Abstract
Adoption of high-content omic technologies in clinical studies, coupled with computational methods, has yielded an abundance of candidate biomarkers. However, translating such findings into bona fide clinical biomarkers remains challenging. To facilitate this process, we introduce Stabl, a general machine learning method that identifies a sparse, reliable set of biomarkers by integrating noise injection and a data-driven signal-to-noise threshold into multivariable predictive modeling. Evaluation of Stabl on synthetic datasets and five independent clinical studies demonstrates improved biomarker sparsity and reliability compared to commonly used sparsity-promoting regularization methods while maintaining predictive performance; it distills datasets containing 1,400–35,000 features down to 4–34 candidate biomarkers. Stabl extends to multi-omic integration tasks, enabling biological interpretation of complex predictive models, as it hones in on a shortlist of proteomic, metabolomic and cytometric events predicting labor onset, microbial biomarkers of pre-term birth and a pre-operative immune signature of post-surgical infections. Stabl is available at https://github.com/gregbellan/Stabl.
Subject terms: Predictive medicine, Computational models
Stabl selects sparse and reliable biomarker candidates from predictive models.
Main
High-content omic technologies, such as transcriptomics, metabolomics or cytometric immunoassays, are increasingly employed in biomarker discovery studies1,2. These technologies allow researchers to measure thousands of molecular features in each biological specimen, offering unprecedented opportunities for advancing precision medicine tools across the spectrum of health and disease. Whether it is personalizing breast cancer diagnostics through multiplex imaging3 or identifying transcriptional signatures governing patient-specific vaccine responses across multiple vaccine types4, omic technologies have also dictated a shift in statistical analysis of biological data. The traditional univariate statistical framework is maladapted to large omic datasets characterized by a high number of molecular features p relative to the available samples n. The p ≫ n scenario reduces the statistical power of univariate analyses, and simply increasing n is often impractical due to cost or sample constraints5,6.
Statistical analysis in biomarker discovery research comprises three distinct tasks, all necessary for clinical translation and impacted by the p ≫ n challenge: (1) predicting clinical endpoints via identification of a multivariable model with high predictive performance (predictivity); (2) selecting a limited number of features as candidate clinical biomarkers (sparsity); and (3) ensuring confidence that the selected features are truly related to the outcome (reliability).
Several machine learning methods, including sparsity-promoting regularization methods (SRMs), such as Lasso7, Elastic Net (EN)8, Adaptive Lasso (AL)9 and sparse group Lasso (SGL)10, provide predictive modeling frameworks adapted to p ≫ n omic datasets. Furthermore, data fusion methods, such as early-fusion and late-fusion Lasso, enable integration of multiple, often heterogeneous, omic datasets11,12. Nevertheless, the challenge of selecting a sparse and reliable set of candidate biomarkers persists. Most SRMs employ ℓ1 regularization to limit the number of features in the final model. However, as the learning phase often relies on a limited number of samples, small perturbations in the training data can yield widely different sets of selected features13–15, undermining confidence in their relevance to the outcome. This inherent limitation hampers sparsity and reliability, impeding the biological interpretation and clinical significance of predictive models. Consequently, few omic biomarker discovery studies progress to later clinical development phases1,2,5,6,16,17.
High-dimensional feature selection methods, such as stability selection (SS), Model-X (MX) knockoff or bootstrap-enhanced Lasso (Bolasso), improve reliability by controlling for false discoveries in the selected feature set18–20. However, these methods often require a priori definition of the feature selection threshold or target false discovery rate (FDR), which decouples feature selection from the multivariable modeling process. Without prior knowledge of the data, this can lead to suboptimal feature selection, requiring multiple iterations to identify a desirable threshold and hindering optimal integration of multiple omic datasets into a unique predictive model, as a single fixed selection threshold may not be suited to the specificities of each dataset.
In this context, we introduce Stabl, a supervised machine learning framework designed to facilitate clinical translation of high-dimensional omic studies by bridging the gap between multivariable predictive modeling and the sparsity and reliability requirements of clinical biomarker discovery. Stabl combines noise injection into the original data, determination of a data-driven signal-to-noise threshold and integration of the selected features into a predictive model. Systematic benchmarking of Stabl against state-of-the-art SRMs, including Lasso, EN, SGL, AL and SS, using synthetic datasets, four existing real-world omic datasets and a newly generated multi-omic clinical dataset demonstrates that Stabl overcomes the shortcomings of current SRMs, thereby enhancing biological interpretation and clinical translation of sparse predictive models. The complete Stabl package is available at https://github.com/gregbellan/Stabl.
Results
Feature selection via false discovery proportion estimate
When applied to a cohort randomly drawn from the population, SRMs will select informative features (that is, truly related to the outcome) with a higher probability, on average, than uninformative features (that is, unrelated to the outcome)7,18. However, as uninformative features typically outnumber informative features in high-dimensional omic datasets1,2,17, the fit of an SRM model on a single cohort can lead to selection of many uninformative features despite their lower probability of selection18,20. To address this challenge, Stabl implements the following strategy (Fig. 1 and Methods):
Stabl fits SRM models (StablSRM), such as Lasso, EN, SGL or AL, on subsamples of the data using a procedure similar to SS18. Subsampling mimics the availability of multiple random cohorts and estimates each feature’s selection frequency across all iterations. However, this procedure lacks an optimal frequency threshold for distinguishing informative from uninformative features objectively.
To define the optimal frequency threshold, Stabl creates artificial features unrelated to the outcome (noise injection) via MX knockoffs19,21,22 or random permutations1–3 (Extended Data Fig. 1), which we assume behave similarly to uninformative features in the original dataset23 (see ‘Theoretical guarantees’ in Methods). The artificial features are used to construct a false discovery proportion surrogate (FDP+). We define the ‘reliability threshold’, θ, as the frequency threshold that minimizes FDP+ across all possible thresholds. This method for determining θ is objective (minimizing a proxy for the FDP) and data driven (tailored to individual omic datasets).
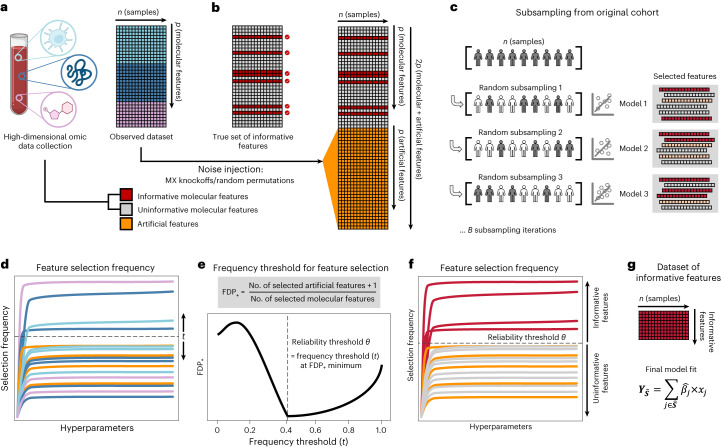
Fig. 1. Overview of the Stabl algorithm.
a, An original dataset of size n × p is obtained from measurement of p molecular features in each of n samples. b, Among the observed features, some are informative (related to the outcome, red), and others are uninformative (unrelated to the outcome, gray). p artificial features (orange), all uninformative by construction, are injected into the original dataset to obtain a new dataset of size n × 2p. Artificial features are constructed using MX knockoffs or random permutations. c, B subsample iterations are performed from the original cohort of size n. At each iteration k, SRM models varying in their regularization parameter(s) λ are fitted on the subsample, resulting in a different set of selected features for each iteration. d, For a given λ, B sets of selected features are generated in total. The proportion of sets in which feature i is present defines the feature selection frequency fi(λ). Plotting fi(λ) against 1/λ yields a stability path graph. Features whose maximum frequency is above a frequency threshold (t) are selected in the final model. e, Stabl uses the reliability threshold (θ), obtained by computing the minimum value of the FDP+ (Methods). f,g, The feature set with a selection frequency larger than θ (that is, reliable features) is included in a final predictive model.
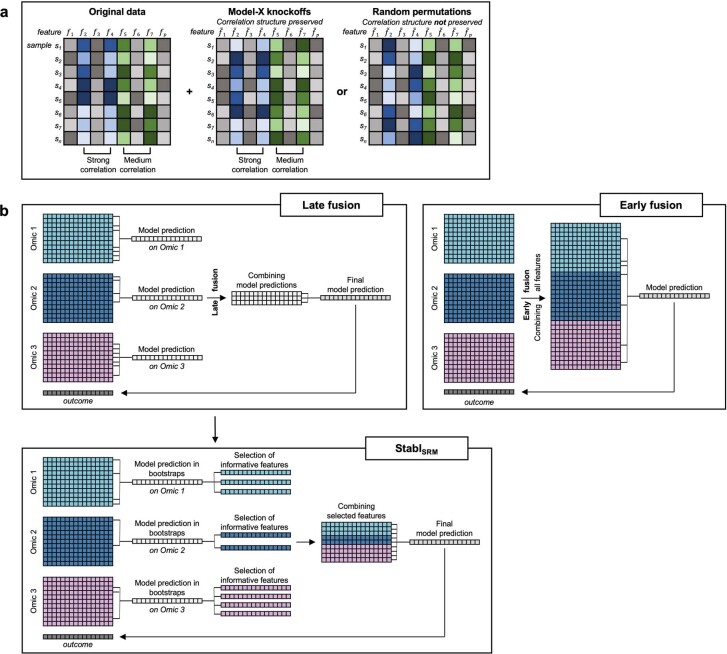
Extended Data Fig. 1. Infographics for noise injection methods and multi-omic data integration with StablSRM.
A. Noise injection methods. Left panel depicting the original dataset with n samples and p features with strong correlation between features f! and f" as well as medium correlation between f# and f$. Middle panel showing MX knockoffs as noise injection method where generated artificial features preserve the original features’ correlation structure. Right panel showing random permutations as alternative noise generation method, which does not preserve the correlation structure. B. Multi-omic data integration with StablSRM. Early fusion approaches of multi-omic data integration combine all features of all omics to a concatenated dataset to derive a multivariate model. Late fusion approaches build predictive models on each omic layer individually, then concatenate the model predictions together and build a predictive model. StablSRM’s method builds models in a bootstrapping fashion on each omic individually to select the informative features, then concatenates all selected (informative) features and builds a final predictive model on all selected features.
As a result, Stabl provides a unifying procedure that selects features above the reliability threshold while building a multivariable predictive model. Stabl is amenable to both classification and regression tasks and can integrate multiple datasets of different dimensions and omic modalities. The complexity of the algorithm is described in Methods, and it allows for a scalable procedure with a runtime of under 1 h on a computer equipped with 32 vCPUs and 128 GB of RAM (Supplementary Table 1).
Improved sparsity and reliability, retained predictivity
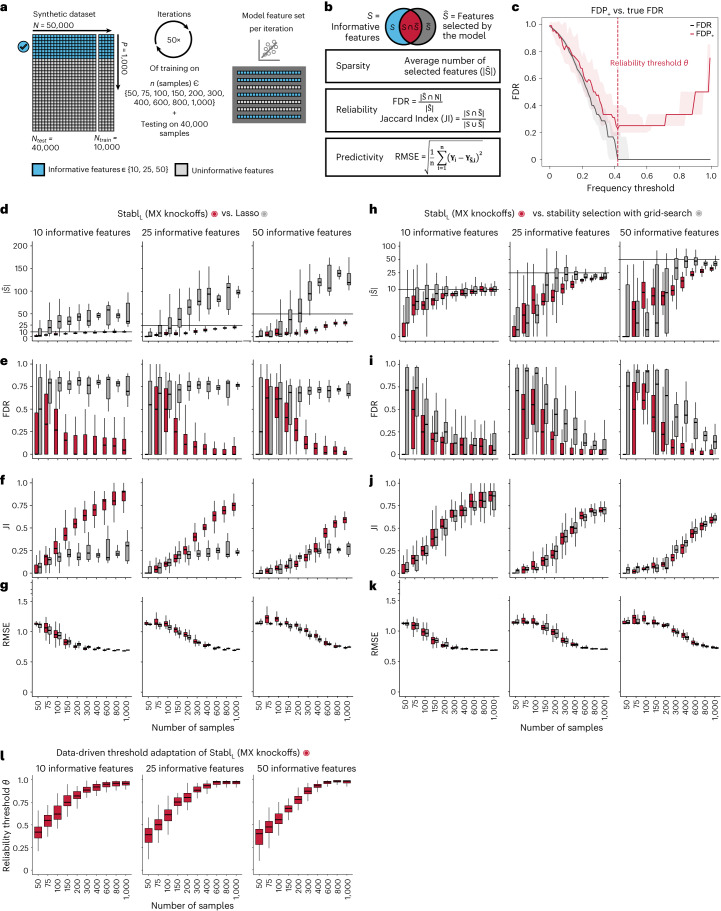
We benchmarked Stabl using synthetic training and validation datasets containing known informative and uninformative features (Fig. 2a). Simulations mimicking real-world scenarios incorporated variations in sample size (n), number of total features (p) and informative features (∣S∣). Three key performance metrics were employed (Fig. 2b and Supplementary Table 2):
Sparsity: measured as the average number of selected features () relative to informative features
Reliability: evaluated through the FDR and Jaccard index (JI), indicating the overlap between algorithm-selected features and true informative features
Predictivity: assessed using root mean square error (RMSE)
Fig. 2. Synthetic dataset benchmarking against Lasso.
a, A synthetic dataset consisting of n = 50,000 samples × p = 1,000 normally distributed features was generated. Some features are correlated with the outcome (informative features, light blue), whereas the others are not (uninformative features, gray). Forty thousand samples are held out for validation. Out of the remaining 10,000, 50 sets of sample sizes n ranging from 50 to 1,000 are drawn randomly to assess model performance. The StablSRM framework is used using Lasso (StablL) with MX knockoffs for noise generation. Performances are tested on continuous outcomes (regression tasks). b, Sparsity (average number of selected features, ), reliability (true FDR and JI) and predictivity (RMSE) metrics used for performance evaluation. c, The FDP+ (red line; 95% CI, red shading) and the true FDR (gray line; 95% CI, gray shading) as a function of the frequency threshold (example shown for n = 150 samples and 25 informative features; see Extended Data Fig. 3 for other conditions). The FDP+ estimate approaches the true FDR around the reliability threshold, θ. d–g, Sparsity (d), reliability (FDR, e; JI, f) and predictivity (RMSE, g) performances of StablL (red box plots) and Lasso (gray box plots) with increasing number of samples (n, x axis) for 10 (left panels), 25 (middle panels) or 50 (right panels) informative features. h–k, Sparsity (h), reliability (i and j) and predictivity (k) performances of models built using a data-driven reliability threshold θ (StablL, red box plots) or grid search-coupled SS (gray box plots). l, The reliability threshold chosen by StablL shown as a function of the sample size (n, x axis) for 10 (left panel), 25 (middle panel) or 50 (right panel) informative features. Boxes indicate median and IQR; whiskers indicate 1.5× IQR.
