Abstract
CHEK2 is considered to be involved in homologous recombination repair (HRR). Individuals who have germline pathogenic variants (gPVs) in CHEK2 are at increased risk to develop breast cancer and likely other primary cancers. PARP inhibitors (PARPi) have been shown to be effective in the treatment of cancers that present with HRR deficiency—for example, caused by inactivation of BRCA1/2. However, clinical trials have shown little to no efficacy of PARPi in patients with CHEK2 gPVs. Here, we show that both breast and non-breast cancers from individuals who have biallelic gPVs in CHEK2 (germline CHEK2 deficiency) do not present with molecular profiles that fit with HRR deficiency. This finding provides a likely explanation why PARPi therapy is not successful in the treatment of CHEK2-deficient cancers.
Checkpoint kinase 2 (CHEK2) is a tumor suppressor gene and is considered a key component of the DNA damage response and the homologous recombination repair (HRR) of DNA double-strand breaks (1). Individuals heterozygous for germline pathogenic variants (gPVs) in CHEK2 have a low-to-moderate risk to develop breast cancer (2,3). Individuals with biallelic gPVs in CHEK2 are, next to breast cancer, likely also at increased risk for other primary cancers (4).
It has been shown that cancers that have a defective HRR (HRD) are sensitive to platinum-based therapies and PARP inhibitors (PARPi) (5). Some PARPi treatment approvals are limited to cancers that have gPVs in BRCA1/2 or genomic instability features indicative of HRD (6). It is thought that cancers that are defective for other components of the HRR pathway, such as ATM, CHEK1, CHEK2, NBN, BRIP1, MRE11, RAD50, RAD51B, RAD51C, RAD51D, RAD54L, PALB2 and BARD1, may also benefit from PARPi (7). However, studies have shown that no clinical benefit is seen in individuals who have ATM or CHEK2 gPVs (8).
Recent molecular studies on breast cancers occurring in individuals who are heterozygous for gPVs in CHEK2 suggest that these cancers do not present with genomic instability features indictive of HRD (9,10). However, these studies focused only on breast cancers from individuals heterozygous for gPVs in CHEK2. The aim of this study was to investigate the molecular profiles of cancers from individuals with biallelic gPVs in CHEK2 (from here on CHEK2-deficient cancers) (11), which may offer explanations why PARPi therapy is ineffective in CHEK2-deficient tumors. We analyzed the genomes, using shallow whole-genome sequencing (sWGS) and whole-exome sequencing (WES), from 16 cancers of 9 individuals homozygous for the most common loss-of-function (LoF) gPV in CHEK2 (NM_007194.4; c.1100del) (12). In total, we analyzed 8 breast cancers and 8 other CHEK2-deficient cancers [colorectal cancer (CRC) (n = 4), thyroid (n = 2), endometrial (n = 1), and urothelial (n = 1) cancer].
We compared our findings to breast (n = 3) and non-breast cancers (n = 25) from individuals heterozygous for CHEK2 gPVs and to breast (n = 7) and non-breast cancers (n = 21) from individuals heterozygous for BRCA1/2 gPVs (13). All, except 2 cancers with a heterozygous CHEK2 gPV, did not harbor a somatic second hit. The cancers from individuals heterozygous for gPVs in BRCA1/2 were selected for having a second somatic hit rendering them BRCA1/2-deficient (Supplementary Table 1, available online). To investigate tumor mutational profiles, we analyzed genomic instability features including large-scale state transitions (LST) and telomeric allelic imbalances (tAI). Furthermore, we analyzed tumor mutational burden (TMB), mutational signatures via SigProfiler (14,15) and putative cancer driver gene mutations (Supplementary Figure 1, available online). To determine the differences in the presence of somatic pathogenic variants (PVs) in driver genes and the presence of mutational signatures between groups, a Fisher exact test was used. For comparisons of more than 2 groups, one-way analysis of variance or Kruskal-Wallis test were applied. All statistical tests are two-sided, and statistical significance level is a P value less than .05. Further details are provided in the Supplementary Methods (available online).
