Abstract
Since the outbreak in humans of an H5N1 avian influenza virus in Hong Kong in 1997, poultry entering the live-bird markets of Hong Kong have been closely monitored for infection with avian influenza. In March 1999, this monitoring system detected geese that were serologically positive for H5N1 avian influenza virus, but the birds were marketed before they could be sampled for virus. However, viral isolates were obtained by swabbing the cages that housed the geese. These samples, known collectively as A/Environment/Hong Kong/437/99 (A/Env/HK/437/99), contained four viral isolates, which were compared to the 1997 H5N1 Hong Kong isolates. Analysis of A/Env/HK/437/99 viruses revealed that the four isolates are nearly identical genetically and are most closely related to A/Goose/Guangdong/1/96. These isolates and the 1997 H5N1 Hong Kong viruses encode common hemagglutinin (H5) genes that have identical hemagglutinin cleavage sites. Thus, the pathogenicity of the A/Env/HK/437/99 viruses was compared in chickens and in mice to evaluate the potential for disease outbreaks in poultry and humans. The A/Env/HK/437/99 isolates were highly pathogenic in chickens but caused a longer mean death time and had altered cell tropism compared to A/Hong Kong/156/97 (A/HK/156/97). Like A/HK/156/97, the A/Env/HK/437/99 viruses replicated in mice and remained localized to the respiratory tract. However, the A/Env/HK/437/99 isolates caused only mild pathological lesions in these tissues and no clinical signs of disease or death. As a measure of the immune response to these viruses, transforming growth factor β levels were determined in the serum of infected mice and showed elevated levels for the A/Env/HK/437/99 viruses compared to the A/HK/156/97 viruses. This study is the first to characterize the A/Env/HK/437/99 viruses in both avian and mammalian species, evaluating the H5 gene from the 1997 Hong Kong H5N1 isolates in a different genetic background. Our findings reveal that at least one of the avian influenza virus genes encoded by the 1997 H5N1 Hong Kong viruses continues to circulate in mainland China and that this gene is important for pathogenesis in chickens but is not the sole determinant of pathogenicity in mice. There is evidence that H9N2 viruses, which have internal genes in common with the 1997 H5N1 Hong Kong isolates, are still circulating in Hong Kong and China as well, providing a heterogeneous gene pool for viral reassortment. The implications of these findings for the potential for human disease are discussed.
In March and April 1997, there were outbreaks of H5N1 avian influenza viruses on several poultry farms in the province of Hong Kong (8, 30). These viruses were highly pathogenic in chickens and resulted in high mortality of infected birds. During this same period, a 3-year-old boy was infected with an influenza virus that was approximately 99% identical by nucleotide sequence to the chicken H5N1 viruses isolated in the poultry outbreak (10, 11, 33, 35). The child subsequently died from complications of the viral infection. Later in the same year, 17 other confirmed cases of H5N1 avian influenza virus infection of humans were reported, 5 of which resulted in death (9, 12, 33, 44). These viruses were also nearly identical at the nucleotide level to viruses isolated from the avian influenza outbreak in chickens (3, 33, 35). Although avian influenza viruses have infected humans previously (13, 22, 39, 42), this was the first report of an avian influenza virus causing severe disease and death in a human host (11, 12, 35). The continuing occurrence of infection of humans with new subtypes of influenza virus led to fears of a deadly influenza pandemic. Fortunately, the H5N1 viruses were poorly spread by human-to-human contact, and each confirmed case was likely transmitted directly from bird to human, with the source being the live-bird markets of Hong Kong (30, 33). In an effort to control the human epidemic of H5N1 infection in Hong Kong, the poultry in Hong Kong were depopulated in December of 1997. No cases of humans infected with these H5N1 viruses were subsequently reported.
Since the outbreak, the 1997 H5N1 Hong Kong isolates from chickens and humans have been studied in both poultry and mice to determine the unique characteristics of these viruses that allowed them to cause lethal infections in both humans and chickens. For the viruses that have been evaluated for chickens by histopathology and immunohistochemistry, the endothelial cells of blood vessels throughout multiple visceral organs are the primary cell type where lesions and antigens are localized (33, 35). These findings have been reported for other highly pathogenic avian influenza (HPAI) viruses that cause peracute death in chickens (6, 20, 36, 41). Several of the 1997 H5N1 Hong Kong isolates have also been inoculated into mice to evaluate these animals as a model system for avian influenza virus pathogenesis in mammals (14, 16, 18, 21, 23, 26, 30). Independent researchers have found that these viruses are able to replicate and cause disease and high mortality in infected mice (14, 16, 18, 21, 23, 26, 30). Severe lesions of the upper and lower respiratory tract, including alveolar edema and alveolitis, were consistently observed in mice infected with the 1997 H5N1 Hong Kong viruses (14, 16, 18, 21, 23, 26, 30). However, there have been conflicting reports of systemic infection of mice by these viruses. Some researchers see no evidence of systemic infection (14), while in a few cases, others have isolated virus from other visceral organs and the brain (16, 18, 21, 23, 26, 30). These conflicting results may be related to the system used to propagate the viruses and to the number of times the viruses were passaged (14, 30).
Since the poultry of Hong Kong were depopulated at the end of 1997, the live-bird markets have been reestablished in Hong Kong, using the same sources of poultry as before the 1997 outbreak. These sources include some birds raised in Hong Kong, but most birds are imported from mainland China, primarily the province of Guangdong. A surveillance system has been set up to detect influenza virus in poultry that are used to stock the poultry markets in Hong Kong. In March 1999, this surveillance system detected antibodies to H5N1 influenza viruses in a shipment of geese from Guangdong province (L. Sims, personal communication). Although the birds had already been marketed when the results were obtained, the cages that housed the birds were sampled for virus isolation, and the viruses obtained are collectively referred to as A/Environment/Hong Kong/437/99 (A/Env/HK/437/99). Since these viruses were of the H5N1 subtype, their genetic relationship to the 1997 H5N1 Hong Kong viruses was determined. The pathogenicity of the A/Env/HK/437/99 isolates was also evaluated in both chickens and mice to ascertain the potential for future outbreaks of these viruses in both poultry and humans.
MATERIALS AND METHODS
Viruses.
A/Env/HK/437-4/99, A/Env/HK/437-6/99, A/Env/HK/437-8/99, A/Env/HK/437-10/99 (all received from Les Sims at the Agriculture and Fisheries Department, Hong Kong), and A/Human/Hong Kong/156/97 (A/HK/156/97) (received from Nancy Cox at the Centers for Disease Control and Prevention, Atlanta, Ga.) were passaged in specific-pathogen-free (SPF) 10-day-embryonated chicken eggs. The allantoic fluid from infected eggs was harvested, aliquoted, and stored at −70°C for use in all the experiments described herein. The 50% embryo lethal dose (ELD50) was determined in SPF 10-day-embryonated chicken eggs for each stock by the method of Reed and Muench (25). These viruses were handled in biosafety level 3 agriculture containment.
