Abstract
Several characteristics make human papillomavirus (HPV) amenable to vaccination. Anti-HPV-directed vaccines are based on the observation that HPV E6 and E7 oncoproteins are constitutively expressed in HPV-positive cervical cancer and may serve as tumor rejection antigens. Five HPV types (16, 18, 31, 33, and 45) account for 80% of cervical cancer. Until now, the type of immune response capable of mediating an effective antitumor response has not been defined. In order to define the anticancer-directed immune response in situ, we characterized CD4+ and CD8+ sorted T cells from peripheral blood lymphocytes, freshly harvested tumor tissue, and tumor-infiltrating lymphocytes (TIL) from a patient with cervical cancer. The HLA-DR-restricted CD4+ T-cell receptor VB16-, VA10-, VA21-, and VA22-positive CD4+ T-cell line derived from TIL recognizes autologous HLA-DR*0402+ (HPV33+) cervical cancer cells, as determined by gamma interferon secretion. Testing of different peptides spanning the E7 gene revealed that the HPV3373–87 peptide ASDLRTIQQLLMGTV represents the immunodominant epitope which can also be presented by the DR*0401 allele to TIL. Such major histocompatibility complex class II-presented peptides represent attractive candidates to augment T-cell responses directed against autologous tumor cells.
Cervical cancer represents a unique tumor entity compared to most other cancer types, since it is associated with a chronic viral infection of epithelial cells with human papillomavirus (HPV), predominantly type 16 (HPV16) and HPV18 (33). Most of the studies examining T-cell responses directed against cervical cancer utilized the HPV E7 gene product as the target, since it is expressed in all disease stages and represents an attractive candidate for immune intervention. The assumption that T cells effectively recognize cancer cells stems from earlier observations that both CD4+ and CD8+ T cells recognize virus-infected cells (32). In contrast to the vigorous cellular immune response to viral antigens in peripheral blood (3, 21), the number of tumor-reactive T cells in patients with cancer appears to be quite low (9, 10).
Several mechanisms may account for the ultimate development of invasive cervical cancer, one of these being the inability of the cellular immune system to effectively eradicate HPV-positive tumor cells. Some reports demonstrated the presence of HPV-specific T cells after in vitro stimulation with peptides (11) or used autologous dendritic cells in order to expand HPV-specific and major histocompatibility complex (MHC) class I-restricted T cells (24). Animal studies suggested that an anti-papillomavirus E7-directed immune response may be effective in eradicating E7-positive autologous cancer cells (4, 12, 22). However, examination of T cells from patients with cervical cancer suggested that cellular immune responses to E7 from HPV16 may be impaired, potentially due to decreased ζ-chain expression, a key signal molecule associated with the T-cell receptor (TCR) (20).
