Abstract
Retrovirus entry into cells follows receptor binding by the surface-exposed envelope glycoprotein (Env) subunit (SU), which triggers the membrane fusion activity of the transmembrane (TM) protein. TM protein fragments expressed in the absence of SU adopt helical hairpin structures comprising a central coiled coil, a region of chain reversal containing a disulfide-bonded loop, and a C-terminal segment that packs onto the exterior of the coiled coil in an antiparallel manner. Here we used in vitro mutagenesis to test the functional role of structural elements observed in a model helical hairpin, gp21 of human T-lymphotropic virus type 1. Membrane fusion activity requires the stabilization of the N and C termini of the central coiled coil by a hydrophobic N cap and a small hydrophobic core, respectively. A conserved Gly-Gly hinge motif preceding the disulfide-bonded loop, a salt bridge that stabilizes the chain reversal region, and interactions between the C-terminal segment and the coiled coil are also critical for fusion activity. Our data support a model whereby the chain reversal region transmits a conformational signal from receptor-bound SU to induce the fusion-activated helical hairpin conformation of the TM protein.
Retrovirus infection is initiated by the viral envelope (Env) glycoproteins that comprise a receptor-binding subunit (SU) associated with a transmembrane (TM) protein. Envelope glycoproteins are synthesized as precursors and are processed in the Golgi apparatus to yield a mature functional SU-TM protein complex. The mature Env proteins are incorporated into budding virions at the plasma membrane. Retrovirus entry into cells follows SU-mediated attachment to cellular receptors and TM protein-mediated fusion between the viral envelope and target cellular membrane. Cell surface-localized Env proteins can also mediate cell-to-cell retrovirus transmission by fusion between infected cells and target cells.
X-ray crystallography and nuclear magnetic resonance studies have revealed structural similarities in TM protein fragments derived from diverse retroviruses (7, 11, 22, 28, 33, 48, 51), orthomyxoviruses (6, 13, 42, 52), a filovirus (Ebola virus) (34, 50), and a paramyxovirus (simian virus 5) (4). Retroviral and filoviral TM protein structures are long rods comprising an N-terminal central triple-stranded coiled coil, a disulfide-bonded loop associated with a chain reversal at the base of the rod, and a structurally diverse C-terminal ectodomain segment that packs against the exterior of the central coiled coil in an antiparallel manner. These structures, referred to as helical hairpins (12, 45), imply that the hydrophobic N-terminal fusion peptide and C-terminal TM sequence are juxtaposed at one end of the rod in a fusion-activated or postfusion conformation.
Retroviral TM protein hairpins resemble the fusion-activated conformation of the influenza virus TM protein, HA2, suggesting a common mechanism for membrane fusion. The conformational changes accompanying HA2 fusion activation, which are induced by low endosomal pH, involve a helical extension at the N terminus of the central coiled coil which would relocate the fusion peptide from the HA2 core to the tip of the rod, allowing insertion into the target cellular membrane. The envelope-proximal portion of the coiled coil refolds, resulting in reversal of the chain direction, packing of the C-terminal segment on the outside of the coiled coil, and placement of the TM sequence close to the N terminus (6, 14). These conformational changes are thought to draw together and destabilize the viral envelope or infected cell membranes and target cell membranes, inducing membrane fusion. The fusion potential of HA2 is thought to result from the low-pH-induced transition from the metastable, prefusogenic conformation to the most stable, fusion-activated conformation (9, 13, 14). Recent biochemical studies with human immunodeficiency virus type 1 (HIV-1) and avian leukosis and sarcoma virus Env indicate that binding between the viral SU and cellular receptor(s) leads to conformational changes in the SU-TM complex that correlate with increased TM protein hydrophobicity and membrane fusion activity (15, 26, 27).
We reported recently the 2.5-Å-resolution crystal structure of the human T-lymphotropic virus type 1 (HTLV-1) gp21 helical hairpin (28) (Fig. 1A). The structure of gp21 encompassed residues Met-338 to Thr-425 and was solved as part of a chimera with Escherichia coli maltose-binding protein (MBP-gp21). Here we describe the functional consequences of mutating residues within the key structural elements of gp21 that are conserved in retroviral and filoviral TM hairpins. We identified functional roles for (i) a hydrophobic structure that caps the N terminus of the central coiled coil (Fig. 1B); (ii) the region of chain reversal comprising a small hydrophobic core containing Phe-386, a conserved Gly-Gly motif preceding the disulfide-bonded loop, the conserved salt bridge formed between Glu-398 (found within the disulfide-bonded loop) and Arg-380 at the base of the coiled coil, and a conserved solvent-exposed phenylalanine residue (Phe-402) located C terminal to the disulfide-bonded loop (Fig. 1D); and (iii) sites of interaction between the C-terminal ectodomain segment and the coiled coil (Fig. 1E).
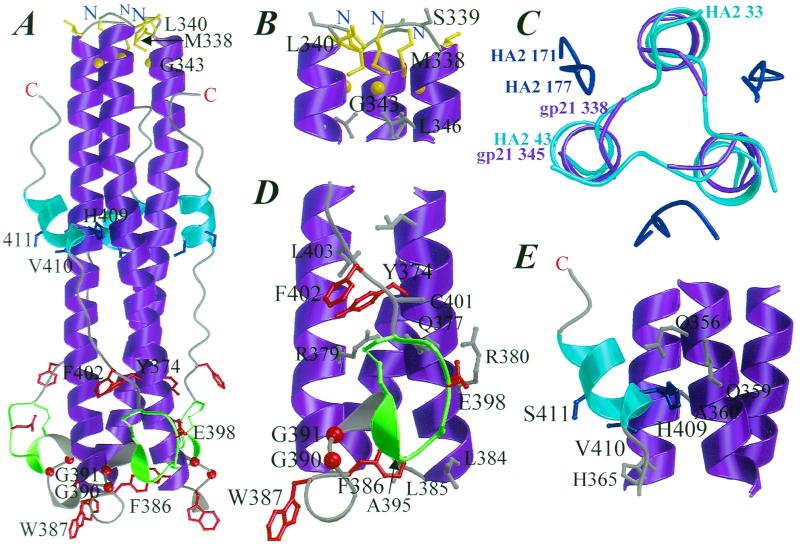
FIG. 1.