Molecular cloning, PCR amplification, and sequencing of viral isolates.
The complete coding sequence of all eight gene segments was determined for all four A/Env/HK/437/99 isolates by methods previously described (33). Briefly, RNA was isolated from allantoic fluid of eggs inoculated with each isolate by using Trizol-LS reagent (Life Technologies, Grand Island, N.Y.). The purified RNA was then used in reverse transcriptase PCR (RT-PCR) to generate cDNA copies for cloning and direct sequencing. The RT step was carried out using Superscript II (Life Technologies) for 1 h at 45°C. Reagents for PCR were added directly to the RT reaction mixtures and amplified for 30 cycles at an annealing temperature of 53°C. The segments encoding the nonstructural (NS), nucleoprotein (NP), and matrix (M) genes were amplified using primers that were complementary to the conserved 5′ 12 bp and 3′ 13 bp of influenza virus gene segments. The primers used to amplify the hemagglutinin (HA) and neuraminidase (NA) gene segments were also complementary to these regions but were longer in order to provide greater specificity for these gene segments. The RT-PCR products were then electrophoresed on a 1.5% agarose gel and purified (Concert matrix extraction system; Life Technologies). The cDNAs were cloned, and colonies were screened by PCR using internal primers for the appropriate gene. Positive clones were grown overnight, and plasmids were extracted using the High Pure plasmid isolation kit (Boehringer Mannheim, Indianapolis, Ind.). cDNAs specific for the three polymerase gene segments (PA, PB1, and PB2) were also generated from viral RNA using RT-PCR. Each segment was amplified in three overlapping pieces, cut from an agarose gel, and purified as described above, except that the PCR annealing temperature was 56°C. The PCR products amplified from the polymerase gene segments were sequenced directly. All plasmids and PCR products were sequenced using a PRISM Ready Reaction Dye Deoxy Terminator cycle-sequencing kit (Perkin-Elmer) and run on a 373A automated sequencer (Perkin-Elmer).
Phylogenetic analysis.
DNASTAR (Madison, Wis.) software was used to create sequence contigs and multiple-sequence alignments of the gene segments from the A/Env/HK/437/99 isolates. Maximum parsimony with 100 bootstrap replicates and a heuristic search method were used with PAUP 3.1 software (37) to generate phylogenetic trees. All phylogenetic trees are midpoint rooted and contain representative influenza virus isolates for each gene segment.
Chicken experiments.
To determine the pathogenicity of the A/Env/HK/437/99 isolates, a modified U.S. Animal Health Association chicken pathogenicity test was performed in 4-week-old SPF white Plymouth Rock chickens (40). Briefly, birds were inoculated via the intravenous route with 0.2 ml of a 10−1 dilution of the bacterium-free virus stock. Based on back titer determination, the doses were 106.6, 106.8, 106.8, and 106.6 ELD50 of A/Env/HK/437-4/99, A/Env/HK/437-6/99, A/Env/HK/437-8/99, and A/Env/HK/437-10/99, respectively per chicken. Eight birds were inoculated per group for pathogenicity testing, using death as the end point. Avian influenza virus is considered to be highly pathogenic when at least 75% of the birds die within 10 days of inoculation. Two additional birds were inoculated per group (using the same route of inoculation and doses as above), and these birds were euthanized with sodium pentobarbital (100 mg/kg of body weight) on day 2 postinoculation. Chickens that died prior to day 2 postinoculation were necropsied on the day of death. The lungs, bursa, kidneys, adrenal gland, thymus, thyroid, brain, liver, heart, pancreas, intestine, spleen, and trachea were taken from these birds and evaluated for gross lesions and by histopathology and immunohistochemistry for influenza virus antigen. In a separate experiment, eight 4-week-old SPF white Plymouth Rock chickens were inoculated with 106 ELD50 of A/Env/HK/437-6/99 per chicken via the intranasal route. Two birds were euthanized on day 3 postinoculation, and tissues were taken and evaluated as described above. The mean death time (MDT) was calculated by determining the sum of the day of death postinoculation for the chickens and dividing by the total number of chickens that died. Chicken experiments were carried out in Horsfal-Bauer stainless steel isolation units ventilated under negative pressure with HEPA-filtered air in biosafety level 3 agriculture facilities (2). Animal care was provided as required by the Institutional Animal Care and Use Committee, based on the Guide for the Care and Use of Agricultural Animals in Agricultural Research and Teaching. Food and water were provided ad libitum.
Mouse experiments.
Seven-week-old male BALB/c mice (Simonsen Laboratories, Inc., Gilroy, Calif.) were anesthetized with ketamine-xylene (1.98 and 0.198 mg per mouse, respectively), and the inoculum was administered intranasally in 50 μl of phosphate-buffered saline (PBS). The data presented below were obtained from two experiments. In experiment 1, virus doses, based on back titers, were 105.5, 106.2, 106.2, and 105.8 ELD50 per mouse for A/Env/HK/437-4/99, A/Env/HK/437-6/99, A/Env/HK/437-8/99, and A/Env/HK/437-10/99, respectively. Eleven mice were inoculated per A/Env/HK/437/99 isolate. Eight mice of the 11 from each group were monitored daily for clinical signs of disease and death. Three mice of the 11 from each experimental group were euthanized on day 4 postinoculation with 50 μl of sodium pentobarbital (100 mg/kg of body weight) administered intraperitoneally. These mice were evaluated for gross lesions, and tissues were taken for virus isolation, histopathology, and immunohistochemistry. The trachea, lungs, and kidneys were removed, using aseptic techniques, for virus isolation. Briefly, tissues were weighed and homogenized in brain heart infusion broth to a 10% slurry. The ELD50 for each tissue was determined in SPF 10-day-embryonated chicken eggs, and the ELD50 per gram of tissue was calculated (14). The sinuses, bone marrow, brain, testes, thymus, kidneys, adrenal gland, lungs, vesicular gland, muscle, heart, liver, spleen, pancreas, intestine, and stomach were taken for evaluation by histopathology and immunohistochemistry for influenza virus antigen. The surviving mice were euthanized on day 14 postinoculation, and their lungs were evaluated by histopathology and immunohistochemistry.
In experiment 2, mice were set up in groups of eight and were anesthetized and inoculated with PBS only as a negative control or as described in experiment 1 above. As determined by back titer determination, mice were inoculated with 105.8, 105.6, or 105.6 ELD50 of A/Env/HK/437-6/99, A/Env/HK/437-10/99, or A/HK/156/97, respectively. The mice were weighed on days 0, 4, 6, 8, and 12 postinoculation. Blood was drawn from the tail vein at 1 and 4 days postinoculation. Sera from mice in the same group and same day postinoculation were pooled and used in experiments described below. Mice were monitored daily for clinical signs of disease and death until day 14 postinoculation, when the surviving mice were euthanized. The MDT was calculated for mice as described above. Mice were housed in standard polypropylene mouse cages and placed inside Horsfal-Bauer stainless steel isolation units that were ventilated with HEPA-filtered air. Experiments were carried out in the facilities and using the guidelines described above.