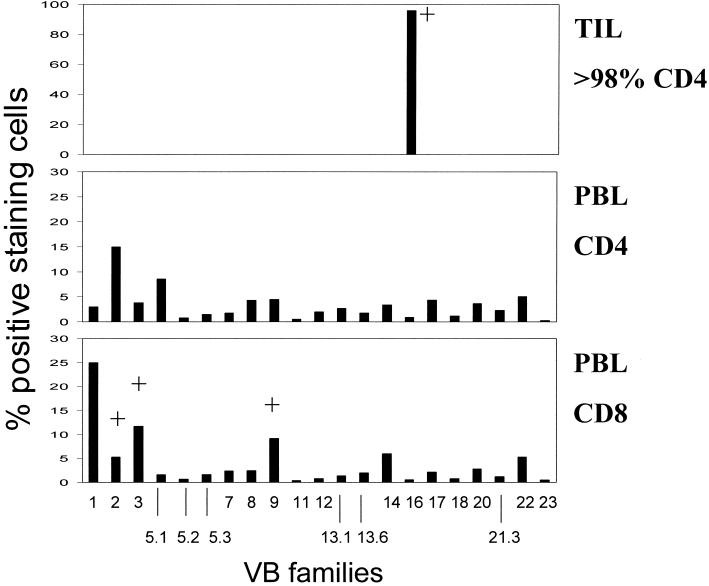
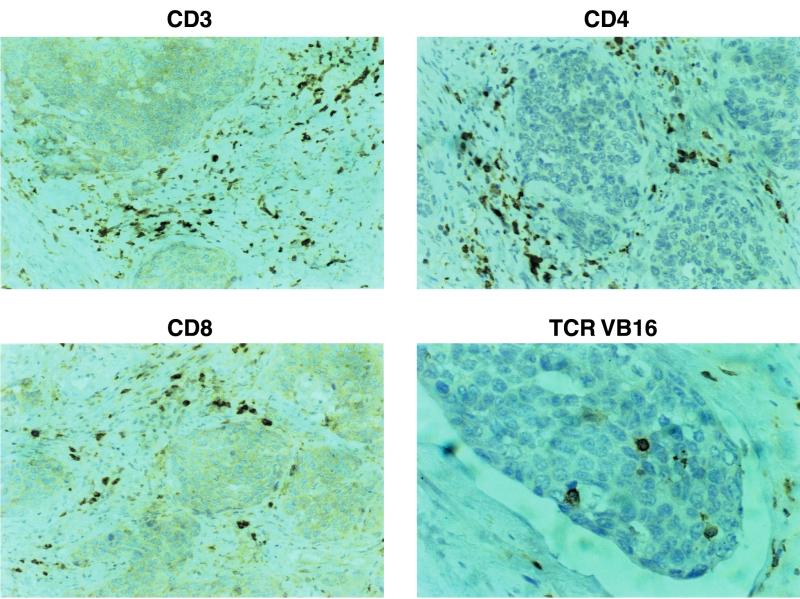
Most of the tumor antigens described thus far have relied on the definition of targets recognized either by T cells infiltrating into cancer tissue or from T cells harvested from peripheral blood (27), and earlier studies of T cells reactive to cervical cancer have predominantly relied on CD8+ or CD4+ T cells from peripheral blood lymphocytes (PBL) (2, 8, 19, 20, 24, 26, 29, 31); CD8+ T cells infiltrating into cancer lesions have recently been described to recognize HLA-A2-binding peptides provided by HPV16 (11). Here, we describe that CD4+ tumor-infiltrating lymphocytes (TIL) expanded in vitro with 100 IU of interleukin-2 (IL-2) and 100 ng of IL-7 per ml, obtained from a patient with cervical cancer, recognize freshly isolated autologous (HPV33+) tumor cells as determined by gamma interferon (IFN-γ) secretion, which could be blocked with a monoclonal antibody (MAb) directed against HLA-DR but not with an MAb directed against MHC class I (Table 1). In order to characterize the structural composition of TIL, we implemented flow cytometry using a panel of 21 different TCR VB-specific MAbs and TCR spectratype analysis of TCR VA and VB chains (18), followed by DNA sequence analysis. The TIL population exhibited less than 2% CD8+ and more than 98% CD4+ staining cells which predominantly express the TCR VB16 chain (Fig. 1), as defined by the MAb TAMAYA1.2 (Beckman/Coulter, Krefeld, Germany). This T-cell expansion could not be detected in T cells obtained from peripheral blood. Immunostaining of the freshly harvested tumor tissue revealed that the TCR VB16+ T-cell population infiltrates into the cancer lesion (Fig. 2).
TABLE 1.
TIL recognize autologous tumor cellsa
| Target | Blocking MAb | IFN-γ (pg/ml/106 cells) |
|---|---|---|
| Tumor | None | 0 |
| TIL | None | 23 |
| Tumor + TIL | None | 400 |
| W6/32 | 400 | |
| L243 | 20 |
Autologous cervical cancer cells harboring HPV33 were tested for T-cell recognition as determined by IFN-γ and IL-2 secretion by ELISA. TIL secreted IFN-γ, but not IL-2 in response to tumor cells, which could be blocked with the anti-DR-directed MAb L243. Blocking with the anti-DR-directed MAb clone B8.12.2 showed a similar result in a parallel experiment.
