Abstract
Norwalk virus (NV) is a causative agent of acute epidemic nonbacterial gastroenteritis in humans. The inability to cultivate NV has required the use of molecular techniques to examine the genome organization and functions of the viral proteins. The function of the NV protein encoded by open reading frame 3 (ORF 3) has been unknown. In this paper, we report the characterization of the NV ORF 3 protein expressed in a cell-free translation system and in insect cells and show its association with recombinant virus-like particles (VLPs) and NV virions. Expression of the ORF 3 coding region in rabbit reticulocyte lysates resulted in the production of a single protein with an apparent molecular weight of 23,000 (23K protein), which is not modified by N-linked glycosylation. The ORF 3 protein was expressed in insect cells by using two different baculovirus recombinants; one recombinant contained the entire 3′ end of the genome beginning with the ORF 2 coding sequences (ORFs 2+3), and the second recombinant contained ORF 3 alone. Expression from the construct containing both ORF 2 and ORF 3 resulted in the expression of a single protein (23K protein) detected by Western blot analysis with ORF 3-specific peptide antisera. However, expression from a construct containing only the ORF 3 coding sequences resulted in the production of multiple forms of the ORF 3 protein ranging in size from 23,000 to 35,000. Indirect-immunofluorescence studies using an ORF 3 peptide antiserum showed that the ORF 3 protein is localized to the cytoplasm of infected insect cells. The 23K ORF 3 protein was consistently associated with recombinant VLPs purified from the media of insect cells infected with a baculovirus recombinant containing the entire 3′ end of the NV genome. Western blot analysis of NV purified from the stools of NV-infected volunteers revealed the presence of a 35K protein as well as multiple higher-molecular-weight bands specifically recognized by an ORF 3 peptide antiserum. These results indicate that the ORF 3 protein is a minor structural protein of the virion.
Norwalk virus (NV) is a prototype strain of human caliciviruses, a group of viruses that are the major pathogens causing epidemic nonbacterial gastroenteritis (14, 49). The NV genome, a positive-sense, single-stranded RNA molecule approximately 7.7 kb in length, is predicted to contain three open reading frames (ORFs) (29). The first and third ORFs are in reading frame 2 of the cDNA, while ORF 2 is in reading frame 3. The first ORF (ORF 1) is predicted to encode the nonstructural proteins. Sequence analysis has identified similarities to the picornavirus 2C helicase, 3C protease, and 3D RNA-dependent RNA polymerase (29). The second ORF (ORF 2) encodes the capsid protein. Expression of the capsid protein in insect cells infected with baculovirus recombinants results in the self-assembly of empty recombinant virus-like particles (rVLPs) (28, 51). The third ORF (ORF 3) is located at the 3′ end of the genome and codes for a 212-amino-acid protein of unknown function. The predicted molecular weight of the NV ORF 3 protein is 22,479. The ORF 3 protein is a basic protein with a predicted isoelectric point of 10.99, which has led to speculation that it may be involved in nucleic acid binding (13).
Purified 38-nm recombinant NV (rNV) VLPs have been characterized antigenically and morphologically (28). Three-dimensional reconstruction studies revealed that these particles fold into T=3 icosahedral structures formed by 180 copies of the capsid protein (40). The finding of virus capsids composed of a single structural protein is a common feature of plant viruses including tomato bushy stunt virus (22) and turnip crinkle virus (25). Caliciviruses and nodaviruses are the only animals viruses described to date with a capsid made of a single structural protein (23, 43). A notable difference between plant virus and NV capsid proteins is that the plant virus capsid proteins have an N-terminal basic domain, which is thought to interact with the RNA during assembly. The NV capsid protein lacks such a basic region, and analysis of the X-ray crystallographic structure of rNV VLPs has revealed that the inner surface of the icosahedral shell is acidic (38). For these reasons, it seems highly possible that the calicivirus ORF 3 protein may aid in RNA encapsidation.
The presence of ORF 3 in the genome is conserved throughout all human and animal caliciviruses, suggesting that it plays a role in replication or assembly. ORF 3 or ORF 3-equivalent proteins were detected in feline calicivirus (FCV)-infected cells (24) and in rabbit hemorrhagic disease virus (RHDV)-infected primary hepatocytes (32) as well as RHDV virions (54). The synthesis of the ORF 3 protein in a cell-free translation system has been reported for FCV and Camberwell virus (24, 54). Considerable sequence variability of ORF 3 among members of the Norwalk-like viruses (NLVs) has been reported and is consistent with the idea that ORF 3 might be a structural protein (42, 45).
The inability to grow NV and a lack of reagents to detect ORF 3 has hampered studies of the ORF 3 protein. In the present paper, we report the characterization of the NV ORF 3 protein synthesized in a cell-free translation system and expressed in insect cells, purified VLPs, and purified NV virions. These studies were facilitated by generation of ORF 3 peptide antisera which are able to recognize the original peptides by enzyme-linked immunosorbent assay (ELISA) and the ORF 3 protein by immunoprecipitation and Western blot analysis.
MATERIALS AND METHODS
Plasmids and recombinant baculoviruses.
The ORF 2 coding region was PCR amplified from pUCNV-4145 (26) using primers NV-110a (5′-GCGGCGAGATCTAATTCGTAAATGATGATGGCG-3′; nucleotides [nt] 5355 to 5374 [BglII site underlined]) and NV-111a (5′-GCGGCGAGATCTAATTGCACCAATTATGGCTT-3′; nt 6953 to 6934 [BglII site underlined]). This PCR product was agarose gel purified, restriction enzyme digested with BglII, and cloned into the pGem-7Zf(+) vector (Promega, Madison, Wis.) and the baculovirus transfer vector pVL1393 (PharMingen, San Diego, Calif.) for the production of pGNV2 and pVLNV2, respectively (White et al., unpublished). A construct containing the entire 3′ end of the genome beginning with the ORF 2 coding sequences was cloned into pSP65 (Promega) following PCR amplification from pSPNV-FL (20) using primers T7NV5 (forward, 5′-CACGTCGACTAATACGACTCACTATAGTGAATGATGATGGCGTCAAAAGACG-3′; nt 1 to 22, similar to 6950 to 6972 [SalI site underlined, T7 promoter sequence in boldface italic type]) and T7NN3 (5′-CACGTCGACCTCGAGT T T T T T T T T T T T T T T T T T T T T T T T T T T TT T-3′ [SalI site underlined; XhoI site in boldface type]) to produce pNV-2/3. The T7NV5 forward primer contains a SalI restriction enzyme site for cloning and the T7 promoter upstream of the NV ORF 2 sequences. The T7NN3 reverse primer contains a SalI restriction enzyme site for cloning and a XhoI restriction enzyme site to linearize the plasmid for cell-free transcription reactions. The ORF 3 coding region was cloned into the pGem-7Zf(+) and pVL1393 vectors following amplification by PCR. ORF 3 was amplified from pUCNV-4145 using primers NV100 (5′-CGGGGATCCATGGCCCAAGCCATA-3′; nt 6950 to 6967 [BamHI site underlined]) and NV101b (5′-CGGGGATCCAATATGATGCCCACA-3′; nt 7618 to 7600 [BamHI site underlined]). The amplified fragment was digested with BamHI and cloned separately to generate pG7BamNV3 and pVLNV3. Additionally, a hemagglutinin (HA) tag was cloned on the 3′ end of the ORF 3 amplified from pUCNV-4145 using primers NV100 and NV120HA (5′-GATCCTCGAGTCATGCGTAGTCCGGTACGTCGTACGGGTATCGCCTATATTTGCG-3′; nt 7570 to 7585, [HA tag italic, XhoI site underlined]). The amplified fragment was digested with BamHI and XhoI for cloning separately to generate pG7NV3HA and pVLNV3HA. All clones were sequenced to ensure authenticity. Baculovirus recombinants Bac-NV2, Bac-NV3, and Bac-NV3HA were generated as previously described (28). The Bac-rNV C8 (ORFs 2 and 3) used in these experiments was generated and described previously (28).
Cell-free translation of ORF 3 in RRLs.
Plasmid DNA was linearized and purified on an agarose gel (Jetsorb, Genomed; PGC, Gaithersburg, Md.). The purified DNA was transcribed in vitro with SP6 or T7 polymerase using the MaxiScript kit (Ambion, Austin, Tex.). The proteins were translated in rabbit reticulocyte lysates (RRLs; Promega) in the presence of Tran35S-label (1,175 Ci/mmol; 43.48 TBq/mmol [ICN, Cosa Mesa, Calif.]). Plasmid pG7BamNV3 was digested with XbaI and transcribed using the SP6 polymerase. Plasmid pNV-2/3 was digested either with XhoI for the generation of RNAs containing both ORFs 2 and 3 or with HindIII for the production of ORF 2 transcripts; both RNAs were transcribed using the T7 polymerase. Translation also was examined in the absence or presence of canine microsomal membranes to determine if the NV ORF 3 protein was modified by N-linked glycosylation. Microsomal membrane quality was evaluated by translation of the rotavirus glycoprotein NSP4 from a plasmid, pG10, which contains the rotavirus gene 10 (2). The pG10 plasmid was digested with XbaI, and RNA was synthesized using the T7 polymerase. XbaI-digested pGem7Zf(+) vector DNA, containing no insert, was included in an SP6 transcription reaction as a negative control. Translated proteins (10 μl of a 25-μl reaction mixture) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (33) on a 15% gel and detected by fluorography (Autofluor [National Diagnostics, Atlanta, Ga.]).
Production of peptide antisera against the ORF 3 protein.