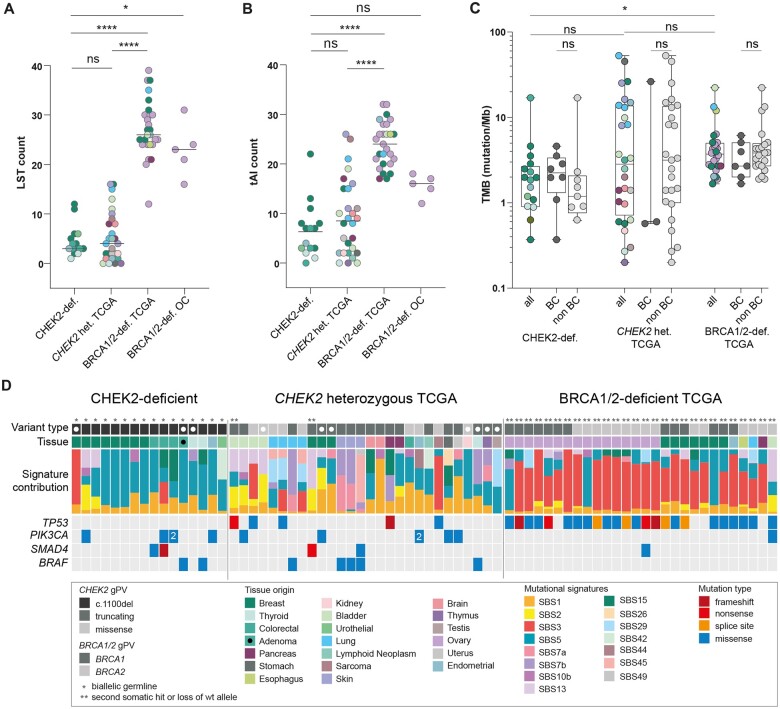
The presence of LSTs and tAIs are genomic instability features indicative of HRD (16). The median number of LSTs and tAIs in CHEK2-deficient cancers was 3 (range = 1-12) and 7 (range = 0-22), respectively (Figure 1, A and B; Supplementary Figure 2, available online). None of the analyzed cancers presented with an LST count of 15 or more, which is considered the cutoff value for HRD. An additional in-house reference set of BRCA1/2-deficient ovarian cancers (n = 5) presented with a median number of LSTs and tAIs of 23 (range = 16-31) and 16 (range = 12-18), respectively (Figure 1, A and B; Supplementary Figure 2, available online). From the Cancer Genome Atlas (TCGA), we also retrieved the LST and tAI counts of cancers from individuals who are heterozygous for CHEK2 gPVs or BRCA1/2 gPVs. The median LST and tAI counts in cancers from individuals heterozygous for CHEK2 gPVs were 4 (range = 0-16) and 8.5 (range = 0-26), respectively, which is comparable to the counts observed in our CHEK2-deficient cancers (Figure 1, A and B; Supplementary Figure 2, available online). BRCA1/2-deficient cancers, as expected, presented with higher LST and tAI counts (median LST count of 26 [range = 12-39], with a median tAI count of 24 [range = 17-32]), (Figure 1, A and B; Supplementary Figure 2, available online). Our findings are in line with literature on breast cancers with heterozygous CHEK2 (g)PVs (10) and show no statistically significant differences in genomic instability features between breast and non-breast cancers of CHEK2-deficient individuals (Supplementary Figure 3, A and B, available online).
Figure 1.