Histopathology, ultrastructural pathology, and immunohistochemistry.
Tissues were fixed in 10% neutral-buffered formalin solution, sectioned, and stained with hematoxylin and eosin. Duplicate sections were stained immunohistochemically to determine influenza virus antigen distribution in individual tissues. A monoclonal antibody against influenza A virus NP (P13C11), developed at Southeast Poultry Research Laboratory, was used as the primary antibody in a streptavidin-biotin-alkaline phosphatase complex immunohistochemical method as previously described (36).
Determination of TGF-β activity in mouse serum.
In mouse experiment 2 described above, sera were pooled from mice within groups on days 1 and 4 postinoculation. Transforming growth factor β (TGF-β) activity in the serum was determined by evaluating colony formation of normal rat kidney (NRK) cells in the presence of epidermal growth factor (EGF) in soft agar as previously described (28, 29). Briefly, 5% Noble agar (Difco, Detroit, Mich.) was diluted to 0.5% in 10% calf serum–Dulbecco's modified Eagle's medium. This solution was added to 24-well tissue culture plates as a base layer and allowed to solidify. Serum samples, known amounts of TGF-β containing 1 ng of EGF, or 1 ng of EGF alone were added to liquid 0.5% agar and 2 × 103 NRK cells, and this mixture was added to the plates containing the agar base layer. Cultures were incubated at 37°C under 5% CO2 for 7 days and then stained with 1% neutral red in PBS. Colonies greater than 62 μm in diameter (containing >8 to 10 cells) were counted. Samples were run in triplicate. The amount of activated TGF-β in mouse serum was estimated by plotting the standard amounts of TGF-β against the number of colonies produced and then determining the linear best fit of the data.
Nucleotide sequence accession numbers.
Sequence data for the A/Env/HK/437/99 isolates have been submitted to GenBank. The accession numbers are as follows: A/Env/HK/437-4/99, AF216710 to AF216717; A/Env/HK/437-6/99, AF216718 to AF216725; A/Env/HK/437-8/99, AF216726 to AF216733; A/Env/HK/437-10/99, AF216734 to AF216741.
RESULTS
Phylogenetic analysis of A/Env/HK/437/99 isolates.
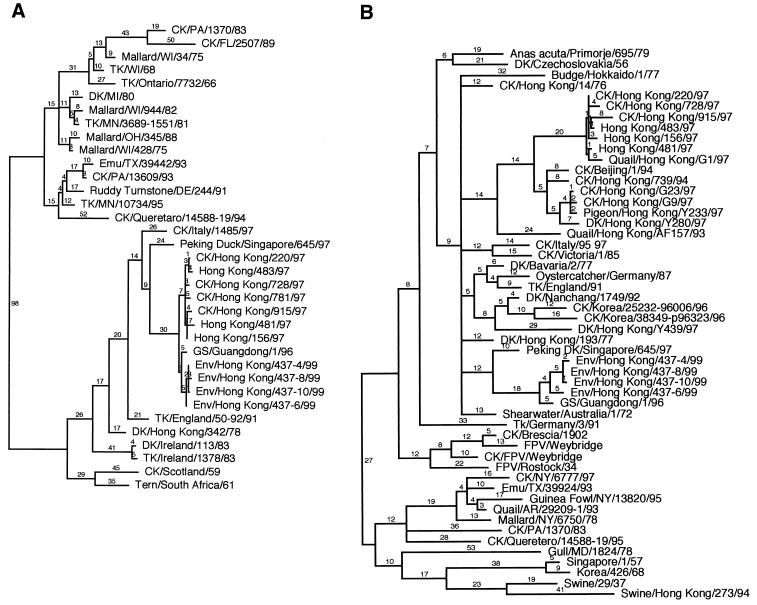
The nucleotide sequences of all eight segments of the four A/Env/HK/437/99 isolates were determined, and phylogenetic trees were constructed using available representative isolates for comparison. Relationships to the H5N1 influenza viruses isolated in Hong Kong in 1997 were of particular interest and are described below. The H5 HA1 phylogenetic tree shown in Fig. 1A is composed of 35 isolates and shows that avian influenza viruses isolated in North America grouped together while those isolated in Europe or Asia grouped together (27). As expected, the A/Env/HK/437/99 isolates grouped with other Eurasian avian influenza viruses. The four A/Env/HK/437/99 viruses were nearly identical in nucleotide sequence (99.5 to 99.9% identical) and were most closely related to the A/Goose/Guangdong/1/96 (A/GS/Guangdong/1/96) isolate, with 98.5 to 99% identity (Fig. 1A). This analysis also showed that the H5 HA genes of the A/Env/HK/437/99 viruses were closely related to those of the H5N1 chicken and human isolates from Hong Kong in 1997, indicating that the H5 HA genes encoded by these viruses are from the same lineage (Fig. 1A) (43). The sequence similarity among the A/Env/HK/437/99 isolates and A/HK/156/97, a representative isolate from Hong Kong in 1997, ranged from 98.2 to 98.5% nucleotide identity.
FIG. 1.
Phylogenetic analysis of the nucleotide sequence of the HA and M genes of the A/Env/HK/437/99 isolates. Trees were generated using parsimony with a heuristic search method by bootstrap analysis with 100 replicates and PAUP 3.1 (37). Branch lengths give the number of nucleotide changes, and trees are midpoint rooted. All isolates are type A influenza viruses. Abbreviations: CK, chicken; DK, duck; Env, environment; FPV, fowl plague virus; GS, goose; TK, turkey; standard two-letter abbreviations are used for states in the United States. (A) HA1 subunit of the H5 HA gene segment. (B) M gene segment.
To provide further evidence of a common lineage for the H5 HA genes of the A/Env/HK/437/99, A/GS/Guangdong/1/96, and 1997 H5N1 Hong Kong isolates, the nucleotides and amino acid residues that are unique to these isolates were determined for the H5 HA1 gene. To be considered a unique change within this lineage, the change had to occur in more than 90% of the A/Env/HK/437/99, A/GS/Guangdong/1/96, and 1997 H5N1 Hong Kong isolates evaluated and in less than 10% of the other isolates used in the phylogenetic analysis. These values were chosen to adjust for the high rate of mutation in these viruses, which could result in the same changes by chance. The isolates used in this comparison are shown in Fig. 1A. Using these criteria, there were 27 unique nucleotide changes and 15 unique amino acid changes. The accumulation of unique mutations within this group over time demonstrates that the H5 HA genes of A/Env/HK/437/99, A/GS/Guangdong/1/96, and 1997 H5N1 Hong Kong isolates are in the same lineage (31, 34). These unique changes resulted in a nonsynonymous-to-synonymous ratio of 0.56, suggesting some positive selection of the H5 HA gene.