Two peptide sequences in ORF 3 (amino acids 70 to 99 and 181 to 203) were selected for the production of antibodies based on analysis of the ORF 3 protein sequence with algorithms that predict surface potential (37), turn potential (9), and amphipathic structure (35), as described previously (4). The ORF 3 C-terminal peptide (amino acids 181 to 203) was synthesized by the University of Pittsburgh Peptide Core Facility with the use of a 9-fluorenylmethyloxycarbonyl chemical strategy and standard protocols as previously described (4). The final peptide product was characterized by reverse-phase high-performance liquid chromatography (Deltapak C4; Waters) and plasma desorption mass spectroscopy (31). Only peptides with the correct theoretical mass and 90% or greater full-length product were used in these studies. The ORF 3 N-terminal peptide (amino acids 70 to 99) was synthesized and characterized by the Texas A&M Peptide Core Facility using the procedure described above. Peptide-specific antiserum was generated in CD-1 mice and New Zealand White rabbits by immunization with peptide cross-linked with glutaraldehyde to the protein carrier keyhole limpet hemocyanin as described previously (4). Female CD-1 mice were given five or six inoculations of 100 nmol of ORF 3 peptide. Primary inoculations were given in Freund's complete adjuvant, and boosts were given in Freund's incomplete adjuvant. Each inoculation was divided between the intraperitoneal and intramuscular routes. Female New Zealand White rabbits were given a total of five inoculations of 200 nmol of ORF 3 peptide. Each inoculation was divided between the subcutaneous and intramuscular routes. Pre- and postimmunization sera were evaluated by peptide ELISAs, immunoprecipitation, and Western blot analyses.
Expression of ORF 3 in insect cells.
Spodoptera frugiperda (Sf9) cells were either mock infected or infected with wild-type baculovirus or the baculovirus recombinants Bac-NV2 (ORF 2), Bac-rNV C8 containing the entire 3′-end of the genome (ORFs 2+3), or Bac-NV3 (ORF 3) at a multiplicity of infection (MOI) of 10 PFU/cell. At various times postinfection, the cells were lysed in 5× SDS-PAGE sample buffer (3 parts disruption buffer [10% SDS, 50% 2-mercaptoethanol, 5 M urea (1:1:1)], 2 parts F/2 [stacking gel buffer (0.5 M Tris-HCl, 0.46% TEMED, pH 6.6-6.8), 80% glycerol (1:1), 0.02 g of phenol red]). An aliquot equivalent to 3 × 105 cells was separated by electrophoresis in an SDS–15% polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane for Western blot analysis with either of the two ORF 3 peptide antisera at a dilution of 1:500 followed by horseradish peroxidase-labeled secondary antibody (goat anti-rabbit immunoglobulin G, 1:5,000 [Cappel, West Chester, Pa.) with detection by enhanced chemiluminescence (ECL) (Amersham, Arlington Heights, Ill.) as previously described (6).
Immunoprecipitation of the ORF 3 protein using the ORF 3 peptide antisera.
A standard immunoprecipitation method was used as previously described (8). Briefly, cell-free translation reaction mixtures (25 μl) were adjusted to 500 μl with RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and preabsorbed using 50 μl of Staphylococcus A protein. The sample was incubated with the ORF 3 C-terminal peptide antiserum at a 1:20 dilution overnight at 4°C. Immune complexes were precipitated with Staphylococcus A protein. The precipitated complexes were washed three times, first with salt wash A (1.0 M NaCl, 0.01 M Tris-HCl [pH 7.2], 0.1% [vol/vol] NP-40), then with SDS-salt wash B (0.1 M NaCl, 1 mM EDTA, 0.01 M Tris-HCl [pH 7.2], 0.1% [vol/vol] NP-40, 0.3% [wt/vol] SDS), and finally with no-salt wash C (0.01 M Tris-HCl [pH 7.2], 0.1% [vol/vol] NP-40). Samples were suspended in 5× SDS-PAGE sample buffer and boiled, the Staphylococcus A protein was pelleted, and the entire sample was loaded for detection by autoradiography of proteins separated by electrophoresis (SDS–15% polyacrylamide gel). For immunoprecipitation from infected insect cells, cells were metabolically labeled with 35S-Promix (Amersham Pharmacia Biotech, Piscataway, N.J.) at 61 h postinfection (p.i.) for 4 h. The monolayers were washed once with phosphate-buffered saline (PBS), after which RIPA buffer (500 μl) was added to lyse the cells directly on the plate. The lysates were collected, clarified, and preabsorbed with Staphylococcus A protein. An aliquot (250 μl) was incubated with either of the two ORF 3 peptide antisera overnight at 4°C. The N-terminal serum was used at a 1:50 dilution, and the C-terminal serum was used at a 1:100 dilution. The immune complexes were precipitated and processed as described above. The entire sample was analyzed by autoradiography.
Immunofluorescent detection of ORF 3 expressed in insect cells.
In 15-ml conical tubes, Sf9 cells were infected with either wild-type baculovirus or baculovirus recombinants Bac-NV2, Bac-rNV C8, or Bac-NV3 at a MOI of 10. Following infection, the cells were seeded onto 96-well plates at a density of 2 × 104 cells/well and were incubated at 27°C. At 32 h p.i., the cells were rinsed once with 0.1 M PBS and fixed in 100% cold methanol for 15 min at room temperature. Next, the cells were rehydrated with 0.1 M PBS and stored at 4°C for later use. Primary antibodies were added to the wells at the appropriate dilution in 0.1 M PBS and incubated at 37°C for 2 h. The wells were washed with 0.1 M PBS, and the diluted fluorochrome-conjugated secondary antibodies (Sigma, St. Louis, Mo.) were added and the plates were incubated at 37°C for 2 to 4 h. Expression of the ORF 3 protein was detected using the rabbit ORF 3 C-terminal peptide antiserum (1:100) as the primary antibody and a fluorescein isothiocyanate-conjugated goat anti-rabbit IgG antibody (1:1,000) as the secondary antibody. Fetal bovine serum (5% final concentration) was added to the diluted secondary antibody to decrease background fluorescence. After the final wash step, fluorescence was detected using an Olympus IX-70 inverted-system microscope. Images were captured using the DC3-30 color camera (Dage-MTI, Michigan City, Ind.) and Image Pro Plus software (Media Cybernetics, Silver Spring, Md.).
Phosphatase treatment of the ORF 3 protein expressed in insect cells.
Infected insect cell pellets (3 × 106 cells) were lysed in 0.5 ml of M-Per buffer (Pierce, Rockford, Ill.) plus protease inhibitors (0.5 μg of aprotinin [Sigma] per ml, 0.7 μg of pepstatin [Calbiochem, La Jolla, Calif.] per ml, 0.5 μg of leupeptin [Sigma] per ml). The lysates were clarified, and an aliquot was removed for Western blot analysis. The remaining sample was adjusted to 1× calf intestinal alkaline phosphatase (CIAP) buffer (Life Technologies, Gaithersburg, Md.) and incubated for 30 min at 37°C in the absence or presence of CIAP enzyme (20 U; Gibco). All samples were analyzed by Western blot using the ORF 3 C-terminal peptide antiserum (1:500).
Western blot analysis to detect ORF 3 in preparations of VLPs.
Recombinant NV particles were produced and purified from the media of Sf9 cell cultures collected 7 days after infection with either the ORF 2 baculovirus recombinant (Bac-NV2) or the ORFs 2+3 baculovirus recombinant (Bac-rNV C8) (28). Briefly, cells were infected at a MOI of 5 and grown in the presence of protease inhibitors (as above), added daily. VLPs were concentrated by ultracentrifugation through a 30% sucrose cushion followed by isopycnic CsCl gradient centrifugation (1.362 g/cm3). The VLP band was concentrated by ultracentrifugation and banded by rate-zonal centrifugation on a discontinuous 10 to 50% (wt/vol) sucrose gradient as previously described (51). The VLP-containing fractions were pooled and pelleted by ultracentrifugation. Aliquots of concentrated VLPs and the soluble protein fractions were analyzed by SDS-PAGE and Western blotting. Western blot analysis for the detection of the ORF 3 protein used either rabbit ORF 3 peptide antiserum as described above. The ORF 2 protein was detected using the rabbit anti-rNV serum at a dilution of 1:5,000 followed by a horseradish peroxidase-conjugated goat anti-rabbit serum (Cappel) as described above. In some experiments, the ORF 2 protein was detected using monoclonal antibody (MAb) 8301 at a dilution of 1:1,000 followed by a horseradish peroxidase-conjugated goat anti-mouse serum (1:5,000; Southern Biotechnology Associates, Inc., Birmingham, Ala.). Protein concentrations were determined using the bicinchoninic acid protein assay kit (Pierce). The presence and integrity of particles were confirmed by electron microscopic (EM) analysis.
Additionally, the ability of the ORF 3 protein to be incorporated into rNV VLPs when ORF 2 and ORF 3 were expressed from different baculovirus recombinants was examined. Sf9 cells were dually infected with Bac-NV2 and Bac-NV3, each at a MOI of 5. Cultures were incubated for 7 days in the presence of protease inhibitors. VLPs were harvested as above with minor modifications. These dual-infection VLP preparations (DI2/3 VLPs) were not subjected to isopycnic CsCl gradient centrifugation; they were concentrated though a 30% sucrose cushion and banded by rate-zonal centrifugation on a discontinuous 10 to 60% (wt/vol) sucrose gradient. The VLP-containing fractions were pooled and concentrated as described above. The purified VLPs were analyzed by SDS-PAGE and Western blotting. Protein concentrations and particle integrity were determined as described above.
