To the Editor:
In adult ALL, 5–10% of patients show KMT2A translocations (KMT2A rearrangements) with only a few secondary alterations, implicating it as a leukemia-initiating factor [1, 2]. Approximately 95% of all fusions in adult ALL are KMT2A::AFF1 or KMT2A::MLLT1 [3]. KMT2A-rearranged adult ALL patients are generally considered high-risk and are treated with intensified therapy, including allogeneic hematopoietic stem cell transplantation (SCT) [4]. Current ALL treatment protocols are often guided by measurable residual disease (MRD)-based risk stratification [4–8], however, limited data are available regarding the prognostic value of MRD in adult ALL with KMT2A rearrangement. In infant KMT2A-rearranged ALL, more reliable MRD data were obtained using the individual KMT2A breakpoints as molecular MRD target as compared to IG/TR [6, 9–11], but no such comparisons have been made in adult ALL. We evaluated the impact of MRD on disease-free survival (DFS) and overall survival (OS) in a cohort of 156 KMT2A-rearranged adult patients and compared IG/TR- and KMT2A-based MRD levels in 46 patients.
In total, 769 bone marrow and/or peripheral blood samples from 193 adult ALL patients with KMT2A rearrangement (175 KMT2A::AFF1, 13 KMT2A::MLLT1, 1 KMT2A::MLLT3, 4 KMT2A+ unspecified) obtained between 2001 and 2021 were available for longitudinal MRD measurements. All patients were treated according to different protocols of the German Multicenter ALL (GMALL) study group and gave their informed consent to further scientific investigations on residual material. Patients with KMT2A::AFF1 aged up to 55 years were assigned to the high-risk group and were candidates for SCT in first CR after consolidation I. Immunophenotyping and MRD measurement with real-time PCR based on KMT2A fusion genes and clonal IG/TR gene rearrangements were performed in central laboratories as previously described [10, 11]. MRD measurements were interpreted according to EuroMRD guidelines [12]. MRD results were considered discordant if positivity/negativity discordance in the same sample was evidenced. For the evaluation of DFS and OS, MRD levels were compared at three different time points: end of induction I, after induction II/ pre-consolidation I, post-consolidation I/pre SCT (around week 16) (Fig. S2) [11]. MRD levels were classified as molecular response (MRD < 10–4 or negative), molecular failure with low MRD (≥10–4 and <10–2), and high MRD (≥10–2). Further statistical details are provided in the online supplement to this letter.
KMT2A-based versus IG/TR-based MRD
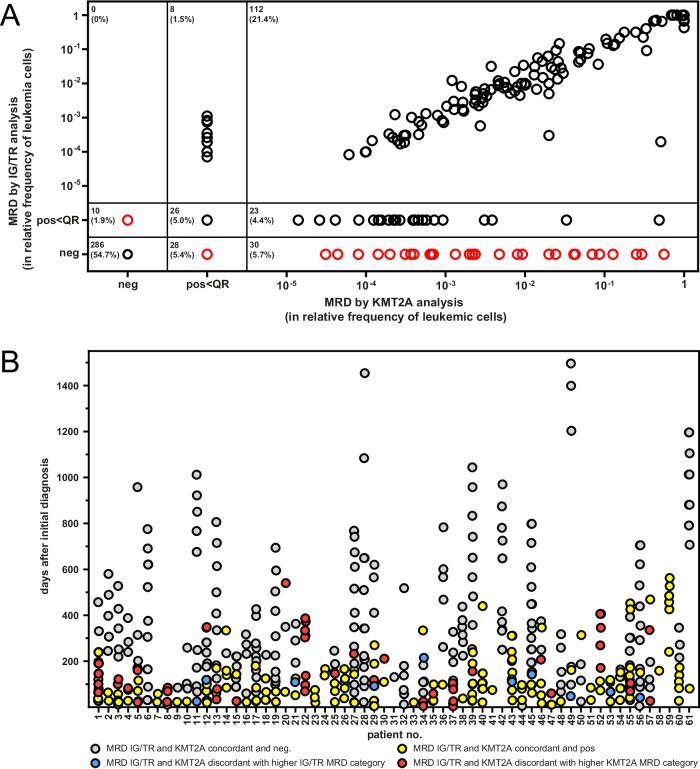