The mutational landscape of CHEK2-deficient cancers. Genomic instability features including A) large-scale state transitions (LST) counts and B) telomeric allelic imbalances (tAI) counts are depicted. Groups (left to right) included in-house generated data for CHEK2-deficient cancers; in-house generated data for BRCA1/2-deficient ovarian cancers; the Cancer Genome Atlas (TCGA) data of cancers from individuals heterozygous for CHEK2 germline pathogenic variants (gPVs); and TCGA data of BRCA1/2-deficient cancers. Cancer origins are represented in different colors. Lines represent median values per group. Groups were compared via nonparametric Kruskal-Wallis test, and Dunn’s multiple comparison correction was applied. *P < .05; ****P < .0001. C) Tumor mutational burden (TMB) of nonsynonymous variants in all cancers and between breast and non-breast cancers are depicted. CHEK2-deficient cancers are compared to the cancers from individuals heterozygous for CHEK2 gPVs from TCGA and BRCA1/2-deficient cancers from TCGA. Lines represent the median value per group. Groups were compared via nonparametric Kruskal-Wallis test, and Dunn’s multiple comparison correction was applied. *P < .05. D) CHEK2-deficient cancers, cancers from individuals heterozygous for CHEK2 gPVs from TCGA, and BRCA1/2-deficient cancers from TCGA were analyzed for single base substitution mutational signature contributions and mutated driver genes. The black dot labels one premalignant adenomatous colonic polyp and white dots indicate cancer samples with <30 single base substitutions, which indicates that mutational signatures from these samples are not completely reliable. gPV = germline pathogenic variant; TCGA = The Cancer Genome Atlas; TMB = tumor mutational burden; LST = large-scale state transitions; tAI = telomeric allelic imbalances; ns = nonsignificant; BC = breast cancer; non-BC = non-breast cancer; OC = ovarian cancer; wt = wild-type; het = heterozygous; def = deficient.
Next, we investigated the nonsynonymous TMB and microsatellite instability (MSI). The median TMB was 1.92 (range = 0.37-16.99 variants/Megabase) in CHEK2-deficient cancers, which is comparable to cancers that developed in individuals heterozygous for CHEK2 gPVs (median TMB of 2.83 [range = 0.2-53 variants/Megabase]; Figure 1, C) and in line with the literature (9,10). BRCA1/2-deficient cancers presented with a significantly higher TMB than CHEK2-deficient cancers (median TMB of 3.72 [range = 1.67-22.27 variants/Megabase]; P < .02; Figure 1, C). Furthermore, the TMB was not significantly different between breast and non-breast CHEK2-deficient cancers (P = .2786; Figure 1, C). One CHEK2-deficient cancer, a colorectal cancer (CRC), showed MSI.
Subsequently, we analyzed mutational signatures. Most CHEK2-deficient cancers presented with the clock-like mutational signatures SBS1 (5/16 cancers; 31%) and/or SBS5 (14/16 cancers; 88%; Figure 1, D, Supplementary Table 1, available online). Cancers from individuals heterozygous for CHEK2 gPVs presented with mutational signatures SBS1 and/or SBS5 in 12/28 (43%; P = 1) and 12/28 (43%; P < .05) cancers, respectively (Figure 1, D). In contrast to the CHEK2-deficient cancers, BRCA1/2-deficient cancers did not present with mutational signature SBS1 (0/28 cancers; 0%; P < .05), and a minority presented with mutational signature SBS5 (11/28 cancers; 39%; P < .05; Figure 1, D). SBS3, the HRD-related mutational signature (17), was observed to some level in 13% (2/16) of CHEK2-deficient cancers, and it was the predominant mutational signature (86%, 24/28) in the BRCA1/2-deficient group (P < .001; Figure 1, D). This finding is in concordance with current literature on breast cancers from individuals heterozygous for CHEK2 PVs (9,10) and indicates that HRD does not seem to be driving the process of tumorigenesis in CHEK2-deficient cancers. In 25% (4/16) of CHEK2-deficient cancers, tissue-specific signatures, including mutational signature SBS15 (defective DNA-mismatch repair) in two CRCs (1 with somatic MSH2 inactivation) and mutational signatures SBS2 and SBS13 (APOBEC-related signatures) in 2 breast cancers, were identified (Figure 1, D). Cancers from individuals heterozygous for CHEK2 gPVs also presented with tissue-specific mutational signatures in 10/28 (36%) cancers, including mutational signatures SBS2 and SBS13 in bladder and breast cancers, and mutational signature SBS29 (tobacco chewing) in lung cancers (Figure 1, D).