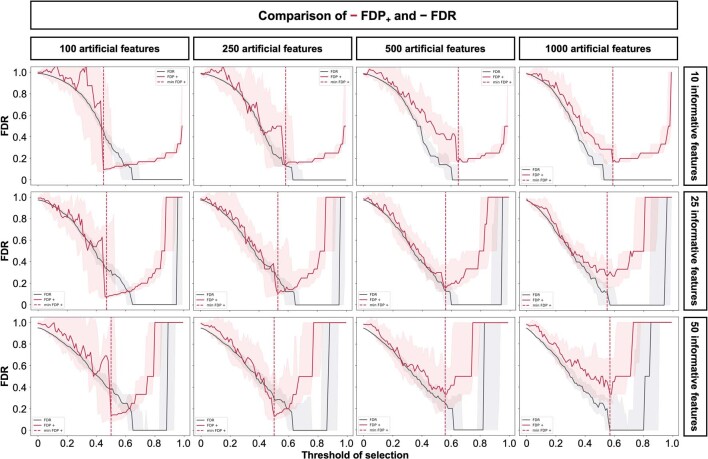
Before benchmarking, we tested whether Stabl’s FDP+ experimentally controls the FDR at the reliability threshold θ, as the actual FDR value is known for synthetic data. We observed that FDP+(θ) consistently exceeded the true FDR value (Fig. 2c and Extended Data Fig. 2). Further experiments explored how the number of artificial features influenced FDP+ computation. Results indicated that increasing artificial features improved FDP+(θ) estimation, notably with more than 500 artificial features (Extended Data Fig. 3). These observations experimentally confirmed Stabl’s validity in optimizing the frequency threshold for feature selection. Furthermore, under the assumption of feature exchangeability between uninformative and artificial features, we bound the probability that FDP exceeds a multiple of the proximity to FDP+(θ), thus providing a theoretical validation of our experimental observations (see ‘Theoretical guarantee’ in Methods).
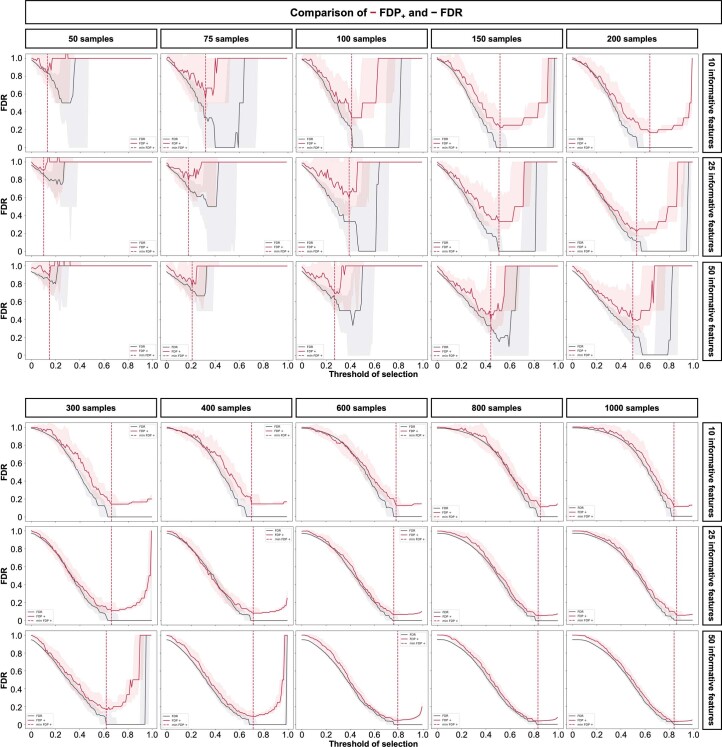
Extended Data Fig. 2. Comparison of FDP+ and FDR in synthetic dataset benchmarking.
On the generated synthetic dataset, the FDP+ and the true FDR were assessed for different dataset sizes ranging from n = 50 to 1000 samples with 10 (upper panels), 25 (middle panels), or 50 (lower panels) informative features. The FDP+ (red line) and the true FDR (black line) are shown as a function of the frequency threshold. The selected reliability threshold (θ, red dotted line) varied across conditions. Boxes in box plots indicate the median and interquartile range (IQR), with whiskers indicating 1.5 × IQR.
Extended Data Fig. 3. Effect of varying numbers of artificial features on the computation of FDP+.
On the generated synthetic dataset, the FDP+ and the true FDR were assessed for a varying number of artificial features on a dataset of n = 200 samples and 10 (upper panels), 25 (middle panels), or 50 (lower panels) informative features within p = 1000 features. The FDP+ (red line) and the true FDR (black line) are shown as a function of the frequency threshold. Increasing the number of artificial features allows for a more accurate estimation of the reliability threshold (θ, red dotted line). Boxes in box plots indicate the median and interquartile range (IQR), with whiskers indicating 1.5 × IQR.
Benchmarking against Lasso and SS
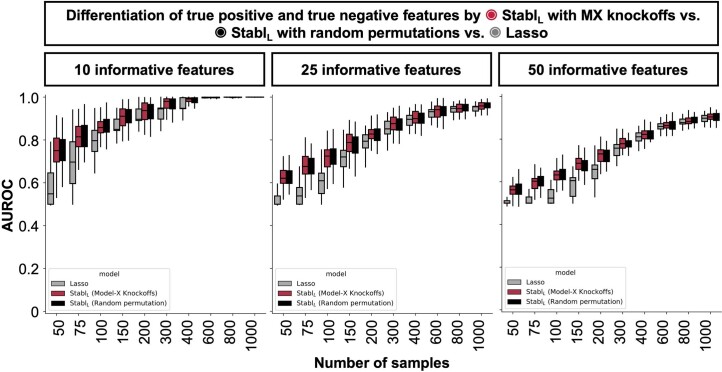
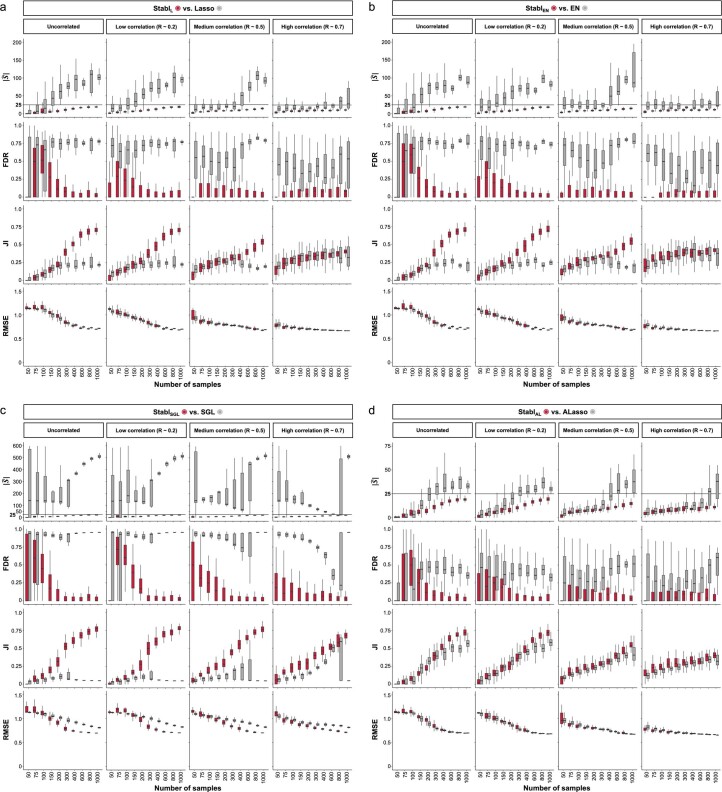
StablSRM was first benchmarked against Lasso using normally distributed, uncorrelated data for regression tasks, incorporating MX knockoffs as artificial features (Fig. 2d–g and Extended Data Fig. 4). StablL consistently achieved greater sparsity compared to Lasso by selecting fewer features across all conditions tested, converging toward the true number of informative features (Fig. 2d). StablL also achieved better reliability compared to Lasso, as evidenced by lower FDR (Fig. 2e) and higher JI (increased overlap with the true informative feature set) (Fig. 2f). Moreover, StablL’s feature selection frequency better distinguished true positives from true negatives, enhancing accuracy, as measured by the area under the receiver operating characteristic (AUROC) curve, compared to Lasso coefficients, thus providing an additional metric for estimating reliability (Extended Data Fig. 5). Notably, StablL and Lasso exhibited similar predictivity (Fig. 2g).
Extended Data Fig. 4. StablL’s performance on synthetic data with varying number of total features compared to Lasso.
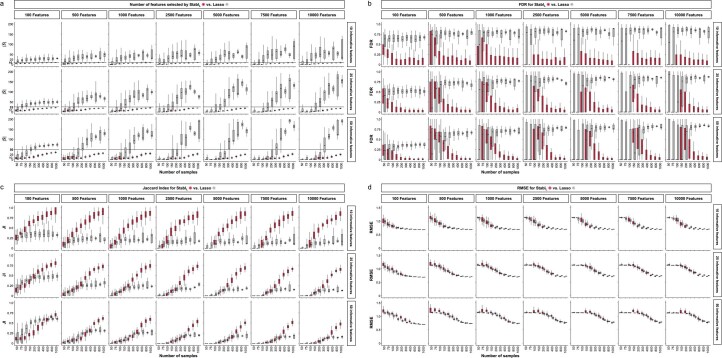
Synthetic datasets differing in the number of features were generated as described in Fig. 2. Sparsity (∣Ŝ∣, a) reliability (FDR, b, and JI, c), and predictivity (RMSE, d) of StablL (red box plots) and Lasso (grey box plots) as a function of the number of samples (n, x-axis) for 10 (left), 25 (middle), or 50 (right) informative features within p = 100, 500, 1000, 2500, 5000, 7500, and 10000 total number of features. Boxes in box plots indicate the median and interquartile range (IQR), with whiskers indicating 1.5 × IQR.
Extended Data Fig. 5. Reliability performance of selection frequency (StablL) and beta coefficients (Lasso) to distinguish true positive and true negative features.
Beta coefficients assigned by Lasso and feature selection frequency assigned by Stabl were used to distinguish true positive and true negative features in a synthetic dataset with p = 1000 total features. The AUROC for this procedure is shown as a function of the number of samples (n, x-axis) for 10 (left panels), 25 (middle panels), or 50 (right panels) informative features. Boxes in box plots indicate the median and interquartile range (IQR), with whiskers indicating 1.5 × IQR.
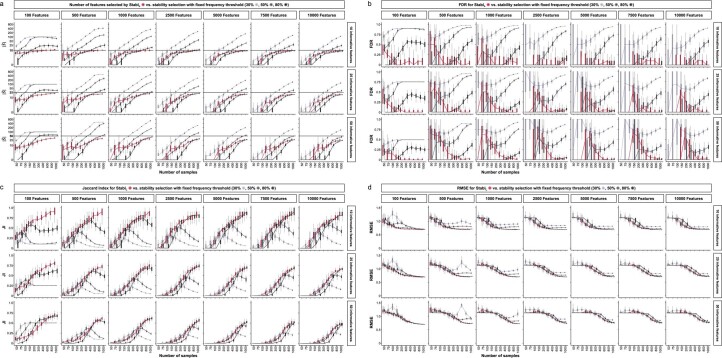
We then assessed the impact of data-driven θ computation in comparison to SS, which relies on a fixed frequency threshold chosen a priori. Three representative frequency thresholds were evaluated: 30%, 50% or 80% (Extended Data Fig. 6). The choice of threshold greatly affected model performance depending on the simulation conditions: the 30% threshold yielded the highest sparsity and reliability with smaller sample sizes (n < 75), whereas the 80% threshold resulted in superior performances with larger sample sizes (n > 500). In contrast, StablL systematically reached optimal sparsity, reliability and predictivity. To generalize the comparative analysis of SS and StablL, we coupled SS with a grid search method to find the optimal feature selection threshold (Fig. 2h–k). The analysis demonstrated that the grid search-coupled SS method produced models with more features and greater variability in feature selection compared to StablL. Furthermore, StablL consistently improved reliability (lower FDR) at similar predictive performance compared to the grid search-coupled SS method. We also show that StablL’s θ varied greatly with sample size (Fig. 2l), illustrating its adaptive ability to identify an optimal frequency threshold solution across datasets of different dimensions.
Extended Data Fig. 6. StablL’s performance on synthetic data with varying number of total features compared to SS with fixed frequency thresholds.
Synthetic datasets differing in the number of total features were generated as described in Fig. 2. Sparsity (∣Ŝ∣, a), reliability (FDR, b, and JI, c), and predictivity (RMSE, d) of StablL (red lines) and stability selection with fixed frequency threshold of 30% (light grey lines), 50% (dark grey lines), or 80% (black lines) as a function of the number of samples (n, x-axis) for 10 (left), 25 (middle), or 50 (right) informative features within p = 100, 500, 1000, 2500, 5000, 7500, and 10000 total number of features. Boxes in box plots indicate the median and interquartile range (IQR), with whiskers indicating 1.5 × IQR.
Extension of StablSRM to multi-omic synthetic datasets
Finally, experiments were performed simulating integration of multiple omic datasets. Unlike the early-fusion method, which concatenates all omic data layers before applying a statistical learner, Stabl adopts an independent analysis approach, fitting specific reliability thresholds for each omic data layer before selecting the most reliable features to merge into a final layer. Consequently, StablL was benchmarked against Lasso using the comparable late-fusion method, wherein a model is trained on each omic dataset independently before merging the predictions into a final dataset (Extended Data Fig. 1)11,12. The results show that StablL improved the sparsity and reliability of integrated multi-omic models compared to late-fusion Lasso at a similar predictive performance (Supplementary Table 3).
In sum, synthetic modeling results show that StablL achieves better sparsity and reliability compared to Lasso while preserving predictivity and that StablL’s feature selection aligns more closely with the true set of informative features. These findings underscore the advantage of data-driven adaptation of the frequency threshold to each dataset’s unique characteristics, as opposed to relying on arbitrarily pre-determined thresholds.
