Abstract
Purpose:
Genetic ancestry influences evolutionary pathways of cancers. However, whether ancestry influences cancer-induced field defects is unknown. The goal of this study was to utilize ancestry-mapped true normal breast tissues as controls to identify cancer-induced field defects in normal tissue adjacent to breast tumors (NATs) in women of African American (AA) and European (EA) ancestry.
Experimental Methods:
A tissue microarray (TMA) comprising breast tissues of ancestry-mapped 100 age-matched healthy women from the Komen Tissue Bank (KTB) at Indiana University and tumor-NAT pairs from 100 women (300 samples total) was analyzed for the levels of ZEB1, an oncogenic transcription factor that is central to cell fate, mature luminal cell enriched estrogen receptor alpha (ERα), GATA3, FOXA1 and for immune cell composition.
Results:
ZEB1+ cells, which were localized surrounding the ductal structures of the normal breast, were enriched in the KTB-normal of AA compared to KTB-normal of EA women. By contrast, in EA women, both NATs and tumors compared to KTB-normal contained higher levels of ZEB1+ cells. FOXA1 levels were lower in NATs compared to KTB-normal in AA but not in EA women. We also noted variations in the levels of GATA3, CD8+ T cells, PD1+ immune cells, and PDL1+ cell but not CD68+ macrophages in NATs of AA and EA women. ERα levels did not change in any of our analyses, pointing to the specificity of ancestry-dependent variations.
Conclusions:
Genetic ancestry-mapped tissues from healthy individuals are required for proper assessment and development of cancer-induced field defects as early cancer detection markers.
Keywords: breast cancer, tumor adjacent normal, ZEB1, FOXA1, early detection
INTRODUCTION:
Recent data demonstrating a correlation between lymph node positivity at the time of detection, and the probability of disease recurrence even decades post detection, only solidifies the principle that detection of breast cancer prior to lymph node metastasis can appreciably improve clinical outcomes (1). Although the last decade witnessed significant improvements in imaging technologies including 3D-mammography, false negatives remain a significant concern (2). One way to overcome these false negatives is to complement radiologic techniques with molecular assays that measure “transcriptomic and epigenetic field effect” of tumors on adjacent “normal” (NATs) tissues. Teschendorff et al demonstrated tumor-induced epigenetic field defects in NATs specifically targeting transcription factor binding sites specifying chromatin architecture and stem cell differentiation pathways (3). These include Wnt and FGF signaling networks. Unfortunately, the Tumor Genome Atlas (TCGA) of breast cancer utilized reduction mammoplasty or NATs as their controls in transcriptome analyses (4). These are often substituted for “normal” controls in comparative analyses with breast cancers. This limitation was highlighted in another study, which compared TCGA “normal” breast transcriptome with the transcriptome of epithelial cells from the breast of healthy women. Significant differences were noted between these two sources of normal tissues (5). Reduction mammoplasty samples are also histologically abnormal compared to breast tissues from healthy women (6).
While molecular markers of cancers, particularly gene expression signatures, are traditionally developed by comparing gene expression between available “normal” and cancer tissues, the possibility of genetic ancestry of samples having an impact on gene expression under normal and abnormal conditions is rarely taken into consideration. The effects of genetic ancestry on tumor evolution and gene expression are just beginning to be recognized (7). This observation is highly relevant in the context of known differences in cancer incidence and/or outcome based on genetic ancestry. For example, women of African American ancestry (AA) suffer higher mortality from the aggressive breast cancer subtype, triple negative breast cancer (TNBC), than women of European Ancestry (EA) (8). By contrast, breast cancer in Hispanic and Native American women is less prevalent and these women have better outcomes (9,10). Whether the worse outcome in AA women is due to an increased incidence of TNBC or unique biological factors that promote aggressive biology is an important but unresolved challenge in cancer disparities research. Dietze et al.(8) recently highlighted that key molecular pathways, including Aurora A-PLK, EZH2, and Wnt-stem cell signaling networks, are significantly upregulated in TNBCs of AA women compared to TNBCs of EA women. The review (8) further emphasized that it remains unknown whether genomic aberrations unique to TNBCs in AA women result in activation of these signaling pathways in tumors or whether the basal activity of these pathways in normal AA women’s breasts is inherently different compared to EA women’s breasts. It remains possible that normal breast biology varies based on genetic ancestry. Evidence for this possibility comes from a recent discovery of breast cancer protective alleles in Latinas (11). Single nucleotide polymorphisms (SNPs) in the protective allele are located on gene regulatory regions affecting the expression of genes linked to differentiation. Our own studies have discovered enrichment of a unique population of cells in the normal breast of AA women (12). Furthermore, a breast cancer susceptibility locus in AA women, potentially altering the expression levels of microRNA miR-3065, has recently been described (13).
Here, we took advantage of genetic ancestry mapped true normal breast tissues to identify differences between true normal and NATs. These differences can potentially be developed into the earliest markers of breast cancer initiation. A tissue microarray (TMA) comprising breast tissues from clinically normal breasts, NATs and tumors were analyzed for markers that are expressed in cells with stem or mature luminal cell properties. We also examined the TMA for CD8+ T cells, CD68+ macrophages, PD1+ immune cells, and PDL1+ epithelial cells to determine whether immune cell composition of tumors and NATs in AA women differ from those of EA women.
Materials and Methods:
Generation of tissue microarray (TMA).
Breast core biopsies from healthy women donated to the Komen Tissue Bank (KTB) at Indiana University and surgical material left over after pathologic assessment as part of a treatment protocol were obtained after informed written consent from the subjects. All experiments were carried out in accordance with the approved guidelines of the Indiana University Institutional Review Board. International Ethical Guidelines for Biomedical Research Involving Human Subjects were followed. We created a tissue microarray comprising healthy breast tissue from the KTB (KTB-normal), matched normal adjacent to tumor (NAT) and tumor tissue of ~50 each of African American and Caucasian women (total ~300 samples). KTB-normal tissues were age- and race-matched to NATs/tumors. BMI of AA women who donated tissues to KTB was 32.3±9, whereas it was 28.3±8.5 in case of Caucasian women. Each sample was spotted in duplicate in cases of NATs and tumors.
Immunohistochemistry and statistical analyses:
TMA was analyzed for ZEB1, MSRB3, estrogen receptor alpha (ERα), FOXA1, and GATA3 expression. All immunohistochemistry (IHC) was done in a CLIA-certified histopathology lab and evaluated by three pathologists in a blinded manner. Quantitative measurements were done using the automated Aperio Imaging system and analysis was done using an FDA approved algorithm. Positivity and H-scores were scored and statistically analyzed as described previously (14,15). With respect to PD1 and PDL1, a tumor proportion score (TPS) was created. The PD1 and PDL1 followed the prescribed FDA reading of TPS <1% positive staining, negative; TPS 2 to 49% tumor cells positive, and TPS greater than 50% tumor cells positive (16,17). Data were analyzed in three different ways: 1) Expression differences between AA and EA KTB-normal; 2) Expression differences between KTB-normal and NATs; and 3) Expression differences between NATs and tumors. The statistical software SAS version 9.4 was used to complete the statistical analyses with p < 0.05 considered significant. Non-parametric Wilcoxon rank-sum tests were used for unpaired analyses, as positivity and H scores were not normally distributed, whereas non-parametric Wilcoxon signed-rank tests were used for paired analyses. The following antibodies were used: CD8 (Dako IR623), CD68 KP1 (Dako IR609), ER clone:EP1 (Dako IR 084), FOXA1 (Santa Cruz sc-6553), GATA3 (Santa Cruz sc-268) MSRB3 (HPA014432, rabbit polyclonal, Sigma), PD1 (Cell Marque 315M-98), PDL1 (Keytruda) (clone 22c3, Dako IHC 22c3) and ZEB1 (3G6, cat no 14-9741-82, eBioscience).