Location of mutated residues in the three-dimensional structure of gp21. (A) Ribbon diagram of the gp21(338–425) trimer. The coiled-coil-forming helices are colored magenta, the disulfide-bonded loop is green, the C-terminal helices are cyan, and the rest of the chain is grey. The side chains of residues mutated in this study are also shown; the mutated residues in the helix cap region are colored yellow, the mutated residues in the chain reversal region are colored red, and the mutated residues in the C-terminal helix region are colored blue. The atomic radii of Cα atoms of glycine residues have been enlarged for clarity. (B) Close-up view of the helix cap region, colored as in panel A. The side chain of Ser-339 and the first heptad repeat residue, Leu-346, are also shown colored in grey. (C) Superimposition of the N-cap regions of gp21 and HA2. The structures have been superimposed using 45 N-terminal residues in the coiled-coil helices. HA2 is colored cyan (residues 33 to 43 of the N-terminal region of the coiled coil) and blue (residues 171 to 177 of the C-terminal segment), and gp21 residues are colored magenta (residues 338 to 345). The structures are viewed down the threefold axis of the coiled coil. (D) Close-up view of the chain reversal region, colored as in panel A. The side chains of some residues interacting with the mutated residues are also shown colored in grey. (E) Close-up view of the C-terminal helix region, colored as in panel A. The side chains of some residues interacting with the mutated residues are also shown colored in grey. The figure was prepared with the programs BOBSCRIPT (20) and RASTER3D (35).
MATERIALS AND METHODS
Cell lines and viruses.
293T and HeLa cells were transfected by the Fugene procedure (Boehringer GmbH, Mannheim, Germany) and maintained in Dulbecco's modification of minimal essential medium with 10% fetal calf serum (complete medium). The recombinant vaccinia virus vTF7.3, which drives expression of bacteriophage T7 polymerase, was obtained from T. M. Fuerst and B. Moss (36).
Expression vector construction.
The cytomegalovirus promoter-driven HTLV-1 Env expression vector pCMV-ENV was a gift of M.-C. Dokhélar (41). This vector was modified to pCELT.1, which expresses HTLV-1 Env, C-terminally tagged with the monoclonal antibody (MAb) C8 epitope (1, 10). Mutations were introduced into a KpnI-NsiI fragment (env nucleotides 939 to 1388) by PCR mutagenesis. The sequences of Env mutants were confirmed by the ABI Prism BigDye terminator system (Applied Biosystems). pMBPL−/gp21(338–425) drives the expression of MBP-gp21 in E. coli (10, 28). Mutations were introduced into the gp21 domain by PCR. The firefly luciferase open reading frame was cloned into the NcoI-StuI sites of pTM.1 (obtained from B. Moss) (36) to yield pTMluc. An HIV-1NL4.3 Env expression vector, pcgp160NL4.3, was constructed by cloning the EcoRI-SalI fragment from pNL4.3 (obtained from M. A. Martin) (2) into the cytomegalovirus-based vector pCDNA3 (Invitrogen).
Antibodies.
MAb C8 (directed against the HIV-1 Env cytoplasmic tail) was obtained from G. Lewis (1), while MAb 46 (directed against HTLV-1 gp46) was a gift of David Tribe, The University of Melbourne, Melbourne, Victoria, Australia. Immunoglobulin G was purified from the plasma of an HTLV-1-infected individual (anti-HTLV) using protein A-Sepharose (Pharmacia Biotech).
Western blotting.
Transfected 293T cells were lysed for 10 min on ice in phosphate-buffered saline containing 1% Triton X-100, 0.02% sodium azide, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 5 μg of aprotinin ml−1, 5 μg of leupeptin ml−1, and 1 mM dithiothreitol. Lysates were clarified by centrifugation at 10,000 × g at 4°C prior to polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE) in 12% polyacrylamide gels under reducing conditions. Proteins were transferred to nitrocellulose prior to Western blotting with MAb C8 using the Boehringer chemiluminescence blotting substrate procedure.
Assessment of cell surface Env expression using an antibody binding assay.
The chloramine-T procedure (24) was used to radioiodinate anti-HTLV. The radioiodinated antibody was precleared with 107 293T cells for 2 h on ice before its addition to transfected 293T cells at 48 h posttransfection. The transfected cells were incubated with 125I-anti-HTLV for 1 h on ice in phosphate-buffered saline containing 10 mg of bovine serum albumin ml−1 with occasional gentle agitation. The cells were then washed four times with the same buffer before counting in a Packard Auto-Gamma counter.
Luciferase reporter assay for cell-to-cell fusion.
293T (effector) cells were cotransfected with the pCELT.1 and pTMluc vectors. Twenty-four hours later, HeLa target cells were infected with vTF7.3 at a multiplicity of infection of 1 PFU per cell. At 16 h postinfection, HeLa cells were resuspended in phosphate-buffered saline containing 50 μM EDTA, washed twice in complete medium, and then cocultured with transfected 293T cells for a further 6 h in the presence of 1 μg of actinomycin D ml−1 and 40 μg of cytosine arabinoside ml−1 at 37°C. Cells were then lysed and assayed for luciferase activity using the Promega (Madison, Wis.) luciferase assay system.