Generalization to other sparse learners and distributions
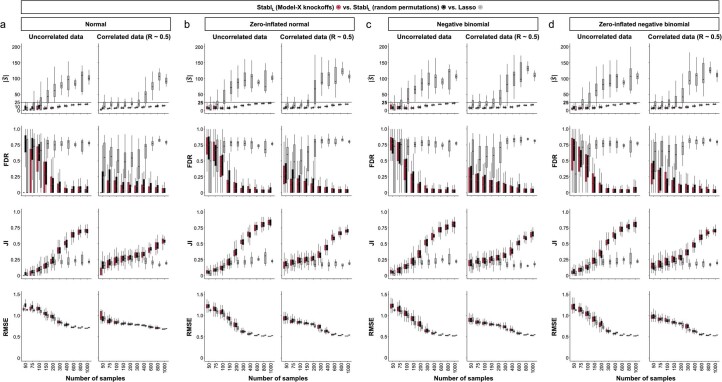
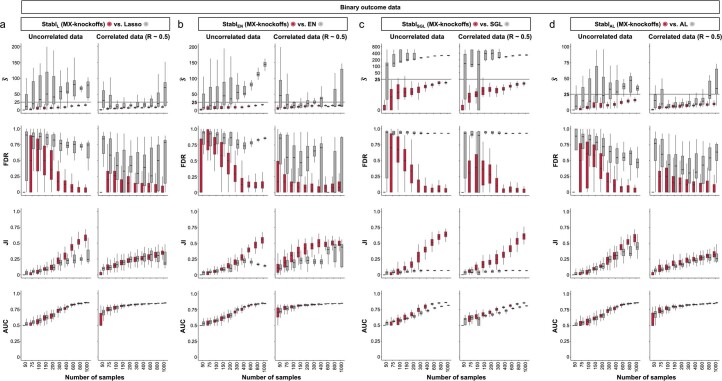
A notable benefit of Stabl is the modularity of the statistical framework, enabling the use of different SRMs as base learners and different noise generation techniques (Methods). This modularity enables customization for datasets with various correlation structures, where specific SRMs may outperform Lasso. We conducted synthetic modeling experiments comparing SRM substitutions within the StablSRM framework to their cognate SRM, including EN, SGL or AL (Fig. 3 and Extended Data Fig. 7). We also explored different feature distributions (normal, zero-inflated normal, negative binomial and zero-inflated negative binomial; Methods and Extended Data Fig. 8) and prediction tasks (regression (Fig. 3 and Extended Data Fig. 7) and classification (Extended Data Fig. 9 and Supplementary Table 2)). Synthetic datasets with S = 25 informative features, p = 1,000 total features and n ranging from 50 to 1,000 samples were used for these experiments.
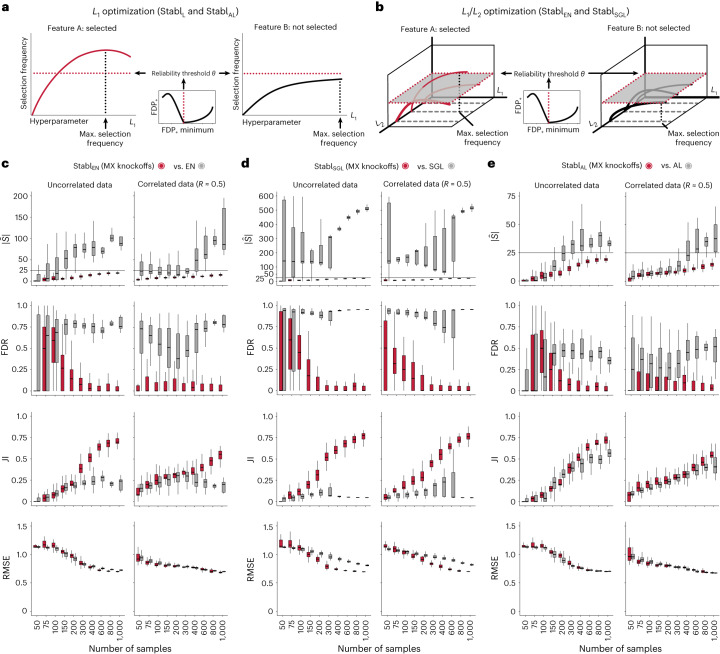
Fig. 3. Extension of the StablSRM framework to EN, SGL and AL: synthetic dataset benchmarking.
The StablSRM framework is benchmarked against various SRMs, including EN (StablEN), SGL (StablSGL) and AL (StablAL), respectively. a,b, Diagrams depict the strategy for identifying the maximum selection frequency for each feature across one (L1 for Lasso and AL, a) or two (L1/L2 for EN and SGL, b) regularization parameters before minimizing the FDP+. c–e, Sparsity (), reliability (FDR and JI) and predictivity (RMSE) performances of StablSRM (red box plots) are compared to their respective SRM (gray box plots) in n = 50 independent experiments for each number of samples for StablEN (c), StablSGL (d) and StablAL (e). Synthetic modeling experiments performed on normally distributed datasets containing S = 25 informative features with uncorrelated (left panels) or intermediate correlation structures (right panels) are shown. For all correlated datasets, the target correlation between informative features is set at a Pearson correlation coefficient, R, of 0.5, yielding a covariance matrix with approximately the target correlation (R ≈ 0.5). Results with low or high correlation structures are shown in Extended Data Fig. 7. Performances are shown for regression tasks. Results for classification tasks are shown in Supplementary Table 10. Box plots indicate median and IQR; whiskers indicate 1.5× IQR.
Extended Data Fig. 7. Stabl’s performance on synthetic data with different correlation structures.
Synthetic datasets differing in correlation structure (low, medium, or high) were generated as described in Fig. 3. Sparsity (∣Ŝ∣, upper panels), reliability (FDR and JI, middle panels), and predictivity performances (AUROC, lower panels) for StablL (a), StablEN (b), StablSGL (c), and StablAL (d) (red box plots) and Lasso (grey box plots) as a function of the number of samples (n, x-axis) for 10 (left panels), 25 (middle panels), or 50 (right panels) informative features. Boxes in box plots indicate the median and interquartile range (IQR), with whiskers indicating 1.5 × IQR.
Extended Data Fig. 8. StablL’s performance with MX knockoffs or random permutations on synthetic data with normal and non-normal distributions compared to Lasso.
Synthetic datasets differing in distribution were generated using the Normal to Anything (NORTA) framework, as described in methods. Sparsity (∣Ŝ∣, upper panels), reliability (FDR and JI, middle panels), and predictivity performances (RMSE, lower panels) of StablL (MX knockoffs, red box plots, or random permutations, black box plots), and Lasso (grey box plots) as a function of the number of samples (n, x-axis) for synthetic data with a normal distribution (a), zero-inflated normal distribution (b), negative binomial distribution (c), or zero-inflated negative binomial distribution (d). The results are shown for datasets with 25 informative features in the context of uncorrelated (left panels) or correlated (right panels, intermediate correlation, R ~ 0.5) data for regression tasks (continuous outcomes). Results obtained for other scenarios, including other SRMs (EN, SGL, and AL), correlation structures (low, R ~ 0.2, high, R ~ 0.7), and classification tasks are listed in Table S2. Boxes in box plots indicate the median and interquartile range (IQR), with whiskers indicating 1.5 × IQR.
Extended Data Fig. 9. Stabl’s performance on synthetic data with binary outcomes.
Synthetic datasets with binary outcome variables were generated as described in Fig. 3. Sparsity (∣Ŝ∣), reliability (FDR and JI), and predictivity (RMSE) performances of StablSRM (red box plots) compared to the respective SRM (grey box plots) as a function of the sample size (n, x-axis) for StablL (a), StablEN (b), StablSGL (c), and StablAL (d). Scenarios with 25 informative features and uncorrelated (left panels) or intermediate feature correlation structures (Spearman R ~ 0.5, right panels) are shown. Boxes in box plots indicate the median and interquartile range (IQR), with whiskers indicating 1.5 × IQR.
Lasso encounters challenges with correlated data structures9,24, often favoring one of two correlated covariates. EN mitigates this by introducing ℓ2 regularization, encouraging consideration of multiple correlated features. Similarly, SGL handles correlated data with known groupings or clusters, by introducing a combination of between-group and within-group sparsity.
To integrate SRMs with multiple regularization hyperparameters (for example, ℓ1/ℓ2 for EN and SGL), StablSRM extends the identification of the maximum selection frequency of each feature to a multi-dimensional space (Fig. 3a,b and Methods). Further simulation experiments benchmarked StablEN against EN across low (R ≈ 0.2), intermediate (R ≈ 0.5) and high (R ≈ 0.7) Spearman correlations and StablSGL against SGL in datasets containing known groups of correlated features (defined in Methods). Here, MX knockoff was used as it preserves the correlation structure of the original dataset (Extended Data Fig. 1)25. For low or intermediate correlation structures, StablEN and StablSGL selected fewer features with improved JI and FDR and similar predictivity compared to EN or SGL (Fig. 3c,d and Extended Data Fig. 7). In highly correlated datasets (Extended Data Fig. 7), the JI for StablEN and StablSGL paralleled that of EN and SGL, respectively, but with lower FDR across all correlation levels. This suggests that, whereas EN or SGL may achieve a similar JI to StablEN or StablSGL, they do so at the expense of selecting more uninformative features.
Other SRMs offer advantages beyond adapting to different correlation structures. For example, AL, an extension of Lasso that demonstrates the oracle property9, ensures accurate identification of informative features as the sample size approaches infinity. Compared to AL, integrating AL within the Stabl framework (StablAL) resulted in fewer selected features, lower FDR and overall improved JI, especially evident with increasing sample sizes (Fig. 3e and Extended Data Fig. 7). For experiments with normally distributed, uncorrelated data, although AL had a higher JI compared to StablAL in two out of 10 cases (sample sizes n = 150 and n = 200), StablAL exhibited lower FDR for these sample sizes and beyond. These findings indicate that StablAL improves the selection of informative features compared to AL, offering an advantageous approach, especially in the context of biomarker discovery studies with large sample sizes.
Stabl enables biomarker discovery in omic studies
We evaluated Stabl’s performance on five distinct clinical omic datasets, encompassing various dimensions, signal-to-noise ratios, data structures, technology-specific pre-processing and predictive performances. Four were previously published with standard SRM analyses, whereas the fifth is a newly generated dataset. These datasets spanned bulk and single-cell omic technologies, including RNA sequencing (RNA-seq) (comprising cell-free RNA (cfRNA) and microbiome datasets), high-content proteomics, untargeted metabolomics and single-cell mass cytometry. To ensure broad applicability, we tested different StablSRM variations using three base SRMs (Lasso, EN and AL) benchmarked against their respective SRM. To preserve the original data’s correlation structure, we primarily employed MX knockoffs for introducing noise across all omic datasets, except for the cfRNA dataset. This dataset exhibited the lowest internal correlation levels (with <1% of features displaying intermediate correlations, R > 0.5; Supplementary Table 4), prompting the use of random permutation as the noise generation approach.
In contrast to synthetic datasets, the true set of informative features is unknown in real-world datasets, precluding an assessment of true reliability performance. Consequently, we employed distinct performance metrics:
Sparsity: representing the average number of features selected throughout the cross-validation (CV) procedure
Predictivity: assessed through the AUROC for classification tasks or the RMSE for regression tasks
Model performances were evaluated over 100 random repetitions using a repeated five-fold or Monte Carlo CV strategy.
Sparse, reliable biomarker discovery from single-omic data
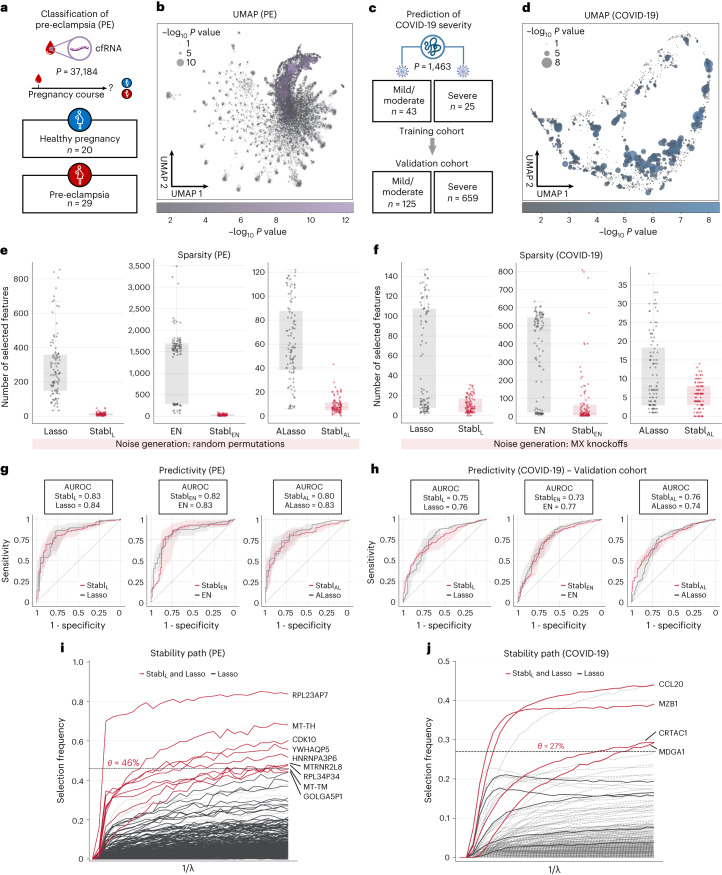
StablSRM was first applied to two single-omic clinical datasets. The first study comprised a large-scale plasma cfRNA dataset (p = 37,184 features) and aimed to classify pregnancies as either normotensive or pre-eclamptic (PE) (Fig. 4a,b)26,27. The second study, involving high-plex plasma proteomics (p = 1,463 features, Olink Explore 1536 assay), aimed to classify coronavirus disease 2019 (COVID-19) severity in two independent cohorts (a training cohort and a validation cohort) of patients positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Fig. 4c,d)28,29. Although both Lasso and EN models achieved very good predictive performance (AUROC = 0.74–0.84) in these examples, suggesting that they have a robust biological signal with diagnostic potential30,31, the lack of model sparsity or reliability hindered the identification of a manageable number of candidate biomarkers, necessitating additional feature selection methods that were decoupled from the predictive modeling process26–29.