RESULTS:
ZEB1+ cells are enriched in the normal breasts of AA women compared to EA women:
In the mouse mammary gland, PROCR+/EpCAM- cells are purported to function as multi-potent stem cells (18). In our previous study focused on evaluating ethnicity-dependent differences in the normal breast, we observed specific enrichment of PROCR+/EpCAM- cells in cultured normal breast epithelial cells from biopsies of healthy AA women compared to EA women (12). These cells are enriched for the expression of stemness-related transcription factor ZEB1 and have enhanced Wnt pathway activity compared to PROCR±/EpCAM+ cells (12). ZEB1 has recently been demonstrated to limit onco-suppressive p53-driven DNA damage response in stem cells and thus increase the stem cells’ intrinsic susceptibility to malignant transformation (19). ZEB1+ cells co-express the methionine sulfoxidase reductase (MSRB3), which protects against DNA damage (19). These observations raised the possibility that PROCR+/ZEB1+ cells are naturally present at a higher levels in the normal breasts of AA women and that failure to consider natural variation in gene expression pattern, influenced at least partially by genetic ancestry, could have an impact on identifying cancer-induced field effect on the adjacent normal breast. Measuring PROCR itself in the breast tissue is complicated because there are four haplotypes of PROCR due to SNPs and only one among them is a cell surface protein (20). Since ZEB1 expression is enriched in PROCR+/EpCAM- cells, we used ZEB1 as a surrogate marker for PROCR+/EpCAM- cells in un-manipulated breast tissues.
Representative IHC staining patterns of ZEB1 in KTB-normal, NATs, and tumors from AA and EA women are shown in Figure 1A and statistical analyses are presented in Figure 1B–D and in Table 1. Descriptive statistics of ethnicity, age, menstrual status, pregnancy and breastfeeding history, hormone replacement therapy and family history of breast cancer for the KTB-normal cohort is shown in Table S1. Highly discriminative 41-ancestry marker profiles of KTB-normal showed >75% African ancestry markers in samples from African American women and >80% European ancestry markers in Caucasian women (Figure S1A) (21). Characteristics of breast cancer in the tumor cohort are shown in Table S2. ZEB1 expressing cells were localized outside the ductal structures of the normal breast and in the stromal part of the tumors (enlarged version on right side of Figure 1A). KTB-normal breast tissue of AA women contained significantly higher levels of ZEB1-positive cells compared to KTB-normal breast of EA women (Figure 1B). NATs of AA women showed a modest increase in ZEB1+ cells compared to those of KTB-normal (Figure 1C and D). The scenario was completely different in EA women; both NATs and tumors contained significantly higher levels of ZEB1+ cells compared to KTB-normal tissue (Figure 1C and D). NAT to tumor differences were noted only in EA women where an increase in ZEB1+ cells was noted predominantly in ERα+ tumors (Table S3). Thus, ZEB1+ cells are intrinsically higher in the normal breasts of AA women, whereas remarkably elevated ZEB1+ cells in the breasts of EA women were observed only in the context of breast cancer. Increases in ZEB1+ cells in KTB-normal tissue of AA women compared to EA women is less likely related to BMI differences. This was demonstrated by subdividing women above and below BMI of 30, irrespective of genetic ancestry; ZEB1 H-score but not positivity showed a marginal relationship (p=0.04) to BMI above and below 30 (Table S4).
Figure 1:
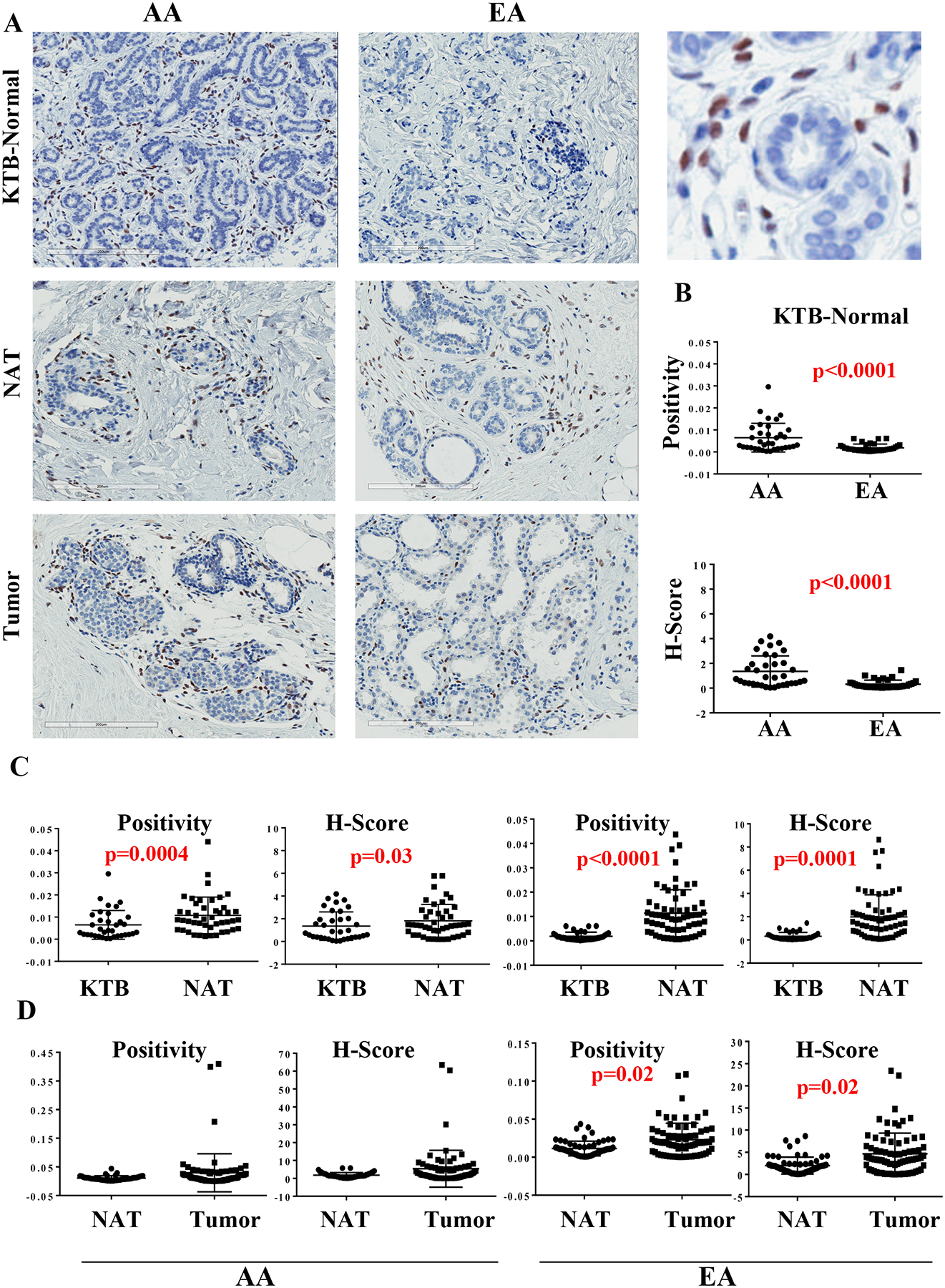
ZEB1 expression pattern in KTB-normal, normal adjacent to tumor (NATs) and in breast tumors. A) Representative immunohistochemistry of KTB-normal, NATs and tumors of women of African American (AA) and European Ancestry (EA). Enlarged view of a KTB-normal is shown on right (top). B) Differences in ZEB1 expression (positivity and H-score) between KTB-normal of AA and EA women. C) Differences between KTB-normal and NATs in AA and EA women. D) Differences between NATs and tumors in AA and EA women.