We further compared mutational signatures occurring in the CHEK2-deficient cancers to those in a cohort of (mostly) sporadic cancers (n = 7515) from TCGA of various tissue origins (Supplementary Figure 4, available online). The spectrum of mutational signatures in CHEK2-deficient cancers resembles sporadic cancers more than what is observed in BRCA1/2-deficient cancers (Supplementary Figure 4, available online), which is also observed when we compare CHEK2-deficient cancers with cancers with heterozygous gPV in BRCA1/2, for which it was not clear if a somatic second hit was present (n = 134 cancers). The latter group has a prominent contribution of mutational signature SBS3, similar to BRCA1/2-deficient cancers, whereas this is absent in CHEK2-deficient cancers (Supplementary Figure 4, available online). Next to the observed tissue-specific mutational signatures, breast and non-breast CHEK2-deficient cancers present with comparable mutational signature profiles (Supplementary Figure 3, C, available online). Our data are not in support of a single base mutational signature that can be linked to CHEK2 deficiency, as has been shown for BRCA1/2 and other genes involved in DNA repair, such as the mismatch repair genes or genes involved in base excision repair (17).
Last, we investigated the mutated cancer driver genes in cancers from CHEK2-deficient individuals. The most frequently mutated gene in CHEK2-deficient cancers was PIK3CA (n = 4 cancers, three cancer types; Figure 1, D). No somatic PVs were observed in TP53 in CHEK2-deficient cancers (Figure 1, D; Supplementary Figure 3, D, available online), in line with Smid et al. (9), whereas this was the most frequently mutated gene in BRCA1/2-deficient cancers (0/16 vs 24/28; P < .001; Figure 1, D). Furthermore, although a complete absence of somatic TP53 PVs was observed in breast cancers with heterozygous CHEK2 gPVs (n = 3), non-breast cancers with heterozygous CHEK2 gPVs did present with somatic TP53 PVs (Figure 1, D; Supplementary Figure 3, D, available online). This finding suggests that either TP53 somatic PVs are not necessary for cancer development in the context of CHEK2-deficiency or that concurrent inactivation of both CHEK2 and TP53 does not confer a growth benefit for cells. In line with this, CHEK2 was identified to cause synthetic lethality in an in silico study on TP53 druggable target partners (18). Although TP53 is one of the most important tumor suppressor genes that also transmits DNA-damage-induced signals received from both ATM and CHEK2, CHEK2 also operates in an independent manner from the HRR pathway in cell-cycle arrest, apoptosis, and senescence (1).
In summary, our data show that CHEK2-deficient cancers are not driven by disruption of HRR as genomic instability features and a mutational signature indicative of HRD are lacking. The absence of a CHEK2-associated mutational signature suggests that CHEK2 does not play a prominent role in DNA repair mechanisms. Its role in regulation of cell division may be more important, which is in line with the apparent mutual exclusivity of CHEK2 deficiency and somatic TP53 mutations.
Our study has a few limitations. We were limited in structural variant size analysis, as the resolution of our sWGS is lower than the WGS approach that was used by Smid et al. and, therefore, does not permit us to investigate structural variants smaller than 1 Mb (9). Furthermore, we were only able to investigate the molecular profile for CHEK2-deficient cancers of 4 origins beyond breast cancer. Ideally, this should be expanded to more cancer types occurring in individuals who have biallelic CHEK2 gPVs and to more cancers per type.
Taken together, our study provides additional evidence that impairment of CHEK2 does not result in HRD, which likely explains the inefficiency of PARPi treatment of CHEK2-deficient cancers.