Further evaluation of the H5 HA1 nucleotide and amino acid sequence showed that the A/Env/HK/437/99 viruses, A/GS/Guangdong/1/96, and the 1997 H5N1 Hong Kong isolates have evolved since the common ancestor to this H5 HA lineage was introduced. For example, nucleotide 159 (numbering scheme for the consensus sequence) of A/Env/HK/437/99 viruses and A/GS/Guangdong/1/96 encoded an adenine while nucleotide 159 of the 1997 H5N1 Hong Kong viruses and other isolates encoded a guanine. Nucleotides 133, 150, and 289 of the A/Env/HK/437/99 isolates encoded changes that are unique to these viruses. Changes specific to this lineage of H5 genes were also noted at amino acid 138, which is a leucine in the 1997 H5N1 Hong Kong isolates, a histidine in the A/Env/HK/437/99 and A/GS/Guangdong/1/96 isolates, and an asparagine in most other H5 isolates that were evaluated.
Amino acid sequence comparison of the H5 HA1 gene from A/GS/Guangdong/1/96, A/Env/HK/437/99 isolates, and representative 1997 H5N1 Hong Kong isolates showed that the unique sequence of the HA cleavage site is conserved among these isolates (data not shown). HA cleavage sites of HPAI viruses are rare and usually unique. The potential glycosylation sites of the H5 HA1 protein of A/GS/Guangdong/1/96 and the A/Env/HK/437/99 isolates were also determined and compared to those of representative 1997 H5N1 Hong Kong isolates. The putative glycosylation sites identified at amino acids 10, 11, 23, 186, 484, and 544 (numbering scheme used for 1997 H5N1 Hong Kong isolates [3]) were conserved in all of the A/Env/HK/437/99 isolates and A/GS/Guangdong/1/96. However, the potential glycosylation site that is found in some of the 1997 H5N1 Hong Kong isolates at position 154 was absent from A/GS/Guangdong/1/96 and all the A/Env/HK/437/99 viruses (3). These additional examples of conservation of functional characteristics also suggest a common ancestor for the H5 HA1 gene of these isolates.
The other seven influenza virus gene segments from the A/Env/HK/437/99 viruses were also evaluated for their genetic relationships to available representative isolates, with particular attention to the H5N1 viruses isolated in Hong Kong in 1997. The phylogenetic tree for the M gene segment showed that A/Env/HK/437/99 viruses clustered with other avian influenza viruses isolated in Europe and Asia, as expected (Fig. 1B). The four A/Env/HK/437/99 isolates were nearly identical to one another, with 98.8 to 99.9% nucleotide sequence identity. These isolates were most closely related to A/GS/Guangdong/1/96, with 98.4 to 98.6% nucleotide sequence identity. However, the M gene segment of A/Env/HK/437/99 isolates did not cluster within the same lineage as that of the 1997 H5N1 Hong Kong viruses (Fig. 1B), as shown by the nucleotide sequence identity, which ranged from 92.2 to 92.6% for A/Env/HK/437/99 compared to A/HK/156/97. Similar results were also observed when phylogenetic trees for the other six gene segments (NS, NA, NP, PA, PB1, and PB2) were generated and nucleotide and amino acid sequence identities were evaluated (data not shown): the isolate most closely related to the A/Env/HK/437/99 viruses for each gene segment was A/GS/Guangdong/1/96; the 1997 H5N1 Hong Kong viruses did not cluster in the same lineage as the A/Env/HK/437/99 isolates for the remaining genes (43). Furthermore, the NS genes of the A/Env/HK/437/99 isolates are of subtype (group) B while those of the 1997 H5N1 Hong Kong isolates are of subtype (group) A (32). This difference results in much lower nucleotide and amino acid sequence identity than observed for the M gene segment. The NA gene of A/Env/HK/437/99 viruses also did not encode the 19-amino-acid deletion in the stalk that was reported for the 1997 H5N1 Hong Kong viruses (reference 3, 11, and 35 and data not shown).
Pathogenicity of A/Env/HK/437/99 isolates in chickens and in mice.
The A/Env/HK/437/99 isolates are closely related to A/GS/Guangdong/1/96. A/GS/Guangdong/1/96 caused an outbreak of disease in domestic geese that was associated with 40% morbidity in the field. This virus was experimentally inoculated into geese and chickens and reported to cause disease and death in these birds (38, 43). However, the details of these experiments are reported in Chinese or are not published. We evaluated the ability of the A/Env/HK/437/99 viruses to grow and cause disease in both chickens and mice. Our study includes detailed gross and histopathological evaluations of pathogenesis for both species and comparison of these findings to those from A/HK/156/97-infected chickens and mice.
To determine the pathogenicity of the A/Env/HK/437/99 isolates in chickens, the U.S. Animal Health Association chicken pathogenicity test was used (40). The birds infected with each of the four isolates exhibited clinical signs of disease that were consistent with an HPAI virus (1, 33, 36). In these experiments, 100% of the infected birds in each group died by day 5 postinoculation, with an average MDT of 3.4 days (Table 1). A virus is classified as highly pathogenic if 75% or more of the birds die within 10 days; thus, these viruses are highly pathogenic in chickens. Similar results were observed when chickens were inoculated intranasally, but the MDT (5.5 days) was longer (Table 1).
TABLE 1.
The A/Env/HK/437/99 viruses and A/HK/156/97 produce high mortality and are highly pathogenic in chickens
| Virus | Mortalitya | MDT (days) | Dose (ELD50) per birdb |
|---|---|---|---|
| A/Env/HK/437-4/99c | 8/8 | 3.4 | 106.6 |
| A/Env/HK/437-6/99c | 8/8 | 3.0 | 106.8 |
| A/Env/HK/437-8/99c | 8/8 | 3.3 | 106.8 |
| A/Env/HK/437-10/99c | 8/8 | 4.1 | 106.6 |
| A/Env/HK/437-6/99d | 8/8 | 5.5 | 106.0 |
| A/HK/156/97e | 8/8 | 2.0 | 106.9 |
Number of chickens that died/number of chickens inoculated.
The titer given was determined by back titer determination of the inoculum.
Pathotyping experiment; birds were inoculated intravenously.
Birds were inoculated intranasally.
Data obtained in reference 33.
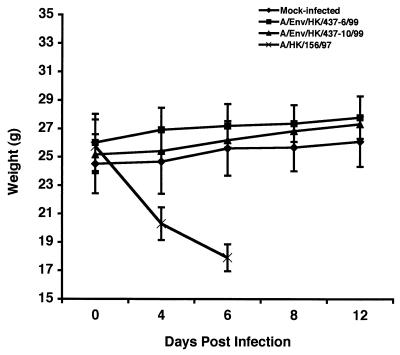
To evaluate the growth and pathogenicity of the A/Env/HK/437/99 viruses in a mammalian host, BALB/c mice were used as a model system as previously described (14). Male BALB/c mice were inoculated intranasally with each of the A/Env/HK/437/99 viruses. These mice displayed no clinical signs of disease and continued to gain weight, like the mock-infected mice, throughout the 14-day experiment (Fig. 2). In contrast, A/HK/156/97 exhibited clinical signs and lost weight until their deaths as previously reported (14) (Fig. 2). All mice that were infected with A/HK/156/97 died by day 8 postinoculation, with an MDT of 6.75 days (14). Although the A/Env/HK/437/99 viruses were not pathogenic to mice, these isolates were able to infect tissues of the upper respiratory tract and replicate to detectable titers (Table 2). Kidneys from the A/Env/HK/437/99-infected mice were negative for virus, suggesting a localized rather than systemic infection (Table 2). These findings are consistent with observations made by Dybing et al. for A/HK/156/97 (14).