Table 1:
Differences in expression levels of ERα, FOXA1, GATA3, MSRB3, and ZEB1 in KTB-normal between women of African American and European ancestry
| African American | European Ancestry | Two-sided Wilcoxon Test p-value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable Name | N | Median | Minimum | Maximum | N | Median | Minimum | Maximum | |
| ER Positivity | 38 | 0.009837 | 0.001769 | 0.085101 | 39 | 0.010387 | 0.000000 | 0.064659 | 0.8345 |
| ER H Score | 38 | 2.165487 | 0.315269 | 21.777591 | 38 | 2.265046 | 0.103586 | 17.165665 | 0.7514 |
| ZEB1 Positivity | 38 | 0.004324 | 0.000316 | 0.025044 | 41 | 0.001224 | 0.000221 | 0.028532 | <0.0001** |
| ZEB H Score | 38 | 0.931903 | 0.045633 | 5.926867 | 41 | 0.157922 | 0.026349 | 3.927299 | <0.0001** |
| FOXA1 Positivity | 42 | 0.037941 | 0.010844 | 0.147725 | 47 | 0.021856 | 0.007987 | 0.171964 | 0.0033** |
| FOXA1 H Score | 42 | 5.108708 | 1.414022 | 20.066698 | 47 | 3.126083 | 1.033697 | 23.666012 | 0.0031** |
| GATA3 Positivity | 27 | 0.009031 | 0.001339 | 0.048353 | 32 | 0.018617 | 0.003970 | 0.067257 | 0.0009** |
| GATA3 H Score | 27 | 1.656681 | 0.170409 | 10.399523 | 32 | 4.020432 | 0.589316 | 17.773060 | 0.0003** |
| MSRB3 Positivity | 29 | 0.006854 | 0.002061 | 0.035347 | 26 | 0.006474 | 0.002085 | 0.034037 | 0.4040 |
MSRB3 has recently been shown to be one of the downstream transcriptional targets of ZEB1 and it cooperates with ZEB1 during transformation of stem-like cells (19). To correlate ZEB1 expression with its activity, we measured the levels of MSRB3 using the same antibody used in the above study. We could measure positivity but not H-score because of low-level expression. The expression pattern was similar to that of ZEB1, as cells surrounding the ducts showed expression (Figure S1B). However, KTB- normal tissues of AA and EA women expressed similar levels of MSRB3 (Table 1 and Figure S1C), which could be due to regulation by other transcription factors or to the low expression levels, making data interpretation difficult. Furthermore, except for a modest change in expression in NATs compared to KTB normal tissues, no other differences were noted (Figure S1C and D).
FOXA1 expression is lower in NATs of only AA women:
FOXA1 serves as a pioneer factor that controls chromatin access of various nuclear receptors including ERα and controls the expression of genes enriched in luminal cells compared to basal cells (22–24). FOXA1 along with another pioneer factor GATA3 and ERα form a lineage restricted hormone-responsive signaling network in the normal breast (25). While higher expression of FOXA1 in the primary tumor is associated with better outcome, its overexpression in metastatic and/or anti-estrogen resistant tumors is associated with rewiring of ERα signaling and poor outcome (26–29). In addition, it is suggested that FOXA1 gene is preferentially methylated in tumors of AA women (30). Because of its relative importance in breast cancer, we assessed our TMA for FOXA1 expression. Representative staining pattern of FOXA1 is shown in Figure 2A and numerical values are presented in Table 1. While FOXA1 levels in KTB-normal tissues of AA women were modestly higher than in EA women, NATs of AA women had lower FOXA1 compared to KTB-normal tissues (Figure 2B). Thus, tumors through their field effect may decrease FOXA1 in the surrounding breast tissues of AA women.
Figure 2:
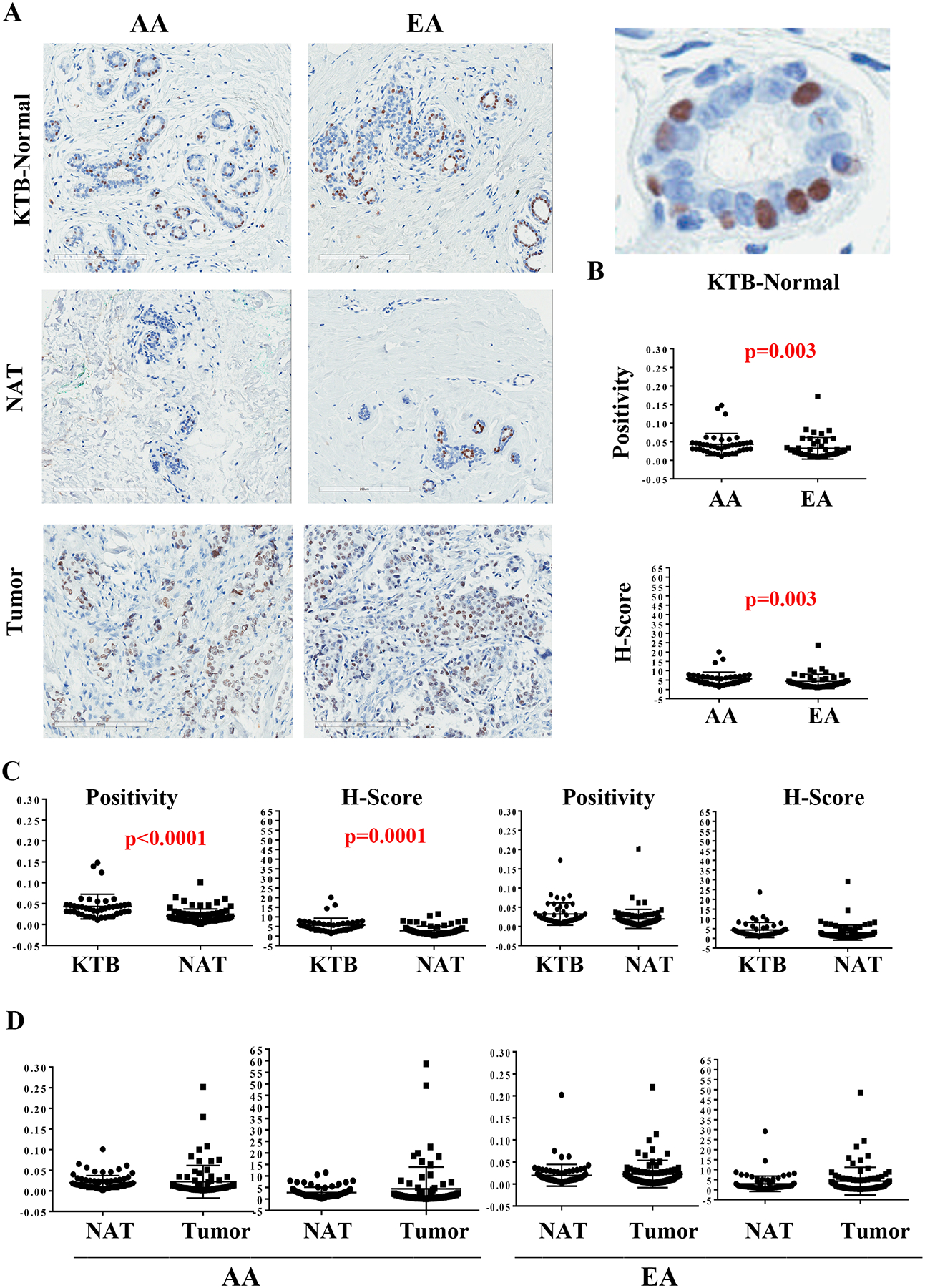
FOXA1 expression pattern in KTB-normal, NATs and in breast tumors. A) Representative immunohistochemistry of KTB-normal, NATs and tumors of AA and EA women. Enlarged view of a KTB-normal is shown on right (top). B) Differences in FOXA1 expression (positivity and H-score) between KTB-normal of AA and EA women. C) Differences between KTB-normal and NATs in AA and EA women. D) Differences between NATs and tumors in AA and EA women.