Biosynthetic labeling and immunoprecipitation.
Transfected 293T cells were first incubated for 30 min in cysteine- and methionine-deficient medium (ICN, Costa Mesa, Calif.) at 27 h posttransfection and then labeled with 120 μCi of [35S]cysteine (ICN) per well for 14 h. Immunoprecipitations were performed as described previously (40).
Expression and purification of MBP-gp21 chimeras.
Fusion proteins were induced in E. coli strain BL21 (DE3/pLysS) cells by the addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and then purified as described previously (10). MBP-gp21 chimera oligomers were isolated by Superdex 200 (HiLoad 26/60) gel filtration chromatography (Pharmacia Biotech) in S buffer as described previously (10). The gel filtration calibration markers, blue dextran, ferritin (440 kDa), catalase (232 kDa), and bovine serum albumin (67 kDa), were obtained from Pharmacia Biotech. Reducing SDS-PAGE (10% polyacrylamide gels) revealed a single ∼45-kDa band for all chimeras. Electrospray mass spectrometry (PE Sciex API III instrument [10]) indicated that the monomer molecular mass of each chimera was within 5 Da of the calculated molecular mass.
RESULTS AND DISCUSSION
Maturation of Env mutants.
The HTLV-1 Env precursor, gp62, is cleaved in the Golgi apparatus to yield the mature SU, gp46, and the TM protein, gp21, which remain noncovalently associated (39). Mutant forms of gp62 were expressed in similar quantities in 293T cells and in most cases processed to yield similar amounts of gp21 (Fig. 2A), indicating intracellular translocation of cleavage-competent Env structures. The F402H, F402L, F402A, and H409P mutants had reduced cleavage, indicative of folding defects that blocked maturation.
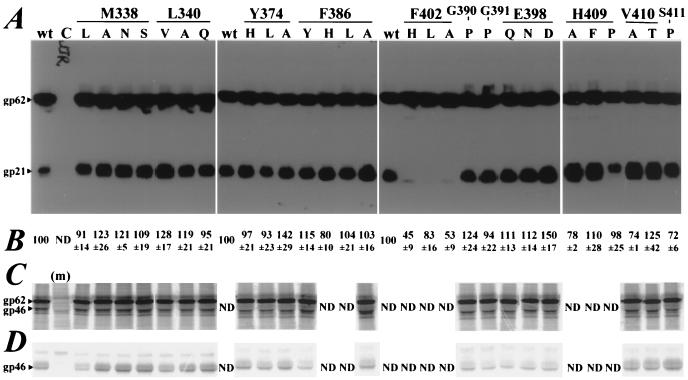
FIG. 2.
(A) Synthesis and processing of HTLV-1 Env glycoprotein mutants. Lysates of Env-expressing 293T cells were subjected to reducing SDS-PAGE in 12% polyacrylamide gels, followed by Western blotting with MAb C8, directed against the C-terminal epitope tag. wt, wild type. Lane C, cells transfected with pCMV-ENV. (B) Cell surface expression of Env glycoprotein mutants. Intact Env-expressing 293T cells were incubated with 125I-anti-HTLV for 1 h on ice and then washed four times with ice-cold phosphate-buffered saline containing 10 mg of bovine serum albumin ml−1. Relative cell surface expression of Env mutants is expressed as follows: ratio of counts per minute bound to cells expressing mutant Env to counts per minute bound to cells expressing wild-type Env × 100. The means ± standard errors of the means from at least three independent transfections are shown. ND, not determined. Relative binding of 125I-anti-HTLV to controls, not shown here, were as follows: for cells transfected with pTM.1, 10 ± 2, and for cells transfected with pcgp160NL4.3, 11 ± 4. (C and D) gp46-anchoring ability of gp21 mutants. (C) Env-expressing 293T cells were labeled with [35S]Cys for 14 h before lysis and immunoprecipitation with a mixture of anti-HTLV and MAb 46. (m), cells transfected with pTM.1. (D) Culture supernatants immunoprecipitated with anti-HTLV MAb 46. gp62 and gp46 were visualized following SDS-PAGE in 5 to 15% gradient gels under reducing conditions and scanning in a PhosphorImager SF. The gp46 bands present in cell lysates were not immunoprecipitated with MAb C8 (data not shown). Panels C and D are composites prepared from scans of multiple gels using PowerPoint software. The differences in the amounts of SU shed into culture supernatants are not reproducible.
The cell surface expression of Env mutants was assessed by using a surface-binding assay employing 125I-radiolabeled anti-HTLV. Anti-HTLV immunoprecipitates only gp62, gp46, and gp21 from [35S]Cys-labeled pCELT.1-transfected 293T cell lysates (data not shown). The levels of cell surface expression were ≥75% of that of the wild type for all but two cleavage-defective mutants, F402H and F402A (Fig. 2B). Background levels of binding to 293T cells transfected with negative control plasmids were 10% ± 2% for pTM.1 and 11% ± 4% for pcgp160NL4.3. The surface expression levels of the cleavage-defective mutants F402L and H409P were >80% of that of wild-type Env. These results suggest that the mutations altered the conformation of the SU-TM protein cleavage site in the absence of major global conformational defects, as has been previously documented for HIV-1 Env mutants (8, 18, 21, 43). By contrast, the surface expression of F402 and F402A mutants was decreased by ∼50% and was consistent with global conformational defects that cause retarded translocation.
The gp46-anchoring ability of gp21 mutants was compared by immunoprecipitation of gp46 shed from [35S]Cys-labeled 293T cells. Similar levels of cell-associated (Fig. 2C) and shed (Fig. 2D) gp46 were observed for wild-type Env and for the mutants tested (i.e., mutants that were cleaved normally but were fusion defective [see Fig. 3]). Thus, all mutants appear to mature normally with the exception of F402H, F402L, F402A, and H409P.