FIG. 1.
TCR VB families in PBL and TIL. PBL were gated on CD4+ or CD8+ T cells and tested for expression of individual TCR VB chains by flow cytometry. No major TCR VB expansion could be detected in PBL. In contrast, the majority of TIL (>98% CD4+) stained positive for the TCR VB16. +, detection of monoclonal TCR VB chains as listed in Table 2.
FIG. 2.
Detection of TCR VB16+ T cells infiltrating into cervical cancer. Serial sections from tumor tissue were stained for CD3, CD4, CD8, and TCR VB16+ T cells. Note that CD4+ T cells represent the majority of T cells and that TCR VB16+ T cells directly infiltrate the tumor lesion.
In order to define the structural anatomy and the magnitude of the T cells in different anatomic compartments, we performed TCR VA/VB spectratype analysis (18) with freshly isolated tumor tissue, in TIL, and in CD4+ and CD8+ sorted PBL. No monoclonal TCRs could be detected in the tumor specimen, but eight monoclonal TCR VA and three monoclonal TCR VB chains were detected in the CD8+ but not in the CD4+ T-cell population in PBL (Table 2). Seven monoclonal TCR VA and one monoclonal TCR VB (VB16) chains could be identified in TIL. DNA sequence analysis of the TCR VA chains in CD8+ PBL revealed that the monoclonal TCR VA3 and VA9 chains are identical to those present in TIL, presumably in the 2% CD8+ T-cell population. Next, we sorted the TCR VB16+ TIL population using the anti-VB16-directed MAb TAMAYA1.2 and anti-murine immunoglobulin G (IgG)-directed immunomagnetic beads. After sorting, three monoclonal TCR VA chains, including TCR VA10, VA21, and VA22, could be detected (Table 2). Thus, other monoclonal TCR VA chains identified in the TIL line may reside in the minority (<4%) of TCR VB16-negative T cells (Fig. 1).
TABLE 2.
Deduced TCR amino acid sequencesa
| T-cell population | TCR chain | Variable region sequence | CDR3 sequence | Sequence of joining region |
|---|---|---|---|---|
| PBL-VA CD8+ | VA2 | DSQPSDSATYLCA | VSG | NDMRFGAG (AJ43) |
| VA3* | ADTASYFCA | SRGGD | NTDKLIFGTG (AJ34) | |
| VA4 | TLRDAAVYYCI | LSFAS | AGGTSYGKLTFGQG (AJ52) | |
| VA9* | FAQEEDSAMYYCA | YGNNRLAFGKG (AJ7) | ||
| VA16 | SALVSDSALYFCA | VRHRI | DTGRRALTFGSG (AJ5) | |
| VA21 | QPGDSAVYFCA | ASES | GTSYGKLTFGQG (AJ52) | |
| VA23 | ASQPGDSATYLCA | VRYLL | SSGSARQLTFGSG (AJ22) | |
| VA25 | STYLCA | VGPS | SSGSARQLTFGSG (AJ22) | |
| PBL-VB CD8+ | VB2 | SAHPEDSSFYIC | SARGGGSG | NQPQHFGDG (BJ1-5) |
| VB3 | SASTNQTSMYLCA | SSLGVG | YEQYFGPG (BJ2-7) | |
| VB9 | LELGDSAVYFCA | SSQLWTSGAPP | STDTQYFGPG (BJ2-3) | |
| CD4+ TIL-VA | VA2 | FNTSCGTDYLCA | MSS | DRGSTLGRLYFGRG (AJ18) |
| VA3* | ADTASYFCA | SRGGD | NTDKLIFGTG (AJ34) | |
| VA6 | ASQLGDSAMYFCA | MREGW | AAGNKLTFGGG (AJ17) | |
| VA9* | FAQEEDSAMYYCA | YGNNRLAFGKG (AJ7) | ||
| VA10 (a) | AAQPGDTGLYLCA | GG | SDGQKLLFARG (AJ16) | |
| VA10 (b) | AAQPGDTGLYLCA | GGP | MDSSYKLIFGSG (AJ12) | |
| VA21 | PSQPGDSAVYFCA | ASGG | KLVFGKG (AJ57) | |
| VA26 | IYLCA | GG | SDGQKLLFARG (AJ16) | |
| CD4+ TIL-VB16 | VB16 | PAELDSGVYFCA | SSRGTSGTLK | QYFGPG (BJ2-7) |
| TIL VB16 sorted | VA10 | AAQPGDTGLYLCA | GGP | MDSSYKLIFGSG (AJ12) |
| VA21 | PSQPGDSAVYFCA | ASGG | KLVFGKG (AJ57) | |