GATA3 levels are higher in KTB-normal of EA compared to AA women:
We also examined expression levels of GATA3 to determine whether hormonal signaling networks show genetic ancestry-dependent variation. Consistent with this possibility, GATA3 H-score and positivity were higher in KTB-normal tissues of EA women compared to those of AA women (Table 1 and Figure S2B). Furthermore, GATA3 is a likely candidate for cancer-induced field defects in EA women as its levels were significantly lower in NATs of EA but not AA women compared to their KTB-normal counterparts (Figure S2C).
ERα+ cells remain stable:
ERα-positive cells in the normal breast are considered to be highly differentiated non-proliferative cells and control proliferation of ERα-negative cells through paracrine mechanisms (31). Representative ERα staining pattern is shown in Figure 3A and statistical analyses are presented in Figure 3B–D and Table 1. Neither KTB normal tissues nor NATs showed genetic ancestry-dependent differences in ERα levels. The results are not only relevant, but also reassure that our TMA detects only specific changes.
Figure 3:
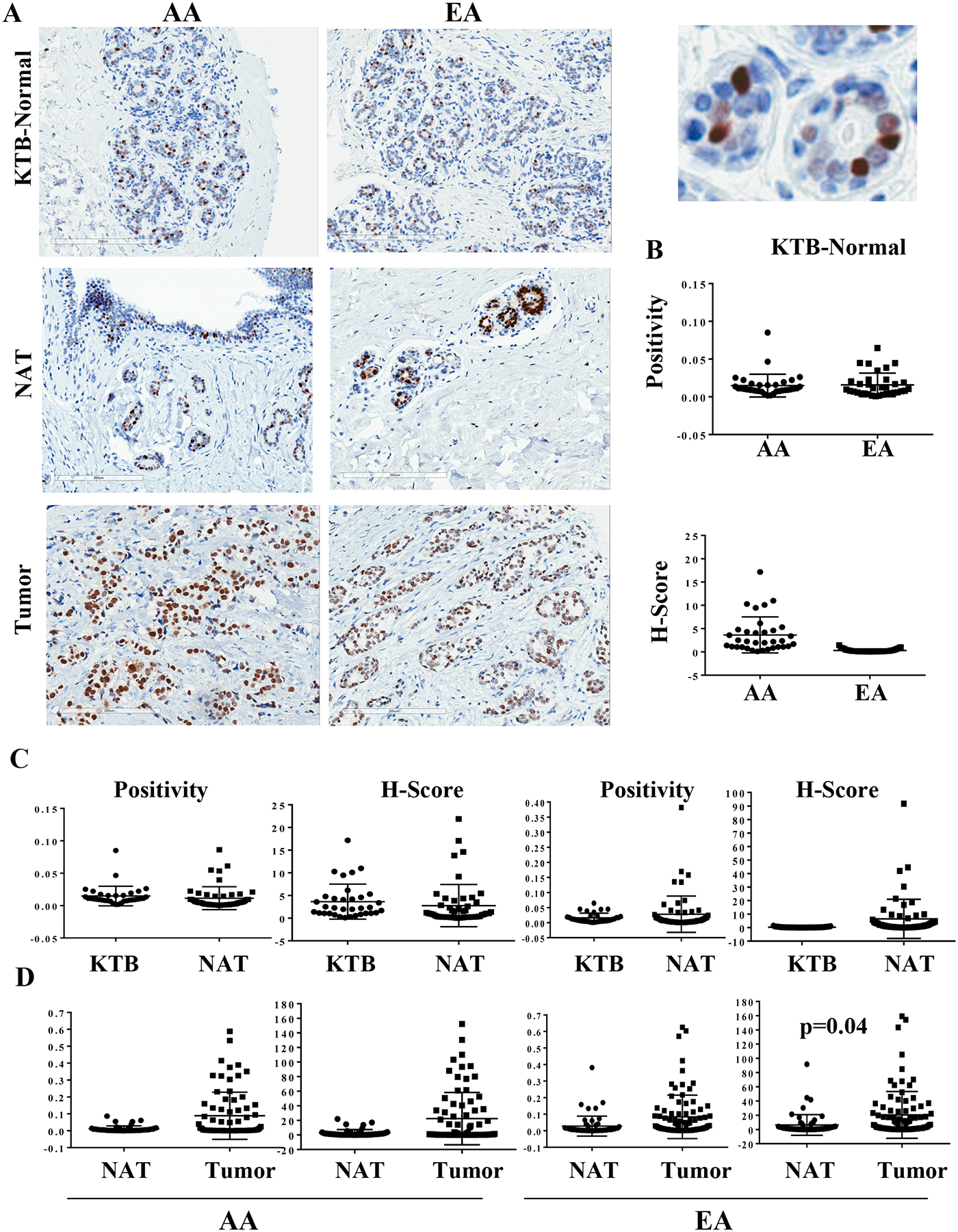
ERα expression pattern in KTB-normal, NATs and in breast tumors. A) Representative immunohistochemistry of KTB-normal, NATs and tumors of AA and EA women. Enlarged view of a KTB-normal is shown on right (top). B) Differences in ERα expression (positivity and H-score) between KTB-normal of AA and EA women. C) Differences between KTB-normal and NATs in AA and EA women. D) Differences between NATs and tumors in AA and EA women.
ERα status in tumors influences differences between NATs and tumors:
Although we observed differences in ZEB1, GATA3, and ERα expression between NATs and tumors (Figures 1D, 2D, 3D and S2D), interpretation of these data is difficult because of differences in characteristics of breast cancer subtypes, particularly ERα-positive and ERα-negative (32). To determine whether ERα-positive and ERα-negative tumors have distinct effects on NATs, FOXA1, GATA3, and ZEB1 expression data in NATs and tumors were subdivided based on ERα status of the tumor and reanalyzed. ZEB1-positivity and H-scores were higher in ERα-positive but not ERα-negative tumors compared to NATs (Table S3). Despite small sample size, these differences were noted only in EA women with ERα+ breast cancers (Table S3). With respect to FOXA1, H-score but not positivity was marginally higher in ERα-positive tumors compared to NATs of EA women (Table S3). ERα-negative tumors of EA but not AA women showed a significant decline in both positivity and H-score of FOXA1 compared to NATs (Table S3). ERα-positive tumors but not ERα-negative tumors showed further increase in GATA3 positivity and H-scores in EA women, which further confirms the role of GATA3 in hormonal regulation of breast cancer (Figure S2D and Table S3). When the analyses was done with paired NAT-tumors, the above noted differences between NATs and tumors in ZEB1, GATA3, and FOXA1 levels remained significant, although sample size was too small to subdivide samples based on genetic ancestry (Table S5).
NATs of AA and EA women show differing levels of CD8, PD1 and PDL1+ cells:
Results thus far point to pro-inflammatory state of NATs of EA women based on the known link between ZEB1 and inflammatory cytokines (33). To address this further, we stained the above TMAs with CD8 for T cells, CD68 for macrophages, and PD1 for immune cells. We also examined epithelial/tumor cells for PDL1. All staining was done in a CLIA-certified lab with FDA-approved antibodies. In KTB-normal TMAs, there was no staining with CD8 and CD68 in either the AA or EA TMAs. Less than 1% of the lymphocytes and macrophages stained and these were considered negative. The same negativity was observed with PD1 and PDL1 immunostains (data not shown). Therefore, we analyzed staining results between NATs of AA and EA women and between NATs and tumors. Representative staining patterns in NATs and tumors are shown in Figure 4.