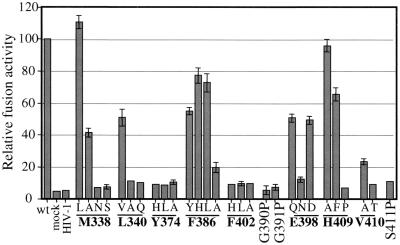
FIG. 3.
Cell-cell fusion activities of Env glycoprotein mutants using a luciferase reporter assay. 293T effector cells, cotransfected with the pCELT.1 and pTMluc vectors, were cocultured with vTF7.3-infected HeLa target cells at 40 h posttransfection. After a 6-h coculture, cells were lysed and assayed for luciferase activity. The relative fusion activities of Env proteins are expressed as follows: ratio of luciferase activity induced by mutant Env to luciferase activity induced by wild-type Env × 100. The means ± standard errors of the means from three independent transfections are shown. wt, wild type; mock, 293T cells transfected with pTM.1; HIV-1, 293T cells transfected with pcgp160NL4.3.
Functional role of the N-terminal helix cap.
The gp21 coiled-coil N terminus is stabilized by contacts between the Cβ atom of Met-338 and the Cβ, Cγ, and Cδ2 atoms of the first helical residue, Leu-340, about the threefold symmetry axis. The remainder of the Met-338 side chain occupies a cavity associated with Gly-343 at the N terminus of the coiled coil (Fig. 1B). This hydrophobic helix-capping structure differs from typical N caps, where the residue immediately preceding the N terminus of an α-helix, hydrogen, bonds with the NH group of the third helical residue; Ser, Thr, Asn, Asp, and Cys are the most favored N-capping residues (17). The side chain of the first nonhelical residue of gp21, Ser-339, points into solvent without making any helix contacts. The gp21 helix cap is an essential functional determinant, because substitutions at either Met-338 or Leu-340 led to decreases in fusion activity correlating inversely with the hydrophobicity of the substituting side chain (Fig. 3). These substitutions did not affect Env maturation (Fig. 2), suggesting that the helix cap may be important in the fusion-activated structure.
Fusion-activated HA2 contains a four-residue (Q34AAD) N cap that is stabilized by a complex network of hydrogen bonds and two nonpolar layers (14). The gp21 and HA2 coiled-coil N caps are superimposable (root mean square distance, 1.0 Å for 21 backbone atoms corresponding to residues 34 to 40 of HA2), with M338S of gp21 and Q34AAD of HA2 exiting the coiled coil laterally in an extended conformation (Fig. 1C). This conservation indicates that the helix caps may have evolved to stabilize the N terminus of the coiled coil in the fusion-activated TM protein structure, linking fusion peptides to the coiled coil through a nonhelical segment (14) (Gly-332 to Ser-337 in gp21). Such a structure positions the fusion peptides to extend away from the threefold symmetry axis of the coiled coil to direct their oblique insertion into the target membrane, as has been inferred from biochemical studies with HA2 model peptides (31, 32). The oblique insertion of fusion peptides may destabilize a larger area of the target membrane than perpendicular insertion. The latter model has been implicated for paramyxovirus TM proteins in which fusion peptides appear to be a helical continuation of the coiled coil (4). However, random insertion of fusion peptides linked to the coiled coil through a flexible linker (45, 51) cannot be ruled out. It is not possible to predict from amino acid alignments whether or not other retrovirus TM proteins have N caps homologous to that of gp21. Because the HA2 and gp21 N caps are stabilized by different types of interactions, other retroviruses may have evolved a variety of structurally related mechanisms to stabilize the N terminus of their central coiled coil.
Functional role of the chain reversal region.
At the base of the gp21 coiled coil, a complex substructure mediates a chain reversal to enable the antiparallel packing of the C-terminal segment against the exterior of the coiled coil. The substructure comprises a 310-helix (Trp-387 to Gln-389), a conserved Gly-390–Gly-391 hinge motif, a short α-helix (Leu-392 to Ala-395), and the disulfide-bonded loop (Cys-393 to Cys-400) (Fig. 1D). Corresponding substructures are present in the murine leukemia virus (MuLV)TM protein (p15E [22]) and in GP2 of Ebola virus (34, 50) (Fig. 4). These substructures pack roughly perpendicular to the coiled coil through hydrophobic and polar interactions.
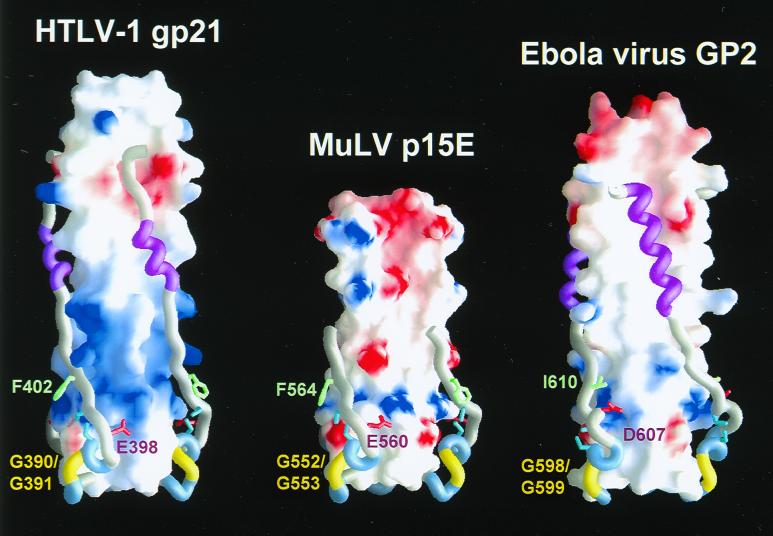
FIG. 4.