We analyzed 193 patients with KMT2A-rearranged ALL with median age at diagnosis of 42.5 years (18.0-76.8), and 63.0% being females. All 187 immunophenotypically characterized patients showed a CD10-negative B cell precursor ALL (146 cyIgneg, 41 cyIgpos). Parallel MRD data of both, KMT2A and IG/TR, were available for 46 patients, totaling 274 MRD data pairs from bone marrow and 99 from peripheral blood. Both methods show good agreement (Table S2; Fig. S1). 197/373 (52.8%) samples were MRD-negative with both methods, 84/373 (22.5%) were congruently positive within quantifiable range (QR), and 22/373 (5.9%) were positive below QR in both MRD targets (Fig. 1A). 18/373 (4.8%) were quantifiable MRD-positive only using KMT2A, whereas IG/TR MRD showed positivity below QR of the method, in 6/373 cases (1.6%) it was the other way around. The remaining 46/373 (13.0%) samples were classified as discordant, with 38/373 (10.2%) being KMT2A-rearranged and IG/TRneg, with 24/46 samples showing quantifiable KMT2A MRD positivity. Only 8/373 (2.1%) were KMT2Aneg and IG/TRpos (P < 0.0001), none of them showing quantifiable IG/TR MRD positivity (Table S1). Discordant samples with the higher KMT2A MRD were detected at least once in 15/46 (32.6%) patients during therapy and follow-up, whereas a higher IG/TR MRD was detected in 8/46 (17.4%) patients (Fig. 1B). These results are consistent with other studies on KMT2A-rearranged ALL, where IG/TR rearrangements at diagnosis were often oligo- or subclonal and underly clonal evolution [9]. Usage of subclonal IG/TR markers or a loss of the MRD marker due to RAG-mediated clonal evolution may lead to false negative results or underestimation of MRD values. In contrast, the KMT2A break cannot get lost because it is an early event and a leukemia-defining molecular hallmark.
Fig. 1. Comparison of KMT2A-based and IG/TR-based MRD measurements.
A Comparison of MRD levels with KMT2A and IG/TR targets in KMT2A-rearranged adult ALL patients. MRD measurements with data on both KMT2A and IG/TR were available from 46 patients totaling 373 sample pairs from peripheral blood or bone marrow aspirates. MRD levels were plotted against each other from negative (neg), positive (pos)< quantifiable range (QR), and quantifiable range in logarithmic format. Black circles represent MRD concordant samples and red circles discordant samples. B Comparison of KMT2A and IG/TR MRD levels over time. All MRD-levels (n = 523) with data on both IG/TR and KMT2A were sorted into four groups (gray color IG/TR and KMT2A MRD level concordant and negative (neg.), blue color IG/TR and KMT2A MRD level discordant with higher IG/TR MRD category, yellow color IG/TR and KMT2A MRD level concordant and positive, and red color IG/TR and KMT2A MRD level discordant with higher KMT2A MRD category) and plotted against days after initial diagnosis (ID).