Figure 4:
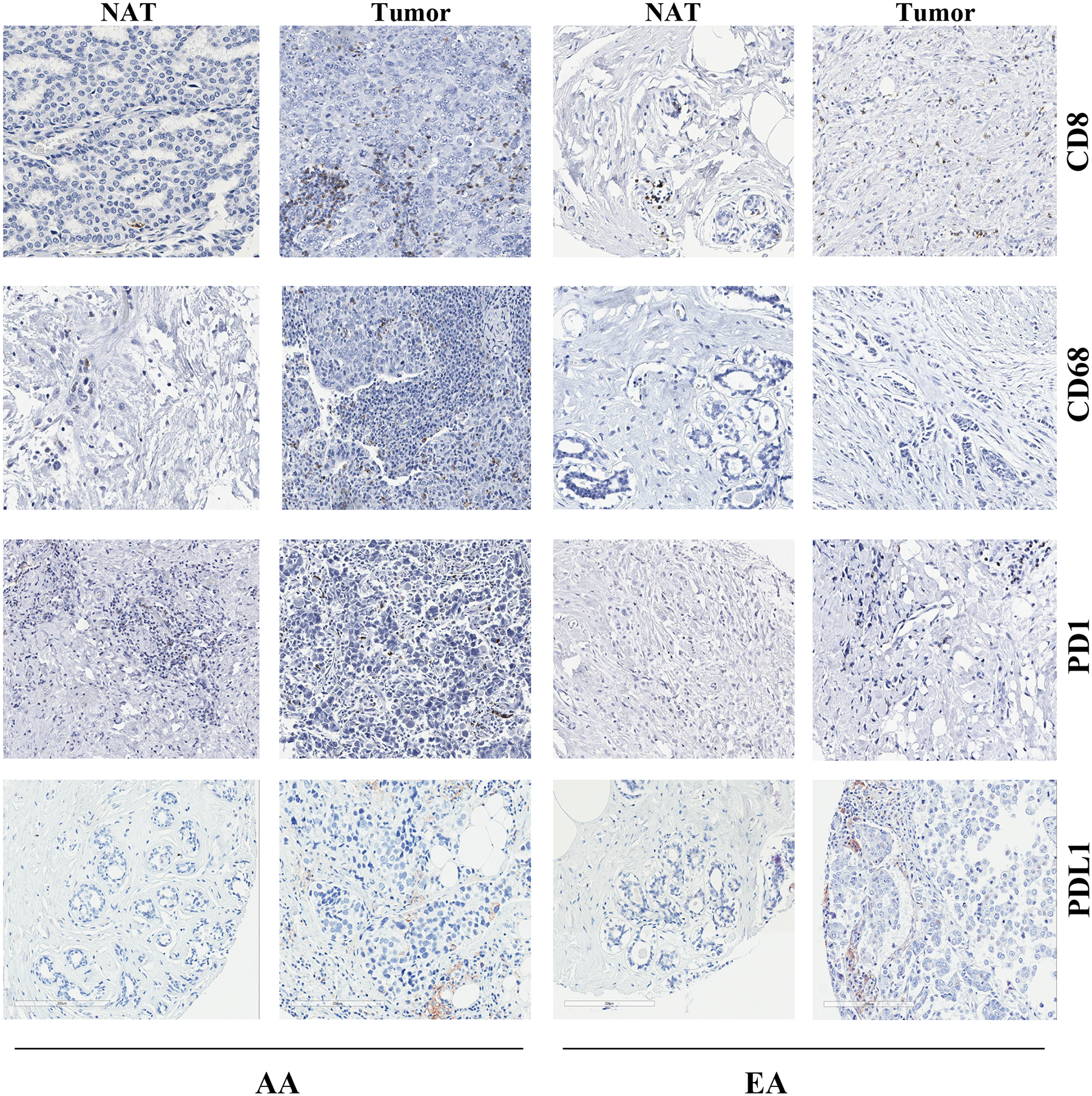
Representative CD8, CD68, PD1, and PDL1 immunohistochemistry of NATs and Tumors of AA and EA women.
CD8 immunostaining was localized to inflammatory cells (T lymphocytes) and not to tumor cells in the breast cancer cores. No background reactivity was observed in any case. NATs of AA women showed statistically significantly higher CD8 positivity compared to EA women (Figure 5 and Table S6). The tumors in EA women had more CD8 immunostaining compared to corresponding NATs but such differences were not seen in the AA women.
Figure 5:
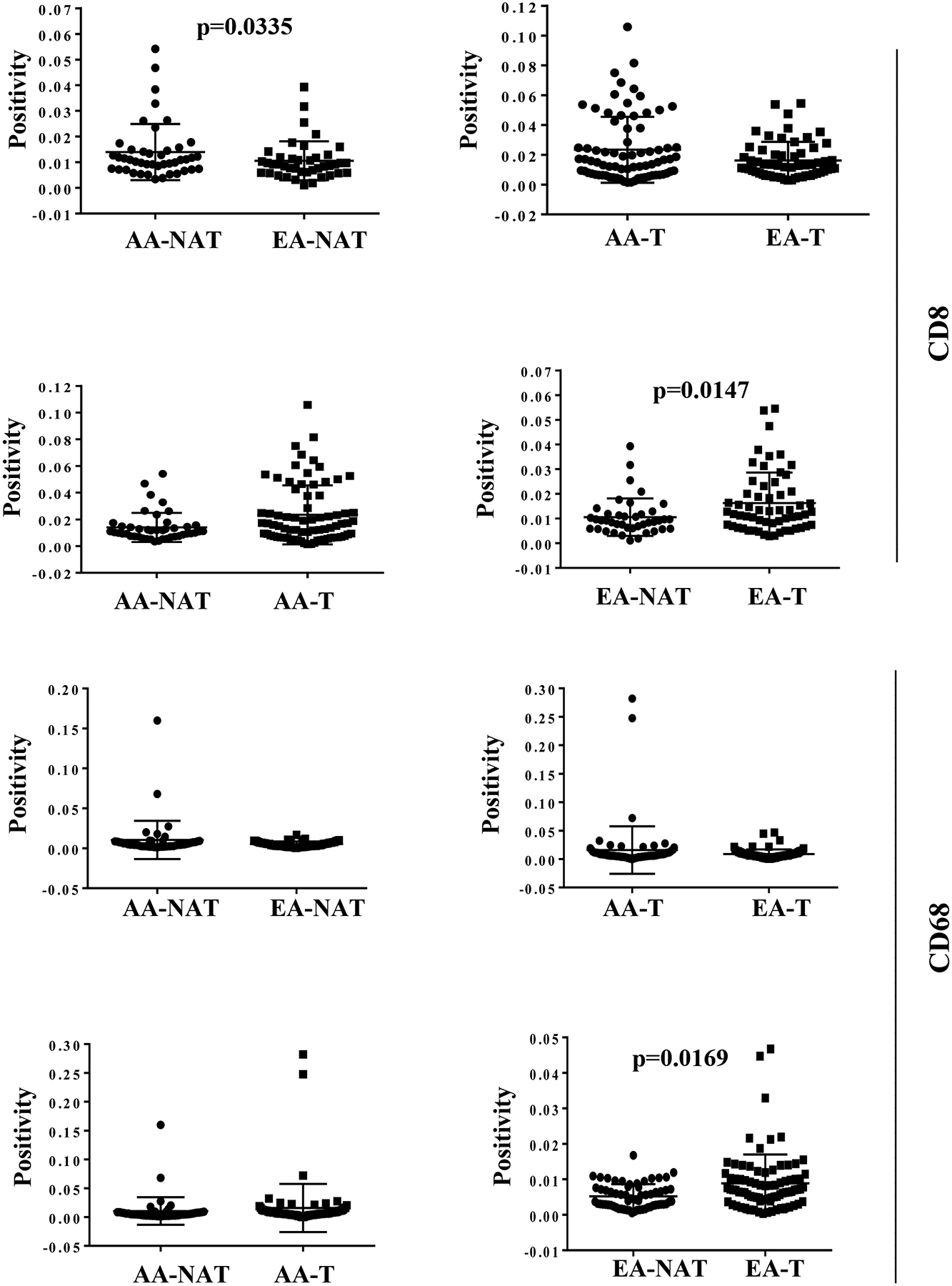
Statistical analyses of CD8 and CD68 positivity in NATs and tumors (T) of AA and EA women. All statistically significant differences are indicated with p values.