| VA22 | AVYFCA | LS | YGCCRLAFGKG (AJ7) |
Monoclonal TCRs in CD8+ or CD4+ T-cell populations were identified by DNA sequence analysis as described previously (18). Exclusively the CDR3 region and the adjacent variable and joining TCR segments are listed. Note that the CD4+ VB16+ population represents 96% of all T cells in TIL (Fig. 1). Sorting of these VB16+ T cells yields a TCR VB16+ VA10+ VA21+ T-cell line. Note that in unsorted TIL, two TCR VA10 chains could be identified, but only one of these was present in the VB16-sorted T-cell population [TCR VA10(b)]. *, identical TCR sequences in PBL and TIL. TCR VA chains combined with VB16 exhibit a common CDR3 motif: TCR VA10 and TCR VA21 share glycine residues (bold), and TCR VA21 shares a serine residue (underlined) with VA22.
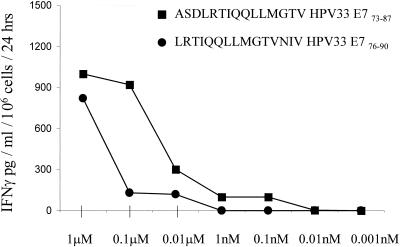
Up till now, most studies dealing with CD4+ cellular immune responses directed against HPV-associated products, particularly E6 and E7, have focused on HPV16 or HPV18 (2, 8). However, HPV33, present in this case, contributes to the high-risk HPV types but represents only a minority of cervical cancer lesions (5). In order to assess immunoreactivity by TIL, we tested a panel of different HPV33 peptides for T-cell recognition as determined in a 24-h IFN-γ secretion assay (Table 3). Based on a computer algorithm (13, 16), eight peptides were selected based on potential binding to DR*0402. Peptides were pulsed onto autologous macrophages, and one peptide (HPV33 E773–87) induced significant IFN-γ secretion in TIL, which could be blocked with the anti-DR-directed MAb but not with control IgG. The same was found to be true for a different peptide (HPV33 E776–90), which induced less IFN-γ secretion in TIL and could also be blocked with the MAb directed against HLA-DR. Titration of these peptides onto autologous cells showed that the peptide derived from HPV33 E773–87 (ASDLRTIQQLLMGTV) represents the dominant epitope defined by TIL (Fig. 3). In contrast, TIL did not secrete significant amounts of IFN-γ in response to a peptide from tetanus toxin.
TABLE 3.
HPV33 E773–87 represents the dominant epitope defined by CD4+ VB16+ TILa
| Peptide | Sequence | IFN-γ secretion (pg/ml per 106 cells per 24 h)
|
||
|---|---|---|---|---|
| Autologous cells (DR*0402) | T2 | T2-DR*0401 | ||
| E7 | 0 | 0 | 0 | |
| E719–3 | PTDLYCYEQLSDSSD | 0 | 0 | 0 |
| E764–78 | TVRLCVNSTASDLRT | 0 | 0 | 0 |
| E773–87 | ASDLRTIQQLLMGTV | 4,000 | 0 | 4,800 |
| E762–76 | NTTVRLCVNSTASDL | 0 | 0 | 0 |
| E75–19 | KPTLKEYVLDLYPEP | 0 | 0 | 0 |
| E79–23 | KEYVLDLYPEPTDLY | 0 | 0 | 0 |
| E776–90 | LRTIQQLLMGTVNIV | 350 | 0 | 0 |
| E779–93 | IQQLLMGTVNIVVPT | 0 | 0 | 0 |
| TT | QYIKANSKFIGITE | 0 | 0 | 0 |
Peptides were pulsed onto autologous antigen-presenting cells (1 μM), T2 cells, or T2 cells expressing the DR*0401 molecule and tested for T-cell recognition by IFN-γ secretion determined by ELISA. The HPV33 E773–87 peptide could be presented by either autologous antigen-presenting cells or T2 cells expressing DR*0401. Nontransfected T2 cells served as a negative control. A tetanus toxin peptide (TT) (28) was not recognized.