Prognostic significance of MRD
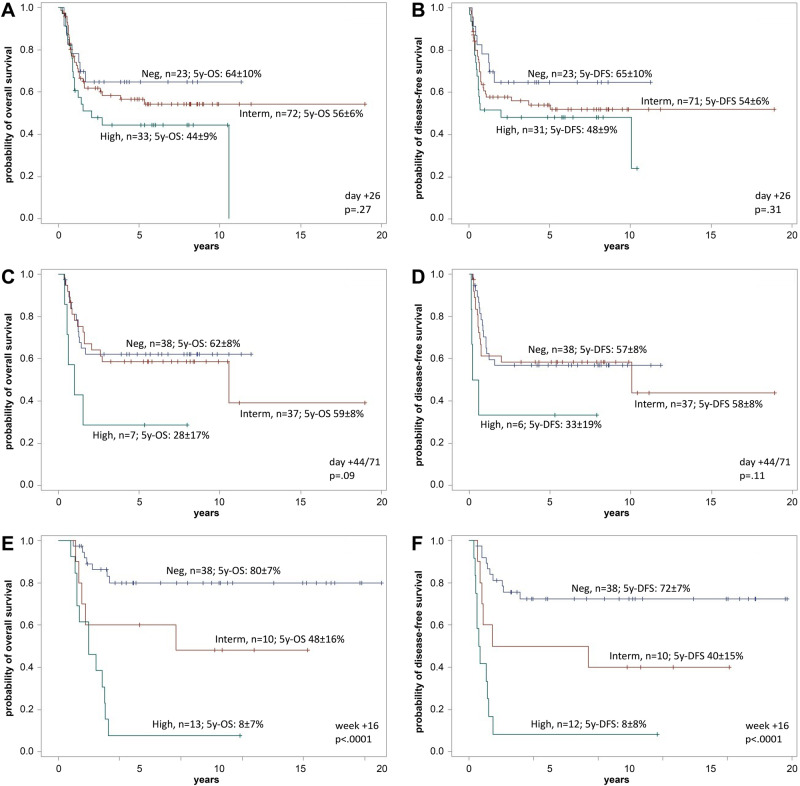
Data for DFS and OS in remission were available for 156/193 patients, with MRD being assessed using IG/TR and/or KMT2A at the end of induction I (n = 140), after induction II/ pre-consolidation I (n = 149) and after consolidation I (n = 68). After induction I, MRD levels did not predict outcome with 5-year DFS and OS (P = 0.31 and P = 0.27) (Fig. 2A+B). At pre-consolidation I, MRD levels did not predict outcome with 5-year DFS but patients with high MRD (≥10–2) levels had a significantly poorer OS. 5-year OS was 62% (95% CI: 54 to 70), 59% (95% CI: 52 to 67), and 28% (95% CI: 11 to 45) for patients with molecular response, and molecular failure with low or high MRD (P = 0.09) (Fig. 2C+D). After consolidation I significant differences were found in both DFS and OS, and MRD levels predicted outcome with 5-year DFS of 72% (95% CI, 64 to 80), 40% (95% CI, 25 to 55), and 8% (95% CI, 0 to 16) and 5-year OS of 80% (95% CI, 73 to 88), 48% (95% CI, 32 to 64), and 8% (95% CI, 1 to 15) for patients with molecular response, and molecular failure with low (MRD ≥ 10–4 and <10–2) and high (≥10–2) MRD levels (P < 0.0001) (Fig. 2E+F). These findings demonstrate that high MRD levels at post-consolidation I in adult KMT2A-rearranged ALL are clearly unfavorable in terms of OS and DFS prior to SCT. Strikingly, early MRD after induction I was not predictive for treatment outcome, which contrasts with published data from other ALL molecular subgroups where early MRD has shown clear prognostic significance [5, 7, 11]. It is possible that this observation reflects the same mechanism that has been described for infant KMT2A-rearranged ALL: In a study by Stutterheim et al. [13], MRD after induction was prognostically relevant only if followed by a lymphoid-style consolidation but not with a myeloid-style type consolidation. In our patient cohort allogeneic SCT was performed in the majority of patients (72%) which may abolish the prognostic effect of postinduction MRD response. However, patients with molecular failure prior to SCT still had poorer outcome. This supports the GMALL approach to offer a targeted therapy with blinatumomab to all patients with molecular failure after consolidation I to eradicate MRD before SCT [14]. However, patients with KMT2A-rearranged ALL occasionally show CD19 antigen loss after blinatumomab and blinatumomab may also be less effective than in non-KMT2A-rearranged ALL, due to lower CD19 expression.
Fig. 2. Overall survival and disease-free survival of patients at different time points.