FIG. 2.
Weights of mice that were mock infected or infected with A/Env/HK/437-6/99, A/Env/HK/437-10/99, or A/HK/156/97. The weights were determined on days 0, 4, 6, 8, and 12 postinoculation. The weights of the mice were averaged for each group on the days indicated, and error bars show the standard deviation of the mean.
TABLE 2.
Viral titers in tissues from mice infected with the A/Env/HK/437/99 viruses
| Virus | Mortalitya | Dose (ELD50) per mouseb | Average tissue titer (ELD50/g of tissue)c in:
|
||
|---|---|---|---|---|---|
| Trachea | Lung | Kidney | |||
| A/Env/HK/437-4/99 | 0/8 | 105.5 | 105.4 (2/3) | 103.6 (3/3) | 0 (0/3) |
| A/Env/HK/437-6/99 | 0/8 | 106.2 | 107.0 (3/3) | 105.4 (3/3) | 0 (0/3) |
| A/Env/HK/437-8/99 | 0/8 | 106.2 | 106.0 (3/3) | 104.9 (3/3) | 0 (0/3) |
| A/Env/HK/437-10/99 | 0/8 | 105.8 | 106.7 (2/3) | 106.0 (1/3)d | 0 (0/3) |
Number of mice that died/number of mice inoculated.
The titer given was determined by back titer determination of the inoculum.
Mice were euthanized on day 4 postinoculation; (number of positive tissues/number of tissues evaluated).
The titer shown is for the one positive sample obtained.
Analysis of tissues from chickens and mice inoculated with A/Env/HK/437/99 by histopathology and immunohistochemistry.
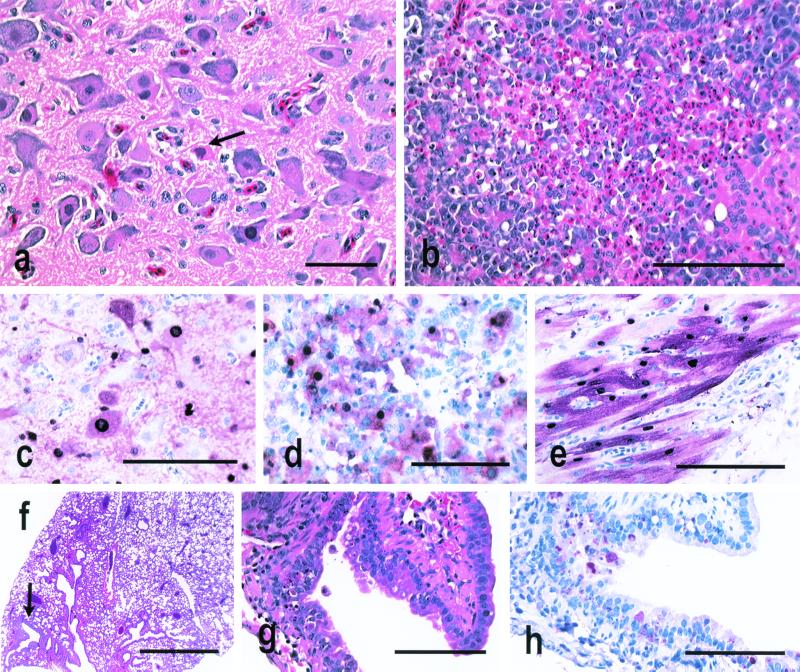
Infection of chickens with the 1997 H5N1 Hong Kong viruses produced severe systemic disease, with the most common lesions being severe pulmonary edema, congestion, and hemorrhage; interstitial pneumonitis with necrosis; necrosis of myocardial cells; and apoptosis of lymphocytes in multiple primary and secondary lymphoid tissues (33, 35). Most frequently, avian influenza virus was localized to the endothelial cells in different-sized blood vessels, vascular sinuses, and endocardium. Influenza virus antigen was also common in cardiac myocytes and in macrophages and heterophils of the lungs (33, 35). Similarly, the A/Env/HK/437/99 H5N1 influenza viruses produced systemic infections, lesions, and death in chickens upon intravenous or intranasal inoculation. After intravenous inoculation, multiple foci of parenchymal cell necrosis in the pancreas, brain, and heart (Fig. 3a and b) and lymphocyte depletion and apoptosis in the spleen, cloacal bursa, thymus, and occasionally the cecal tonsil were the most consistent lesions. Influenza virus NP was demonstrated most consistently in necrotic parenchymal cells of the brain, pancreas, and heart (Fig. 3c to e); sporadically in adrenal corticotrophic cells and kidney tubule epithelium; and only rarely in vascular endothelial cells. After intranasal inoculation, lymphocytic meningoencephalitis and myocarditis were consistent lesions, and apoptosis and lymphocyte depletion were detected in primary and secondary lymphoid tissues. Influenza virus NP was identified in the brain neurons and ependymal cells and in cardiac myocytes (data not shown).
FIG. 3.
Experimental studies in 4-week-old chickens and 7-week-old mice inoculated with A/Env/HK/437-4/99, A/Env/HK/437-6/99, A/Env/HK/437-8/99, and A/Env/HK/437-10/99. Photomicrographs of hematoxylin and eosin-stained tissue sections (a, b, f, and g) and photomicrographs of tissue sections stained immunohistochemically to demonstrate avian influenza virus NP (c to e and h) are shown. (a) Neuronal degeneration and necrosis in the medulla of a chicken euthanized on day 2 after intravenous inoculation with A/Env/HK/437-6/99. Bar, 25 μm. (b) Severe widespread necrosis of pancreatic acinar epithelium from a chicken euthanized on day 2 after intravenous inoculation with A/Env/HK/437-8/99. Bar, 20 μm. (c) Intranuclear and intracytoplasmic avian influenza virus antigen in neurons and glial cells from the chicken in panel a. Bar, 50 μm. (d) Intense staining of pancreatic acinar epithelium and debris for avian influenza virus antigen from the chicken in panel b. Bar, 50 μm. (e) Intranuclear and intracytoplasmic avian influenza virus antigen in cardiac myocytes of a chicken that died on day 2 after intravenous inoculation with A/Env/HK/437-10/99. Bar, 50 μm. (f) Single focal area of bronchitis in a normal lung from a mouse euthanized on day 4 after intranasal inoculation with A/Env/HK/437-4/99. Bar, 500 μm. (g) Higher magnification of panel f, showing focal necrosis of respiratory epithelium from a bronchus. Bar, 50 μm. (h) Avian influenza virus antigen in bronchial respiratory epithelium of the mouse in panel f. Bar, 50 μm.