Fig. 4. Stabl’s performance on transcriptomic and proteomic data.
a, Clinical case study 1: classification of individuals with normotensive pregnancy or PE from the analysis of circulating cfRNA sequencing data. The number of samples (n) and features (p) are indicated. b, UMAP visualization of the cfRNA transcriptomic features; node size and color are proportional to the strength of the association with the outcome. c, Clinical case study 2: classification of mild versus severe COVID-19 in two independent patient cohorts from the analysis of plasma proteomic data (Olink). d, UMAP visualization of the proteomic data. Node characteristics as in b. e,f, Sparsity performances (the number of features selected across n = 100 CV iterations, median and IQR) on the PE (e) and COVID-19 (f) datasets for StablL (left), StablEN (middle) and StablAL (right). g,h, Predictivity performances (AUROC, median and IQR) on the PE (g) and COVID-19 (h, validation set; training set shown in Supplementary Table 5) datasets for StablL (left), StablEN (middle) and StablAL (right). StablSRM performances are shown using random permutations for the PE dataset and MX knockoffs for the COVID-19 dataset. Median and IQR values comparing StablSGL performances to the cognate SRM are listed numerically in Supplementary Table 5. Results in the COVID-19 dataset using random permutations are also shown for StablL in Supplementary Table 5. i,j, StablL stability path graphs depicting the relationship between the regularization parameter and the selection frequency for the PE (i) and COVID-19 (j) datasets. The reliability threshold (θ) is indicated (dotted line). Features selected by StablL (red lines) or Lasso (black lines) are shown. Significance between outcome groups was calculated using a two-sided Mann–Whitney test. Box plots indicate median and IQR; whiskers indicate 1.5× IQR.
Consistent with the results obtained using synthetic data, StablL, StablEN and StablAL demonstrated improved sparsity compared to Lasso, EN and AL, respectively (Fig. 4e,f and Supplementary Table 5). For the PE dataset, StablSRM selected over 20-fold fewer features compared to Lasso or EN and eight-fold fewer compared to AL (Fig. 4e). For COVID-19 classification, StablSRM reduced the number of features by factors of 1.9, >20 and 1.25 for Lasso, EN and AL, respectively (Fig. 4f). Remarkably, StablL, StablEN and StablAL maintained similar predictive performance to their respective SRMs on both datasets (Fig. 4g,h) despite this favorable feature reduction.
Comparing StablL to SS using fixed frequency thresholds (30%, 50% and 80%; Supplementary Table 6) revealed that SS’s predictivity and sparsity performances varied widely based on the chosen threshold, consistent with synthetic modeling findings, whereas StablL consistently optimized sparsity while preserving predictive performance. For example, using SS with a 30% versus a 50% threshold resulted in a 42% decrease in predictivity for the COVID-19 dataset (AUROC30% = 0.85 versus AUROC50% = 0.49), with a model selecting no features. Conversely, for the PE dataset, fixing the frequency threshold at 30% versus 50% yielded a 5.3-fold improvement in sparsity with only a 6% decrease in predictivity (AUROC30% = 0.83 versus AUROC50% = 0.78).
Stabl’s ability to identify fewer, more reliable features streamlined biomarker discovery, pinpointing the most informative biological features associated with the clinical outcome. For simplicity, biological interpretation of predictive model features is provided in the context of the StablL analyses (Fig. 4i,j and Supplementary Tables 7 and 8). For example, the StablL model comprised nine features, including cfRNAs encoding proteins with fundamental cellular function (for example, CDK10 (ref. 32)), providing biologically plausible biomarker candidates. Other features included non-coding RNAs and pseudogenes with yet unknown functions (Fig. 4i). For the COVID-19 dataset, StablL identified features that echoed key pathobiological mechanisms of the host’s inflammatory response, such as CCL20, a known element of the COVID-19 cytokine storm33,34; CRTAC1, a newly identified marker of lung function35–37; and MZB1, a protein associated with high neutralization antibody titers after COVID-19 infection (Fig. 4j)28. The StablL model also selected MDGA1, a previously unknown candidate biomarker of COVID-19 severity.
Application of StablSRM to multi-omic clinical datasets
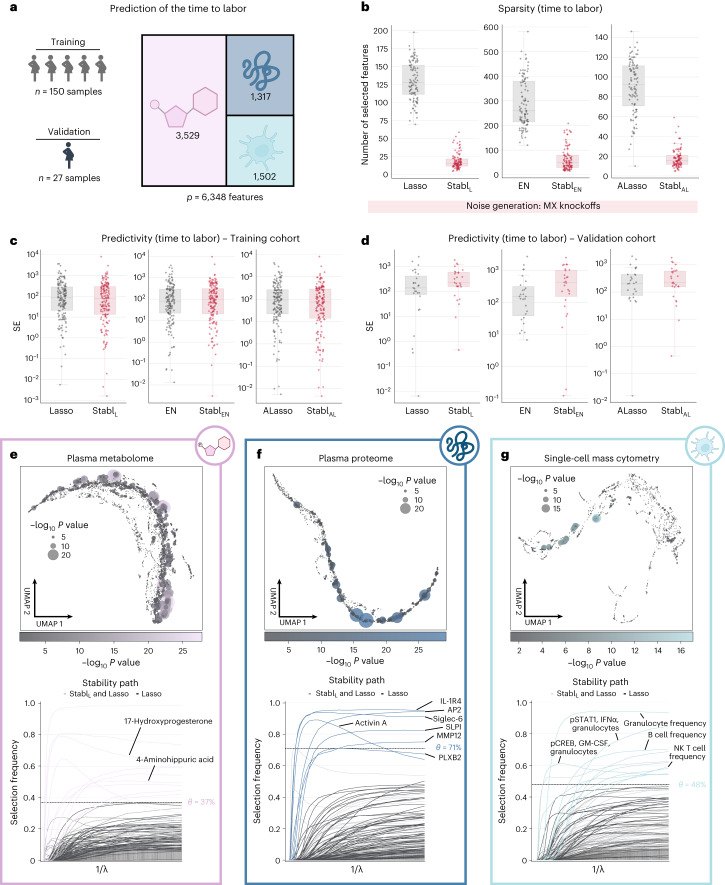
We extended the assessment of Stabl to complex clinical datasets combining multiple omic technologies, comparing StablL, StablEN and StablAL to late-fusion Lasso, EN and AL, respectively, for predicting a continuous outcome variable from a triple-omic dataset and a binary outcome variable from a double-omic dataset.
The first analysis leveraged a unique longitudinal biological dataset collected in independent training and validation cohorts of pregnant individuals (Fig. 5a)38, aiming to predict the time to labor onset, an important clinical need39,40. The triple-omic dataset included plasma proteomics (p = 1,317 features, SomaLogic), metabolomics (p = 3,529 untargeted mass spectrometry features) and single-cell mass cytometry (p = 1,502 immune cell features) (Methods). Relative to late-fusion Lasso, EN or AL, the StablL, StablEN and StablAL models selected fewer features (Fig. 5b) while estimating the time to labor with similar predictivity (training and validation cohorts; Fig. 5c,d). StablSRM calculated a unique reliability threshold for each omic layer (for example, θ[Proteomics] = 71%, θ[Metabolomics] = 37% and θ[mass cytometry] = 48%, for StablL; Fig. 5e–g). These results emphasize the advantage of data-driven thresholds, as a fixed, common frequency threshold across all omic layers would have been suboptimal, risking over-selecting or under-selecting features in each omic dataset for integration into the final predictive model.
Fig. 5. Stabl’s performance on a triple-omic data integration task.
a, Clinical case study 3: prediction of the time to labor from longitudinal assessment of plasma proteomic (SomaLogic), metabolomic (untargeted mass spectrometry) and single-cell mass cytometry data in two independent cohorts of pregnant individuals. b, Sparsity performances (number of features selected across CV iterations, median and IQR) for StablL (left), StablEN (middle) and StablAL (right) compared to their respective SRM (late-fusion data integration method) across n = 100 CV iterations. c,d, Predictivity performances as squared error (SE) on the training (n = 150 samples, c) and validation (n = 27 samples, d) datasets for StablL (left), StablEN (middle) and StablAL (right). StablSRM performances are shown using MX knockoffs. Results using random permutations are shown for StablL in Supplementary Table 5. Median and IQR values comparing StablSRM performances to their cognate SRMs are listed in Supplementary Table 5. e–g, UMAP visualization (upper) and stability path (lower) of the metabolomic (e), plasma proteomic (f) and single-cell mass cytometry (g) datasets. UMAP node size and color are proportional to the strength of association with the outcome. Stability path graphs denote features selected by StablL. The data-driven reliability threshold θ is computed for each individual omic dataset and is indicated by a dotted line. Significance of the association with the outcome was calculated using Pearson’s correlation. Box plots indicate median and IQR; whiskers indicate 1.5× IQR.
From a biological perspective, Stabl streamlined the interpretation of our previous multivariable analyses38, honing in on sentinel elements of a systemic biological signature predicting labor onset, valuable for developing a blood-based diagnostic test. The Stabl model highlighted dynamic changes in 10 metabolomic, seven proteomic and 10 immune cell features with approaching labor (Fig. 5e–g and Supplementary Table 9), including a regulated decrease in innate immune cell frequencies (for example, neutrophils) and their responsiveness to inflammatory stimulation (for example, the pSTAT1 signaling response to IFNα in natural killer (NK) cells41,42), along with a synchronized increase in pregnancy-associated hormones (for example, 17-hydroxyprogesterone43), placental-derived proteins (for example, Siglec-6 (ref. 44) and angiopoietin 2/sTie2 (ref. 45)) and immune regulatory plasma proteins (for example, IL-1R4 (ref. 46) and SLPI47 (ref. 47)).
The use cases provided thus far featured models with good to excellent predictive performance. Stabl was also tested on a dataset where previous models did not perform as well (AUROC < 0.7). The Microbiome Preterm Birth DREAM challenge aimed to classify pre-term (PT) and term (T) labor pregnancies using nine publicly available vaginal microbiome (phylotypic and taxonomic) datasets48,49. The top 20 models submitted by 318 participating analysis teams achieved AUROC scores between 0.59 and 0.69 for the task of predicting PT delivery. When applied to a subset of this dataset (n = 1,569 samples, 609 T and 960 PT deliveries), StablL and StablEN achieved better sparsity at similar predictive performance compared to late-fusion Lasso and EN (Supplementary Table 5).
Identifying promising candidate biomarkers from a new multi-omic dataset
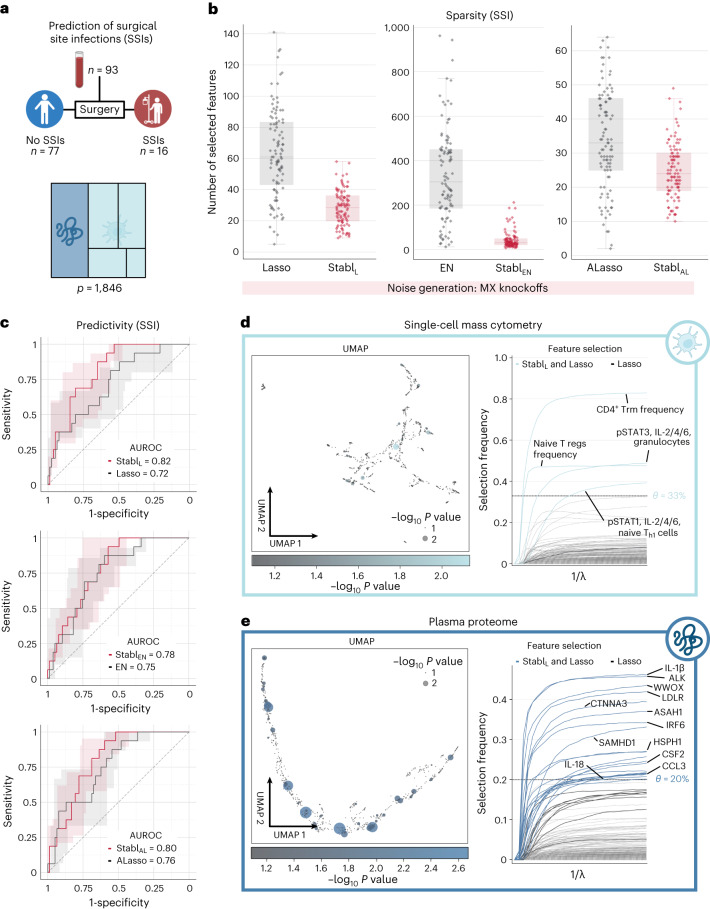
Application of Stabl to the four existing omic datasets demonstrated the algorithm’s performance in biomarker discovery studies with known biological signal. To complete its systematic evaluation, Stabl was applied to our multi-omic clinical study performing an unbiased biomarker discovery task. The aim was to develop a predictive model for identifying patients at risk for post-operative surgical site infection (SSI) from analysis of pre-operative blood samples collected from 274 enrolled patients (Fig. 6a). Using a matched, nested case–control design, 93 patients were selected from the larger cohort to minimize the influence of clinical or demographic confounders on identified predictive models (Supplementary Table 10). These samples were analyzed using a combined single-cell mass cytometry (Extended Data Fig. 10 and Supplementary Table 11) and plasma proteomics (SomaLogic) approach.
Fig. 6. Candidate biomarker identification using Stabl for analysis of a newly generated multi-omic clinical dataset.
a, Clinical case study 5: prediction of post-operative SSIs from combined plasma proteomic and single-cell mass cytometry assessment of pre-operative blood samples in patients undergoing abdominal surgery. b, Sparsity performances (the number of features selected across n = 100 CV iterations) for StablL (left), StablEN (middle) and StablAL (right) compared to their respective SRMs (late-fusion data integration method). c, Predictivity performances (AUROC) for StablL (upper), StablEN (middle) and StablAL (lower). StablSRM performances are shown using MX knockoffs. Results using random permutations are shown in Supplementary Table 5. Median and IQR values comparing StablSRM performances to their cognate SRMs are listed in Supplementary Table 5. d,e. UMAP visualization (left) and stability path (right) of the mass cytometry (d) and plasma proteomic (e) datasets. UMAP node size and color are proportional to the strength of association with the outcome. Stability path graphs denote features selected by StablL. The data-driven reliability threshold θ is computed for individual omic datasets and indicated by a dotted line. Significance of the association with the outcome was calculated using a two-sided Mann–Whitney test. Box plots indicate median and IQR; whiskers indicate 1.5× IQR.
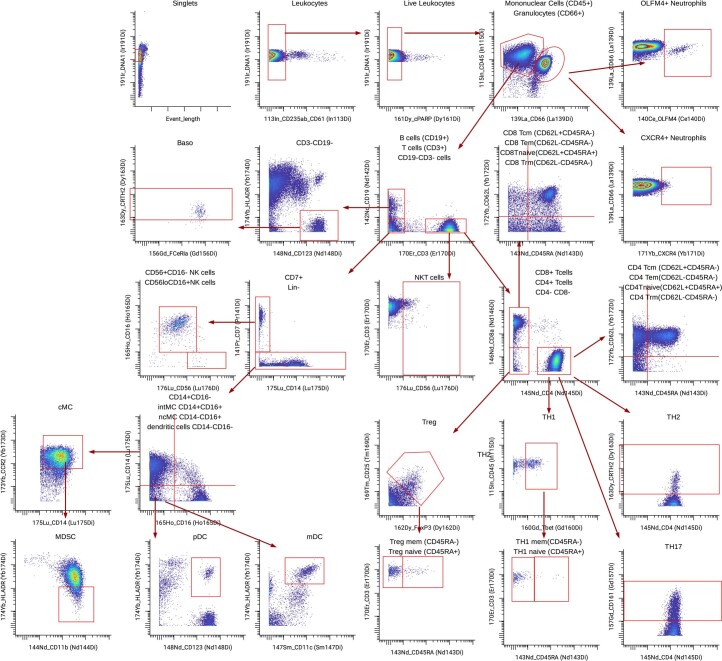
Extended Data Fig. 10. Gating strategy for mass cytometry analyses (SSI dataset).