Molecular surface of gp21, p15E (22), and GP2 (50) coiled coils, color coded according to electrostatic potential (continuous coloring from blue [+10 kTe−1] to red [−10 kTe−1]). The region of chain reversal and C-terminal ectodomain segment are drawn as tubes. The glycine hinges are colored yellow, cysteines are cyan, and C-terminal α-helices are magenta. The figure was drawn using GRASP (37).
(i) A hydrophobic core at the base of the coiled coil.
A small hydrophobic core is formed at the base of the coiled coil where Phe-386 packs with Ala-395 within the disulfide-bonded loop and with Leu-384 and Leu-385 of an adjacent monomer's central α-helix (Fig. 1D). Whereas replacement of Phe-386 with other bulky residues (His and Ile) was tolerated, an F386A substitution reduced fusion activity to 20% of that of the wild type (Fig. 3). This effect is most likely due to a decrease in protein stability resulting from the creation of a cavity in a hydrophobic core, as has been observed in other systems (19). Replacement of Phe-386 with Tyr decreased fusion activity by approximately 50%, probably due to steric problems associated with the burial of the larger and more polar side chain of tyrosine. None of these substitutions affected Env maturation, suggesting that the contacts mediated by Phe-386 are most critical in the context of a fusion-activated structure. Thus, hydrophobic interactions appear to play an important role in stabilizing both ends of the central coiled coil in a fusion-competent gp21 structure. Hydrophobic clusters are present at similar positions in MuLV p15E (22) and Ebola virus GP2 (34, 50), highlighting their general importance in stabilizing the base of the central coiled coil of fusion-competent Env.
(ii) The conserved diglycine motif preceding the disulfide-bonded loop.
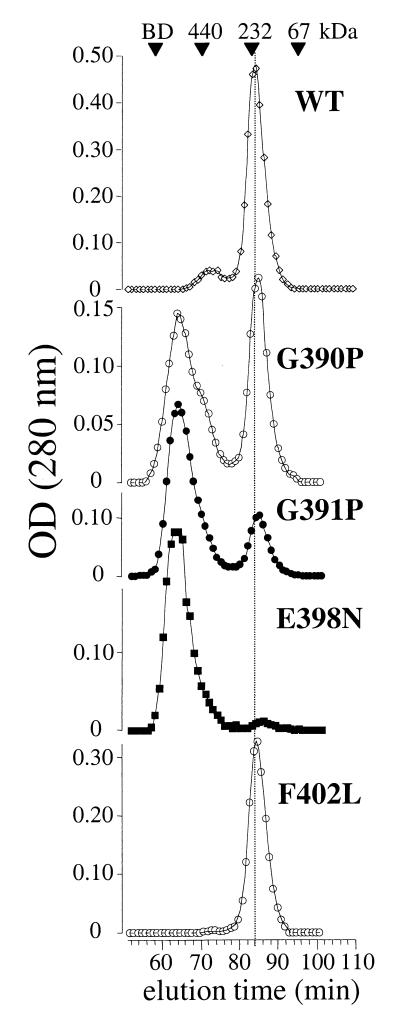
Most retroviruses (excluding type B retroviruses and lentiviruses of ungulates) and filoviruses contain one or more glycines immediately N terminal to the disulfide-bonded loop of the TM protein. The gp21, p15E, and GP2 crystal structures reveal that the Gly-Gly hinge motif precedes a short α-helix (Leu-392 to Ala-395 in gp21) that contains the first cysteine of the disulfide bridge (Fig. 4). Replacement of either Gly-390 or Gly-391 of gp21 with Pro, a residue that severely limits the flexibility of protein backbones by restricting rotation about the N-Cα bond (46), led to the complete abolition of Env fusion function (Fig. 3) without affecting Env maturation (Fig. 2). We tested the effect of the proline mutations on the trimerization of the MBP-gp21 chimera, which contains the gp21 helical hairpin (28). We assumed that MBP-gp21 trimerization indicated the formation of the gp21 hairpin, whereas aggregation indicated misfolding. MBP containing three alanines fused to its C terminus has a monomeric structure (10). Superdex-200 gel filtration chromatography shows that both the G390P and G391P mutations induced aggregation of the MBP-gp21 chimera (Fig. 5). The block in fusogenicity may therefore be due to an inability of gp21 with a Pro substitution to fold into a helical hairpin. Because the Gly-to-Pro substitutions did not affect Env maturation, the conserved Gly-Gly motif appears to be essential for the formation of the helical hairpin structure, perhaps by functioning as a flexible hinge to facilitate the conversion of prefusogenic Env to the fusion-active form.
FIG. 5.
Gel filtration of MBP-gp21 chimeras. MBP-gp21 chimeras bearing fusion-inhibiting mutations were purified from the soluble fraction of E. coli lysates using amylose-agarose. The oligomerization of wild-type (WT) and mutant chimeras was assessed by Superdex 200 gel filtration chromatography. BD, blue dextran; OD, optical density.
(iii) The Arg-380–Glu-398 salt bridge.
Glutamic acid-398 within the gp21 disulfide-bonded loop forms a salt bridge with Arg-380 or a hydrogen bond with Gln-377, located close to the base of the coiled-coil-forming sequence of an adjacent monomer. Fusion activity (Fig. 3) and MBP-gp21 trimerization (Fig. 5), but not Env maturation (Fig. 2), were abolished by an E398N substitution that precludes the electrostatic interaction with Arg-380, creating a small cavity at this site. The proteins with the more conservative substitution mutations E398Q (geometry) and E398D (charge) retained approximately 50% of wild-type fusion activity. These intermediate phenotypes may be due to the Gln side chain maintaining a hydrogen bond with Gln-377 while an Asp maintains a less favorable electrostatic contact with Arg-380. The electrostatic contact is conserved in the helical hairpin structures of MuLV p15E (Arg-542 to Glu-560) and Ebola virus GP2 (Lys-588 to Asp-607) (Fig. 4); structure-based alignments of retroviral TM protein sequences suggest that these electrostatic contacts are also likely to be important features in TM proteins of type D and avian leukosis and sarcoma retroviruses (28). Our observations suggest that these conserved salt bridges serve to stabilize or position the region of chain reversal against the base of the coiled coil in the helical hairpin.