A/HK/156/97 virus in experimentally inoculated mice caused high mortality, which resulted from infection and frequent, severe lesions in the upper and lower respiratory tract; especially prominent were alveolar edema and alveolitis (14). Influenza virus antigen was localized to tracheal and bronchial epithelium and, in the most severe cases, the alveolar epithelium (14). By contrast, none of the mice inoculated with the A/Env/HK/437/99 influenza viruses died, but the viruses replicated in the respiratory tract and caused infrequent, mild lesions in the trachea and bronchi, with occasional associated mild foci of acute to chronic alveolitis (Fig. 3f and g). Influenza virus was localized to the tracheal and bronchial respiratory epithelium (Fig. 3h). The viruses did not undergo significant systemic spread, since NP was not detected by immunohistochemistry in visceral organs or the brain (data not shown). The A/Env/HK/437/99 viruses were most similar pathogenically to experimental infections of mice with two other HPAI viruses, A/CK/Queretaro/7653-20/95 (H5N2) and A/CK/Scotland/59 (H5N1) (14).
Comparison of levels of activated TGF-β in serum in A/Env/HK/437/99- and A/HK/156/97-infected mice.
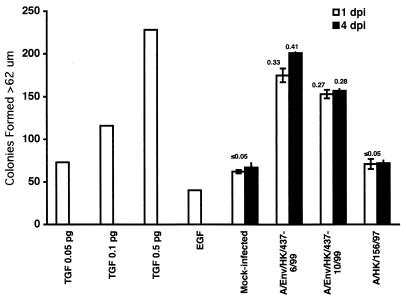
To evaluate the host response to A/Env/HK/437/99 viruses, we examined the levels of TGF-β activity in the sera of infected mice. TGF-β activity is elevated in influenza virus-infected mice early in the course of infection (14, 28). Studies in vitro show that the influenza virus NA protein activates latent TGF-β, suggesting that the increase in TGF-β activity in the serum of influenza virus-infected mice is due to the in vivo activation of latent TGF-β by influenza virus NA (28). Therefore, blood was obtained on days 1 and 4 postinoculation from mice inoculated with PBS only (negative control), A/Env/HK/437-6/99, A/Env/HK/437-10/99, or A/HK/156/97 to evaluate TGF-β activity in serum by using the NRK soft agar assay (28, 29). Control mice showed low levels of TGF-β activity that were slightly above background (EGF alone) on both days 1 and 4 postinoculation (Fig. 4). Figure 4 shows that A/Env/HK/437-6/99- and A/Env/HK/437-10/99-infected mice had levels of TGF-β activity much higher than those in control mice. As shown previously and in Fig. 4, A/HK/156/97-infected mice exhibited low levels of TGF-β activity, similar to the levels observed in control mice (14). These results showed that there is an increase in the level of activated TGF-β in the sera of mice infected with A/Env/HK/437-6/99 and A/Env/HK/437-10/99 over that in the sera of mice infected with A/HK/156/97. These findings suggest that the A/Env/HK/437/99 viruses, like many other avian influenza viruses, elevate the levels of activated TGF-β in vivo (14, 28). In contrast, activated TGF-β levels in A/HK/156/97-infected mouse serum are similar to those in the negative control (14).
FIG. 4.
TGF-β activity levels in the serum of mock- and virus-infected mice. TGF-β activity levels were determined using an NRK soft-agar assay and are reported in number of colonies formed. A standard curve was derived from known levels of TGF-β. EGF alone represents the background level of colony formation in this assay. Mice were mock infected or infected with A/Env/HK/437-6/99, A/Env/HK/437-10/99, or A/HK/156/97, and serum was evaluated on days 1 and 4 postinoculation (dpi). Numbers above the experimental samples represent the amount (in picograms) of activated TGF-β per 50 μl of serum as determined using the standard curve generated from known amounts of TGF-β. The value could not be accurately estimated for mock-infected or A/HK/156/97-infected mouse serum because the number of colonies formed was smaller than that of the lowest TGF-β value determined for the standard curve. Experiments were performed in triplicate and are represented as the mean of the samples and the standard error of the mean.
DISCUSSION
Genetic evaluation of the A/Env/HK/437/99 viruses clearly showed that these isolates have an HA gene in common with the 1997 H5N1 Hong Kong viruses and that all eight segments are closely related to A/GS/Guangdong/1/96. These findings show that these avian influenza virus genes have been in circulation for at least 3 years and that these viruses continue to circulate even after the poultry depopulation in Hong Kong. Other researchers have shown that H9N2 viruses (A/quail/HK/G1/97 and A/CK/HK/G9/97) isolated from Hong Kong in 1997 encode internal genes that are closely related to those of the 1997 H5N1 Hong Kong viruses (17). Since the depopulation of poultry in Hong Kong, H9N2 viruses have also been isolated from pigs, humans, and poultry in mainland China (references 19 and 24 and A. Hay, personal communication). It is clear from the data summarized here that the 1997 H5N1 Hong Kong viruses were derived as a result of reassortment of cocirculating viruses. Thus, the continued presence of genetically distinct avian influenza viruses in mainland China provides a varied pool of genes for generation of reassortant viruses. The presence of multiple genetically different viruses provides the potential for future outbreaks of human disease from avian influenza, especially with the cocirculation of viruses containing genes that contributed to the H5N1 outbreak in Hong Kong in 1997.
The pathogenicity of the HA gene associated with the 1997 H5N1 outbreak in Hong Kong was evaluated in chickens and mice infected with A/Env/HK/437/99 isolates. These studies provided the opportunity to examine the effects of the H5 gene on disease without the contribution of the other 1997 H5N1 Hong Kong virus genes. The 1997 H5N1 Hong Kong viruses caused severe disease in poultry (30, 33). Since HPAI viruses in poultry are generally characterized by H5 or H7 genes that encode multiple basic amino acids at the HA cleavage site (4, 5), it is not surprising that the A/Env/HK/437/99 isolates are also highly pathogenic in chickens. Although the HA gene made a major contribution to the virulence of A/HK/156/97 and the A/Env/HK/437/99 virus infection of chickens, other genes also had an effect on pathogenesis, since the main sites of infection and MDT values were different for these viruses (Fig. 3 and Table 1). These differences may be related to differences in the abilities of these viruses to infect different cell types or the rate of replication and/or dissemination of the viruses in the host.