Live, non-erythroid cell populations were used for analysis.
Stabl merged all omic datasets into a final model that accurately classified patients with and without SSI (StablL: AUROC = 0.82 (0.71, 0.90); StablEN: AUROC = 0.78 (0.68, 0.88); and StablAL: AUROC = 0.80 (0.70, 0.89)). Compared to late-fusion Lasso, EN and AL, StablL, StablEN and StablAL had superior sparsity performances (Fig. 6b) yet similar predictive performances (Fig. 6c). The frequency-matching procedure ensured that major demographic and clinical variables did not differ significantly between patient groups, suggesting that model predictions were primarily driven by pre-operative biological differences in patients’ SSI susceptibility.
StablL selected four mass cytometry and 21 plasma proteomic features, combined into a biologically interpretable immune signature predictive of SSI. Examination of StablL features unveiled cell-type-specific immune signaling responses associated with SSI (Fig. 6d), which resonated with circulating inflammatory mediators (Fig. 6e and Supplementary Table 12). Notably, the model revealed elevated STAT3 signaling response to IL-6 in neutrophils before surgery in patients predisposed to SSI. Correspondingly, patients with SSI had increased plasma levels of IL-1β and IL-18, potent inducers of IL-6 production in response to inflammatory stress50,51. Other selected proteomic features included CCL3, which coordinates recruitment and activation of neutrophils, and the canonical stress response protein HSPH1. These findings concur with previous studies indicating that heightened innate immune cell responses to inflammatory stress, such as surgical trauma52,53, can result in diminished defensive responses to bacterial pathogens39, increasing susceptibility to subsequent infection.
Altogether, application of Stabl in a biomarker discovery study provided a manageable number of candidate SSI biomarkers, pointing at plausible biological mechanisms that can be targeted for further diagnostic or therapeutic development.
Discussion
Stabl is a machine learning framework developed to facilitate clinical translation of high-dimensional omic biomarker studies. Through artificial noise injection and minimization of a proxy for FDP, Stabl enables data-driven selection of sparse and reliable biomarker candidates within a multivariable predictive modeling architecture. The modular framework of Stabl allows for customization across various SRMs and noise injection techniques, catering to the specific requirements of individual studies. When applied to real-world biomarker discovery tasks spanning different omic technologies, single-omic and multi-omic datasets and clinical endpoints, Stabl consistently demonstrates its adaptability and effectiveness in reliable selection of biologically interpretable biomarker candidates conducive to further clinical translation.
Stabl builds upon earlier methodologies, including SS and MX knockoff. These approaches aim to improve reliability of sparse learning algorithms by incorporating bootstrapping or artificial features7,18,20,22. However, they typically rely on fixed or user-defined frequency thresholds to distinguish informative from uninformative features. In practical scenarios where p ≫ n, determining the optimal frequency threshold without prior data knowledge is challenging, as illustrated by our synthetic modeling results. This reliance on prior knowledge limits these methods to feature selection only.
Stabl improves on these methodologies by experimentally and, under certain assumptions, theoretically extending FDR control techniques devised for MX knockoff and random permutation noise19,54,55. Minimizing the FDP+ offers two key advantages: it balances the tradeoff between reliability and sparsity by combining an increasing and decreasing function of the threshold, and, assuming exchangeability between artificial and uninformative features, it guarantees a stochastic upper bound on FDP using the reliability threshold, ensuring reliability during the optimization procedure. By minimizing this function ex-ante, Stabl objectively defines a model fit without requiring prior data knowledge.
Experimental results on synthetic datasets demonstrate Stabl’s ability to select an optimal reliability threshold by minimizing FDP+, leading to improved reliability and sparsity compared to popular SRMs such as Lasso, EN, SGL or AL, all while maintaining similar predictivity performance. These findings hold across different data distributions, correlation structures and prediction tasks. When applied to real-world omic studies, Stabl consistently performs favorably compared to other SRMs. In each case study, identification of a manageable number of reliable biomarkers facilitated the interpretation of the multivariable predictive models. Stabl embeds the discovery of reliable candidate biomarkers within the predictive modeling process, eliminating the need for separate analyses that risk overfitting, such as post hoc analyses with user-defined cutoffs after the initial model fitting or the selection of clinical endpoint-associated features before modeling.
Stabl’s versatility extends to multi-omic datasets, offering an alternative that avoids the potential shortcomings of early-fusion and late-fusion strategies. Although early fusion combines all omic data layers for joint optimization, regardless of each dataset’s unique properties, and late fusion independently fits models for each omic before integrating predictions without weighing features from different omics against each other11,12, Stabl computes a distinct reliability threshold for each omic layer, tailoring its approach to the specific dataset. This enables integration of selected features into a final modeling layer, a capability that was particularly useful for analysis of our dataset involving patients undergoing surgery. Stabl identified a patient-specific immune signature spanning both plasma and single-cell datasets that appears to be programmed before surgery and predictive of SSIs.
Our study has limitations. The assumption of exchangeability between artificial and uninformative features underpins our theoretical guarantee, which builds on a recent line of research focused on constructing artificial features to establish control over the FDR19,21,23,54–56. Hence, Stabl’s validity hinges on the accuracy of the artificial feature generation technique. Future efforts will investigate relaxing the exchangeability assumption by exploring pairwise exchangeability settings to accommodate a wider range of data scenarios where complete exchangeability may not hold19. Additionally, improving knockoff generation methods, such as deep knockoff57 and metropolized knockoff25, may enhance the robustness and flexibility of our approach in handling diverse data distributions and structures. We also observed that Stabl can be overly conservative. However, Stabl is designed to optimize reliability, sparsity and predictivity performances simultaneously, which can result in feature under-selection when only a subset of informative features is sufficient for optimal predictive performance. Other algorithms addressing these performance tasks individually, such as double machine learning58 for reliability, Boruta59 for sparsity and random forest60 or gradient boosting61 for predictivity, warrant further evaluation to systematically investigate each method’s performance in comparison to, or integrated with, the Stabl statistical framework. Finally, integrating emerging algorithms for multi-omic data, such as cooperative multiview learning11, may further enhance Stabl’s capabilities in multi-omic modeling tasks.
Analysis of high-dimensional omic data has transformed biomarker discovery, necessitating adjustments to machine learning methods to facilitate clinical translation. Stabl addresses key requirements of an effective biomarker discovery pipeline by offering a unified supervised learning framework that bridges the gap between predictive modeling of clinical endpoints and selection of reliable candidate biomarkers. Across diverse real-world single-omic and multi-omic datasets, Stabl identified biologically meaningful biomarker candidates, providing a robust machine learning pipeline that holds promise for generalization across all omic data.
Methods
Notations
Given a vector of outcomes and a matrix of covariates , where n denotes the number of observations (sample size) and p denotes the number of covariates (features) in each sample. We are interested in estimating parameters , within the linear model:
Here, ε is an unknown noise vector, which is centered and independent of X.
We denote the columns of X by and the entries of Y by y1, …, yn. We denote by S ≔ {i ∈ [p]: βi ≠ 0} the set of informative features and by N ≔ {i ∈ [p]: βi = 0} the set of uninformative features. Throughout [m] ≔ {1, …, m} is the set of first m integers, and ∣A∣ is the cardinality of a set A.
Our main objective is to estimate S, and we will generally denote by an estimator of this set. Given coefficient estimates , an estimate of S can be constructed using the support of . We will denote this by .
Lasso, EN, AL and SGL
Motivated by omic application, our main focus is on the high-dimensional regime p ≫ n. Lasso is a regression method that uses an ℓ1-regularization penalty to yield sparse solutions7. Denoting with λ the regularization parameter, Lasso estimate is defined by:
For sparse linear models, and under suitable conditions on the design matrix X (for example, restricted isometry or restricted eigenvalue conditions), the Lasso is known to provide consistent estimates of β, for certain choices of λ (refs. 62–64). It is also known that the Lasso can yield consistent variable selection—that is, (refs. 65,66). However, variable selection consistency requires stronger conditions on X, such as the irrepresentability or the generalized irrepresentability condition65,67.
EN is a regression method that combines ℓ1-regularization and ℓ2-regularization penalties8. Denoting by λ1 and λ2 the regularization parameters of these two penalties, the EN estimate is defined by:
Although we will mostly focus on Lasso as our basic estimator, this can be replaced by EN or other sparse regression methods without much change to our overall methodology.
AL is a regression method based on the Lasso with adaptive weights to penalize different coefficients in the ℓ1 penalty differently. To define the model, first we need , a root-n-consistent estimator to β, and we can consider βOLS. Then, choose a γ > 0 and define . As such, denoting with λ the regularization parameter, the AL estimate is defined by:
The weighted Lasso above can be solved with the same algorithm used to solve the Lasso. With well-chosen weights and regularization parameters, AL also enjoys the oracle property9:
and , with Σ* the covariance matrix of the true subset model.
SGL extends the concept of sparse regression methods for problems with group covariates, with sparsity on both within and between groups in high-dimensional settings10. The SGL penalty can be formulated as:
where G is the number of groups, and pg denotes the number of covariates in group g. The first term in the objective function measures the data-fitting loss, whereas the second and third terms enforce sparsity at the individual feature level and group level, respectively.
SS
SS18 is a technique to improve variable selection in high-dimensional methods, including the Lasso. The algorithm uses Lasso on subsamples of the original data . At each iteration k ∈ {1, …, B}, a different subsample (Y, X)k of size ⌊n/2⌋ × (p + 1) is selected. Lasso is used to fit a linear model on (Y, X)k over a range of regularization parameters . This yields an estimate that we denote by:
After B iterations, it is possible, for any feature i and regularization parameter λ, to define a ‘frequency of selection’ fi measuring how often feature i was selected by Lasso:
Plotting fi as a function of 1/λ yields a ‘stability path’ for feature i. Plotting all stability paths on the same graph yields a ‘stability graph’. Denoting the ‘selection threshold’ by t ∈ (0, 1), selected features are those whose stability path fi(λ) crosses the line y = t. In other words, the set of stable features is defined as:
Notice that, in SS, t is arbitrary in that it has to be defined ex-ante. The threshold value is a tuning parameter whose influence is very small18. However, we observe that, in some cases, the results are sensitive to the chosen threshold, thereby motivating the development of a data-driven threshold optimization.
Stabl framework
Preliminaries
Our algorithm builds upon the framework of SS and provides a way to define a data-driven threshold by optimizing a surrogate for the FDP. We construct such a surrogate by introducing artificially generated features in the Lasso regression. We thus build upon a recent fruitful line of work that develops several constructions of such artificial features and establishes control of the FDR under varying assumptions23,54–56.
The general Stabl procedure can accommodate a variety of feature-generating procedures. In our implementation, we experimented with two specific constructions:
Stabl algorithm
The initial step of the Stabl procedure involves selecting a base SRM (for example, Lasso, AL, EN and SGL), in which case the procedure is denoted StablSRM. It runs as follows:
From the original matrix , we generate a matrix of artificial features of the same dimensions as the original matrix.
- We concatenate the original matrix X and the artificial matrix , and define:
All the following steps run using the matrix as input. We denote by the set of artificial features and by the set of original features. In the context of SGL, an extra layer of information regarding feature groupings is needed. Specifically, each feature requires supplementary information about its respective group assignment. To adapt the procedure to this requirement, each artificial feature is linked to the group of its original feature source. We fix B the number of subsampling iterations. At each iteration , a subsample of size ⌊n/2⌋ is drawn without replacement from , denoted by . The size of subsamples could be ⌊αn⌋ with α ∈ (0, 1). Selecting subsamples of size ⌊n/2⌋ most closely resembles the bootstrap while allowing computationally efficient implementation18.
- We use the base SRM to fit a model on data for different values of regularization parameters λ ∈ Λ. For models with only one penalization (Lasso and AL), . For models with two penalizations (EN and SGL), . For each set of hyperparameters λ (in the context of EN), beyond the conventional pursuit of the ℓ1-regularization parameter, we introduce three distinct options for determining the parameter that governs the equilibrium between ℓ1 and ℓ2 regularization. This results in the creation of a hyperparameter set, within which the maximum value is selected for each feature; this yields an estimate defined as:
- For each feature j, the maximum frequency of selection over Λ is computed. In the case of models with two hyperparameters (EN and SGL), this leads to a two-dimensional optimization.
- For a given frequency threshold t ∈ [0, 1], a feature j is selected if fj ≥ t. We define the augmented FDP at t by
The set of selected features at t is . - We define the reliability threshold as:
which results in a selected feature set . When multiple minimizers exist, we select one arbitrarily (but, in practice, we always found a unique minimizer). At θ, we achieve the following augmented FDP:1 2 - We obtain the final estimate for the Stabl model using:
Link with FDP and FDR
FDP and FDR68 are classical metrics to assess the quality of a model selection method. Consider a general method parameterized by a threshold t ∈ (0, 1) (for example, the stability threshold in our approach). For any fixed t, the method returns a selected subset of features , resulting in the FDP
Several approaches use a threshold that is dependent on the data. The resulting FDP is a random quantity at fixed and is also random because it is evaluated at a random threshold. An important goal of a model selection procedure is to achieve a small with as large a probability as possible. Often the distribution of is summarized via the FDR
Because is not observed, several methods estimate it by constructing a set of artificial features that share common behavior with the uninformative features19,21,23,54,56. In all of these cases, the artificial features are used to construct a surrogate of FDP(t) that we denoted in the previous section by FDP+(t).
The key distinction between Stabl and previous work is in the selection of the threshold . Previous approaches start by fixing a target FDR, denoted by q ∈ (0, 1), and then set
| 3 |
In contrast, we choose the Stabl threshold θ by minimizing FDP+(t) over t ∈ (0, 1) as per equation (1). The resulting observed FDP surrogate q+, defined in equation (2), is now a random variable.