FIG. 3.
Titration of HPV33 E773–87 peptides onto autologous antigen-presenting cells. Peptides were serially diluted and pulsed onto antigen-presenting cells, incubated for 2 h at room temperature, and tested for CD4+ VB16+ T-cell recognition, as determined by IFN-γ secretion. The peptide ASDLRTIQQLLMGTV represents the dominant epitope.
In addition to autologous antigen-presenting cells, peptides were also pulsed onto T2 cells transfected with HLA-DR*0401. In contrast to autologous antigen-presenting cells, TIL could exclusively recognize the HPV33 E773–87 peptide (Table 4). Similarly to autologous antigen-presenting cells, T-cell recognition could be blocked with the anti-DR-specific MAb but not with control IgG. At least two possibilities may account for this observation. (i) The HPV33 E773–87 but not the HPV33 E776–90 peptide may bind to HLA-DR*0401 and to the autologous HLA-DR*0402 allele expressed by tumor cells. (ii) The HPV33 E776–90 peptide may not be presented directly but may first be engulfed (e.g., by macrophages), degraded, transported to the cell surface, and presented by MHC class II molecules. The latter possibility is unlikely, since not only DR*0401-transfected T2 cells but also DR*0401-positive macrophages are unable to present the HPV33 E776–90 peptide to TIL. Additionally, different TCR VA chains, either VA10, VA21, or VA22, paired to VB16+ T cells may account for the differential recognition of the closely related HPV33 E7 epitopes.
TABLE 4.
HLA-DR-restricted recognition of HPV33 E773–87 epitope by CD4+ TILa
| Target cells | Blocking MAb | IFN-γ secretion (pg/ml/2 × 106 cells per 24 h) |
|---|---|---|
| Autologous macrophages | IgG | 4,800 |
| Anti-DR | 400 | |
| T2-DR4 | IgG | 3,800 |
| Anti-DR | 0 |
The HPV33 E773–87 peptide was pulsed onto autologous antigen-presenting cells or T2 cells expressing HLA-DR*0401. Antigen-presenting cells were incubated with IgG or anti-DR-directed MAbs (5 μg/well), and an IFN-γ release assay was performed. Different anti-DR-directed MAbs (clone L243 and clone B8.12.2) exhibited similar results. Data are from a blocking experiment with the L243 MAb.
The significance of CD4+-mediated recognition of immune responses directed against cervical cancer has already been suggested in earlier studies. CD4+ T-cell clones obtained from peripheral blood from asymptomatic HPV16+ individuals responded to different amino acid stretches of the HPV E7 protein (2), predominantly to amino acids 67 to 98 of the HPV16 E7 protein (8), to HPV16 L1 peptides (29), or to peptides provided by the less prevalent HPV59 and HPV68 in the context of HLA-DR4 (17). The role of CD4+ T cells in immune surveillance is further underscored by the observation that CD4+ T cells dominate in T-cell infiltrates in clinically regressing genital warts (16). Additionally, decreased CD4+ T-cell numbers enhance progression of HPV-positive cervical lesions to invasive cancer. This may be due to infection with human immunodeficiency virus or to iatrogenically decreased CD4+ T-cell numbers (25). Women diagnosed with cervical cancer appear to show lower CD4 counts than women with preinvasive lesions or healthy controls (14).