Supplementary Material
Acknowledgments
We thank all the patients who were a part of the study. We thank clinical geneticists Wendy A.G. van Zelst-Stams, C. Marleen Kets, Maaike Haadsma, Liesbeth Spruijt, and Marijke R. Wevers (Department of Human Genetics, Radboudumc, Nijmegen, the Netherlands) for their cooperation. We thank Daniel von Rhein for help with WES and sWGS data analysis and Samhita Pamidimarri Naga (Department of Human Genetics, Radboudumc, Nijmegen, the Netherlands) for help in sWGS data analysis. We thank Lisa Elze (Department of Human Genetics, Radboudumc, Nijmegen, the Netherlands) for providing control data for this study. We thank Christian Gilissen (Radboudumc, Nijmegen, the Netherlands) for the use of the variant calling and annotation pipelines. We thank Central Genome Analysis Laboratory (CGAL; Radboudumc, Nijmegen, the Netherlands) for the whole-exome and shallow whole-genome sequencing of cancer DNA. The funder did not play a role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Ethics approval: Written informed consent was obtained, and this study was approved by the local medical ethics committee of the Radboud University Medical Center (CMO; study numbers 2019-5082, 2019-5738, and 2020-7036). This study was performed in accordance with the ethical standards of the Helsinki Declaration.
Contributor Information
Snežana Hinić, Department of Human Genetics, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Rachel S van der Post, Department of Pathology, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Lilian Vreede, Department of Human Genetics, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Janneke Schuurs-Hoeijmakers, Department of Human Genetics, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Saskia Koene, Department of Human Genetics, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Erik A M Jansen, Department of Human Genetics, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Franziska Bervoets-Metge, Department of Pathology, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Arjen R Mensenkamp, Department of Human Genetics, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Nicoline Hoogerbrugge, Department of Human Genetics, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Marjolijn J L Ligtenberg, Department of Human Genetics, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands; Department of Pathology, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Richarda M de Voer, Department of Human Genetics, Radboud University Medical Center, Research Institute for Medical Innovation, Nijmegen, The Netherlands.
Data availability
All WES and sWGS data generated in this study are available from European Genome-Phenome Archive (EGA) numbers EGAS50000000080 and EGAS00001007258 via https://ega-archive.org/. Public data are available through https://www.cbioportal.org/.
Author contributions
Snežana Hinić, MSc (Conceptualization; Data curation; Formal analysis; Project administration; Software; Visualization; Writing—original draft; Writing—review & editing), Rachel S. van der Post, MD, PhD (Conceptualization; Formal analysis; Resources; Supervision; Writing—review & editing), Lilian Vreede, BSc (Data curation; Investigation; Validation; Writing—review & editing), Janneke Schuurs-Hoeijmakers, MD, PhD (Resources; Writing—review & editing), Saskia Koene, MD, PhD (Resources; Writing—review & editing), Erik A.M. Jansen, BSc (Data curation; Formal analysis; Writing—review & editing), Franziska Bervoets-Metge, PhD (Data curation; Formal analysis; Writing—review & editing), Arjen R. Mensenkamp, PhD (Resources; Validation; Writing—review & editing), Nicoline Hoogerbrugge, MD, PhD (Conceptualization; Funding acquisition; Resources; Supervision; Writing—review & editing), Marjolijn J.L. Ligtenberg, PhD (Conceptualization; Funding acquisition; Resources; Supervision; Writing—review & editing), Richarda de Voer, PhD (Conceptualization; Data curation; Formal analysis; Funding acquisition; Project administration; Resources; Supervision; Writing—original draft; Writing—review & editing).
Funding
This work was supported by a grant from the Dutch Cancer Society (KWF 12174).
Conflict of interests
All authors declare no financial or nonfinancial conflict of interests.