Preparation of radiolabeled rNV VLPs to determine the number of molecules of ORF 3 per VLP.
Recombinant NV particles were metabolically labeled as previously described (50) with slight modifications. Briefly, at 28 h p.i., Sf9 cells infected with the ORFs 2+3 baculovirus recombinant were starved for 45 min in methionine-free medium and then labeled with [35S]Met label (>1,000 Ci/mmol; 185 MBq/mmol [Amersham Pharmacia Biotech]) at a final concentration of 20 μCi/ml. Both 4 and 24 h after the initial labeling, an additional 0.25 μCi of [35S]Met per ml was added to the culture. Protease inhibitors were added daily as described above. The radiolabeled VLPs were purified on both CsCl and sucrose gradients from the media of Sf9 cell cultures collected 5 days p.i. The total protein concentration was determined as described above, and the specific activity was determined by liquid scintillation counting. An aliquot (35 μg) was separated by SDS-PAGE, transferred to nitrocellulose to allow for both PhosphorImager analysis using the Molecular Dynamics Storm system and Western blot analysis with the rabbit ORF 3 C-terminal peptide antiserum. ImageQuantNT software was used for PhosphorImager data analysis. The obtained quantitative representation of the sample was used to calculated the number of ORF 3 molecules per VLP.
Western blot analysis to detect the ORF 3 protein in virus purified from stools of NV-infected volunteers.
Virus particles were partially purified from the stools of volunteers experimentally infected with NV (16; I. LeParc-Goffart and M. K. Estes, unpublished data). Briefly, stool samples diluted in 0.1 M PBS–5% Zwittergent detergent (Calbiochem, La Jolla, Calif.) were extracted with 1,1,2-trichloro-1,2,2-trifluoroethane (Freon; Fisher Scientific, Pittsburgh, Pa.) and centrifuged at 12,400 × g for 10 min. The supernatant was collected, and virus was precipitated with polyethylene glycol (molecular weight, 6,000 [BDH Laboratory Supplies, Poole, England]) and NaCl for 30 min at room temperature (1). The precipitated virus was pelleted at 10,000 × g for 15 min, and the pellet was suspended in Tris-buffered saline (pH 7.4 to 7.6) (0.14 M NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 5.5 mM dextrose, 25 mM Tris, 0.5 mM MgCl2·6H2O). The virus suspension was pelleted through a 30% sucrose cushion and further purified on a CsCl gradient. The gradient was fractionated by bottom puncture, and each fraction was tested for the presence of virus-specific RNA by dot blot analysis using a 32P-labeled ORF 3 antisense probe. Positive fractions were pooled, diluted, and pelleted at 120,000 × g for 3 h. The purified virus was suspended in Milli-Q water and separated on SDS-polyacrylamide gels under reducing conditions. The presence of the ORF 3 protein was determined by Western blot analysis as described above.
RESULTS
Production and characterization of peptide antisera against the ORF 3 protein.
Characterization of the product of the NV ORF 3 has been hampered by a lack of reagents to directly detect the ORF 3 protein. ORF 3 amino acid residues 181 to 203 and 70 to 99 were chosen for the production of antisera due to their high amphipathic score and high surface potential. The peptide sequences were synthesized, coupled to keyhole limpet hemacyanin and used to generate antisera in mice and rabbits. The titers against the original peptide varied between 1:250 and 1:10,240 for individual mouse sera. The titer of the rabbit antiserum against the homologous peptide was 1:10,240 for the C-terminal peptide serum and 1:327,680 for the N-terminal peptide serum as detected by ELISA (data not shown). No reactivity was detected by ELISA against heterologous peptides of unrelated sequence. By Western blot analysis, all sera, at a dilution of 1:500, were able to detect a glutathione S-transferase–ORF 3 fusion protein expressed in Escherichia coli (data not shown) or the ORF 3 protein expressed in insect cells (see below).
The ORF 3 protein is expressed in a cell-free translation system.
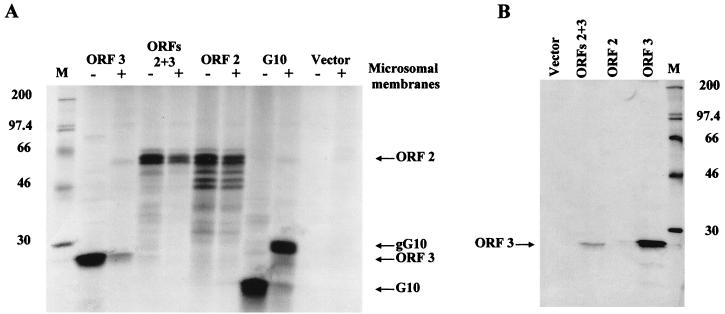
First, we examined the expression of the ORF 3 protein by cell-free translation in RRLs. Transcripts were made from linearized DNA transcribed using either the SP6 or T7 polymerase and translated in RRLs. Translated proteins were radiolabeled with Tran35S-label in the absence or presence of microsomal membranes. Translation in the presence of microsomal membranes was tested because the ORF 3 protein contains one predicted site for N-linked glycosylation. The ORF 3 protein product migrated with a molecular weight of approximately 25,000 (Fig. 1A). Further analysis of the cell-free translated ORF 3 protein revealed that it comigrated with the 23,000-molecular-weight ORF 3 protein (23K protein) produced in insect cells (data not shown). The difference in apparent size was due to the use of different molecular weight marker preparations on different gels. No modification of the ORF 3 protein was detected in the presence of microsomes. Rotavirus gene 10 was included as a positive control for glycosylation. Expression of gene 10 in the absence of microsomal membranes resulted in the synthesis of a 20K protein, and addition of microsomal membranes resulted in a glycosylated product of the expected apparent molecular weight (ca. 29,000) (12). The NV capsid protein was included as a negative control for glycosylation. In the absence or presence of microsomal membranes, translation of the ORF 2 coding region yielded the major capsid protein with a molecular weight of 58,000 (58K protein) and a few faster-migrating ORF 2-related bands that represent proteins translated from alternate initiation sites or degradation products (28). Translation of an RNA containing the entire 3′ end of the genome (ORFs 2+3) yielded the 58K major capsid protein product as well as the lower-molecular-weight bands. It was not clear whether the ORF 3 protein was synthesized from the ORFs 2+3 RNA. Immunoprecipitation using the ORF 3 C-terminal peptide antiserum confirmed that the ORF 3 protein was indeed generated from the ORFs 2+3 RNA (Fig. 1B). No corresponding protein was immunoprecipitated from either the vector or ORF 2 translation reactions. Thus, the ORF 3 protein was produced from RNAs containing both ORFs 2+3 as well as from RNA containing ORF 3 alone, and this protein was not modified by N-linked glycosylation.
FIG. 1.
The ORF 3 protein is produced by cell-free translation of ORF 3 and ORFs 2+3 transcripts. (A) Analysis of proteins produced by translation of synthetic mRNAs using RRLs in the absence (−) or presence (+) of canine pancreatic microsomal membranes. The translated proteins were labeled using Tran35S-label. An aliquot (10 μl/25 μl) of the translation reaction mixture was analyzed on an SDS–15% polyacrylamide gel. (B) Immunoprecipitation of the ORF 3 protein from cell-free translation reaction mixtures using the ORF 3 C-terminal peptide antiserum. The remaining aliquot of the translation reaction mixtures in panel A was immunoprecipitated in RIPA buffer, and the entire sample was analyzed by autoradiography of an SDS–15% polyacrylamide gel. Lane M contains 14C-methylated protein molecular weight markers (Amersham); lane ORFs 2+3 contains the entire 3′ end of the genome beginning with the ORF 2 coding sequences; lane G10 contains rotavirus gene 10; lane Vector contains pGem7Zf(+) vector with no insert. The ORF 2, ORF 3, G10, and glycosylated G10 (gG10) protein products are designated.
ORF 3 expressed in insect cells has multiple forms.
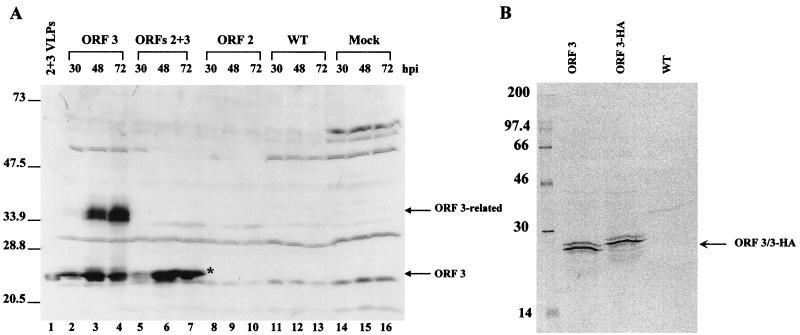
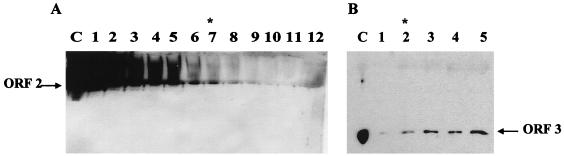
We next examined the expression of the ORF 3 protein in insect cells infected with baculovirus recombinants by Western blot analysis using the ORF 3 peptide antisera. The same results were obtained with either serum. The ORF 3 protein was present in insect cell lysates infected with baculovirus recombinants that contained either ORF 3 or ORFs 2+3 (Fig. 2A). In insect cells infected with the ORF 3 baculovirus recombinant, two main forms of the ORF 3 protein were detected. The molecular weights of these ORF 3-related proteins were calculated to be 23,000 and 35,000, migrating as a lower doublet and an upper doublet (Fig. 2A, lanes 2 to 4). At 30 h p.i., the predominant form was the lower doublet. The amount of this lower form of the ORF 3 protein increased over the first 48 h p.i. and decreased slightly by 72 h p.i. The ORF 3 upper-doublet form (35K protein) appeared to increase in concentration over time. In insect cells infected with the ORFs 2+3 baculovirus recombinant, only the lower-23K doublet protein was detected (lanes 5 to 7). No corresponding protein was detected in cells that were either mock infected or infected with wild-type baculovirus or with a baculovirus recombinant that expressed ORF 2 (lanes 8 to 16). Only the 23K protein was incorporated into VLPs (lane 1) (see Fig. 4).