IFN-γ secretion by tumor- or peptide-reactive T cells served as the marker for immune recognition in this study. However, other Th1-associated cytokines, e.g., IL-2, appear to represent a good marker for T-helper responses directed to different 20-mer peptides spanning the HPV16 E7 protein, as reported in a different study (8). Interestingly, T-helper reactivity was restricted to patients infected with HPV and associated with viral persistence and disease progression (8). However, these data have been generated by utilizing CD4 T cells from peripheral blood, which is easily accessible for screening but may not necessarily reflect the in situ situation. Analysis of T cells infiltrating into cancer lesions suggested that the expression of HLA-DR by keratinocytes and an increased CD4 T-cell infiltrate of the T-helper 2 type is correlated with high-grade squamous intraepithelial lesions (1).
The biological role of CD4 T cells in orchestrating a cellular immune response may be quite diverse. T-helper cells may provide T-cell help for B-cell responses or cytolytic CD8+ T cells. Alternatively, CD4+ T cells may themselves also be able to mediate tumor regression (15, 30), even if tumor cells lack MHC class II expression (23). This may be important since MHC class I antigen processing or presentation defects appear to be a common event associated with progression of cervical cancer lesions (7). These data suggest that targeting MHC class II responses may benefit patients suffering from cervical cancer and represent an attractive approach to activate or to expand T cells recognizing tumor cells in an MHC class II-restricted fashion.
Acknowledgments
We are grateful to Edgar Hilmes and W. E. Hitzler, Blood Bank, University of Mainz, for MHC typing.
This work was supported by the German Research Foundation (SFB 432/A9).
REFERENCES
- 1.Al-Saleh W, Giannini S L, Jacobs N, Moutschen M, Doyen J, Boniver J, Delvenne P. Correlation of T helper secretory differentiation and types of antigen-presenting cells in squamous intraepithelial lesions of the uterine cervix. J Pathol. 1998;184:283–290. doi: 10.1002/(SICI)1096-9896(199803)184:3<283::AID-PATH25>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 2.Altmann A, Jochmus-Kudielka I, Frank R, Gausepohl H, Moebius U, Gissmann L, Meuer S C. Definition of immunogenic determinants of the human papillomavirus type 16 nucleoprotein E7. Eur J Cancer. 1992;28:26–33. doi: 10.1016/s0959-8049(05)80047-4. [DOI] [PubMed] [Google Scholar]
- 3.Callan M F, Tan L, Annels N, Ogg G S, Wilson J D, O'Callaghan C A, Steven N, McMichael A J, Rickinson A B. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus in vivo. J Exp Med. 1998;187:1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L P, Thomas E K, Hu S L, Hellström I, Hellström K E. Human papillomavirus type 16 nucleoprotein E7 is a tumor rejection antigen. Proc Natl Acad Sci USA. 1991;88:110–114. doi: 10.1073/pnas.88.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole S T, Streeck R E. Genome organization and nucleotide sequence of human papillomavirus 33, which is associated with cervical cancer. J Virol. 1986;58:991–995. doi: 10.1128/jvi.58.3.991-995.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman N, Birley H D, Renton A M, Hanna N F, Ryait B K, Byrne M, Taylor-Robinson D, Stanley M A. Immunological events in regressing genital warts. Am J Clin Pathol. 1994;102:768–774. doi: 10.1093/ajcp/102.6.768. [DOI] [PubMed] [Google Scholar]