Although the idea of minimizing FDP+(t) over t is very natural from an empirical viewpoint, it is less natural mathematically. Indeed, earlier work exploits in a crucial way the fact that defined via equation (3) is a stopping time for a suitably defined filtration, to conclude that
| 4 |
In contrast, our threshold θ is not a stopping time, and, therefore, a similarly simple argument is not available. Related to this is the fact that q+ is itself random.
We carried out numerical simulations on synthetic data (compare to Section 4.6). We observe empirically that often
| 5 |
In the next section, we will provide mathematical support for this finding.
Theoretical guarantees
We will establish two bounds on the FDP achieved by Stabl, under the following exchangeability assumption.
Assumption 1
Exchangeability of the extended null set. Denote by the covariates in the informative set and by the covariates in the null set or in the artificial set. We assume that XN∪A is exchangeable. Namely, for any permutation π of the set N ∪ A, we have
| 6 |
(Here, is the matrix obtained by permuting the columns of XN∪A using π, and denotes equality in distribution.)
Our first result establishes that the true FDP(θ) cannot be much larger than the minimum value of the FDP surrogate, , with large probability. We defer proofs to Section 11.4.5.
Proposition 1
Under Assumption 1, we have, for any Δ > 0,
| 7 |
Although reassuring, Lemma 1 exhibits only a slow decrease of the probability that FDP(θ) ≥ (1 + Δ)q+ with Δ. A sharper result can be obtained when the optimal threshold is not too high.
Theorem 1
Under Assumption 1, further assume ∣S∣ ≤ p/2. Let be the total number of false discoveries (including those among artificial features). Then, there exist constants c*, C* > 0 such that, for any ,
| 8 |
This result gives a tighter control of the excess of FDP(θ) over the surrogate q+. It implies that, in the event that the number of false discoveries is at least m, we have
| 9 |
As should be clear from the proof, the assumption ∣S∣ ≤ p/2 could be replaced by ∣S∣ ≤ (1 − c)p for any strictly positive constant c.
Proofs
Throughout this appendix, will be used to denote absolute constants whose value might change from line to line. We begin by defining the stopping time:
| 10 |
In words, tk is the threshold for the k-th-to-last false discovery. We will assume the tk to be distinct: 0 = t0 < t1 < ⋯ < t∣A∣+∣N∣ < 1. Indeed, we can always reduce the problem to this case by a perturbation argument. We define . We let , , and define .
Lemma 1
Under Assumption 1, for any , we have
| 11 |
Proof
By definition (recalling that O = S ∪ N is the set of original features):
The claim follows.
We next define and
| 12 |
The next result is standard, but we provide a proof for the reader’s convenience.
Lemma 2
Under Assumption 1, the process is a supermartingale with respect to the filtration , and is a martingale. Finally, for all k.
Proof
By exchangeability, the (k + 1)-th false discovery is equally likely to be among any of the nk + ak nulls that have not yet been rejected. Hence, conditional joint distribution of nk+1, ak+1 is (for ):
Hence, in the event {k < k0} (in which case ak > 0)
On the other hand, in the event {k ≥ k0}:
Hence, is a supermartingale. The same calculation implies that Zk is a martingale. The inequality follows from the fact that is decreasing in k for k ≥ k0.
We are now in a position to prove Proposition 1 and Theorem 1.
Proof
Proof of Proposition 1 by Lemma 1
where (a) follows from Lemma 2 and (b) from Doob’s maximal inequality. Because (Zk) is a martingale, following the above:
This proves the claim.
Proof
Proof of Theorem 1. By the same argument as in Lemma 1 (and adopting the standard notation ):
where (a) follows from Lemma 2.
Letting , for any non-negative, non-decreasing convex function
| 13 |
where (a) follows from Doob’s inequality for the submartingale .
Recalling the definition , we estimate the last expectation by
Here, , , and δ is a small constant.
Let X ~ Binom(∣N∣, ρ), Y ~ Binom(∣A∣, ρ) be independent binomial random variables. Then, it is easy to see that, for any ρ ∈ (0, 1),
| 14 |
| 15 |
In particular, because ∣A∣ = p and, by assumption, ∣N∣ ≥ p/2, we have , . Further choosing ρ = m/(∣A∣ + ∣N∣),
| 16 |
| 17 |
| 18 |
where the first inequality follows by the local central limit theorem and the second by Bernstein inequality. Of course, a similar bound holds for aK.
Substituting above, we get, for δ < 1,
| 19 |
Let and . For , , δ ≤ 1/4, we have
We next choose for some ℓ ≥ 1 (this function is monotone and convex as required). We thus get, from (19), fixing δ = 1/4,
where the last inequality holds for ℓ < m/C with C a sufficiently large constant. If we further choose , then we get
Substituting in equation (13), we get, for any :
Choosing ℓ = c0Δ2m for a sufficiently small constant c0 implies the claim.
Comparison of algorithmic complexity
We compare the algorithmic complexity of the Lasso, EN, SS and Stabl algorithms:
Lasso, EN and AL: Given the number of samples (n) and the number of features (p), the time complexity of the Lasso, EN or AL algorithm is (refs. 18,69).
SGL: Given the number of groups (g) and the average number of features in a group (m), the time complexity of the SGL would be .
SS: SS’s complexity depends on the number of subsamples (B) and the number of regularization parameters (R) considered. Assuming Lasso or EN is used as the base model, the time complexity of SS would be .
Stabl: Stabl’s complexity is driven by the base model (Lasso, EN, SGL or AL) and the additional steps introduced by the method. The time complexity of Stabl would be or , where represents the number of artificial features introduced by Stabl’s method.
Synthetic datasets
Gaussian models without correlation
We use a standard Gaussian covariates model70,71. Denoting the rows of X by x1, …xn, and the responses by y1, …, yn, we let the samples (yi, xi) be i.i.d. with:
| 20 |
| 21 |
We use the following covariance and coefficients
| 22 |
| 23 |
| 24 |
This structure was also used in ref. 11.
Note that the above can also be written in the standard form as
where
| 25 |
| 26 |
This distribution is parametrized by:
Number of features p, number of informative features k and sample size n
Variance parameters s, t
β coefficients
Note that, for a binary outcome, we can use the new response . S being the sigmoid function:
Gaussian models with correlation
Following the procedure devised in the previous section, we simulate the Gaussian model with three levels of correlations. In this case, we use the same model as the previous section, but ΣZ is a k × k matrix that captures the correlation among the informative features. We can define ΣZ as:
| 27 |
where ρ1, …, ρk−1 are the correlation parameters for the informative features.
The coefficients β and the covariance matrix Σ used to generate the covariates xi can be defined as before.
Non-Gaussian models
Although previous simulations were based on normally distributed data, omic data, such as bulk or single-cell RNA-seq datasets, often follow negative binomial and zero-inflated negative binomial distributions. The MX knockoff framework, despite its inherent adaptability to non-normal distributions, often requires modification based on the dataset’s specific nature and any existing model that describes the joint distribution of feature covariates19,25. For scenarios governed by a known data generation process, the MX knockoff framework was adjusted to generate artificial features. These features mirrored the marginal covariate distribution and correlation structure of non-normally distributed datasets. For these scenarios, the StablSRM framework combined with MX knockoffs consistently enhanced sparsity and reliability in both regression and classification tasks (Extended Data Fig. 8 and Supplementary Table 2). In cases with undisclosed joint distribution, random permutations offer a viable option for generation of artificial features. Although ensuring genuine marginal distributions, this technique might not retain the dataset’s original correlation structure. However, the StablSRM’s results using random permutations paralleled those achieved with MX knockoffs on non-normally distributed datasets of varying correlation structures (Extended Data Fig. 8).
Collectively, synthetic modeling experiments underscore that the choice of base SRM and noise generation techniques within the StablSRM framework can influence feature selection and model performance. Ideally, the correlation structure and data distribution should dictate this choice, but real-world datasets often have unknown true distributions. Therefore, selecting between MX knockoff or random permutation for artificial feature generation within the Stabl framework hinges on knowledge of covariate distribution and the analyst’s priority-preserving the original dataset’s correlation structure or its distribution.
Normal to Anything framework
Normal to Anything (NORTA) was designed to synthesize high-dimensional multivariate datasets72–75. This method can be used to generate random variables with arbitrary marginal distributions and correlation matrix from a multivariate normal distribution. In essence, the problem boils down to finding the pairwise correlations between the normal vectors that yield the desired correlation between the vectors of the non-normal distribution. In practice, this can be achieved using quantile functions. Using this method, we created correlated vectors following either a zero-inflated negative binomial model or a standard negative binomial model. Our simulations harnessed the capabilities of the Julia package Bigsimr, which implements this framework. This package enables data generation via the Gaussian copula (a joint distribution of the multivariate uniform vector obtained from the multivariate normal vector), facilitating the creation of datasets with targeted correlations and specified marginal distributions, such as Gaussian and negative binomials.
Negative binomial models
To generate the synthetic negative binomial models, we initially create a correlation matrix ΣZ for the multivariate normal from which the copula is computed, and we verify that the informative features match the desired level of correlation (low (ρ = 0.2), intermediate (ρ = 0.5) and high (ρ = 0.7)).
We constructed zi using this strategy and used the following parameters for the marginal distributions: NB(μ = 2, ϕ = 0.1).
Similar to the Gaussian cases, we then use the generated data to create the response with the following procedure:
| 2 |
Zero-inflated negative binomial and normal models
To generate zero-inflated (ZI) covariates in our models, we follow a similar process as described earlier for the non-zero values in a negative binomial distribution or Gaussian distribution. Let xij represent the j-th covariate of the i-th observation. The ZI covariate can be generated as follows:
where π is the probability of observing a zero and is fixed in our examples at 0.1.
Adaptation of the MX knockoff with Gaussian copulas
In situations where the quantile–quantile transformation is available, we can easily adapt the MX knockoff procedure to generate knockoffs tailored to the chosen distribution. Specifically, from the synthetic data, we can estimate ΣZ and generate MX knockoffs, thereby establishing the correspondence to the chosen distribution. For the sake of comparison with the random permutation procedure, we use this modified version of the knockoffs when we considered synthetic non-normal distributions.
Synthetic data for SGL
To apply SGL, the creation of predefined feature groups for analysis is needed. This was achieved through the construction of sets of five correlated covariates (Xi). This was accomplished by generating a block diagonal correlation matrix (ΣZ) where, apart from the diagonal entries, all other elements were set to zero. This matrix was formulated to encapsulate the interrelationships solely within each covariate group. Specifically, the diagonal blocks, each of size five, represented distinct groups. By adopting this approach, we explicitly defined the covariate groups to be considered during the optimization process of the algorithm. This methodology remained consistent across various scenarios involving correlation structures and data distributions.
Let Xi denote the i-th group of correlated covariates, where i = 1, 2, …, m is the index of the group. The block diagonal correlation matrix ΣZ is given by:
Here, each ΣZi represents the correlation matrix among the covariates within group Xi. By structuring ΣZ in this way, we intentionally limit the relationships within each group and disregard correlations between different groups.
SS coupled with grid search
In this approach, we combined SS with grid search to optimize the threshold used to select the features. The procedure was as follows:
We used a grid search method with a predefined number of possible thresholds ranging from 0% to 100%, evenly spaced across the range. This allowed us to test the sensitivity of SS performance to different thresholds.
For each threshold, we applied the SS algorithm with the chosen threshold to select a subset of features. We then used this subset of features to train a logistic regression model.
We used a CV method to compute the R2 score of the model for each threshold in the grid search on the training set.
We selected the threshold that resulted in the highest R2 score as the optimal threshold.
Finally, we used the selected threshold to predict the outcome variable on the test set using the logistic regression model trained on the full dataset with the selected subset of features.
Computational framework and pre-processing
Stabl was designed and executed using the Python packages ‘scikit-learn’ (version 1.1.2), ‘joblib’ (version 1.1.0) and ‘knockpy’ (version 1.2) (for the knockoff sampling generation). The Lasso algorithm fed into the Stabl subsampling process was executed using ‘scikit-learn’ (version 1.1.2) using the default threshold for feature selection at 10−5 in absolute value. The synthetic data generation was done using the Python package ‘numpy’ (version 1.23.1). Basic pre-processing steps, including variance thresholds and standardization, were executed using the Python packages ‘scikit-learn’ (version 1.1.2), ‘pandas’ (version 1.4.2) and ‘numpy’ (version 1.23.1). Visualization functions to plot stability path and FDR curves were executed using ‘seaborn’ (version 0.12.0) and ‘matplotlib’ (version 3.5.2).
Metrics on synthetic datasets
Predictive performance for binary classification
To evaluate our models, we use the AUROC and the area under the precision-recall curve (AUPRC) in the case of binary classification.
A common scale of performance was used to refer to the AUROC:
0.5–0.7 AUROC: modest performance
0.7–0.8 AUROC: good performance
0.8–0.9 AUROC: very good performance
0.9–1 AUROC: excellent performance
Predictive performance for regression
For regression tasks, the coefficient of determination R2, the RMSE and the mean absolute error (MAE) were used conventionally.
As for the AUROC, an arbitrary but common scale of performances was used in terms of R2 score:
0.0–0.3: No linear relationship
0.3–0.5: A weak linear relationship
0.5–0.7: A moderate linear relationship
0.7–0.9: A strong linear relationship
0.9–1.0: A very strong linear relationship
Note that, in some specific situations, the R2 score can be negative when the predictions are arbitrarily worse than using a constant value.
To assess the statistical significance of our results, we always performed a statistical test (two-sided Pearsonʼs r). To compare between methods, a two-sided Mann–Whitney rank-sum test was performed on the distribution of the repetition of the training for a given n.
Sparsity
Our measure of sparsity is the number of features that are selected in the final model. On the synthetic dataset, random samples are generated many times, so the average size of the set of selected features serves as our metric. To compare between methods, a two-sided Mann–Whitney rank-sum test was performed on the distribution of the repetition of the training for a given n.
Reliability
On the synthetic dataset, as we can sort out informative from uninformative features, we are able to compute the JI and the FDR, which are defined as:
The JI ranges from 0 (if no informative features are selected) to 1 (if the selected set comprises all informative features). To compare between methods, a two-sided Mann–Whitney rank-sum test was performed on the distribution of the repetition of the training for a given n.
Benchmark on real-world datasets
Description of the datasets
PE dataset
The PE dataset contained cfRNA data previously collected as part of a prospective study of 49 pregnant women (29 with PE, 20 normotensive) receiving routine antenatal care at Lucile Packard Children’s Hospital at Stanford University. Blood samples were collected three times in pregnancy (early, mid and late pregnancy). Women were diagnosed as having PE following American College of Obstetrics and Gynecology76 guidelines. Women in the control group had uncomplicated term pregnancies. Samples collected from women who developed PE were collected before clinical diagnosis. The study was reviewed and approved by the institutional review board (IRB) at Stanford University (no. 21956). The details of the study design and the cfRNA sample preparation and data quality assessment were previously described26,27.