(iv) The conserved, solvent-exposed Phe-402.
Phenylalanine-402 occupies a solvent-exposed position on the C-terminal side of the disulfide-bonded loop, adjacent to the unpaired Cys-401. Mutation of Phe-402 to Leu, His, or Ala led to the abolition of Env maturation (Fig. 2). The inhibition of processing with mutants of Phe-402 suggests that it plays a structural role in gp62, implying that Phe-402 may be associated with the core of prefusogenic Env, becoming solvent exposed during the formation of the helical hairpin. Phenylalanine-402 does not, however, appear to be essential for the acquisition of the helical hairpin conformation, because the F402L mutation was tolerated in the MBP-gp21 trimer (Fig. 5).
Phenylalanine residues that are C terminal to the disulfide-bonded loop are conserved in the TM proteins of type C (e.g., MuLV) and type D retroviruses (28). The solvent-exposed Phe-564 of MuLV p15E (Fig. 4) is adjacent to the unpaired Cys-563, which forms a labile disulfide bond with SU. Proteolysis experiments have suggested that the SU sequence C312WLCL, which closely resembles the active-site sequences of thiol-disulfide exchange enzymes, is responsible for the labile disulfide bond with Cys-563 (38). Because this SU sequence is rich in hydrophobic residues, it may be a contact site for Phe-564 in the SU-TM protein interface. Similar CXXCX motifs (where X is a hydrophobic residue) are present in the SUs of HTLV-1 (C226IVCI), HTLV-2, bovine leukemia virus, and type C and type D retroviruses, providing potential docking sites for the Phe-564 homologs of these viruses. A possible mechanism for TM fusion activation may require SU-receptor binding to trigger the expulsion of Phe-402 from the core of prefusogenic SU-TM protein complex to become solvent exposed in the fusion-activated helical hairpin. It should be noted that the receptor-binding GP1 subunit of filoviruses and the SU of avian leukosis and sarcoma retroviruses do not contain thiol-disulfide exchange CXXCX motifs, which may account for the stable covalent associations between GP1-GP2 (44) and SU-TM protein complex (29) in these viruses. Nevertheless, Ile-610 of Ebola virus GP2 occupies a solvent-exposed position homologous to Phe-402 of gp21 (Fig. 4), while avian leukosis and sarcoma virus TM proteins also contain gp21 Phe-402 homologs, suggesting conserved functional roles for these hydrophobic residues.
Interactions between the coiled coil and the C-terminal ectodomain segment.
The C-terminal ectodomain segment of gp21 (Leu-403 to Asn-421) packs into a groove on the surface of the coiled coil, burying a large complementary surface area (28). This packing is mediated in part by the C-terminal α-helix residues, His-409, Val-410, Ile-412, Leu-413, and Gln-414, contacting the coiled-coil residues. The results presented in Fig. 3 indicate that polar interactions mediated by His-409 (Fig. 1E) are not important for fusogenicity, whereas hydrophobic interactions between Val-410 and His-365 are essential. The C-terminal α-helical structure may also be important, because fusion activity is abolished by the S411P mutation, which was designed to introduce a kink into the α-helical element. Tyrosine-374 is located on the surface of the coiled coil, making hydrophobic contacts with Cys-401 and Leu-403 of the C-terminal segment and a hydrogen bond with Arg-379 of an adjacent monomer's coiled-coil-forming sequence (Fig. 1D). These contacts are also essential for Env fusion competence, as mutation of Tyr-374 to His, Leu, or Ala completely abolished fusogenicity (Fig. 3). The V410A, S411P, and Y374A substitutions did not affect MBP-gp21 trimerization (data not shown), and V410A and Y374A MBP-gp21 mutants were recognized by conformational MAbs, HT2 and HT48, raised against MBP-gp21 (H. E. Drummer and P. Poumbourios, unpublished observation). The ability of these mutants to retain a global helical hairpin conformation despite the loss of fusion activity may be due to multiple contacts being used to pack the C-terminal ectodomain segment into the groove on the surface of the coiled coil (28).
Implications for membrane fusion.
Recent models of retrovirus fusion (12, 45) are based on the influenza virus HA2 fusion process, in which endosomal pH induces refolding of the metastable prefusogenic HA2 structure into a thermostable, ∼110-Å-long fusion-active helical rod (6, 14). Whereas the receptor for HTLV-1 is not known, studies with HIV-1 (23, 27, 47), simian immunodeficiency virus (SIV) (3), and avian leukosis and sarcoma virus (15, 26) indicate that receptor binding by SU is the fusion activation trigger for retrovirus TM proteins. TM protein fragments derived from MuLV, HIV-1, SIV, Ebola virus, simian virus 5, and influenza virus also adopt thermostable helical hairpin structures analogous to fusion-activated HA2 when expressed in E. coli in the absence of receptor-binding subunits (4, 6, 11, 22, 33, 34, 48, 50, 51). These structures imply that the N-terminal fusion peptide and C-terminal TM sequence are positioned at the same end of the rod, consistent with a structure that closely apposes viral and target cellular membranes in order that they may fuse.