FIG. 2.
Expression of the ORF 3 protein in infected insect cells. Sf9 cells were either mock infected (Mock) or infected with wild-type baculovirus (WT), the ORF 2 baculovirus recombinant, the ORFs 2+3 baculovirus recombinant, or the ORF 3 baculovirus recombinant. (A) Kinetics of ORF 3 expression in infected insect cells. Cells were harvested at 30, 48, and 72 h p.i. by lysis in SDS-PAGE sample buffer. The samples were separated by SDS-PAGE in a 15% polyacrylamide gel, and Western blot analysis was performed using the rabbit ORF 3 C-terminal peptide antiserum at a dilution of 1:500. The asterisk highlights the 23K ORF 3 protein band that separates into two bands on some gels. (B) Immunoprecipitation of the ORF 3 protein from infected insect cells. Cells infected with wild-type baculovirus (WT), the ORF 3 or the HA-tagged ORF 3 (ORF 3-HA) baculovirus recombinants were harvested in RIPA buffer at 65 h p.i. and immunoprecipitated using the ORF 3 C-terminal peptide antiserum.
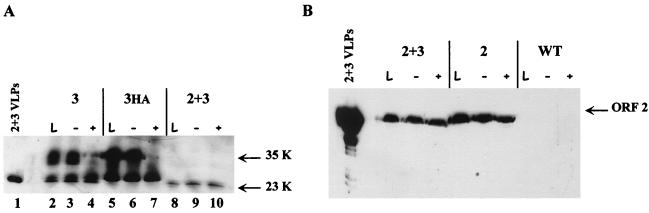
FIG. 4.
The 35K ORF 3 protein is phosphorylated. Sf9 cells were infected with wild-type baculovirus (WT) or the ORF 2, ORFs 2+3, ORF 3, or ORF 3HA baculovirus recombinants. Cells were harvested at 65 h p.i. in M-Per buffer plus protease inhibitors. An aliquot of the clarified lysate was removed for analysis (lanes L). The remaining sample was adjusted to 1× CIAP buffer and incubated in the absence (−) or presence (+) of CIAP (20 U). Western blot analysis was performed using the ORF 3 C-terminal peptide antiserum (1:500) (A) or the anti-rNV serum (1:1,000) (B). rNV 2+3 VLPs were included as a positive control for Western blot analysis.
The ORF 3 protein was immunoprecipitated from infected insect cell lysates by using either the ORF 3 C-terminal or N-terminal peptide antiserum. Infected cells were metabolically labeled with 35S-Promix, and lysates were harvested in RIPA buffer. Both the ORF 3 peptide antisera efficiently immunoprecipitated the 23K ORF 3 protein from insect cells infected with either the ORF 3, another ORF 3 construct containing an engineered HA tag (ORF 3HA [Fig. 2B]) or ORFs 2+3 (data not shown) baculovirus recombinants; the 35K ORF 3 protein was not immunoprecipitated.
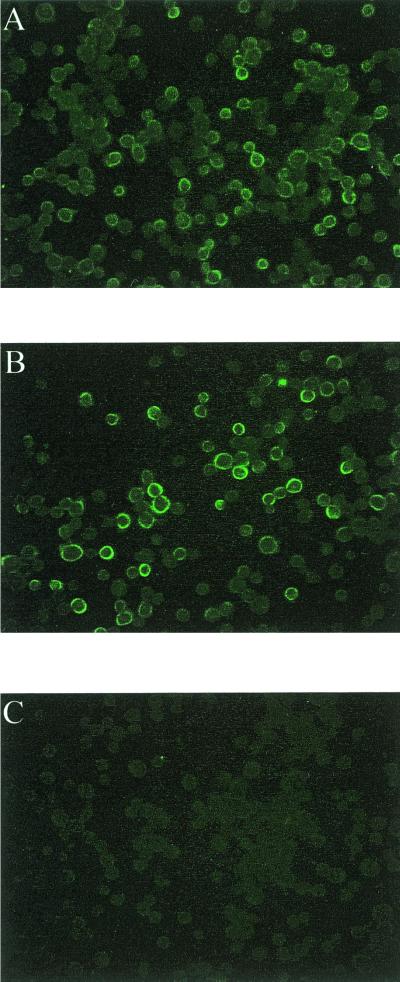
Since the ORF 3 protein is predicted to be a highly basic protein, the possibility existed that it might be localized to the nucleus; therefore, infected insect cells were examined by indirect immunofluorescence to localize the site of expression of the ORF 3 protein. The ORF 3 protein was detected in the cytoplasm of cells infected with either the ORFs 2+3 or the ORF 3 baculovirus recombinant (Fig. 3A and B); only background fluorescence was seen in wild-type-baculovirus-infected Sf9 cells (Fig. 3C). As expected, a mouse anti-ORF 2 antiserum detected the ORF 2 protein in the cytoplasm of cells infected with either the ORF 2 or the ORFs 2+3 baculovirus recombinant but not in cells expressing only ORF 3 (data not shown).
FIG. 3.
Immunofluorescence analysis of ORF 3 protein expression in infected insect cells. Sf9 cells were infected with the ORF 3 baculovirus recombinant (A), the ORFs 2+3 baculovirus recombinant (B), or wild-type baculovirus (C) and seeded onto 96-well plates. At 32 h p.i., the cells were fixed in methanol and rehydrated in PBS. The ORF 3 protein was detected using the ORF 3 C-terminal peptide antiserum (dilution of 1:100) followed by a fluorescein isothiocyanate-conjugated goat anti-rabbit serum (dilution of 1:1,000). Fluorescence was detected using an Olympus IX-70 inverted-system microscope. Images were captured using the DC3-30 color camera and Image Pro Plus software.
Additionally, infected insect cell cultures were metabolically labeled with orthophosphate to examine whether the ORF 3 protein was phosphorylated, since this protein contains nine potential sites of phosphorylation. At 72 h p.i., a large number of proteins were labeled; however, no proteins were immunoprecipitated with either ORF 3 peptide antiserum (data not shown). Recombinant NV VLPs were purified from insect cells infected with the ORFs 2+3 that were metabolically labeled with orthophosphate. This analysis revealed that the ORF 3 protein present in the rNV VLPs did not incorporate orthophosphate (data not shown); however, this did not rule out the possibility that ORF 3 protein is phosphorylated in infected cells. Therefore, further experiments were done to examine whether the ORF 3 protein is phosphorylated in insect cells. Infected insect cell cultures were lysed in M-Per buffer plus protease inhibitors. This buffer was chosen due to its high efficiency of lysis and mild composition, which allows downstream assays to be performed without the removal of the detergent. Clarified lysates were adjusted to 1× CIAP buffer and incubated in the absence or presence of CIAP. Western blot analysis revealed that when no CIAP was added, no change was seen in the migration of the ORF 3 proteins (Fig. 4A, lanes 2 and 3 and lanes 5 and 6). With the addition of 20 U of CIAP enzyme, the 35K ORF 3 protein was no longer detected in either the ORF 3- or the ORF 3-HA-infected cell lysates by Western blot analysis and a slight increase in the amount of the 23K protein was detected (lanes 4 and 7). No change in the 23K protein was detected in ORFs 2+3-infected cell lysates (lane 10). Additionally, analysis of these same samples by Western blot using the anti-rNV serum (Fig. 4B) showed that the ORF 2 protein was not affected by the addition of the CIAP enzyme.
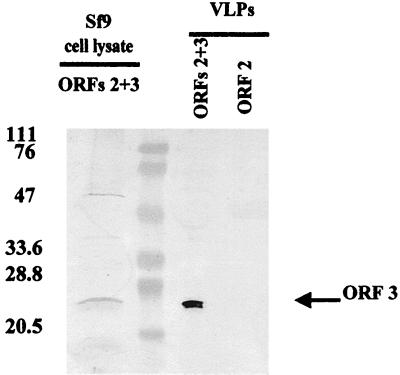
ORF 3 is specifically associated with rNV VLPs.
Since the ORF 3 protein was expressed by the ORFs 2+3 baculovirus recombinant, we examined whether this protein was present in purified VLPs. VLPs were purified first through a CsCl gradient to band NV VLPs and component proteins and then through a sucrose gradient to separate proteins in the form of VLPs from soluble proteins. Western blot analysis of sucrose gradient-purified ORF 2 and ORFs 2+3 VLPs (20 μg) revealed that the ORF 3 protein was present in VLPs purified from supernatants of cells infected with the ORFs 2+3 baculovirus recombinant (Fig. 5). The ORF 3 protein was also detected in the soluble protein fractions when equal amounts of protein (>10 μg) were analyzed (data not shown). No corresponding protein was detected in the soluble fractions or VLPs purified from cell cultures infected with the baculovirus recombinant encoding only the ORF 2 protein (Fig. 5). Examination of over 10 preparations of ORFs 2+3 VLPs showed that the ORF 3 protein was always present in VLPs whether they were purified by banding in CsCl gradients alone, sucrose gradients alone, or a combination of CsCl gradients followed by sucrose gradients. The same results were obtained with the ORF 3 N-terminal peptide antiserum (data not shown).