Prognostic impact of measurable residual disease (MRD) levels at the end of induction I (A + B), pre-consolidation I (C + D), and post-consolidation I (E + F). Data shown by Kaplan–Meier estimates of overall survival (A, C, E) and disease-free survival (B, D, F). MRD results were classified as low (blue color; neg; <10–4), intermediate (red color; interm; <10–2), and high MRD levels (green color; ≥10–2). Information on DFS was not available for some patients (day+26: 3 pts., day+44/71: 1 pt., w +16: 1 pt.).
Myeloid coexpression and MRD response
In the Interfant-06 study, patients with myeloid coexpression had significantly higher MRD levels at the end of induction and benefitted from subsequent myeloid-style consolidation [13]. In our cohort data were available in 96 patients for both, detailed immunophenotype and MRD. Expression of at least one myeloid marker (CD13, CD15, CD65s, CD33) was detected in 77 (80.2%) patients. We observed no significant differences in MRD response at end of induction I, pre- or post-consolidation in patients with or without myeloid co-expression (Fig. S3; Table S3).
In conclusion, our study suggests that in adult KMT2A-rearranged ALL the KMT2A genomic fusion breakpoint has clear technical advantages as MRD target, as has also recently been reported by Kim et al. [15]. However, patient numbers in our study were too small to prove a clinical impact of MRD discordance between these two methods. The MRD status has a very strong prognostic value in DFS and OS post-consolidation I in a transplant-oriented regimen. The optimal therapy of patients with treatment failure or MRD persistence is under investigation. Particularly the term “myeloid-style therapy” needs to be defined more precisely, since most relevant compounds are also part of ALL therapy. More promise probably lies in the use of immunotherapies directed to lymphoid surface markers like CD19.
Supplementary information
Acknowledgements
We thank all participating clinics and patients of the GMALL study group. The technical assistance of Mara Molkentin, Daniela Gröger and Maike Ipsen is highly appreciated. TB was supported by Deutsche José Carreras Leukämie-Stiftung grants 13 R/2016 and R10/37 f. This study was in part funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—“Clinician Scientist Program in Evolutionary Medicine” (GEPRIS project 4134905, to A-SS), and DFG project number 444949889 (KFO 5010/1 Clinical Research Unit ‘CATCH ALL’ to CB and MB).
Author contributions
TB, HT, CM, RM, SS, and MB performed experiments. A-SS, TB, BK, MN, NG, and MB analyzed results. NG, CM, RM, and SS provided relevant patient information for the study. TB, NG, and MB designed the research. A-SS drafted the first version of manuscript. All authors discussed the results and contributed to the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Competing interests
MB is contracted to carry out research for Affimed, Amgen, Regeneron, the advisory board of Amgen, Incyte, Speaker bureau of Amgen, Janssen, Pfizer, Roche. SS is advisory board member of AMGEN and received honoraria as a speaker bureau member of AMGEN. CB is contracted to carry out research for Novartis, the advisory board of Amgen. TB received speakers’ honoraria from Novartis and Pfizer. The other authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Thomas Burmeister, Aeint-Steffen Ströh.
These authors jointly supervised this work: Nicola Gökbuget, Monika Brüggemann.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-024-02209-7.