COVID-19 dataset
The analysis leveraged existing plasma proteomics data collected from 68 adults with a positive SARS-CoV-2 test (qRT–PCR on a nasopharyngeal swab specimen29). Publicly available plasma proteomic data using 784 SARS-CoV-2 samples from 306 positive patients was used for independent validation of the findings28. In the first study, 30 individuals reported having mild COVID-19 disease—that is, asymptomatic or various mild symptoms (for example, cough, fever, sore throat and loss of smell and taste) without any breathing issues. Thirteen individuals reported having moderate disease—that is, evidence of lower respiratory tract disease but with oxygen saturation (SpO2) above 94%. Twenty-five individuals were hospitalized with severe disease due to respiratory distress (SpO2 % ≥94%, respiratory frequency ≤30 breaths per minute, PaO2/FiO2 ≤300 mmHg or lung infiltrates ≥50%). For modeling purposes, COVID-19 severity was dummy-coded as follows: mild or moderate = 1 and severe = 2. The validation cohort consisted of 125 samples from patients with mild or moderate COVID-19 and 659 samples from patients with severe COVID-19. For both training and validation datasets, the Olink proximity extension assay (PEA, Olink Proteomics, Explore panel) was used to measure the plasma protein levels of 1,472 proteins77. Plasma was pre-treated with 1% Triton X-100 for 2 h at room temperature to inactivate the virus before freezing at −80 °C and shipping. The arbitrary unit normalized protein expression (NPX) is used to express the raw expression values obtained with the Olink assay, where high NPX values represent high protein concentration. Values were log2 transformed to account for heteroskedasticity.
Time-to-labor dataset
This dataset consisted of existing single-cell proteomic (mass cytometry), plasma proteomic and metabolomic data derived from the analysis of samples collected in a longitudinal cohort of pregnant women receiving routine antepartum and postpartum care at the Lucile Packard Children’s Hospital at Stanford University, as previously described38. The study was approved by the IRB of Stanford University (no. 40105), and all participants signed an informed consent form.
In brief, n = 63 study participants were enrolled in their second or third trimester of an uncomplicated, singleton pregnancy. Serial peripheral blood samples were collected at one to three times throughout pregnancy before the onset of spontaneous labor (the median sample size per patient is three).
In plasma, high-throughput untargeted mass spectrometry and an aptamer-based proteomic platform were used to quantify the concentration of 3,529 metabolites and 1,317 proteins, respectively. In whole blood, a 46-parameter mass cytometry assay measured a total of 1,502 single-cell immune features in each sample. These included the frequencies of 41 immune cell subsets (major innate and adaptive populations), their endogenous intracellular activities (phosphorylation states of 11 signaling proteins) and the capacities of each cell subset to respond to receptor-specific immune challenges (lipopolysaccharide (LPS), interferon-α (IFN-α), granulocyte macrophage colony-stimulating factor (GM-CSF) and a combination of IL-2, IL-4 and IL-6).
The original model to predict the time to onset of labor was trained on a cohort of n = 53 women with n = 150 samples. The independent validation of the model was performed on n = 10 additional pregnancies with n = 27 samples. A total of 6,348 immune, metabolite and protein features were included per sample. In this specific dataset, to account for the longitudinal nature of the data, we performed a patient shuffle split (PSS) method to assess the generalizability of our models. Specifically, we divided the dataset into two subsets and used one subset for training and the other for testing. Each subset contains all data from an individual patient (that is, for a given patient, its data are either in the training subset or the testing subset). We repeated this process n times, leaving out different patients (that is, all their data) each time. This approach allowed us to evaluate the performance of our models in predicting time to labor for patients not included in the training data. The dataset obtained was first z-scored, and the knockoff method was used for Stabl modeling experiments.
DREAM challenge dataset
The DREAM challenge study aimed at classifying PT and T labor pregnancies from vaginal microbiome data48. The DREAM challenge dataset contains nine publicly available and curated microbiome datasets with 1,569 samples, across 580 individuals (336 individuals delivered at T and 244 delivered PT). The DREAM challenge included 318 teams who submitted results for the classification of PT versus T pregnancies.
The MaLiAmPi pipeline was used to process all the data48,49. Essentially, DADA2 was used to assemble each project’s raw reads into approximate sequence variants (ASVs). These ASVs were then employed to recruit complete 16S rRNA gene alleles from a repository based on sequence similarity. The recruits were then assembled into a maximum-likelihood phylogeny using RAxM78, and the ASVs were placed onto this common phylogenetic tree through EPA-ng79. The final step was to use these placements to determine community alpha-diversity, phylogenetic (KR) distance between communities and taxonomic assignments for each ASV and to cluster ASVs into phylotypes based on their phylogenetic distance. Moreover, VALENCIA was used to identify each sample’s community state type (CST)80. MaLiAmPi is accessible as a nextflow workflow and is containerized at 100%, enabling it to be used on multiple high-performance computing resources.
Following the description for pre-processing of the best-performing team on the first challenge, we use specimens collected no later than 32 weeks of gestation to develop the prediction model. We extract microbiome data from phylotype_nreads.5e_1.csv, phylotype_nreads.1e0.csv, taxonomy_nreads.species.csv, taxonomy_nreads.genus.csv and taxonomy_nreads.family.csv tables. The phylotype_nreads.1e_1.csv table is not used because its number of columns (9,718) is overwhelming compared to the sample size.
We apply the centered log-ratio (clr) transformation81 on microbiome data to obtain scale-invariant values. In clr transformation, given a D-dimensional input x,
where is the geometric mean of x.
In this dataset, to account for the longitudinal nature of the data, we performed a PSS method to assess the generalizability of our models on the time-to-labor dataset.
SSI dataset
Patients undergoing non-urgent major abdominal colorectal surgery were prospectively enrolled between 11 July 2018 and 11 November 2020 at Stanford University Hospital after approval by the IRB of Stanford University and the obtaining of written informed consent (IRB-46978). Inclusion criteria were patients over 18 years of age who were willing and able to sign a written consent. Exclusion criteria were a history of inflammatory/autoimmune conditions not related to the indication for colorectal surgery as well as undergoing surgery that did not include resection of the bowel.
A nested case–control study was designed to identify pre-operative immunological factors predictive of the occurrence of an SSI. The study protocol was designed following the STROBE guidelines. The primary clinical endpoint was the occurrence of an SSI within 30 d of surgery, defined as superficial, deep or organ space SSI, anastomotic leak or dehiscence of the surgical incision. The primary clinical endpoint and all clinical variables were independently curated and validated by a colorectal surgeon and a practicing anesthesiologist. To minimize the effect of clinical and demographic variables potentially associated with the development of an SSI, patients who developed an SSI were matched to a control group of patients who did not develop an SSI. Patient characteristics and types of surgical procedures are provided in Supplementary Table 7. We performed a power analysis82 to determine the minimum required sample size of 80 patients to achieve an expected AUROC of 0.8, with a maximum 95% confidence interval (CI) of 0.25 and an expected SSI incidence of 25%. After conducting a frequency-matching procedure, we included a total of 93 patients, which reduced the expected confidence interval range to 0.23.
Whole blood and plasma samples were collected on the day of surgery (DOS) before induction of anesthesia, processed and analyzed following a similar workflow as previously described53. In brief, whole blood samples were either left unstimulated (to quantify cell frequency and endogenous cellular activities) or stimulated with a series of receptor-specific ligands eliciting key intracellular signaling responses implicated in the host’s immune response to trauma/injury, including LPS, TNFα and a combination of IL-2, IL-4 and IL-6. From each sample, 1,134 single-cell proteomic features were extracted using a 41-parameter single-cell mass cytometry immunoassay (Supplementary Table 11), including the frequency of 35 major innate and adaptive immune cells (Extended Data Fig. 10) and their intracellular signaling activities (for example, the phosphorylation state of 11 proteins). In addition, the plasma concentrations of 712 inflammatory proteins were quantified using the SOMAscan manual assay for human plasma83,84. SOMAscan kits were run in a SomaLogic trained and certified assay site. Mass cytometry data were collected using the default software for the CyTOF 3.0 Helios instrument (Helios CyTOF software, version 7.0.5189, Standard BioTools) and then gated using CellEngine (CellCarta).
Stabl analysis of real-world datasets
For each real-world dataset, the dataset obtained was first z-scored, and the StablSRM method was applied using Lasso, EN or AL as the base SRM (hyperparameters listed in Supplementary Table 13). To preserve the correlation structure of synthetic features, MX knockoffs served as the primary method for introducing noise in all omics datasets, except for the PE dataset (cfRNA). This dataset demonstrated the lowest internal correlation level (≤1% of features with intermediate or high correlations, R ≥ 0.5), and, therefore, random permutations were employed as the noise generation approach.
Metrics on real-world datasets
Monte Carlo CV
The Monte Carlo CV is done as follows. At each fold, the dataset is split randomly into training and testing sets, and the model is then trained and evaluated using the training and testing sets, respectively:
In the COVID-19 and SSI datasets, we executed Monte Carlo CV using the RepeatedStratifiedKFold class of ‘scikit-learn’ (version 1.1.2), which repeats the multiple K-fold CV scheme. We then take the median of the predictions to obtain the final predictions. This technique ensures that all samples are evaluated the same number of times. We used stratified five-fold CV (20% of the data are tested at each fold) to ensure that the class repartition was preserved among all the folds.
In the time-to-labor, PE and DREAM datasets, we used the Monte Carlo CV with the GroupShuffleSplit class of ‘scikit-learn’ (version 1.1.2), allowing us to preserve the patients’ repartition between the training and testing sets as no patient’s samples are split into both sets. As before, the final predictions are obtained by taking the median of the predictions for each sample. The testing proportion was set at 20% at each fold.
Predictive performance
The predictive performance was measured using the same metrics as in the artificial datasets. The values were computed using the median from the Monte Carlo CV procedure for all the training cohorts. For the validation, the predictions from the final models were applied to compute the relevant metrics. When comparing predictive performance between methods, a two-sided bootstrap test was performed on the distribution of the CV folds.
Sparsity
Sparsity was defined as the average number of features selected in the model during the CV procedure. When comparing sparsity performance between methods, a two-sided Mann–Whitney rank-sum test was performed on the distribution of the CV folds.
Multi-omic modeling using Stabl and late-fusion Lasso
In early fusion, the features from different omics data are combined into a single feature set before training a model. This means that the model sees the combined feature set as a single input and learns a single set of weights for all the features. In contrast, late fusion involves training separate models on each omics data and then combining their predictions at the end. This can be done by taking the average of the predictions or by training a final model to combine the predictions from the separate models. Late fusion can be more flexible, allowing the different models to learn different weights for the features from each data source. Similarly to late fusion, Stabl adopts an independent analysis approach for each omic data layer by fitting specific reliability thresholds before selecting the most reliable features to be merged into a final layer. However, in contrast to late fusion, Stabl computes a specific reliability threshold for each omic data layer, allowing for the integration of the features selected from each omic data layer into a final modeling layer.
Visualization
Uniform manifold approximation and projection
Uniform manifold approximation and projection (UMAP) is a dimensionality reduction technique that can be used to reduce the number of dimensions in a dataset while preserving the global structure of the data. UMAPs were plotted using the ‘umap-learn’ library and default parameters. The two first UMAP supports were used to represent all the molecular features in two-dimensional plots for all omics. The node sizes and colors were then calculated based on the intensity of the association with the outcome as the −log10 Pvalue.
Stability paths
The stability path is used to visualize how the features are selected as the regularization parameter is varied. The stability path is a curve that plots the mean stability of each feature as a function of the regularization parameter. The stability of a feature is defined as the proportion of times that the feature is selected by the model when trained on different subsets of the data. The stability path can identify a range of regularization parameters that result in a stable set of features being selected.
Box plots
Throughout the figures, the box plots show the three-quartile values of the distribution along with extreme values. The whiskers extend to points that lie within 1.5× interquartile range (IQR) of the lower and upper quartile, and observations that fall outside this range are displayed independently.
ROC and PR curves
In the figures, the ROC and PR curves are displayed along with their CIs. The 95% CIs are computed with 2,000 stratified bootstrap replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41587-023-02033-x.
Supplementary information
Supplementary Tables 1–13.
Acknowledgements
We thank R. Tibshirani for thorough and critical reading of the manuscript. This work was supported by the National Institutes of Health (NIH): R35GM137936 (B.G.), P01HD106414 (N.A., D.K.S. and B.G.), 1K99HD105016-01 (I.A.S.), R35GM138353 (N.A.), R01HL139844 (B.G., N.A. and M.S.A.); the Center for Human Systems Immunology at Stanford (B.G.); the German Research Foundation (J.E.); the March of Dimes Prematurity Research Center at Stanford University (D.F., D.K.S., B.G., N.A. and M.S.A.); the March of Dimes Prematurity Research Center at the University of California, San Francisco (T.T.O., A.R., J.L.G. and M. Sirota); the Burroughs Welcome Fund (N.A.); the Alfred E. Mann Foundation (N.A.); the Stanford Maternal and Child Health Research Institute (D.F., D.K.S., B.G., N.A. and M.S.A.); and the Charles and Mary Robertson Foundation (B.G., N.A. and D.K.S.).
Extended data
Author contributions
Conceptualization: J.H., I.M., G.B., J.E., A.M. and B.G.; data collection: F.V., I.A.S., D.F., A.S.T., E.A.G., A.C., T.T.O., A.R., J.L.G., T.A.B., M. Sato and M.D.; formal analysis: J.H., I.M., G.B., J.E., M. Sabayev, J.G., J.A. and X.D.; investigation/data acquisition: E.A.G., I.A.S., D.F., A.C., M. Sato, M.D., T.T.O., A.R., J.L.G. and M. Sirota; methodology: J.H., I.M., G.B., N.A., A.M. and B.G.; software: J.H., G.B., M. Sabayev and J.A.; supervision: M.S.A., D.K.S., N.A., A.M. and B.G.; visualization: J.H., G.B. and J.E.; mathematical proof: A.M.; writing—original draft: J.H., I.M., J.E., D.K.G., F.X.L., D.K.S., M.S.A., N.A., A.M. and B.G.; writing—review and editing: all authors.
Peer review
Peer review information
Nature Biotechnology thanks Arnaud Droit, Hakim Benkirane and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editor: Anne Doerr, in collaboration with the Nature Biotechnology team.
Data availability
The datasets generated and/or analyzed during the current study are available on GitHub (https://github.com/gregbellan/Stabl/tree/main/Sample%20Data) and Dryad (10.5061/dryad.stqjq2c7d).