FIG. 5.
The ORF 3 protein is associated with rNV VLPs. Recombinant rNV VLPs were purified from supernatants of Sf9 cells infected with the ORF 2 (rNV 2 VLPs) or the ORFs 2+3 (rNV 2+3 VLPs) baculovirus recombinants. VLPs sequentially purified over both CsCl and sucrose gradients were analyzed by SDS-PAGE and Western blotting using the rabbit ORF 3 C-terminal peptide antiserum (1:500).
Next, we determined the minimal amount of VLPs that had to be analyzed to detect the ORF 3 protein. This was done by Western blot analysis of serial dilutions of ORFs 2+3 VLPs using MAb 8301 to the 58K capsid protein (21) and the ORF 3 C-terminal peptide antiserum. This analysis showed consistent detection of the ORF 2 and ORF 3 proteins in three preparations of VLPs. The ORF 2 protein was detected in VLP protein amounts between 8 and 15 ng. Initially, the ORF 3 protein was not detected in the largest amount of VLPs tested (2 μg); further studies showed that 10 to 17 μg of protein in the form of VLPs must be analyzed to detect the ORF 3 protein by Western blotting with the ORF 3 peptide antiserum. A representative VLP preparation is shown in Fig. 6. In this sample, the ORF 2 protein was clearly detectable at ≥15 ng of VLPs (Fig. 6A, lane 7) while the ORF 3 protein was detected at ≥17 μg of VLPs (Fig. 6B, lane 2).
FIG. 6.
Western blot analysis of the ORF 2 protein and the ORF 3 protein in an ORFs 2+3 VLP preparation. (A) Detection of the ORF 2 protein. Purified VLPs were diluted to a loading concentration of 2 μg and serially diluted twofold for separation on an SDS–15% polyacrylamide gel. Western blot analysis was performed using MAb 8301 at a dilution of 1:1000. (B) Detection of the ORF 3 protein. A higher concentration of purified VLPs was loaded, and ORF 3 was detected using the rabbit ORF 3 peptide antiserum (1:500). Asterisks denote the lowest level at which each protein was clearly detectable. The lanes designated C are ORFs 2+3 VLPs run as a control for ORF 2 and ORF 3 detection. (A) Lanes: 1, 2 μg; 2, 1 μg; 3, 0.5 μg; 4, 0.25 μg; 5, 0.125 μg; 6, 0.0625 μg; 7, 0.03125 μg; 8, 0.015625 μg; 9, 0.0078 μg; 10, 0.0039 μg; 11, 0.00195 μg; 12, 0.00097 μg. (B) Lanes: 1, 15 μg; 2, 17 μg; 3, 20 μg; 4, 22 μg; 5, 25 μg.
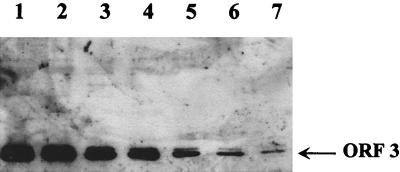
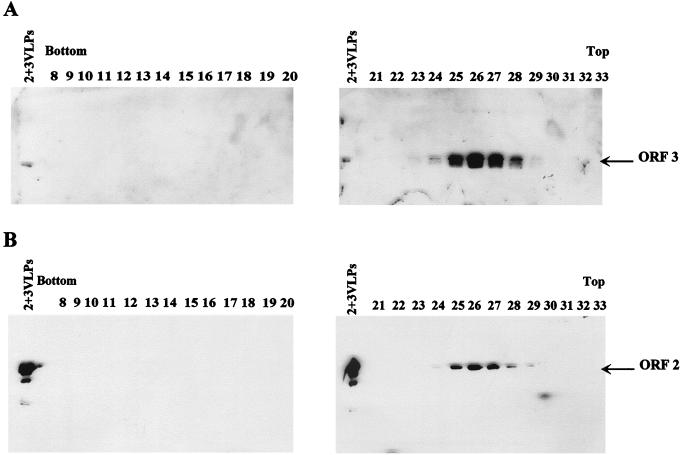
VLPs harvested from insect cells dually infected with both the ORF 2 and the ORF 3 baculovirus recombinants (DI2/3 VLPs) were analyzed by Western blotting. This analysis revealed that the ORF 3 protein was detected in the DI2/3 VLPs (Fig. 7). Examination of cell lysates showed that both the 23K and 35K ORF 3 protein forms were generated (data not shown); however, only the 23K protein was incorporated into the particles (Fig. 7). These particles were less stable than the ORFs 2+3 particles. The DI2/3 VLPs were unable to withstand banding through a CsCl gradient and were purified directly on a sucrose gradient. It also appeared that more molecules of ORF 3 were incorporated into these particles, since the ORF 3 protein could be detected when as little as 15 ng of protein in the form of VLPs was analyzed. Aliquots (15 μl/1,000 μl) of the sucrose gradient fractions were analyzed by Western blotting using either the ORF 3 C-terminal peptide antiserum or a MAb to the ORF 2 capsid protein to show that the increased amount of the ORF 3 protein in the DI2/3 VLPs was not due to an overall increase in the amount of soluble ORF 3 protein throughout the gradient. In this analysis, the ORF 3 protein was detectable only in the VLP-containing fractions, as indicated by the detection of the ORF 2 protein in the same fractions (Fig. 8).
FIG. 7.
Western blot analysis of DI2/3 VLPs using the ORF 3 C-terminal peptide antiserum. VLPs were diluted to a loading concentration of 1 μg and serially diluted twofold for separation on an SDS-polyacrylamide gel. Western blot analysis was performed using the rabbit ORF 3 C-terminal peptide antiserum (1:500). Lanes: 1, 1 μg; 2, 0.5 μg; 3, 0.25 μg; 4, 0.125 μg; 5, 0.0625 μg; 6, 0.03125 μg; 7, 0.015625 μg. The DI2/3 particles were as pure as previously described VLP preparations (3).
FIG. 8.
The ORF 3 protein comigrates with the ORF 2 protein in VLPs in sucrose gradients. Western blot analysis of the sucrose gradient fractions from the DI2/3 VLP purification is shown. Proteins were detected using the rabbit ORF 3 C-terminal peptide antiserum (A) or the ORF 2 MAb 3912 (B). Fractions numbers and gradient orientation are indicated. Each gel contained a VLP control, 2+3VLPs (left lanes).
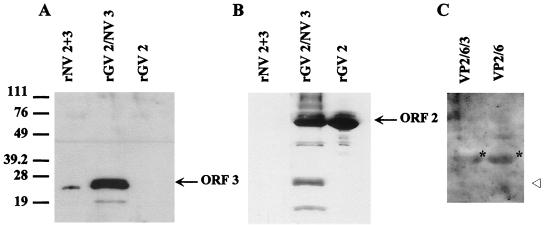
Additionally, dual-infection experiments were performed using another “Norwalk-like virus” capsid protein, the genogroup II Grimsby virus (GV) ORF 2 protein (18). Cells were dually infected with baculovirus recombinants expressing the GV ORF 2 protein and the NV ORF 3 protein. The recombinant GV (rGV) VLPs were purified directly on a sucrose gradient. VLP-containing fractions were pooled, pelleted, and analyzed by Western blotting with the ORF 3 C-terminal peptide antiserum or anti-rGV antiserum. The rGV ORF 2-NV ORF 3 VLPs contained the NV ORF 3 protein; no corresponding protein was detected in rGV ORF 2 VLPs purified from cultures infected with only the GV ORF 2 baculovirus recombinant (Fig. 9A). Reactivity with the anti-rGV serum confirmed the presence of the ORF 2 protein in these VLPs and the lack of cross-reactivity between the ORF 2 proteins of rNV and rGV (Fig. 9B).
FIG. 9.
The ORF 3 protein associates with GV ORF 2 VLPs but not rotavirus VP2/6 VLPs. (A) GV VLPs (25 μg) were analyzed in an SDS–15% polyacrylamide gel. Western blot analysis was first performed using the rabbit ORF 3 C-terminal peptide antiserum. (B) The blot was then stripped in ECL stripping buffer as specified by the manufacturer (Amersham) and reprobed with the rabbit anti-rGV serum (1:500); the rGV ORF 2 and degradation products are detected by this serum. (C) Rotavirus VLPs (25 μg) were loaded for separation in an SDS–15% polyacrylamide gel and analyzed by Western blotting using the ORF 3 C-terminal peptide antiserum. Lane designations are as follows: rGV 2, GV ORF 2 VLPs; rGV 2/NV 3, dually infected GV ORF 2 and NV ORF 3 VLPs; rNV 2+3, NV ORFs 2+3 VLPs; VP2/6/3, dually infected rotavirus 2/6- and NV ORF 3 VLPs; VP2/6, rotavirus 2/6-VLPs. The open arrowhead designates where the ORF 3 protein would migrate if present. The asterisks show the location of rotavirus VP6 that is detected nonspecifically by the secondary antibody because of the large amount of protein on the gel.
To show that the ORF 3 protein does not associate with any heterologous VLP, insect cells were dually infected with a rotavirus VP2/6 baculovirus recombinant (A. F. Bertolotti-Ciarlet, unpublished data) and the NV ORF 3 baculovirus recombinant as described above. Rotavirus 2/6-VLPs were purified as previously described (10), and Western blot analysis confirmed that the ORF 3 protein was not present in the rotavirus particles (Fig. 9C). Taken together, these results suggest that the ORF 3 protein is a minor structural protein of the NV virion.