CD68 staining was localized to macrophages in the breast cancer cores (Figure 4). CD68 had lower positivity compared to CD8 by both visual and the Aperio positive pixel reads. CD68 positivity was higher in tumors compared to their NATs (p=0.02) in EA women but no such differences were noted in AA women (Figure 5).
PD1 immunostaining was localized to immune cells only and no background staining was observed (Figure 4). There was no staining of tumor cells. NATs of AA women contained significantly higher PD1+ cells, similar to CD8+ cells, compared to NATs of EA women (Figure 6 and Table S6). PD1 staining did not show any differences between NATs and tumors in both groups (Table S3).
Figure 6:
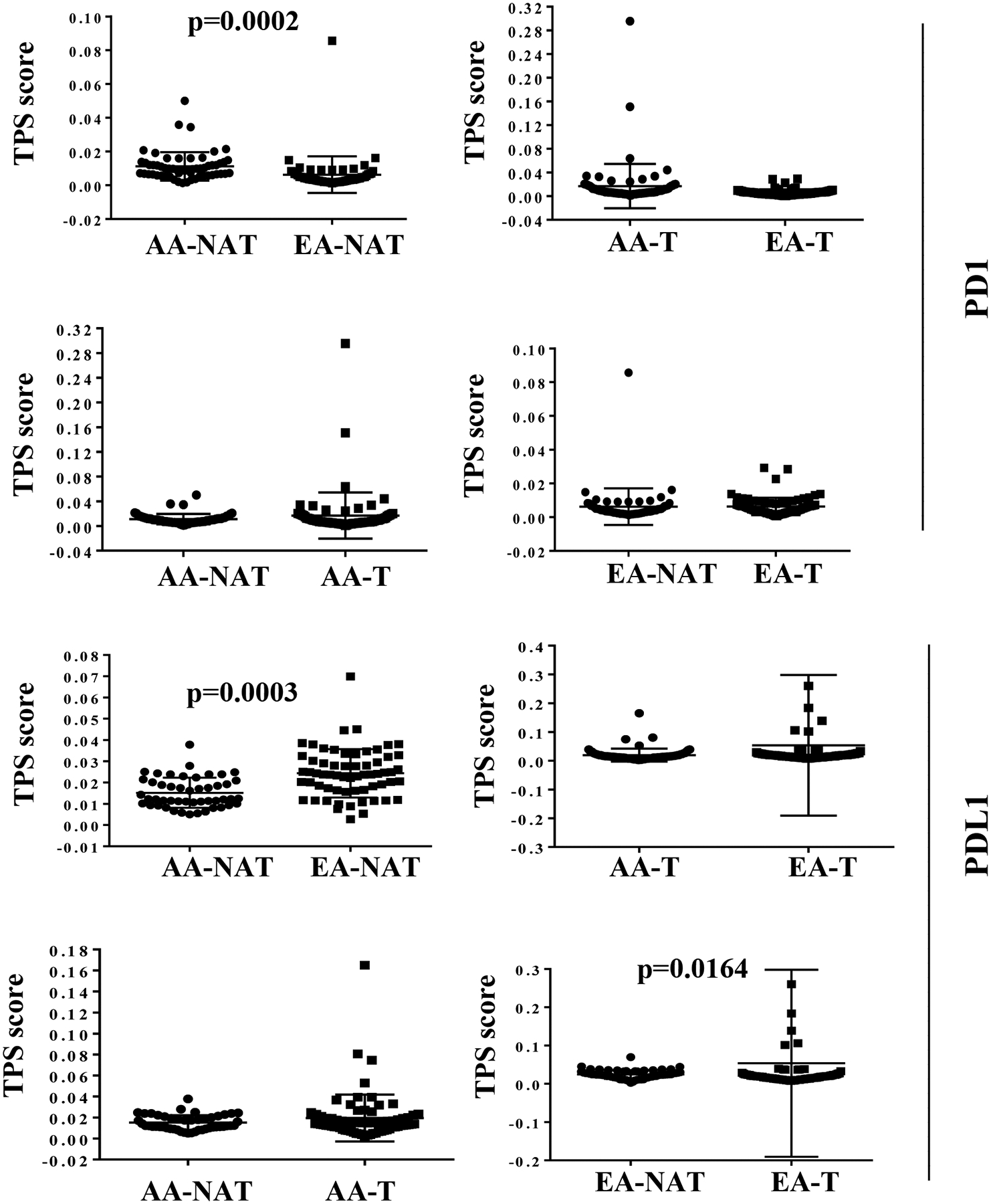
Statistical analyses of PD1 and PDL1 TPS scores in NATs and tumors (T) of AA and EA women. All statistically significant differences are indicated with p values.
PDL1 immunostaining was seen localized in the tumor cell cytoplasm and cell membrane (Figure 4). In a few EA cases, only lymphocytes were stained. PDL1 staining of NATs of AA women was significantly lower than EA cases (Figure 6 and Table S6). Although PDL1 staining did not differ between NATs and tumors of AA women, its levels were marginally lower in ER+ tumors but not ER- tumors compared to NATs in case of EA women (Figure 6 and Table S3). It is interesting that PD1 and PDL1 staining scores in NATs of AA is the reverse of the patterns seen in EA women. In summation, the immune environment in NATs is different from that in KTB-normal tissue with further differences between NATs and tumors, showing variations based on genetic ancestry.
Discussion:
Recent studies have shown cancer-induced field defects influencing gene expression patterns in histologically normal tissues surrounding cancer (3,34,35). These observations raise a concern as well as provide an opportunity for further investigation. The concern is the use of tumor adjacent normal as a “normal” control, whereas the opportunity pertains to the development of cancer-induced field defects in the adjacent normal as early markers of cancer. However, recent discovery of inter-individual differences in gene expression patterns due to SNPs in gene regulatory regions and genetic ancestry-dependent enrichment of SNPs with breast cancer protective or elevated risk characteristics necessitate the use of ancestry-matched control samples from healthy individuals to develop molecular features of tumor adjacent normal as cancer-initiation or progression markers (11,13,36,37). Ethnicity contributing to inter-individual differences in normal biology is just beginning to be explored, as evident from a recent study that demonstrated distinct gut microbiota in different ethnic groups with shared geography (38). Furthermore, genetic ancestry has been shown to influence mutation patterns in cancer (7). Resources available at the Komen Normal Tissue Bank at Indiana University, namely ancestry-mapped breast tissues from >5000 healthy women, should enable us to take these factors into consideration as we develop molecular features of NATs as cancer detection markers. Utilizing a small fraction of those tissues, we provide evidence for ancestry-dependent differences in the number of ZEB1-positive and GATA3-positive cells in the normal breast as well as cancer-induced field effects on ZEB1, GATA3, and FOXA-positive cells in the tumor-adjacent normal tissue.