- 7.Cromme F V, Airey J, Heemels M T, Ploegh H L, Keating P J, Stern P L, Meijer C J, Walboomers J M. Loss of transporter protein, encoded by the TAP-1 gene, is highly correlated with loss of HLA expression in cervical carcinomas. J Exp Med. 1994;179:335–340. doi: 10.1084/jem.179.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeGruijl T D, Bontkes H J, Walboomers J M, Stukart M J, Doekhie F S, Remmink A J, Helmerhorst T J, Verheijen R H, Duggan-Keen M F, Stern P L, Meijer C J, Scheper R J. Differential T helper cell responses to human papilloma virus type 16 E7 related to viral clearance or persistence in patients with cervical neoplasia: a longitudinal study. Cancer Res. 1998;58:1700–1706. [PubMed] [Google Scholar]
- 9.Dunbar P R, Chen J L, Chao D, Rust N, Teisserenc H, Ogg G S, Romero P, Weynants P, Cerundolo V. Rapid cloning of tumor-specific CTL suitable for adoptive immunotherapy of melanoma. J Immunol. 1999;162:6959–6962. [PubMed] [Google Scholar]
- 10.Dunbar P R, Ogg G S, Chen J, Rust N, van der Bruggen P, Cerundolo V. Direct isolation, phenotyping and cloning of low-frequency antigen-specific cytotoxic T lymphocytes from peripheral blood. Curr Biol. 1998;8:413–427. doi: 10.1016/s0960-9822(98)70161-7. [DOI] [PubMed] [Google Scholar]
- 11.Evans E M, Man S, Evans A S, Borysiewicz L K. Infiltration of cervical cancer tissue with human papillomavirus-specific cytotoxic T-lymphocytes. Cancer Res. 1997;57:2943–2950. [PubMed] [Google Scholar]
- 12.Feltkamp M C W, Vreugdenhil G R, Vierboom M P, Ras E, van der Burg S H, ter Schegget J, Melief C J, Kast W M. Cytotoxic T lymphocytes raised against a subdominant epitope offered as a synthetic peptide eradicate human papillomavirus type 16-induced tumors. Eur J Immunol. 1995;25:2638–2642. doi: 10.1002/eji.1830250935. [DOI] [PubMed] [Google Scholar]
- 13.Friede T, Gnau V, Jung G, Keilholz W, Stevanovic S, Rammensee H G. Natural ligand motifs of closely related HLA-DR4 molecules predict features of rheumatoid arthritis associated peptides. Biochim Biophys Acta. 1996;1316:85–101. doi: 10.1016/0925-4439(96)00010-5. [DOI] [PubMed] [Google Scholar]
- 14.Gemignani M, Maiman M, Fruchter R G, Arrastia C D, Gibbon D, Ellison T. CD4 lymphocytes in women with invasive and preinvasive cervical neoplasia. Gynecol Oncol. 1995;59:364–369. doi: 10.1006/gyno.1995.9961. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg P D. Adoptive T cell therapy of tumors: mechanisms operative in the recognition and elimination of tumor cells. Adv Immunol. 1991;49:281–355. doi: 10.1016/s0065-2776(08)60778-6. [DOI] [PubMed] [Google Scholar]
- 16.Hammer J, Galazzi F, Bono E, Karr R W, Guenot J, Valsasnini P, Nagy Z A, Sinigaglia F. Peptide binding specificity of HLA-DR4 molecules: correlation with rheumatoid arthritis association. J Exp Med. 1995;181:1847–1855. doi: 10.1084/jem.181.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Höhn H, Pilch H, Günzel S, Neukirch C, Hilmes C, Kaufmann A, Seliger B, Maeurer M. CD4+ tumor infiltrating lymphocytes (TIL) in cervical cancer recognize HLA-DR-restricted peptides provided by HPV-E7. J Immunol. 1999;163:5715–5722. [PubMed] [Google Scholar]
- 18.Höhn H, Reichert T, Neukirch C, Pilch H, Maeurer M J. Monoclonal TCR mRNA transcripts are preferentially detected in the TCR variable alpha chain in CD8+ lymphocytes: implications for immunomonitoring. Int J Mol Med. 1996;3:139–144. doi: 10.3892/ijmm.3.2.139. [DOI] [PubMed] [Google Scholar]
