 1999;73:9555–9567. doi: 10.1128/jvi.73.11.9555-9567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin M T, Cervera M T, Garcia J A, Bonay P. Properties of the active plum pox potyvirus RNA polymerase complex in defined glycerol gradient fractions. Virus Res. 1995;37:127–137. doi: 10.1016/0168-1702(95)00028-o. [DOI] [PubMed] [Google Scholar]
- 28.Mas P, Beachy R N. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J Cell Biol. 1999;147:945–958. doi: 10.1083/jcb.147.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maynell L A, Kirkegaard K, Klymkowsky M W. Inhibition of poliovirus RNA synthesis by brefeldin A. J Virol. 1992;66:1985–1994. doi: 10.1128/jvi.66.4.1985-1994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyawaki A, Llopis J, Heim R, McCaffery J M, Adams J A, Ikura M, Tsien R Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 31.Moche M, Schneider G, Edwards P, Dehesh K, Lindqvist Y. Structure of the complex between the antibiotic cerulenin and its target, beta-ketoacyl-acyl carrier protein synthase. J Biol Chem. 1999;274:6031–6034. doi: 10.1074/jbc.274.10.6031. [DOI] [PubMed] [Google Scholar]
- 32.Molla A, Paul A V, Wimmer E. Effects of temperature and lipophilic agents on poliovirus formation and RNA synthesis in a cell-free system. J Virol. 1993;67:5932–5938. doi: 10.1128/jvi.67.10.5932-5938.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohkuma M, Park S M, Zimmer T, Menzel R, Vogel F, Schunck W H, Ohta A, Takagi M. Proliferation of intracellular membrane structures upon homologous overproduction of cytochrome P-450 in Candida maltosa. Biochim Biophys Acta. 1995;1236:163–169. doi: 10.1016/0005-2736(95)00040-a. [DOI] [PubMed] [Google Scholar]
- 34.Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Packter N M, Stumpf P K. Fat metabolism in higher plants. The effect of cerulenin on the synthesis of medium- and long-chain acids in leaf tissue. Arch Biochem Biophys. 1975;167:655–667. doi: 10.1016/0003-9861(75)90509-3. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen K W, van der Meer Y, Roos N, Snijder E J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichel C, Beachy R N. Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc Natl Acad Sci USA. 1998;95:11169–11174. doi: 10.1073/pnas.95.19.11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Restrepo-Hartwig M, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J Virol. 1999;73:10303–10309. doi: 10.1128/jvi.73.12.10303-10309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Restrepo-Hartwig M A, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rezelman G, Franssen H J, Goldbach R, Ie T S, van Kammen A. Limits to the independence of bottom component RNA of cowpea mosaic virus. J Gen Virol. 1982;60:335–342. [Google Scholar]
- 41.Ruiz M T, Voinnet O, Baulcombe D C. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandoval I V, Carrasco L. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J Virol. 1997;71:4679–4693. doi: 10.1128/jvi.71.6.4679-4693.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaad M C, Jensen P E, Carrington J C. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlegel A, Giddings T H, Jr, Ladinsky M S, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider F, Lessire R, Bessoule J J, Juguelin H, Cassagne C. Effect of cerulenin on the synthesis of very-long-chain fatty acids in microsomes from leek seedlings. Biochim Biophys Acta. 1993;1152:243–252. doi: 10.1016/0005-2736(93)90255-x. [DOI] [PubMed] [Google Scholar]
- 46.Sidrauski C, Chapman R, Walter P. The unfolded protein response: an intracellular signalling pathway with many surprising features. Trends Cell Biol. 1998;8:245–249. doi: 10.1016/s0962-8924(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 47.Siemering K R, Golbik R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- 48.Staehelin L A. The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 1997;11:1151–1165. doi: 10.1046/j.1365-313x.1997.11061151.x. [DOI] [PubMed] [Google Scholar]
- 49.Ushiyama R, Matthews R E. The significance of chloroplast abnormalities associated with infection by turnip yellow mosaic virus. Virology. 1970;42:293–303. doi: 10.1016/0042-6822(70)90273-4. [DOI] [PubMed] [Google Scholar]
- 50.van Bokhoven H, Le Gall O, Kasteel D, Verver J, Wellink J, van Kammen A B. Cis- and trans-acting elements in cowpea mosaic virus RNA replication. Virology. 1993;195:377–386. doi: 10.1006/viro.1993.1387. [DOI] [PubMed] [Google Scholar]
- 51.van Bokhoven H, van Lent J W, Custers R, Vlak J M, Wellink J, van Kammen A. Synthesis of the complete 200K polyprotein encoded by cowpea mosaic virus B-RNA in insect cells. J Gen Virol. 1992;73:2775–2784. doi: 10.1099/0022-1317-73-11-2775. [DOI] [PubMed] [Google Scholar]
- 52.van Bokhoven H, Verver J, Wellink J, van Kammen A. Protoplasts transiently expressing the 200K coding sequence of cowpea mosaic virus B-RNA support replication of M-RNA. J Gen Virol. 1993;74:2233–2241. doi: 10.1099/0022-1317-74-10-2233. [DOI] [PubMed] [Google Scholar]
- 53.Vance D E, Trip E M, Paddon H B. Poliovirus increases phosphatidylcholine biosynthesis in HeLa cells by stimulation of the rate-limiting reaction catalyzed by CTP: phosphocholine cytidylyltransferase. J Biol Chem. 1980;255:1064–1069. [PubMed] [Google Scholar]
- 54.van der Meer Y, Snijder E J, Dobbe J C, Schleich S, Denison M R, Spaan W J, Locker J K. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Lent J W, Groenen J T, Klinge-Roode E C, Rohrmann G F, Zuidema D, Vlak J M. Localization of the 34 kDa polyhedron envelope protein in Spodoptera frugiperda cells infected with Autographa californica nuclear polyhedrosis virus. Arch Virol. 1990;111:103–114. doi: 10.1007/BF01310508. [DOI] [PubMed] [Google Scholar]
- 56.Verver J, Wellink J, Van Lent J, Gopinath K, van Kammen A. Studies on the movement of cowpea mosaic virus using the jellyfish green fluorescent protein. Virology. 1998;242:22–27. doi: 10.1006/viro.1997.8982. [DOI] [PubMed] [Google Scholar]
- 57.Wellink J, van Lent J, Goldbach R. Detection of viral proteins in cytopathic structures in cowpea protoplasts infected with cowpea mosaic virus. J Gen Virol. 1988;69:751–755. [Google Scholar]
- 58.Wellink J, van Lent J W, Verver J, Sijen T, Goldbach R W, van Kammen A. The cowpea mosaic virus M RNA-encoded 48-kilodalton protein is responsible for induction of tubular structures in protoplasts. J Virol. 1993;67:3660–3664. doi: 10.1128/jvi.67.6.3660-3664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu S X, Ahlquist P, Kaesberg P. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc Natl Acad Sci USA. 1992;89:11136–11140. doi: 10.1073/pnas.89.23.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zabel P, Weenen-Swaans H, van Kammen A. In vitro replication of cowpea mosaic virus RNA. I. Isolation and properties of the membrane-bound replicase. J Virol. 1974;14:1049–1055. doi: 10.1128/jvi.14.5.1049-1055.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]