Recently discovered functions of ZEB1 have raised considerable interest in this molecule within the oncology field. The regulatory regions of this gene remain in a bivalent state, enabling the regulatory regions to respond readily to the tumor microenvironment and increase breast cancer plasticity and tumorigencity (39). Another study showed elevated ZEB1 expression in normal breast stem cells and it functionally protects stem cells from p53-mediated cell death in response to oncogene activation-induced DNA damage and promotes tumorigenecity with limited genomic instability (19). It was also reported that ZEB1 is expressed in both tumor and stromal cells of the breast (40). ZEB1 directly increases the expression of pro-inflammatory cytokines such as IL-6 and IL-8, and it promotes vascular mimicry of breast cancer cells by remodeling extracellular matrix (33,41). We had previously demonstrated that cytokines such as tumor necrosis factor induce the expression of ZEB1 (42). These observations along with our unique observations of genetic ancestry-dependent differences in ZEB1-positive cells in the normal breast, elevated number of ZEB1+ cells in NATs compared to healthy breast tissues of women of European ancestry, and its localization outside the ductal structures raise several questions about the function of ZEB1+ cells in the normal and tumor adjacent normal breast. We have shown previously that cytokeratin-positive, PROCR+/EpCAM- cells of the normal breast, which are enriched in the normal breast of AA women compared to EA women, express 50-fold higher ZEB1 compared to cytokeratin-positive, PROCR-/EpCAM+ cells of the breast (12,43). Thus, we suspect that ZEB1+ cells in the normal breast correspond to PROCR+/EpCAM- cells and that cancer-induced field effect leads to expansion/proliferation of such cells in the breast of EA women. Signaling pathways leading to proliferation of ZEB1+ cells in NATs of EA women are unknown, but the Wnt pathway is the prime suspect as it is activated in cells surrounding cancer due to altered DNA methylation (3). In this respect, Wnt and ZEB1 constitute a reciprocal feed-forward signaling loop where ZEB1 enhances TCF4/β-Catenin-mediated transcription and Wnt signaling converts ZEB1 from a transcription repressor to an activator (44).
The reason for an intrinsically higher number of ZEB1+ cells in AA women is unknown. TNBCs in AA compared to EA women display elevated Wnt pathway activation and it could be that Wnt pathway activity is intrinsically higher in AA women leading to elevated ZEB1 expression (8). It has also been demonstrated that vitamin D through Vitamin D Receptor (VDR) represses ZEB1 expression and serum vitamin D levels are significantly lower in AA than EA individuals (45,46). Therefore, lower VDR activity and resulting increase in the activity of pro-inflammatory cytokines could be responsible for higher number of ZEB1+ cells in the normal breast of AA women, which needs further investigation.
In contrast to stemness-associated ZEB1, FOXA1 and GATA3, which are expressed predominantly in differentiated luminal cells, showed opposite pattern in AA women. While the normal breasts of AA women had higher number of FOXA1-positve cells compared to EA women, a decline in FOXA1-positive cells in NATs as a consequence of cancer field effect was observed only in AA women. How tumors cause down regulation of FOXA1 in NATs is unknown but could involve inflammatory cytokines, as cytokine inducible transcription repressors such as TWIST1 repress FOXA1 expression (47,48). In this regard, we observed genetic ancestry-dependent differences in the levels of immune cells in NATs; NATs of AA women contained an elevated number of CD8+ T cells and PD1+ immune cells compared to NATs of EA women. In addition, FOXA1 regulatory regions are highly susceptible for DNA methylation and transcriptional repression, particularly in the context of BRCA1 deficiency (49). Furthermore, ER-negative tumors in AA women show elevated FOXA1 DNA methylation compared to ER-negative tumors of EA women (30). Recent studies have also demonstrated racial differences in plasma levels of cytokines with CCL2, CCL11, IL4, and IL10 being higher in EA women, and IL1RA and IFNα2 being higher in AA women (50).
Differential expression of GATA3 in the normal breasts of AA and EA women is intriguing, as GATA3 is one of the major signaling molecules required for hormonal response and differentiation of normal breast epithelial cells (25). Our results suggest that hormonal- and differentiation-signaling networks show genetic ancestry-dependent differences and it is likely that ERα:GATA3-dependent transcriptional program is more active in the normal breast of EA compared to AA women. Whether such difference between EA and AA persist in ERα-positive tumors is unknown and potentially worth investigating as it is relevant for response to antiestrogen therapy.
Collectively, data presented in this study suggest the need to consider the following aspects for cancer biomarker discovery: 1) NATs are molecularly abnormal and thus are not suitable as controls; 2) These abnormalities can be detected only when true normal breast tissues are used as controls and differences in normal gene expression attributable to genetic ancestry are taken into consideration; 3) ZEB1 and GATA3 show unique expression pattern in the normal breast, which is influenced by the genetic ancestry and could potentially be developed as biomarkers of breast cancer initiation of women of European Ancestry; and 4) Genetic ancestry has an influence on the immune environment of tumors as well as NATs.
Supplementary Material
Translational Relevance:
Breast cancer diagnosis prior to lymph node metastasis can appreciably improve clinical outcomes. While radiologic techniques have improved early diagnosis, molecular markers that can complement radiologic techniques are needed to improve specificity. This study aimed to investigate how both genetic ancestry and appropriate control tissues influence detection of cancer-induced changes in the breast. We show that alterations in ZEB1+ cells in tissues surrounding tumors are observed predominantly in women of European ancestry, whereas FOXA1+ cells were altered in normal tissues adjacent to tumors of women of African American Ancestry. Immune cell activation in tumors as well as surrounding tissue showed genetic ancestry-dependent variations as evident from differences in PD1+ and PDL1+ cells in the normal tissue adjacent to tumors of women of African American and European Ancestry. Thus, biomarker discovery needs to consider not only sample size and statistical methods but also genetic ancestry and true normal control tissues.
Statement of Significance:
We demonstrate that genetic ancestry mapped tissues from healthy individuals are required as controls to identify cancer-induced field defects in tumor-adjacent normal tissues as well as defects in tumors. This finding is significant in light of recent discoveries of influence of genetic ancestry on both normal biology and tumor evolution.
Acknowledgement:
We thank the members of IU Simon Cancer Center tissue procurement facility, IU School of Medicine immunohistochemistry core, Komen Tissue Bank for their service and countless number of women who donated their breast tissue for research purpose as well as volunteers who facilitated tissue collection. Department of Defense DOD-W81XWH-15-1-0707, Susan G Komen for the Cure SAC110025, 100 Voices of Hope (to HN), and Catherine Peachy Foundation (HN and CDS) funded this study. Susan G. Komen for the Cure, Breast cancer Research Foundation and Vera Bradley Foundation for Breast Cancer Research provide funding support to the Komen Normal Tissue Bank.
Footnotes
Conflict of Interest: Authors have no conflict of interest to declare.