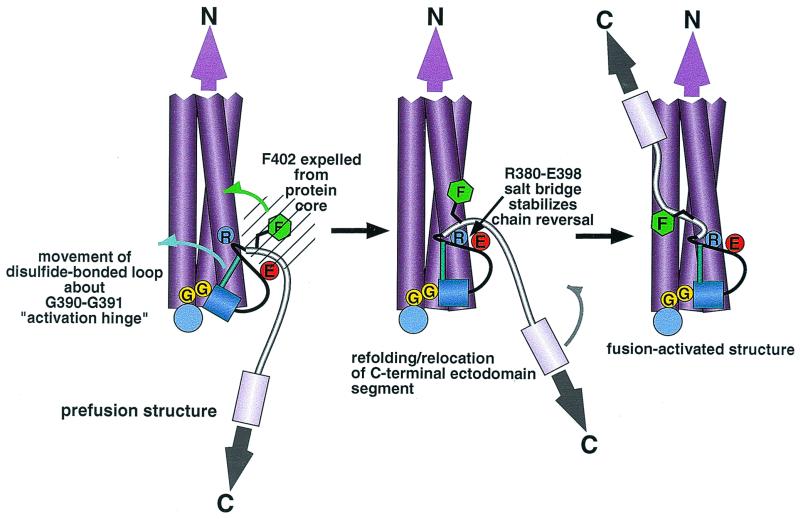
The data presented here indicate that retrovirus Env fusion competence requires that the central coiled coil of the fusion-activated TM protein structure be stabilized by an N-terminal helix cap and C-terminal hydrophobic cluster. These interactions may help to lock the central coiled coil into a stable membrane-embedded rod about which the fusion-inducing conformational change occurs. Our data are also consistent with the idea that the region of chain reversal plays a conserved role in transmitting a conformational signal from receptor-bound SU to the TM protein so that the fusion-activated helical hairpin structure can be induced. A hypothetical pathway for the conformational change is summarized in Fig. 6, using HTLV-1 Env as an example. Receptor binding by SU induces the expulsion of Phe-402 from the core of prefusogenic Env to a solvent-exposed location. Refolding of the TM protein to a more stable conformation involves movement of the disulfide-bonded loop about the Gly-Gly motif to allow formation of the Arg-380–Glu-398 salt bridge.
FIG. 6.
Fusion activation model for retrovirus TMs. Receptor-SU binding induces the expulsion of Phe-402 (green hexagon) from a hydrophobic environment in the Env core (hatched circle). The disulfide-bonded loop rotates about the Gly-Gly hinge (yellow circles), resulting in stabilization of the region of chain reversal against the base of the central coiled coil (purple cylinders) through the Arg-380–Glu-398 salt bridge (blue and red circles). These events may enable the antiparallel packing of the C-terminal segment against the exterior of the coiled coil to provide energy for the relocation and juxtaposition of C-terminal TM anchors with N-terminal fusion peptides (not shown) to induce virus-cell membrane fusion.
The available HIV-1 and SIV gp41 helical hairpin structures lack an intact chain-reversal region (7, 11, 33, 48, 51). However, sequence comparisons reveal that various features of the HIV and SIV chain reversal region may have parallels in other retroviruses, including HTLV-1 and MuLV (28). These features include the hydrophobic sequence immediately C terminal to the coiled coil, the conserved glycine preceding the disulfide-bonded loop, the conserved basic residue in the center of the disulfide-bonded loop that could make ionic contacts with conserved acidic residues close to the base of the coiled coil, and a conserved hydrophobic sequence, Val-X-Trp, located C terminally to the disulfide bridge (28). The results of in vitro mutagenesis experiments have revealed that residues C terminal to the gp41 disulfide-bonded loop are in intimate contact with the C terminus of gp120 (5) and that the N- and C-terminal regions of gp120 provide a hydrophobic binding site for gp41 (25). It is therefore important to determine if the Val-X-Trp motif of gp41 contacts gp120, perhaps acting as a functional analog of the solvent-exposed Phe residue of gp21 and p15E.
Mutations of the ectodomain cysteines of MuLV p15E and HIV-1 gp41 result in Env maturation defects (16, 49), suggesting that the disulfide-bonded loop is already formed in prefusogenic retroviral Env proteins, functioning as a module. Stabilization of the region of chain reversal against the base of the coiled coil may therefore involve repositioning of the disulfide-bonded loop rather than extensive refolding. This scenario is in contrast to the case with influenza virus HA2, in which fusion activation requires refolding of the C-terminal one-third of the central coiled coil, creating a new hydrophobic core that stabilizes the chain reversal (6). These conformational events at the base of the rod may trigger the refolding and relocation of the C-terminal ectodomain segment into a groove on the surface of the coiled coil that, in the case of gp21, buries approximately 2,630 Å2 in the interface (28). Similar or larger surface areas are buried in the interfaces formed between the central coiled coil and C-terminal segments of HA2 (5,351 Å2), GP2 (2,840 Å2), and SIV gp41 (3,648 Å2) helical hairpins, although the structures of the C-terminal ectodomain segments vary considerably. The gain in free energy associated with the refolding and burial of these large surface areas (30) may help drive membrane destabilization and fusion when TM anchors become juxtaposed with fusion peptides.
ACKNOWLEDGMENTS
We thank Ken Mitchellhill and Kirilee Wilson for mass spectrometry and the NIH AIDS Research and Reference Reagent Program for the supply of vTF7.3, pNL4.3, and MAb C8.
This work was supported by grants from the NHMRC, NIH, and Wellcome Trust. B.K. is a Wellcome Senior Research Fellow in Medical Science in Australia. B.E.K. is an NHMRC Fellow.