Evaluation of A/Env/HK/437/99 isolates in mice revealed that these viruses, like A/HK/156/97, remained confined to the respiratory tract, where they replicated to detectable titers (Table 2) (14). However, A/Env/HK/437/99 did not cause clinical signs of disease in mice and histologically caused only mild pathological lesions in the upper respiratory tract. In contrast, A/HK/156/97 caused severe disease and pathological lesions in the upper and lower respiratory tract and resulted in death by 8 days postinoculation. These findings demonstrate that the HA gene of the 1997 H5N1 Hong Kong isolates is not sufficient alone to cause disease in a mammalian host and thus is not likely to be the major determinant of virulence in mammals as it is in poultry. Pathogenicity studies of A/quail/HK/G1/97 and A/CK/HK/G9/97 in chickens and mice confirm this data. These H9N2 isolates replicated in chickens, but the birds did not display any clinical signs of disease. However, these viruses caused severe disease in mice, killing three of eight and two of eight mice, respectively, without prior adaptation to the host (19).
The data from this study, as well as the evaluation of H9N2 Hong Kong isolates (19) in mice, provide further evidence that the ability of the 1997 H5N1 Hong Kong viruses to cause disease in mammals is specified by multiple genes. The HA gene alone is not the sole determinant of pathogenesis in mice, and the internal genes encoded by A/quail/HK/G1/97 do not produce the same degree of virulence in these animals as seen with A/HK/156/97. Thus, some combination of the internal genes and the HA and/or NA of the 1997 H5N1 Hong Kong isolates is likely responsible for the lethal phenotype. The NA gene has not been evaluated in another genetic background; thus, its contribution to pathogenesis is unknown. However, the NA gene of the 1997 H5N1 Hong Kong viruses encodes a 19-amino-acid deletion, and others have shown that NA stalk deletions can affect the ability of the NA to free virions from red blood cells and to cause disease in mice (7, 15). These findings suggest that stalk length is important for NA function and pathogenesis. Influenza virus NA is also known to activate latent TGF-β in vitro, by a direct or indirect mechanism (28, 29), and data presented here and by Dybing et al. (14) show that latent TGF-β can also be activated in influenza virus-infected mice as well. Unlike other influenza viruses, the 1997 H5N1 Hong Kong isolates tested do not produce elevated levels of activated TGF-β in the sera of infected mice. Conversely, mice infected with A/Env/HK/437/99 viruses have high levels of activated TGF-β. Activated TGF-β levels in the sera of H9N2-infected mice have not been determined. Unfortunately, the role of TGF-β in influenza virus pathogenesis is not understood. However, TGF-β is known to mediate both proinflammatory and immunosuppresive activities, depending on the levels produced. If activation of TGF-β by influenza virus infection is important for a proper immune response, it is possible that the 1997 H5N1 Hong Kong viruses are more virulent, in part, because they lack the ability to activate TGF-β to high levels. The reduced ability to activate TGF-β may produce greater inflammation at the site of infection and thus cause more severe disease. Alternatively, the low levels of activated TGF-β in the sera of A/HK/156/97-infected mice may allow the viruses to replicate and spread unchecked in the respiratory tracts of the mice, causing more severe disease. Studies are under way to explore these possibilities.
This study shows that the A/Env/HK/437/99 isolates are highly pathogenic in chickens but cause no clinical signs of disease in mice, suggesting that the H5 gene of the 1997 H5N1 viruses is important for virulence in poultry but is not the sole determinant of pathogenicity in mammals. When these data are reviewed along with those from other current literature, it becomes clear that the pathogenicity of the 1997 H5N1 Hong Kong viruses is multigenic. These results also show that the H5 gene of the 1997 H5N1 Hong Kong isolates is still circulating in HPAI viruses in China, even after poultry depopulation. This finding, in combination with the continued circulation of H9N2 viruses containing the internal genes of the 1997 H5N1 Hong Kong isolates, indicates that there is a pool of avian influenza virus genes that could cause disease in humans and poultry if reassortment occurs among the right group of viruses.
ACKNOWLEDGMENTS
We thank Joan Beck, Patsy Decker, Suzanne DeBlois, Liz Turpin, John Latimer, and Roger Brock for technical assistance. We thank Les Sims and Kitman Dyrting at the Agriculture and Fisheries Department in Hong Kong for providing the A/Env/HK/437/99 isolates and Robert Webster at St. Jude Children's Research Hospital, Memphis, Tenn., for transporting these isolates from Hong Kong. We thank Nancy Cox at the Centers for Disease Control and Prevention, Atlanta, Ga., for providing A/HK/156/97.
This work was supported by USDA/ARS Cris project number 6612-32000-022-93.
REFERENCES
- 1.Alexander D J. Avian influenza. Recent developments. Vet Bull. 1982;52:341–359. [Google Scholar]
- 2.Barbeito M S, Abraham G, Best M, Cairns P, Langevin P, Sterritt W G, Barr D, Meulepas W, Sanchez-Vizcaino J M, Saraza M, Requena E, Collado M, Mani P, Breeze R, Brunner H, Mebus C A, Morgan R L, Rusk S, Siegfried L M, Thompson L H. Recommended biocontainment features for research and diagnostic facilities where animal pathogens are used. Rev Sci Tech Off Int Epizoot. 1995;14:873–887. doi: 10.20506/rst.14.3.880. [DOI] [PubMed] [Google Scholar]
- 3.Bender C, Hall H, Huang J, Klimov A, Cox N, Hay A, Gregory V, Cameron K, Lim W, Subbarao K. Characterization of the surface proteins of influenza A (H5N1) viruses isolated from humans in 1997–1998. Virology. 1999;254:115–123. doi: 10.1006/viro.1998.9529. [DOI] [PubMed] [Google Scholar]
- 4.Bosch F X, Garten W, Klenk H D, Rott R. Cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza virus. Virology. 1981;113:725–735. doi: 10.1016/0042-6822(81)90201-4. [DOI] [PubMed] [Google Scholar]
- 5.Bosch F X, Orlich M, Klenk H D, Rott R. The structure of the hemagglutinin. A determinant for the pathogenicity of influenza viruses. Virology. 1979;95:197–207. doi: 10.1016/0042-6822(79)90414-8. [DOI] [PubMed] [Google Scholar]
- 6.Brown C C, Olander H J, Senne D A. A pathogenesis study of highly pathogenic avian influenza virus H5N2 in chickens, using immunohistochemistry. J Comp Pathol. 1992;107:341–348. doi: 10.1016/0021-9975(92)90009-j. [DOI] [PubMed] [Google Scholar]
- 7.Castrucci M R, Kawaoka Y. Biologic importance of neuraminidase stalk length in influenza A virus. J Virol. 1993;67:759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. Isolation of avian influenza A (H5N1) from humans—Hong Kong, May–December, 1997. Morbid Mortal Weekly Rep. 1997;46:1204–1207. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. Update: isolation of avian influenza A (H5N1) viruses from humans-Hong Kong, 1997–1998. Morbid Mortal Weekly Rep. 1998;46:1245–1247. [PubMed] [Google Scholar]