References
- 1. Bartek J, Lukas J.. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3(5):421-429. doi: 10.1016/S1535-6108(03)00110-7 [DOI] [PubMed] [Google Scholar]
- 2. Dorling L, Carvalho S, Allen J, et al. ; Breast Cancer Association Consortium. Breast cancer risk genes—association analysis in more than 113,000 women. N Engl J Med. 2021;384(5):428-439. doi: 10.1056/NEJMoa1913948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dorling L, Carvalho S, Allen J, et al. SGBCC Investigators. Breast cancer risks associated with missense variants in breast cancer susceptibility genes. Genome Med. 2022;14(1):51. doi: 10.1186/s13073-022-01052-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rainville I, Hatcher S, Rosenthal E, et al. High risk of breast cancer in women with biallelic pathogenic variants in CHEK2. Breast Cancer Res Treat. 2020;180(2):503-509. doi: 10.1007/s10549-020-05543-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keung MYT, Wu Y, Vadgama JV.. PARP inhibitors as a therapeutic agent for homologous recombination deficiency in breast cancers. J Clin Med. 2019;8(4):435. doi: 10.3390/jcm8040435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ragupathi A, Singh M, Perez AM, Zhang D.. Targeting the BRCA1/2 deficient cancer with PARP inhibitors: clinical outcomes and mechanistic insights. Front Cell Dev Biol. 2023;11:1133472. doi: 10.3389/fcell.2023.1133472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Risdon EN, Chau CH, Price DK, Sartor O, Figg WD.. PARP inhibitors and prostate cancer: to infinity and beyond BRCA. Oncologist. 2021;26(1):e115-e129. doi: 10.1634/theoncologist.2020-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tung NM, Robson ME, Ventz S, et al. TBCRC 048: phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J Clin Oncol. 2020;38(36):4274-4282. doi: 10.1200/jco.20.02151 [DOI] [PubMed] [Google Scholar]
- 9. Smid M, Schmidt MK, Prager-van der Smissen WJC, et al. Breast cancer genomes from CHEK2 c.1100delC mutation carriers lack somatic TP53 mutations and display a unique structural variant size distribution profile. Breast Cancer Res. 2023;25(1):53. doi: 10.1186/s13058-023-01653-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mandelker D, Kumar R, Pei X, et al. The landscape of somatic genetic alterations in breast cancers from CHEK2 germline mutation carriers. JNCI Cancer Spectr. 2019;3(2):pkz027. doi: 10.1093/jncics/pkz027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bell DW, Kim SH, Godwin AK, et al. Genetic and functional analysis of CHEK2 (CHK2) variants in multiethnic cohorts. Int J Cancer. 2007;121(12):2661-2667. doi: 10.1002/ijc.23026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huijts PE, Hollestelle A, Balliu B, et al. CHEK21100delC homozygosity in the Netherlands—prevalence and risk of breast and lung cancer. Eur J Hum Genet. 2014;22(1):46-51. doi: 10.1038/ejhg.2013.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. The Cancer Genome Atlas (TCGA). https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga. Accessed September 26, 2023.
- 14. Bergstrom EN, Huang MN, Mahto U, et al. SigProfilerMatrixGenerator: a tool for visualizing and exploring patterns of small mutational events. BMC Genomics. 2019;20(1):685. doi: 10.1186/s12864-019-6041-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Islam SMA, Díaz-Gay M, Wu Y, et al. Uncovering novel mutational signatures by de novo extraction with SigProfilerExtractor. Cell Genomics. 2022;2(11). doi: 10.1016/j.xgen.2022.100179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Watkins JA, Irshad S, Grigoriadis A, Tutt ANJ.. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16(3):211. doi: 10.1186/bcr3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alexandrov LB, Kim J, Haradhvala NJ, et al. ; PCAWG Consortium. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94-101. doi: 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X, Simon R.. Identification of potential synthetic lethal genes to p53 using a computational biology approach. BMC Med Genomics. 2013;6(30). doi: 10.1186/1755-8794-6-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All WES and sWGS data generated in this study are available from European Genome-Phenome Archive (EGA) numbers EGAS50000000080 and EGAS00001007258 via https://ega-archive.org/. Public data are available through https://www.cbioportal.org/.