The number of ORF 3 molecules per VLP was determined by analysis of the stoichiometric ratio of the two proteins in VLPs by using PhosphorImager analysis. The Storm system captures images from both strong and weak signals in a single exposure, and the linear dynamic range is 1,000 times greater than that of film, allowing the quantitation of ORF 3 without overexposure of ORF 2. The specific activity of the purified rNV VLPs metabolically labeled with [35S]Met was 5.34 × 104 cpm/μg. These VLPs were separated by SDS-PAGE, transferred to nitrocellulose, and exposed to the phosphor screen. The quantitative representation of the ORF 2 and ORF 3 proteins was determined using ImageQuantNT software. The number of molecules of ORF 3 per VLP was calculated based on knowing the quantity of VLPs loaded (35 μg), the number of VLPs per microgram (5.78 × 1010) (52), and the number of capsid molecules per VLP (180 copies). After the data were scanned from the membrane, Western blot analysis was performed using the rabbit ORF 3 peptide antiserum to confirm that the ORF 3 band was detected properly. Several assumptions were made in the analysis of these data. First, production of the VLPs in the presence of protease inhibitors still yielded several ORF 2 degradation products. These were included in the calculations and assumed to be originally incorporated as full-length ORF 2, since these VLPs were purified through both CsCl and sucrose gradients to remove soluble protein. Second, an efficiency of labeling of 100% was assumed, since no cold methionine was added to the cultures. Based on these assumptions, it was calculated that ∼1.5 molecules of ORF 3 are present per VLP.
ORF 3 is associated with native NV purified from stool.
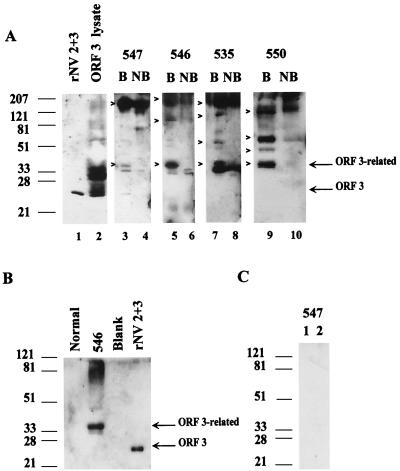
We have shown that the ORF 3 protein is a component of the rNV VLPs. We next examined whether this protein is a component of native NV virions. Due to the lack of a cell culture system or animal model for NV, the only available source of virus is stools from infected individuals. Virus was purified from the stools of NV-infected volunteers (I. LeParc-Goffart and M. K. Estes, unpublished). Purified virions were detected by EM in preparations from five volunteers. An aliquot (5 or 10 μl) from each of these virus preparations was examined by Western blot analysis using the ORF 3 peptide antiserum. The results of analysis of virus from the stools of four volunteers are shown in Fig. 10A. The partially purified virions isolated from the stool of volunteer 547 contained the 23K and 35K ORF 3 protein forms, as well as higher-molecular-weight forms (Fig. 10A, lane 3). Samples from volunteers 546, 535, and 550 contained only the 35K protein and higher-molecular-weight forms (lanes 5, 7, and 9). EM analysis of the sample from volunteer 547 revealed that it contained more empty particles than the other purified samples, which may account for the detection of the 23K protein. The apparent molecular weight of the detected larger forms varied from sample to sample. Neither the 23K nor the 35K ORF 3 protein reacted with the rabbit anti-rNV ORF 2 serum, as expected (data not shown). The higher-molecular-weight forms probably represent multimers of the ORF 2 and/or ORF 3 proteins. If the virion samples were not boiled prior to separation by SDS-PAGE, the 23K and 35K proteins were not detected and only higher-molecular-weight forms of ORF 3 were detected (lanes 4, 6, 8, and 10). A similar pattern, consisting of detection of only higher-molecular-weight forms, was seen for ORF 2 when the samples were not boiled (data not shown). No proteins were detected in an identically prepared stool sample from an uninfected volunteer (Fig. 10B). No proteins were detected when the partially purified virions were analyzed by Western blotting with rabbit preimmune sera (Fig. 10C). Taken together, these data suggest that the ORF 3 protein is a structural protein of NV.
FIG. 10.
Western blot analysis of partially purified virions from NV-infected volunteers using the ORF 3 peptide antiserum. (A) Analysis of samples from volunteers 547, 546, 535, and 550. Boiled (B) and not-boiled (NB) samples were analyzed to examine both continuous and discontinuous epitopes. Controls included ORFs 2+3 VLPs and ORF 3 baculovirus recombinant-infected cell lysate. The ORF 3-related proteins are designated on the right and by arrowheads. (B) Partially purified sample from an uninfected volunteer (Normal) prepared identically to the infected volunteer samples in panel A, partially purified NV from volunteer 546 (independent purification), and rNV 2+3 VLPs analyzed by Western blot analysis using the ORF 3 C-terminal peptide antiserum. (C) Two independent partial purifications of NV from stools of volunteer 547, analyzed by Western blotting using a rabbit preimmune serum (1:500).
DISCUSSION
Studies of the expression of the proteins of the human caliciviruses remain limited due to a lack of tissue culture system and reagents to detect many of the proteins. However, there has been considerable progress in understanding the structural and antigenic characteristics of the viral capsid protein (21, 38–40, 51). Baculovirus recombinants expressing either ORF 2 alone or ORFs 2+3 are able to produce rNV VLPs, indicating that the ORF 3 protein is not essential for the formation of empty VLPs (15, 17, 18, 27, 28, 30, 34, 51). However, the conservation of ORF 3 in the genomes of all caliciviruses suggests that this protein performs an important function in the virus life cycle. Studies by Wirblich et al. (54) showed that the ORF 3-equivalent protein of an animal calicivirus, RHDV, is a minor structural component of purified virions. In this paper, we provide the first evidence that the ORF 3 protein is a minor structural protein of the Norwalk-like human caliciviruses.
Previous characterization of the translation products of an RNA encoding the ORF 2 and ORF 3 proteins or of the proteins produced in insect cells infected with the ORFs 2+3 baculovirus recombinant concluded that the ORF 3 protein was not expressed or was expressed insufficiently to allow the detection of an 35S-labeled ORF 3 protein (28). This may have been because the ORF 3 protein lacks cysteines and contains only three methionines, so that radiolabeling with methionine was inefficient as a detection method. The present studies showed that the ORF 3 protein was efficiently detected in a cell-free translation system with Tran35S-label only if the entire reaction was analyzed. To examine the expression of ORF 3, we first prepared two antisera in mice and rabbits to ORF 3 peptides spanning amino acids 70 to 99 or 181 to 203.
Expression of ORF 3 in a cell-free translation system from RNAs containing either ORF 3 or ORFs 2+3 resulted in the production of a protein with an apparent molecular weight of 23,000. This size is consistent with the predicted molecular weight based on the amino acid sequence. Additionally, these experiments showed that the NV ORF 3 protein was not modified by N-linked glycosylation. The detection of an ORF 3 protein with an apparent size similar to that predicted by the sequence is in agreement with previously published results for the detection of the ORF 3 protein of FCV and RHDV (24, 32, 54) and recently for the human calicivirus Camberwell virus (45) expressed in a cell-free translation system.
Expression of ORF 3 in insect cells from the ORFs 2+3 baculovirus recombinant resulted in the production of a protein doublet with an apparent molecular weight of 23,000. This result is consistent with previously published studies of FCV and RHDV, which show detection of the ORF 3 protein in infected cells (24, 32). When ORF 3 was expressed in insect cells from a baculovirus recombinant containing only the ORF 3 sequences, two forms of the ORF 3 protein were produced. The ORF 3 protein was not modified by N-linked glycosylation, and a [32P]orthophosphate-labeled 35K protein was not incorporated into VLPs. Additionally, analysis of infected insect cell lysates subjected to strong denaturants (urea or guanidinium isothiocynate) prior to electrophoresis resulted in no change in migration of the proteins (data not shown). Further experiments revealed that the 35K ORF 3 protein is phosphorylated (perhaps by a kinase reaction), since treatment of insect cell lysates with CIAP resulted in the disappearance of the 35K ORF 3 protein. We hypothesize that the phosphorylated 35K ORF 3 protein is involved in encapsidation of genomic RNA.
The expression of the ORF 3 protein in RRLs from the ORFs 2+3 RNA and in insect cells infected with the ORFs 2+3 baculovirus recombinant was surprising because the ORF 2 and ORF 3 proteins are encoded in different reading frames and the only known promoter driving the transcription of these constructs lies upstream of the ORF 2 coding sequences. These results suggest that the ORF 3 protein is expressed from an mRNA encoding ORFs 2 and 3. It is possible that the ORF 3 protein is made by utilizing mechanisms of ribosomal frameshifting, internal initiation, or termination-reinitiation. Previously, Neill (36) suggested that ORF 3 might be expressed by a −1 frameshift. Sequence analysis has revealed a lack of known stem-loop structures or heptapeptide sequences seen in other viruses (retroviruses, astroviruses, and coronaviruses) which utilize a mechanism of ribosomal frameshifting for the production of some of their proteins (5). Additionally, ribosomal frameshifting would result in the production of an ORF 2-ORF 3 fusion protein, which was not detected in our studies for NV or by Herbert et al. for FCV (24). A mechanism of termination-reinitiation or internal initiation has been suggested for synthesis of the ORF 3 protein of FCV based on in vitro studies with RRLs (24). Termination-reinitiation seems the most likely mechanism utilized by NV since there is no nontranslated region between the ORF 2 and ORF 3 coding sequences and no apparent internal ribosome entry site element contained within the ORF 2 coding region.