Code availability
The Stabl framework and custom computer code used in this study can be accessed on GitHub (https://github.com/gregbellan/Stabl) and Zenodo (10.5281/zenodo.8406758).
Competing interests
J.H., B.G., D.K.G. and F.V. are advisory board members; G.B. and X.D. are employed; and E.A.G. is a consultant at SurgeCare. N.A. is a member of the scientific advisory boards of January AI, Parallel Bio, Celine Therapeutics and WellSim Biomedical Technologies, is a paid consultant for MARAbio Systems and is a cofounder of Takeoff AI. Part of this work was carried out while A.M. was on partial leave from Stanford University and was Chief Scientist at nData, Inc. dba, Project N. The present research is unrelated to A.M.’s activity while on leave. J.H., N.A., M.S.A. and B.G. are listed as inventors on a patent application (PCT/US22/71226). The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Julien Hédou, Ivana Marić, Grégoire Bellan, Jakob Einhaus.
Extended data
is available for this paper at 10.1038/s41587-023-02033-x.
Supplementary information
The online version contains supplementary material available at 10.1038/s41587-023-02033-x.
References
- 1.Subramanian, I., Verma, S., Kumar, S., Jere, A. & Anamika, K. Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights14, 1177932219899051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wafi, A. & Mirnezami, R. Translational -omics: future potential and current challenges in precision medicine. Methods151, 3–11 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature578, 615–620 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Fourati, S. et al. Pan-vaccine analysis reveals innate immune endotypes predictive of antibody responses to vaccination. Nat. Immunol.23, 1777–1787 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunkler, D., Sánchez-Cabo, F. & Heinze, G. Statistical analysis principles for omics data. Methods Mol. Biol.719, 113–131 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Ghosh, D. & Poisson, L. M. ‘omics’ data and levels of evidence for biomarker discovery. Genomics93, 13–16 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Series B Methodol.58, 267–288 (1996). [Google Scholar]
- 8.Zou, H. & Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol.67, 301–320 (2005). [Google Scholar]
- 9.Zou, H. The adaptive lasso and its oracle properties. J. Am. Stat. Assoc.101, 1418–1429 (2006). [Google Scholar]
- 10.Simon, N., Friedman, J., Hastie, T. & Tibshirani, R. A sparse-group lasso. J. Comput. Graph. Stat.22, 231–245 (2013). [Google Scholar]
- 11.Ding, D. Y., Li, S., Narasimhan, B. & Tibshirani, R. Cooperative learning for multiview analysis. Proc. Natl Acad. Sci. USA119, e2202113119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang, P., Yang, J., Zhou, B. & Zomaya, A. A review of ensemble methods in bioinformatics. Curr. Bioinform.5, 296–308 (2010). [Google Scholar]
- 13.Huan, X., Caramanis, C. & Mannor, S. Sparse algorithms are not stable: a no-free-lunch theorem. IEEE Trans. Pattern Anal. Mach. Intell.34, 187–193 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Roberts, S. & Nowak, G. Stabilizing the lasso against cross-validation variability. Comput. Stat. Data Anal.70, 198–211 (2014). [Google Scholar]
- 15.Homrighausen, D. & McDonald, D. The lasso, persistence, and cross-validation. Proc. of the30th International Conference on Machine Learning 2068–2076 (PMLR, 2013).
- 16.Olivier, M., Asmis, R., Hawkins, G. A., Howard, T. D. & Cox, L. A. The need for multi-omics biomarker signatures in precision medicine. Int. J. Mol. Sci.20, 4781 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarazona, S., Arzalluz-Luque, A. & Conesa, A. Undisclosed, unmet and neglected challenges in multi-omics studies. Nat. Comput. Sci.1, 395–402 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Meinshausen, N. & Bühlmann, P. Stability selection. J. R. Stat. Soc. Series B Stat. Methodol.72, 417–473 (2010). [Google Scholar]
- 19.Candès, E., Fan, Y., Janson, L. & Lv, J. Panning for gold: ‘model-X’ knockoffs for high dimensional controlled variable selection. J. R. Stat. Soc. Series B Stat. Methodol.80, 551–577 (2018). [Google Scholar]
- 20.Bach, F. Bolasso: model consistent lasso estimation through the bootstrap. Proc. of the 25th International Conference on Machine Learning 33–40 (PMLR, 2008).
- 21.Barber, R. F. & Candès, E. J. Controlling the false discovery rate via knockoffs. Ann. Stat.43, 2055–2085 (2015). [Google Scholar]
- 22.Ren, Z., Wei, Y. & Candès, E. Derandomizing knockoffs. J. Am. Stat. Assoc.118, 948–958 (2023). [Google Scholar]
- 23.Weinstein, A., Barber, R. & Candès, E. A power and prediction analysis for knockoffs with lasso statistics. Preprint at 10.48550/arXiv.1712.06465 (2017).
- 24.Bondell, H. D. & Reich, B. J. Simultaneous regression shrinkage, variable selection, and supervised clustering of predictors with OSCAR. Biometrics64, 115–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bates, S., Candès, E., Janson, L. & Wang, W. Metropolized knockoff sampling. J. Am. Stat. Assoc.116, 1413–1427 (2020). [Google Scholar]
- 26.Moufarrej, M. N. et al. Early prediction of preeclampsia in pregnancy with cell-free RNA. Nature602, 689–694 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marić, I. et al. Early prediction and longitudinal modeling of preeclampsia from multiomics. Patterns (N Y)3, 100655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filbin, M. R. et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell–cell interactions. Cell Rep. Med.2, 100287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feyaerts, D. et al. Integrated plasma proteomic and single-cell immune signaling network signatures demarcate mild, moderate, and severe COVID-19. Cell Rep. Med.3, 100680 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosmer, D. & Lemeshow, S. Applied Logistic Regression 376–383 (Wiley, 2000).
- 31.Davis, K. D. et al. Discovery and validation of biomarkers to aid the development of safe and effective pain therapeutics: challenges and opportunities. Nat. Rev. Neurol.16, 381–400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasten, M. & Giordano, A. Cdk10, a Cdc2-related kinase, associates with the Ets2 transcription factor and modulates its transactivation activity. Oncogene20, 1832–1838 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Markovic, S. S. et al. Galectin-1 as the new player in staging and prognosis of COVID-19. Sci. Rep.12, 1272 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.COvid-19 Multi-omics Blood ATlas (COMBAT) Consortium. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell185, 916–938 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayr, C. H. et al. Integrative analysis of cell state changes in lung fibrosis with peripheral protein biomarkers. EMBO Mol. Med.13, e12871 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Overmyer, K. A. et al. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst.12, 23–40 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohammed, Y. et al. Longitudinal plasma proteomics analysis reveals novel candidate biomarkers in acute COVID-19. J. Proteome Res.21, 975–992 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Stelzer, I. A. et al. Integrated trajectories of the maternal metabolome, proteome, and immunome predict labor onset. Sci. Transl. Med.13, eabd9898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suff, N., Story, L. & Shennan, A. The prediction of preterm delivery: what is new? Semin. Fetal Neonatal Med.24, 27–32 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Marquette, G. P., Hutcheon, J. A. & Lee, L. Predicting the spontaneous onset of labour in post-date pregnancies: a population-based retrospective cohort study. J. Obstet. Gynaecol. Can.36, 391–399 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Shah, N. et al. Changes in T cell and dendritic cell phenotype from mid to late pregnancy are indicative of a shift from immune tolerance to immune activation. Front. Immunol.8, 1138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraus, T. A. et al. Characterizing the pregnancy immune phenotype: results of the viral immunity and pregnancy (VIP) study. J. Clin. Immunol.32, 300–311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah, N. M., Lai, P. F., Imami, N. & Johnson, M. R. Progesterone-related immune modulation of pregnancy and labor. Front. Endocrinol.10, 198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brinkman-Van der Linden, E. C. M. et al. Human-specific expression of Siglec-6 in the placenta. Glycobiology17, 922–931 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Kappou, D., Sifakis, S., Konstantinidou, A., Papantoniou, N. & Spandidos, D. A. Role of the angiopoietin/tie system in pregnancy (Review). Exp. Ther. Med.9, 1091–1096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang, B. et al. Interleukin-33-induced expression of PIBF1 by decidual B cells protects against preterm labor. Nat. Med.23, 128–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, A., Lee, R. H., Felix, J. C., Minoo, P. & Goodwin, T. M. Alteration of secretory leukocyte protease inhibitor in human myometrium during labor. Am. J. Obstet. Gynecol.200, 311.e1–311.e10 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Golob, J. L. et al. Microbiome preterm birth dream challenge: crowdsourcing machine learning approaches to advance preterm birth research. Preprint at medRxiv10.1101/2023.03.07.23286920 (2023). [DOI] [PMC free article] [PubMed]
- 49.Minot, S. S. et al. Robust harmonization of microbiome studies by phylogenetic scaffolding with MaLiAmPi. Cell Rep. Methods3, 100639 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tosato, G. & Jones, K. D. Interleukin-1 induces interleukin-6 production in peripheral blood monocytes. Blood75, 1305–1310 (1990). [PubMed] [Google Scholar]
- 51.Lee, J.-K. et al. Differences in signaling pathways by IL-1β and IL-18. Proc. Natl Acad. Sci. USA101, 8815–8820 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fong, T. G. et al. Identification of plasma proteome signatures associated with surgery using SOMAscan. Ann. Surg.273, 732–742 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rumer, K. K. et al. Integrated single-cell and plasma proteomic modeling to predict surgical site complications: a prospective cohort study. Ann. Surg.275, 582–590 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He, K. et al. A theoretical foundation of the target-decoy search strategy for false discovery rate control in proteomics. Preprint at 10.48550/arXiv.1501.00537 (2015).
- 55.He, K., Li, M.-J., Fu, Y., Gong, F.-Z. & Sun, X.-M. Null-free false discovery rate control using decoy permutations. Acta Math. Appl. Sin.38, 235–253 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinstein, A., Su, W. J., Bogdan, M., Barber, R. F. & Candès, E. J. A power analysis for Model-X knockoffs with ℓp-regularized statistics. Preprint at 10.48550/arXiv.2007.15346 (2020).
- 57.Romano, Y., Sesia, M. & Candès, E. Deep knockoffs. J. Am. Stat. Assoc.115, 1861–1872 (2019). [Google Scholar]
- 58.Chernozhukov, V. et al. Double/debiased machine learning for treatment and structural parameters. Econometrics J.21, C1–C68 (2018). [Google Scholar]
- 59.Kursa, M. B. & Rudnicki, W. R. Feature selection with the Boruta package. J. Stat. Softw.36, 1–13 (2010). [Google Scholar]
- 60.Breiman, L. Random forests. Mach. Learn.45, 5–32 (2001). [Google Scholar]
- 61.Friedman, J. Stochastic gradient boosting. Comput. Stat. Data Anal.38, 367–378 (2002). [Google Scholar]
- 62.Candes, E. & Tao, T. The Dantzig selector: statistical estimation when p is much larger than n. Ann. Stat.35, 2313–2351 (2007). [Google Scholar]
- 63.Bickel, P. J., Ritov, Y. & Tsybakov, A. B. Simultaneous analysis of Lasso and Dantzig selector. Ann. Stat.37, 1705–1732 (2009). [Google Scholar]
- 64.Bühlmann, P. & Van De Geer, S. Statistics for High-Dimensional Data: Methods, Theory and Applications (Springer, 2011).
- 65.Zhao, P. & Yu, B. On model selection consistency of Lasso. J. Mach. Learn. Res.7, 2541–2563 (2006). [Google Scholar]
- 66.Zhang, C.-H. & Huang, J. The sparsity and bias of the lasso selection in high-dimensional linear regression. Ann. Stat.36, 1567–1594 (2008). [Google Scholar]
- 67.Javanmard, A. & Montanari, A. Model selection for high-dimensional regression under the generalized irrepresentability condition. Proc. of the 26th International Conference on Neural Information Processing Systems 3012–3020 (Curran Associates, 2013).
- 68.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Methodol.57, 289–300 (1995). [Google Scholar]
- 69.Efron, B., Hastie, T., Johnstone, I. & Tibshirani, R. Least angle regression. Ann. Stat.32, 407–499 (2004). [Google Scholar]
- 70.Meinshausen, N. & Bühlmann, P. High-dimensional graphs and variable selection with the Lasso. Ann. Stat.34, 1436–1462 (2006). [Google Scholar]
- 71.Celentano, M., Montanari, A. & Wei, Y. The Lasso with general Gaussian designs with applications to hypothesis testing. Preprint at 10.48550/arXiv.2007.13716 (2020).
- 72.Cario, M. C. & Nelson, B. L. Modeling and generating random vectors with arbitrary marginal distributions and correlation matrix. http://www.ressources-actuarielles.net/EXT/ISFA/1226.nsf/769998e0a65ea348c1257052003eb94f/5d499a3efc8ae4dfc125756c00391ca6/$FILE/NORTA.pdf (1997).
- 73.Kurtz, Z. D. et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol.11, e1004226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McGregor, K., Labbe, A. & Greenwood, C. M. MDiNE: a model to estimate differential co-occurrence networks in microbiome studies. Bioinformatics36, 1840–1847 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, Y. & Lê Cao, K.-A. PLSDA-batch: a multivariate framework to correct for batch effects in microbiome data. Brief. Bioinformatics24, bbac622 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.American College of Obstetricians and Gynecologists. Gestational hypertension and preeclampsia: ACOG practice bulletin, number 222. Obstet. Gynecol. 135, e237–e260 (2020). [DOI] [PubMed]
- 77.Assarsson, E. et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE9, e95192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barbera, P. et al. EPA-ng: massively parallel evolutionary placement of genetic sequences. Syst. Biol.68, 365–369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.France, M. T. et al. VALENCIA: a nearest centroid classification method for vaginal microbial communities based on composition. Microbiome8, 166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aitchison, J. The statistical analysis of compositional data. J. R. Stat. Soc. Series B Methodol.44, 139–177 (1982). [Google Scholar]
- 82.Hanley, J. A. & McNeil, B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology143, 29–36 (1982). [DOI] [PubMed] [Google Scholar]
- 83.Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. Nat. Prec.10.1038/npre.2010.4538.1 (2010). [DOI] [PMC free article] [PubMed]
- 84.Rohloff, J. C. et al. Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol. Ther. Nucleic Acids3, e201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1–13.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available on GitHub (https://github.com/gregbellan/Stabl/tree/main/Sample%20Data) and Dryad (10.5061/dryad.stqjq2c7d).
The Stabl framework and custom computer code used in this study can be accessed on GitHub (https://github.com/gregbellan/Stabl) and Zenodo (10.5281/zenodo.8406758).