References
- 1.Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–7. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki Y, Watanabe T, Saito Y, Kuroki Y, Hijikata A, Takagi M, et al. Identification of CD34+ and CD34- leukemia-initiating cells in MLL-rearranged human acute lymphoblastic leukemia. Blood. 2015;125:967–80. doi: 10.1182/blood-2014-03-563304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burmeister T, Meyer C, Schwartz S, Hofmann J, Molkentin M, Kowarz E, et al. The MLL recombinome of adult CD10-negative B-cell precursor acute lymphoblastic leukemia: results from the GMALL study group. Blood. 2009;113:4011–5. doi: 10.1182/blood-2008-10-183483. [DOI] [PubMed] [Google Scholar]
- 4.Gökbuget N, Stelljes M, Viardot A, Nachtkamp K, Steffen B, Schneller F, et al. First results of the risk-adapted, MRD-stratified GMALL trial 08/2013 in 705 adults with newly diagnosed acute lymphoblastic leukemia/lymphoma (ALL/LBL) Blood. 2021;138:362. doi: 10.1182/blood-2021-146306. [DOI] [Google Scholar]
- 5.Beldjord K, Chevret S, Asnafi V, Huguet F, Boulland ML, Leguay T, et al. Oncogenetics and minimal residual disease are independent outcome predictors in adult patients with acute lymphoblastic leukemia. Blood. 2014;123:3739–49. doi: 10.1182/blood-2014-01-547695. [DOI] [PubMed] [Google Scholar]
- 6.van Dongen JJ, van der Velden VH, Brüggemann M, Orfao A. Minimal residual disease diagnostics in acute lymphoblastic leukemia: need for sensitive, fast, and standardized technologies. Blood. 2015;125:3996–4009. doi: 10.1182/blood-2015-03-580027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gökbuget N, Kneba M, Raff T, Trautmann H, Bartram CR, Arnold R, et al. Adult patients with acute lymphoblastic leukemia and molecular failure display a poor prognosis and are candidates for stem cell transplantation and targeted therapies. Blood. 2012;120:1868–76. doi: 10.1182/blood-2011-09-377713. [DOI] [PubMed] [Google Scholar]
- 8.Bassan R, Spinelli O, Oldani E, Intermesoli T, Tosi M, Peruta B, et al. Improved risk classification for risk-specific therapy based on the molecular study of minimal residual disease (MRD) in adult acute lymphoblastic leukemia (ALL) Blood. 2009;113:4153–62. doi: 10.1182/blood-2008-11-185132. [DOI] [PubMed] [Google Scholar]
- 9.van der Velden VH, Corral L, Valsecchi MG, Jansen MW, De Lorenzo P, Cazzaniga G, et al. Prognostic significance of minimal residual disease in infants with acute lymphoblastic leukemia treated within the Interfant-99 protocol. Leukemia. 2009;23:1073–9. doi: 10.1038/leu.2009.17. [DOI] [PubMed] [Google Scholar]
- 10.Burmeister T, Marschalek R, Schneider B, Meyer C, Gökbuget N, Schwartz S, et al. Monitoring minimal residual disease by quantification of genomic chromosomal breakpoint sequences in acute leukemias with MLL aberrations. Leukemia. 2006;20:451–7. doi: 10.1038/sj.leu.2404082. [DOI] [PubMed] [Google Scholar]
- 11.Brüggemann M, Raff T, Flohr T, Gökbuget N, Nakao M, Droese J, et al. Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukemia. Blood. 2006;107:1116–23. doi: 10.1182/blood-2005-07-2708. [DOI] [PubMed] [Google Scholar]
- 12.van der Velden VH, Cazzaniga G, Schrauder A, Hancock J, Bader P, Panzer-Grümayer ER, et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21:604–11. doi: 10.1038/sj.leu.2404586. [DOI] [PubMed] [Google Scholar]
- 13.Stutterheim J, van der Sluis IM, de Lorenzo P, Alten J, Ancliffe P, Attarbaschi A, et al. Clinical implications of minimal residual disease detection in infants with KMT2A-rearranged acute lymphoblastic leukemia treated on the interfant-06 protocol. J Clin Oncol. 2021;39:652–62. doi: 10.1200/JCO.20.02333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gökbuget N. MRD in adult Ph/BCR-ABL-negative ALL: how best to eradicate? Hematology Am Soc Hematol Educ Program. 2021;2021:718–25. doi: 10.1182/hematology.2021000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim R, Bergugnat H, Pastoret C, Pasquier F, Raffoux E, Larcher L, et al. Genetic alterations and MRD refine risk assessment for KMT2A-rearranged B-cell precursor ALL in adults: a GRAALL study. Blood. 2023;142:1806–17. doi: 10.1182/blood.2023021501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.