References:
- 1.Pan H, Gray R, Braybrooke J, Davies C, Taylor C, McGale P, et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N Engl J Med 2017;377(19):1836–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzaghal AA, DiPiro PJ. Applications of Advanced Breast Imaging Modalities. Curr Oncol Rep 2018;20(7):57. [DOI] [PubMed] [Google Scholar]
- 3.Teschendorff AE, Gao Y, Jones A, Ruebner M, Beckmann MW, Wachter DL, et al. DNA methylation outliers in normal breast tissue identify field defects that are enriched in cancer. Nature communications 2016;7:10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radovich M, Clare SE, Atale R, Pardo I, Hancock BA, Solzak JP, et al. Characterizing the heterogeneity of triple-negative breast cancers using microdissected normal ductal epithelium and RNA-sequencing. Breast Cancer Res Treat 2014;143(1):57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Degnim AC, Visscher DW, Hoskin TL, Frost MH, Vierkant RA, Vachon CM, et al. Histologic findings in normal breast tissues: comparison to reduction mammaplasty and benign breast disease tissues. Breast Cancer Res Treat 2012;133(1):169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan J, Hu Z, Mahal BA, Zhao SD, Kensler KH, Pi J, et al. Integrated Analysis of Genetic Ancestry and Genomic Alterations across Cancers. Cancer Cell 2018;34(4):549–60 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietze EC, Sistrunk C, Miranda-Carboni G, O’Regan R, Seewaldt VL. Triple-negative breast cancer in African-American women: disparities versus biology. Nat Rev Cancer 2015;15(4):248–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sineshaw HM, Gaudet M, Ward EM, Flanders WD, Desantis C, Lin CC, et al. Association of race/ethnicity, socioeconomic status, and breast cancer subtypes in the National Cancer Data Base (2010–2011). Breast Cancer Res Treat 2014;145(3):753–63. [DOI] [PubMed] [Google Scholar]
- 10.DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin 2016;66(1):31–42. [DOI] [PubMed] [Google Scholar]
- 11.Fejerman L, Ahmadiyeh N, Hu D, Huntsman S, Beckman KB, Caswell JL, et al. Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nature communications 2014;5:5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakshatri H, Anjanappa M, Bhat-Nakshatri P. Ethnicity-Dependent and -Independent Heterogeneity in Healthy Normal Breast Hierarchy Impacts Tumor Characterization. Scientific reports 2015;5:13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bensen JT, Graff M, Young KL, Sethupathy P, Parker J, Pecot CV, et al. A survey of microRNA single nucleotide polymorphisms identifies novel breast cancer susceptibility loci in a case-control, population-based study of African-American women. Breast Cancer Res 2018;20(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perkins SM, Bales C, Vladislav T, Althouse S, Miller KD, Sandusky G, et al. TFAP2C expression in breast cancer: correlation with overall survival beyond 10 years of initial diagnosis. Breast Cancer Res Treat 2015;152(3):519–31. [DOI] [PubMed] [Google Scholar]
- 15.Padua MB, Bhat-Nakshatri P, Anjanappa M, Prasad MS, Hao Y, Rao X, et al. Dependence receptor UNC5A restricts luminal to basal breast cancer plasticity and metastasis. Breast Cancer Res 2018;20(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandusky GE, Mintze KS, Pratt SE, Dantzig AH. Expression of multidrug resistance-associated protein 2 (MRP2) in normal human tissues and carcinomas using tissue microarrays. Histopathology 2002;41(1):65–74. [DOI] [PubMed] [Google Scholar]
- 17.NSCLC-Keytruda. PsGtP-LTi. Dako Denmark A/S PD-L1 IHC 22C3 pharmDX Interpretation Manual 10. [Google Scholar]
- 18.Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L, et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature 2014;517:81–84. [DOI] [PubMed] [Google Scholar]
- 19.Morel AP, Ginestier C, Pommier RM, Cabaud O, Ruiz E, Wicinski J, et al. A stemness-related ZEB1-MSRB3 axis governs cellular pliancy and breast cancer genome stability. Nat Med 2017;23(5):568–78. [DOI] [PubMed] [Google Scholar]
- 20.Medina P, Navarro S, Bonet E, Martos L, Estelles A, Bertina RM, et al. Functional analysis of two haplotypes of the human endothelial protein C receptor gene. Arteriosclerosis, thrombosis, and vascular biology 2014;34(3):684–90. [DOI] [PubMed] [Google Scholar]
- 21.Nievergelt CM, Maihofer AX, Shekhtman T, Libiger O, Wang X, Kidd KK, et al. Inference of human continental origin and admixture proportions using a highly discriminative ancestry informative 41-SNP panel. Investig Genet 2013;4(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 2005;122(1):33–43. [DOI] [PubMed] [Google Scholar]
- 23.Bernardo GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, et al. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development 2010;137(12):2045–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernardo GM, Bebek G, Ginther CL, Sizemore ST, Lozada KL, Miedler JD, et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene 2013;32(5):554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res 2007;67(13):6477–83. [DOI] [PubMed] [Google Scholar]
- 26.Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, et al. FOXA1 expression in breast cancer--correlation with luminal subtype A and survival. Clin Cancer Res 2007;13(15 Pt 1):4415–21. [DOI] [PubMed] [Google Scholar]
- 27.Thorat MA, Marchio C, Morimiya A, Savage K, Nakshatri H, Reis-Filho JS, et al. Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J Clin Pathol 2008;61(3):327–32. [DOI] [PubMed] [Google Scholar]
- 28.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 2011;43(1):27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu X, Jeselsohn R, Pereira R, Hollingsworth EF, Creighton CJ, Li F, et al. FOXA1 overexpression mediates endocrine resistance by altering the ER transcriptome and IL-8 expression in ER-positive breast cancer. Proc Natl Acad Sci U S A 2016;113(43):E6600–E09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espinal AC, Buas MF, Wang D, Cheng DT, Sucheston-Campbell L, Hu Q, et al. FOXA1 hypermethylation: link between parity and ER-negative breast cancer in African American women? Breast Cancer Res Treat 2017;166(2):559–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke RB, Howell A, Potten CS, Anderson E. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res 1997;57(22):4987–91. [PubMed] [Google Scholar]
- 32.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98(19):10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katsura A, Tamura Y, Hokari S, Harada M, Morikawa M, Sakurai T, et al. ZEB1-regulated inflammatory phenotype in breast cancer cells. Molecular oncology 2017;11(9):1241–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan JS, Tan MJ, Sng MK, Teo Z, Phua T, Choo CC, et al. Cancer-associated fibroblasts enact field cancerization by promoting extratumoral oxidative stress. Cell death & disease 2017;8(1):e2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moller M, Strand SH, Mundbjerg K, Liang G, Gill I, Haldrup C, et al. Heterogeneous patterns of DNA methylation-based field effects in histologically normal prostate tissue from cancer patients. Scientific reports 2017;7:40636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC, et al. Genetic effects on gene expression across human tissues. Nature 2017;550(7675):204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013;501(7468):506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, Herrema H, et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat Med 2018;24:1526–31. [DOI] [PubMed] [Google Scholar]
- 39.Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 2013;154(1):61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jang MH, Kim HJ, Kim EJ, Chung YR, Park SY. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum Pathol 2015;46(9):1267–74. [DOI] [PubMed] [Google Scholar]
- 41.Langer EM, Kendsersky ND, Daniel CJ, Kuziel GM, Pelz C, Murphy KM, et al. ZEB1-repressed microRNAs inhibit autocrine signaling that promotes vascular mimicry of breast cancer cells. Oncogene 2018;37(8):1005–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene 2007;26(5):711–24. [DOI] [PubMed] [Google Scholar]
- 43.Kumar B, Prasad MS, Bhat-Nakshatri P, Anjanappa M, Kalra M, Marino N, et al. Normal breast-derived epithelial cells with luminal and intrinsic subtype-enriched gene expression document inter-individual differences in their differentiation cascade. Cancer Res 2018;78(17):5107–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez-Tillo E, de Barrios O, Valls E, Darling DS, Castells A, Postigo A. ZEB1 and TCF4 reciprocally modulate their transcriptional activities to regulate Wnt target gene expression. Oncogene 2015;34(46):5760–70. [DOI] [PubMed] [Google Scholar]
- 45.Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest 2016;126(9):3219–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, Bartz TM, Chittoor G, Eiriksdottir G, Manichaikul AW, Sun F, et al. Meta-analysis across Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium provides evidence for an association of serum vitamin D with pulmonary function. Br J Nutr 2018:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y, Qin L, Sun T, Wu H, He T, Yang Z, et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene 2017;36(8):1157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li CW, Xia W, Huo L, Lim SO, Wu Y, Hsu JL, et al. Epithelial-mesenchymal transition induced by TNF-alpha requires NF-kappaB-mediated transcriptional upregulation of Twist1. Cancer Res 2012;72(5):1290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong C, Fujino K, Monteiro LJ, Gomes AR, Drost R, Davidson-Smith H, et al. FOXA1 repression is associated with loss of BRCA1 and increased promoter methylation and chromatin silencing in breast cancer. Oncogene 2015;34(39):5012–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao S, Hong CC, Ruiz-Narvaez EA, Evans SS, Zhu Q, Schaefer BA, et al. Genetic ancestry and population differences in levels of inflammatory cytokines in women: Role for evolutionary selection and environmental factors. PLoS genetics 2018;14(6):e1007368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