REFERENCES
- 1.Abacioglu Y H, Fouts T R, Laman J D, Claassen E, Pincus S H, Moore J P, Roby C A, Kamin-Lewis R, Lewis G K. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retrovir. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 2.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allan J S, Strauss J, Buck D W. Enhancement of SIV infection with soluble receptor molecules. Science. 1990;247:1084–1088. doi: 10.1126/science.2309120. [DOI] [PubMed] [Google Scholar]
- 4.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 5.Binley J M, Sanders R W, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma D J, Maddon P J, Olson W C, Moore J P. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 7.Caffrey M, Cai M, Kaufman J, Stahl S J, Wingfield P T, Covell D G, Gronenborn A M, Clore G M. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 10.Center R J, Kobe B, Wilson K A, Teh T, Howlett G J, Kemp B E, Poumbourios P. Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein. Protein Sci. 1998;7:1612–1619. doi: 10.1002/pro.5560070715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 12.Chan D C, Kim P S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Lee K H, Steinhauer D A, Stevens D J, Skehel J J, Wiley D C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Skehel J J, Wiley D C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA2 subunit to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci USA. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damico R L, Crane J, Bates P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA. 1998;95:2580–2585. doi: 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dedera D, Gu R, Ratner L. Conserved cysteine residues in the human immunodeficiency virus type 1 transmembrane envelope protein are essential for precursor envelope cleavage. J Virol. 1992;66:1207–1209. doi: 10.1128/jvi.66.2.1207-1209.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doig A J, MacArthur M W, Stapley B J, Thornton J M. Structures of N-termini of helices in proteins. Protein Sci. 1997;6:147–155. doi: 10.1002/pro.5560060117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubay J W, Dubay S R, Shin H-J, Hunter E. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J Virol. 1995;69:4675–4682. doi: 10.1128/jvi.69.8.4675-4682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eriksson A E, Baase W A, Zhang X J, Heinz D W, Blaber M, Baldwin E P, Matthews B W. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 20.Esnouf R M. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 21.Farzan M, Choe H, Desjardins E, Sun Y, Kuhn J, Cao J, Archambault D, Kolchinsky P, Koch M, Wyatt R, Sodroski J. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J Virol. 1998;72:7620–7625. doi: 10.1128/jvi.72.9.7620-7625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 Å resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 23.Furuta R A, Wild C T, Weng Y, Weiss C D. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 24.Greenwood F C, Hunter W M, Glover J S. The preparation of 131I-labelled human growth hormone of high specific radioactivity. Biochem J. 1963;89:114–123. doi: 10.1042/bj0890114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez L D, Peters R J, Delos S E, Young J A, Agard D A, White J M. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J Cell Biol. 1997;139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones P L, Korte T, Blumenthal R. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J Biol Chem. 1998;273:404–409. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- 28.Kobe B, Center R J, Kemp B E, Poumbourios P. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc Natl Acad Sci USA. 1999;96:4319–4324. doi: 10.1073/pnas.96.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leamnson R N, Halpern M S. Subunit structure of the glycoprotein complex of avian tumor virus. J Virol. 1976;18:956–968. doi: 10.1128/jvi.18.3.956-968.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 31.Luneberg J, Martin I, Nussler F, Ruysschaert J M, Herrmann A. Structure and topology of the influenza virus fusion peptide in lipid bilayers. J Biol Chem. 1995;270:27606–27614. doi: 10.1074/jbc.270.46.27606. [DOI] [PubMed] [Google Scholar]
- 32.Macosko J C, Kim C H, Shin Y K. The membrane topology of the fusion peptide region of influenza hemagglutinin determined by spin-labeling EPR. J Mol Biol. 1997;267:1139–1148. doi: 10.1006/jmbi.1997.0931. [DOI] [PubMed] [Google Scholar]
- 33.Malashkevich V N, Chan D C, Chutkowski C T, Kim P S. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc Natl Acad Sci USA. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malashkevich V N, Schneider B J, McNally M L, Milhollen M A, Pang J X, Kim P S. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merritt E A, Bacon D J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–525. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 36.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 37.Nicholls A, Sharp K A, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 38.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pique C, Pham D, Tursz T, Dokhélar M-C. Human T-cell leukemia virus type I envelope protein maturation process: requirements for syncytium formation. J Virol. 1992;66:906–913. doi: 10.1128/jvi.66.2.906-913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poumbourios P, Wilson K A, Center R J, El Ahmar W, Kemp B E. Human immunodeficiency virus type 1 envelope glycoprotein oligomerization requires the gp41 amphipathic α-helical/leucine zipper-like sequence. J Virol. 1997;71:2041–2049. doi: 10.1128/jvi.71.3.2041-2049.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg A R, Delamarre L, Pique C, Pham D, Dokhélar M-C. The ectodomain of the human T-cell leukemia virus type 1 TM glycoprotein is involved in postfusion events. J Virol. 1997;71:7180–7186. doi: 10.1128/jvi.71.10.7180-7186.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenthal P B, Zhang X, Formanowski F, Fitz W, Wong C H, Meier-Ewert H, Skehel J J, Wiley D C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salzwedel K, West J T, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez A, Yang Z-Y, Xu L, Nabel G J, Crews T, Peters C J. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J Virol. 1998;72:6442–6447. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skehel J J, Wiley D C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 46.Stites W E, Pranata J. Empirical evaluation of the influence of side chains on the conformational entropy of the polypeptide backbone. Proteins. 1995;22:132–140. doi: 10.1002/prot.340220206. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan K, Liu J, Wang J, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas A, Roth M J. Analysis of cysteine mutations on the transmembrane protein of Moloney murine leukemia virus. Virology. 1995;211:285–289. doi: 10.1006/viro.1995.1402. [DOI] [PubMed] [Google Scholar]
- 50.Weissenhorn W, Carfi A, Lee K H, Skehel J J, Wiley D C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 51.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 52.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]