- 10.Claas E C J, de Jong J C, van Beek R, Rimmelzwaan G F, Osterhaus A D M E. Human influenza virus A/Hong Kong/156/97 (H5N1) infection. Vaccine. 1998;16:977–978. doi: 10.1016/s0264-410x(98)00005-x. [DOI] [PubMed] [Google Scholar]
- 11.Claas E C J, Osterhaus A D M E, van Beek R, De Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 12.De Jong J C, Claas E C J, Osterhaus A D M E. A pandemic warning? Nature. 1997;389:554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLay P D, Casey H L, Tubiash H S. Comparative study of fowl plaque virus and a virus isolated from man. Pub Health Rep. 1967;82:615–620. [PMC free article] [PubMed] [Google Scholar]
- 14.Dybing J K, Schultz-Cherry S, Swayne D E, Suarez D L, Perdue M L. Distinct pathogenesis of Hong Kong-origin H5N1 viruses in mice compared to that of other highly pathogenic H5 avian influenza viruses. J Virol. 2000;74:1443–1450. doi: 10.1128/jvi.74.3.1443-1450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Els M C, Air G M, Murti K G, Webster R G, Laver W G. An 18 amino acid deletion in an influenza neruaminidase. Virology. 1985;142:241–248. doi: 10.1016/0042-6822(85)90332-0. [DOI] [PubMed] [Google Scholar]
- 16.Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley A J, Kawaoka Y. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol. 1999;73:3184–3189. doi: 10.1128/jvi.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan Y, Shortridge K F, Krauss S, Webster R G. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gubareva L V, McCullers J A, Bethell R C, Webster R G. Characterization of influenza A/Hong Kong/156/97 (H5N1) virus in a mouse model and protective effect of zanamivir on H5N1 infection in mice. J Infect Dis. 1998;178:1592–1596. doi: 10.1086/314515. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y J, Krauss S, Senne D A, Mo I P, Lo K S, Xiong X P, Norwood M, Shortridge K F, Webster R G, Guan Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi Y, Horimoto T, Kawaoka Y, Alexander D J, Itakura C. Pathological studies of chickens experimentally infected with two highly pathogenic avian influenza viruses. Avian Pathol. 1996;25:285–304. doi: 10.1080/03079459608419142. [DOI] [PubMed] [Google Scholar]
- 21.Kodihalli S, Goto H, Kobasa D L, Krauss S, Kawaoka Y, Webster R G. DNA vaccine encoding hemagglutinin provides protective immunity against H5N1 influenza virus infection in mice. J Virol. 1999;73:2094–2098. doi: 10.1128/jvi.73.3.2094-2098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtz J, Manvell R, Banks J. Avian influenza virus isolated from a woman with conjunctivitis. Lancet. 1996;348:901–902. doi: 10.1016/S0140-6736(05)64783-6. [DOI] [PubMed] [Google Scholar]
- 23.Lu X, Tumpey T M, Morken T, Zaki S R, Cox N J, Katz J M. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J Virol. 1999;73:5903–5911. doi: 10.1128/jvi.73.7.5903-5911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peiris M, Yuen K Y, Leung C W, Chan K H, Ip P L S, Lai R W M, Orr W K, Shortridge K F. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 25.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 26.Riberdy J M, Flynn K J, Stech J, Webster R G, Altman J D, Doherty P C. Protection against a lethal avian influenza A virus in a mammalian system. J Virol. 1999;73:1453–1459. doi: 10.1128/jvi.73.2.1453-1459.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohm C, Horimoto T, Kawaoka Y, Suss J, Webster R G. Do hemagglutinin genes of highly pathogenic avian influenza viruses constitute unique phylogenetic lineages? Virology. 1995;209:664–670. doi: 10.1006/viro.1995.1301. [DOI] [PubMed] [Google Scholar]
- 28.Schultz-Cherry S, Hinshaw V S. Influenza virus neuraminidase activates latent transforming growth factor-β. J Virol. 1996;70:8624–8629. doi: 10.1128/jvi.70.12.8624-8629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz-Cherry S, Murphy-Ullrich J E. Thrombospondin causes activation of latent transforming growth factor-β secreted by endothelial cells by a novel mechanism. J Cell Biol. 1993;122:923–932. doi: 10.1083/jcb.122.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shortridge K R, Zhou N N, Guan Y, Gao P, Ito T, Kawaoka Y, Kodihalli S, Krauss S, Markwell D, Murti K G, Norwood M, Senne D, Sims L, Takada A, Webster R G. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- 31.Suarez D L, García M, Latimer J, Senne D, Perdue M. Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the Northeast United States. J Virol. 1999;73:3567–3573. doi: 10.1128/jvi.73.5.3567-3573.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suarez D L, Perdue M L. Multiple alignment comparison of the non-structural genes of influenza A viruses. Virus Res. 1998;54:59–69. doi: 10.1016/s0168-1702(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 33.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suarez D L, Senne D A. Sequence analysis of related low pathogenic and highly pathogenic H5N2 avian influenza isolates from the United States live bird markets and poultry farms from 1983–1989. Avian Dis. 1999;44:356–364. [PubMed] [Google Scholar]
- 35.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 36.Swayne D E. Pathobiology of H5N2 Mexican avian influenza virus infections of chickens. Vet Pathol. 1997;34:557–567. doi: 10.1177/030098589703400603. [DOI] [PubMed] [Google Scholar]
- 37.Swofford D. PAUP: phylogenetic analysis using parsimony, version 3 ed. Champaign: Illinois Natural History Survey; 1989. [Google Scholar]
- 38.Tang X, Tian G, Zhao C, Zhou J, Y K. Isolation and characterization of prevalent strains of avian influenza viruses in China. Chin J Anim Poult Infect Dis. 1998;20:1–5. . (In Chinese.) [Google Scholar]
- 39.Taylor H R, Turner A J. A case report of fowl plaque keratoconjunctivitis. Br J Ophthalmol. 1977;61:86–88. doi: 10.1136/bjo.61.2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.U.S. Animal Health Association. Proceedings of the 98th Annual Meeting of the U.S. Grand Rapids, Mich: Animal Health Association. U.S. Animal Health Association; 1994. Report of the committee on transmissible diseases of poultry and other avian species. Criteria for determining that an AI virus isolation causing an outbreak must be considered for eradication; p. 522. [Google Scholar]
- 41.Van Campen H, Easterday B C, Hinshaw V S. Virulent avian influenza A viruses: Their effect on avian lymphocytes and macrophages in vivo and in vitro. J Gen Virol. 1989;70:2887–2895. doi: 10.1099/0022-1317-70-11-2887. [DOI] [PubMed] [Google Scholar]
- 42.Webster R G, Geraci J, Petursson G, Skirnisson K. Conjunctivitis in human beings caused by influenza A virus of seals. N Engl J Med. 1981;304:911. doi: 10.1056/NEJM198104093041515. [DOI] [PubMed] [Google Scholar]
- 43.Xu S, Subbarao K, Cox N J, Guo Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology. 1999;261:15–19. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- 44.Yuen K Y, Chan P K S, Peiris M, Tsang D N C, Que T L, Shortridge K F, Cheung P T, To W K, Ho E T F, Sung R, Cheng A F B Members of the H5N1 Study Group. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]