Using the ORF 3 peptide antisera, we have shown that the 23K ORF 3 protein is associated with ORFs 2+3 rNV VLPs and is consistently present in every ORFs 2+3 VLP preparation tested. We determined that there are ∼1.5 molecules of ORF 3 per VLP, as calculated from experiments with radiolabeled particles. This interaction is specific based on the ORF 3 protein being present in VLPs purified through both CsCl and sucrose gradients, the ORF 3 protein associating with VLPs when ORF 2 and ORF 3 are expressed from different baculovirus recombinants, and the ORF 3 protein not associating with heterologous rotavirus 2/6-VLPs. Somewhat unexpectedly, VLPs purified from the media of insect cells dually infected with ORF 2 and ORF 3 baculovirus recombinants contained a greater amount of only the 23K ORF 3 protein compared to VLPs purified from cultures infected with the ORFs 2+3 baculovirus recombinant. A possible explanation for these data is that the ORF 2 and ORF 3 proteins interact with each other prior to assembly and an increase in ORF 3 expression increases the number of ORF 2-ORF 3 multimers formed in the infected insect cells. Expression of the ORF 3 protein from the ORFs 2+3 construct resulted in low levels of the ORF 3 protein being produced; consequently, fewer ORF 2-ORF 3 multimers were available for VLP formation. The increase in the amount of ORF 3 in the DI2/3 VLPs affected their stability. This may be analogous to experiments with some plant virus systems aimed at creating designer virions carrying either labels, such as green fluorescent protein, or therapeutic peptides using a capsid-fusion protein, which require the production of both capsid and capsid-fusion protein to generate detectable virions (11, 19). While the DI2/3 particles are able to form, it is likely that only a limited number of ORF 3 molecules can be incorporated into stable particles.
The association of the NV ORF 3 protein with the rGV VLPs was somewhat unexpected, since these two viruses are classified in different genogroups within the NLVs and are antigenically distinct (18). This result suggests that regions common to the ORF 2 and ORF 3 proteins are involved in this protein-protein interaction. Sequence analysis of the viral capsid proteins did not reveal any obvious regions that might be responsible for incorporation the ORF 3 protein into VLPs. This analysis obviously does not rule out the possibility that a common structural motif may be involved.
Structural studies to date have not detected the ORF 3 protein in three-dimensional reconstructions (39) or in the high-resolution structure (38). These studies were conducted on rNV VLPs prepared from supernatants of insect cells infected with the ORFs 2+3 baculovirus recombinant. The methods used to obtain the three-dimensional reconstruction and the high-resolution structure require that ≥60 copies of a protein be present with icosahedral symmetry for detection. Our findings indicate that the ORF 3 protein is not present in large enough quantities to be detected.
Caliciviruses are structurally similar to many plant viruses. Plant virus capsids are composed of 180 copies of a single structural protein which assembles into a T=3 structure (22, 25, 47, 52, 53). In plant viruses such as brome mosaic virus (41, 44), tomato bushy stunt virus (22), and turnip crinkle virus (25), RNA packaging occurs through an interaction of the viral RNA with the basic N-terminal domain of the capsid protein. The capsid protein of caliciviruses is acidic (pI = 5.64) and lacks a basic N-terminal arm. It has been speculated that the ORF 3 protein may function in viral RNA encapsidation in caliciviruses. Our results, along with those of others, are consistent with this hypothesis (54).
Multiple forms of the ORF 3 protein were detected in the native NV virions, including the 23K and 35K proteins and higher-molecular-weight forms. The detection of some higher-molecular-weight bands with both the ORF 3 peptide antiserum and an anti-rNV serum suggests that multimeric forms of ORF 2 and ORF 3 are present in these samples. In partially purified virion samples that were not boiled, only the higher-molecular-weight forms of the ORF 3 protein were detected. In only one sample, from volunteer 547, was the 23K ORF 3 protein detected. EM analysis revealed that this sample appeared to contain more empty particles than did the other samples. It may be that the 23K protein is associated with empty virions and empty VLPs while the 35K protein is associated with RNA-containing virions. One interpretation of our data is that the 35K protein detected in NV virions represents a dimeric or modified (phosphorylated?) form of the ORF 3 protein necessary for the encapsidation of RNA into the virions. Future experiments will test this hypothesis.
Another interesting observation was that the NV virion samples appear to contain more ORF 3 protein than was detected in the rNV VLPs. Several possibilities may explain this observation. First, an aliquot of partially purified sample was analyzed; therefore, it is not clear exactly how many virions are present per sample. Since these preparations are not completely pure, the protein concentration could not be used to calculate the number of virus particles. An ELISA was unable to determine the number of virions present due to the small amounts of sample available. Second, it is possible that there are more molecules of ORF 3 in native virions than are incorporated into empty rNV VLPs. If the ORF 3 protein is involved in the encapsidation of the viral RNA, it may require more copies of protein for this function.
Other systems provide examples of viral capsids composed of both a major and a minor structural protein. Similar to NV, the capsid of single-stranded RNA bacteriophages (group I leviviruses) is composed of 180 copies of a coat protein and exhibits T=3 icosahedral symmetry. The levivirus virion also contains one molecule of an additional virus-encoded polypeptide, the maturation or A protein, which appears to be similar in abundance and possibly in function to the ORF 3 protein of NV. During infection, the A protein is cleaved to yield 15- and 24-kDa fragments (46). It is thought that this event liberates the 5′ end of the viral RNA. There is no evidence to suggest that the ORF 3 protein is cleaved in insect cells, yet it may function similarly. Sequence analysis of the amino acid composition predicts that the A protein is basic (pI = 9.66). Further studies showed the A protein interacts with several different sites on the RNA and is partially exposed on the surface of the virion and that bacteriophages lacking the A protein contain RNA with the 5′ terminus missing (46). Therefore, it is assumed that the A protein prevents the 5′ terminus of the RNA from protruding from the viral capsid, suggesting that the A protein might help alter the tertiary structure of the RNA for packaging (7, 43, 48). The ORF 3 protein is not essential for VLP formation, but it may be required for specificity and proper conformation of the encapsidated viral RNA. The fact that the VLPs produced in this system are empty suggests that the elements necessary for encapsidation are masked or, more likely, not present on the ORFs 2+3 RNA. Our current hypothesis is that the ORF 3 protein is located on the interior of the virion and that it functions in encapsidation of RNA and may aid in regulating assembly of the calicivirus virion.
ACKNOWLEDGMENTS
This work was supported by funding from the U.S. Public Health Service (grants AI38036 and AI46581), and P.J.G. was funded by a training fellowship (grant AI07471) from the National Institutes of Health.
We thank Andrea Bertolotti-Ciarlet and Brenda Hogue for critical comments and suggestions in the preparation of the manuscript.
REFERENCES
- 1.Atmar R L, Metcalf T G, Neill F H, Estes M K. Detection of enteric viruses in oysters by using the polymerase chain reaction. Appl Environ Microbiol. 1993;59:631–635. doi: 10.1128/aem.59.2.631-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au K-S, Mattion N M, Estes M K. A subviral particle binding domain on the rotavirus nonstructural glycoprotein NS28. Virology. 1993;194:665–673. doi: 10.1006/viro.1993.1306. [DOI] [PubMed] [Google Scholar]
- 3.Ball J M, Hardy M E, Atmar R L, Conner M E, Estes M K. Oral immunization with recombinant Norwalk virus-like particles induces a systemic and mucosal immune response in mice. J Virol. 1998;72:1345–1353. doi: 10.1128/jvi.72.2.1345-1353.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball J M, Tian P, Zeng C Q-Y, Morris A, Estes M K. Age-dependent diarrhea is induced by a viral nonstructural glycoprotein. Science. 1996;272:101–104. doi: 10.1126/science.272.5258.101. [DOI] [PubMed] [Google Scholar]
- 5.Brierley I. Ribosomal frameshifting viral RNAs. J Gen Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 6.Burns J W, Chen D, Estes M K, Ramig R F. Biological and immunological characterization of a simian rotavirus SA11 variant with an altered genome segment 4. Virology. 1989;169:427–435. doi: 10.1016/0042-6822(89)90168-2. [DOI] [PubMed] [Google Scholar]
- 7.Casjens S. Nucleic acid packaging by viruses. In: Casjens S, editor. Virus structure and assembly. Portola Valley, Calif: Jones and Bartlett Publishers, Inc.; 1985. pp. 75–147. [Google Scholar]
- 8.Chan W K, Penaranda M E, Crawford S E, Estes M K. Two glycoproteins are produced from the rotavirus neutralization gene. Virology. 1986;151:243–252. doi: 10.1016/0042-6822(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 9.Chou P Y, Fassman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol. 1978;47:45–147. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 10.Crawford S E, Labbe M, Cohen J, Burroughs M H, Zhou Y-J, Estes M K. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994;68:5945–5952. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz S S, Chapman S, Roberts A G, Roberts I M, Prior D A, Oparka K J. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc Natl Acad Sci USA. 1996;93:6286–6290. doi: 10.1073/pnas.93.13.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ericson B L, Graham D Y, Mason B B, Estes M K. Identification, synthesis, and modifications of simian rotavirus SA11 polypeptides in infected cells. J Virol. 1982;42:825–839. doi: 10.1128/jvi.42.3.825-839.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estes M K, Hardy M E. Norwalk virus and other enteric caliciviruses. In: Blaser M, Smith P, Ravdin J, Greenberg H B, Guerrant R, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 1009–1034. [Google Scholar]
- 14.Fankhauser R L, Noel J S, Monroe S S, Ando T, Glass R I. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J Infect Dis. 1998;178:1571–1578. doi: 10.1086/314525. [DOI] [PubMed] [Google Scholar]
- 15.Geissler K, Schneider K, Fleuchaus A, Parrish C R, Sutter G, Truyen U. Feline calicivirus capsid protein expression and capsid assembly in cultured feline cells. J Virol. 1999;73:834–838. doi: 10.1128/jvi.73.1.834-838.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham D Y, Jiang X, Tanaka T, Opekun A R, Madore H P, Estes M K. Norwalk virus infection of volunteers: new insights based on improved assays. J Infect Dis. 1994;170:34–43. doi: 10.1093/infdis/170.1.34. [DOI] [PubMed] [Google Scholar]
- 17.Green K Y, Kapikian A Z, Valdesuso J, Sosnovtsev S, Treanor J J, Lew J F. Expression and self-assembly of recombinant capsid protein from the antigenically distinct Hawaii human calicivirus. J Clin Microbiol. 1997;35:1909–1914. doi: 10.1128/jcm.35.7.1909-1914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale A D, Crawford S E, Ciarlet M, Green J, Gallimore C, Brown D W, Jiang X, Estes M K. Expression and self-assembly of Grimsby virus: antigenic distinction from Norwalk and Mexico viruses. Clin Diagn Lab Immunol. 1999;6:142–145. doi: 10.1128/cdli.6.1.142-145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamamoto H, Sugiyama Y, Nakagawa N, Hashida E, Matsunaga Y, Takemoto S, Watanabe Y, Okada Y. A new tobacco mosaic virus vector and its use for the systemic production of angiotensin-I-converting enzyme inhibitor in transgenic tobacco and tomato. Bio/Technology. 1993;11:930–932. doi: 10.1038/nbt0893-930. [DOI] [PubMed] [Google Scholar]
- 20.Hardy M E, Estes M K. Completion of the Norwalk virus genome sequence. Virus Genes. 1996;12:289–292. doi: 10.1007/BF00284649. [DOI] [PubMed] [Google Scholar]
- 21.Hardy M E, Tanaka T N, Kitamoto N, White L J, Ball J M, Jiang X, Estes M K. Antigenic mapping of the recombinant Norwalk virus capsid protein using monoclonal antibodies. Virology. 1996;217:252–261. doi: 10.1006/viro.1996.0112. [DOI] [PubMed] [Google Scholar]
- 22.Harrison S C, Olson A, Schutt C E, Winkler F K, Bricogne G. Tomato bushy stunt virus at 2.9 angstrom resolution. Nature. 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 23.Harrison S C, Skehel J J, Wiley D C. Virus structure. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 59–99. [Google Scholar]
- 24.Herbert T P, Brierley I, Brown T D K. Detection of the ORF 3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J Gen Virol. 1996;77:123–127. doi: 10.1099/0022-1317-77-1-123. [DOI] [PubMed] [Google Scholar]
- 25.Hogle J M, Maeda A, Harrison S C. Structure and assembly of turnip crinkle virus. I. X-ray crystallographic structure analysis at 3.2 angstrom resolution. J Mol Biol. 1986;191:625–638. doi: 10.1016/0022-2836(86)90450-x. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X, Graham D Y, Wang K N, Estes M K. Norwalk virus genome cloning and characterization. Science. 1990;250:1580–1583. doi: 10.1126/science.2177224. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X, Matson D O, Ruiz-Palacios G M, Hu J, Treanor J, Pickering L K. Expression, self-assembly, and antigenicity of a Snow Mountain agent-like calicivirus capsid protein. J Clin Microbiol. 1995;33:1452–1455. doi: 10.1128/jcm.33.6.1452-1455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X, Wang M, Graham D Y, Estes M K. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X, Wang M, Wang K, Estes M K. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X, Zhong W, Kaplan M, Pickering L K, Matson D O. Expression and characterization of Sapporo-like human calicivirus capsid proteins in baculovirus. J Virol Methods. 1999;78:81–91. doi: 10.1016/s0166-0934(98)00169-4. [DOI] [PubMed] [Google Scholar]
- 31.Jonsson G, Hedin A, Hakansson P, Sundqvist B U. Competition between protein/protein interactions and protein/substrate interactions studied by plasma desorption mass spectrometry. Rapid Commun Mass Spectrom. 1988;2:154–156. doi: 10.1002/rcm.1290020803. [DOI] [PubMed] [Google Scholar]
- 32.König M, Thiel H-J, Meyers G. Detection of viral proteins after infection of cultured hepatocytes with rabbit hemorrhagic disease virus. J Virol. 1998;72:4492–4497. doi: 10.1128/jvi.72.5.4492-4497.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli U K, Beguin F, Gujer-Kellenberger G. A factor preventing the major head protein of bacteriophage T4 from random aggregation. J Mol Biol. 1970;47:69–85. doi: 10.1016/0022-2836(70)90402-x. [DOI] [PubMed] [Google Scholar]
- 34.Leite J P, Ando T, Noel J S, Jiang B, Humphrey C D, Lew J F, Green K Y, Glass R I, Monroe S S. Characterization of Toronto virus capsid protein expressed in baculovirus. Arch Virol. 1996;141:865–875. doi: 10.1007/BF01718161. [DOI] [PubMed] [Google Scholar]
- 35.Margalit H, Spouge J L, Cornette J L, Cease K B, Delisi C, Berzofsky J A. Prediction of immunodominant helper T cell antigenic sites from the primary sequence. J Immunol. 1987;138:2213–2229. [PubMed] [Google Scholar]
- 36.Neill J D. Nucleotide sequence of the capsid protein gene of two serotypes of San Miguel sea lion virus: identification of conserved and non-conserved amino acid sequences among calicivirus capsid proteins. Virus Res. 1992;24:211–222. doi: 10.1016/0168-1702(92)90008-w. [DOI] [PubMed] [Google Scholar]
- 37.Parker J M R, Guo D, Hodges R S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenticity and x-ray-derived accessible sites. Biochemistry. 1986;25:5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 38.Prasad B V, Hardy M E, Dokland T, Bella J, Rossmann M G, Estes M K. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 39.Prasad B V, Matson D O, Smith A W. Three-dimensional structure of calicivirus. J Mol Biol. 1994;240:256–264. doi: 10.1006/jmbi.1994.1439. [DOI] [PubMed] [Google Scholar]
- 40.Prasad B V V, Rothnagel R, Jiang X, Estes M K. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J Virol. 1994;68:5117–5125. doi: 10.1128/jvi.68.8.5117-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao A L, Grantham G L. Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology. 1996;226:294–305. doi: 10.1006/viro.1996.0657. [DOI] [PubMed] [Google Scholar]
- 42.Roerink F, Hashimoto M, Tohya Y, Mochizuki M. Organization of the canine calicivirus genome from the RNA polymerase gene to the poly(A) tail. J Gen Virol. 1999;80:929–935. doi: 10.1099/0022-1317-80-4-929. [DOI] [PubMed] [Google Scholar]
- 43.Rossmann M G, Johnson J E. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 44.Sacher R, Ahlquist P. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J Virol. 1989;63:4545–4552. doi: 10.1128/jvi.63.11.4545-4552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seah E L, Gunesekere I C, Marshall J A, Wright P J. Variation in ORF3 of genogroup 2 Norwalk-like viruses. Arch Virol. 1999;144:1–8. doi: 10.1007/s007050050563. [DOI] [PubMed] [Google Scholar]
- 46.Shiba T, Suzuki Y. Localization of A protein in the RNA-A protein complex of RNA phage MS2. Biochim Biophys Acta. 1981;654:249–255. doi: 10.1016/0005-2787(81)90179-9. [DOI] [PubMed] [Google Scholar]
- 47.Silva A M, Rossmann M G. Refined structure of southern bean mosaic virus at 2.9 A resolution. J Mol Biol. 1987;197:69–87. doi: 10.1016/0022-2836(87)90610-3. [DOI] [PubMed] [Google Scholar]
- 48.van Duin J. The single-stranded RNA bacteriophages. In: Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. pp. 117–161. [Google Scholar]
- 49.Vinje J, Altena S A, Koopmans M P. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J Infect Dis. 1997;176:1374–1378. doi: 10.1086/517325. [DOI] [PubMed] [Google Scholar]
- 50.White L J, Ball J M, Hardy M E, Tanaka T N, Kitamoto N, Estes M K. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J Virol. 1996;70:6589–6597. doi: 10.1128/jvi.70.10.6589-6597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White L J, Hardy M E, Estes M K. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J Virol. 1997;71:8066–8072. doi: 10.1128/jvi.71.10.8066-8072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wikoff W R, Tsai C J, Wang G, Baker T S, Johnson J E. The structure of cucumber mosaic virus: cryoelectron microscopy, X-ray crystallography, and sequence analysis. Virology. 1997;232:91–97. doi: 10.1006/viro.1997.8543. [DOI] [PubMed] [Google Scholar]
- 53.Winkler F K, Schutt C E, Harrison S C, Bricogne G. Tomato bushy stunt virus at 5.5-A resolution. Nature. 1977;265:509–513. doi: 10.1038/265509a0. [DOI] [PubMed] [Google Scholar]
- 54.Wirblich C, Theil H-J, Meyers G. Genetic map of the calicivirus hemorrhagic disease virus as deduced from in vitro translation studies. J Virol. 1996;70:7974–7983. doi: 10.1128/jvi.70.11.7974-7983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]