- 19.Kadish A S, Ho G Y, Burk R D, Wang Y, Romney S L, Ledwige R, Angeletti R H. Lymphoproliferative responses to human papillomavirus (HPV) type 16 proteins E6 and E7: outcome of HPV infection and associated neoplasia. J Natl Cancer Inst. 1997;89:1285–1293. doi: 10.1093/jnci/89.17.1285. [DOI] [PubMed] [Google Scholar]
- 20.Kono K, Bessing M E, Brandt R M P, Melief C J M, Potkul R K, Andersson B, Petersson M, Kast W M, Kiessling R. Decreased expression of signal-transducing chain in peripheral T cells and natural killer cells in patients with cervical cancer. Clin Cancer Res. 1996;2:1825–1828. [PubMed] [Google Scholar]
- 21.Kuroda M J. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meneguzzi G, Cerni C, Kieny M P, Lathe R. Immunization against human papillomavirus type 16 tumor cells with recombinant vaccinia viruses expressing E6 and E7. Virology. 1991;181:62–69. doi: 10.1016/0042-6822(91)90470-v. [DOI] [PubMed] [Google Scholar]
- 23.Mumberg D, Monach P A, Wanderling S, Philip M, Todedano A Y, Schreiber R D, Schreiber H. CD4+ T cells eliminate MHC class II-negative cancer cells in vivo by indirect effects of IFN-γ. Proc Natl Acad Sci USA. 1999;96:8633–8638. doi: 10.1073/pnas.96.15.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami M, Gurski K J, Marincola F M, Ackland J, Steller M A. Induction of specific CD8+ T lymphocyte responses using a human papillomavirus-16 E6/E7 fusion protein and autologous dendritic cells. Cancer Res. 1999;59:1184–1187. [PubMed] [Google Scholar]
- 25.Petry K U, Scheffel D, Bode U, Gabrysiak T, Kochel H, Kupsch E, Glaubitz M, Niesert S, Kuhnle H, Schedel I. Cellular immunodeficiency enhances the progression of human papillomavirus-associated cervical lesions. Int J Cancer. 1994;57:836–840. doi: 10.1002/ijc.2910570612. [DOI] [PubMed] [Google Scholar]
- 26.Ressing M E, van Driel W J, Celis E, Sette A, Brandt M P, Hartmann M, Anholts J D, Schreuder G M, ter Harmsel W B, Fleuren G J, Trimbos B J, Kast W M, Melief C J. Occasional memory cytotoxic T-cell responses of patients with human papillomavirus type-16 positive cervical lesions against a human leukocyte antigen-A*0201-restricted E7-encoded epitope. Cancer Res. 1996;56:582–588. [PubMed] [Google Scholar]
- 27.Rosenberg S A. A new era for cancer immunotherapy based on the genes that encode cancer antigens. Immunity. 1999;10:281–287. doi: 10.1016/s1074-7613(00)80028-x. [DOI] [PubMed] [Google Scholar]
- 28.Sette A, Sidney J, Oseroff C, del Guercio M F, Southwood S, Arrhenius T, Powell M F, Colon S M, Gaeta F C, Grey H M. HLA DR4w4-binding motifs illustrate the biochemical basis of degeneracy and specificity in peptide-DR interactions. J Immunol. 1993;151:3163–3170. [PubMed] [Google Scholar]
- 29.Shepherd P S, Rowe A J, Cridland J C, Coletart T, Wilson P, Luxton J C. Proliferative T cell responses to human papillomavirus type 16 L1 peptides in patients with cervical dysplasia. J Gen Virol. 1996;77:593–602. doi: 10.1099/0022-1317-77-4-593. [DOI] [PubMed] [Google Scholar]
- 30.Toes R E M, Ossendorf F, Offringa R, Melief C J M. CD4 T cells and their role in antitumor immune responses. J Exp Med. 1999;189:753–756. doi: 10.1084/jem.189.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsukui T, Hildesheim A, Schiffman M H, Lucci J, Contois D, Lawler P, Rush B B, Lorincz A T, Corrigan A, Burk R D, Qu W, Marshall M A, Mann D, Carrington M, Clerici M, Shearer G M, Carbone D P, Scott D R, Houghten R A, Berzofsky J A. Interleukin-2 production in vitro by peripheral lymphocytes in response to human papillomavirus-derived peptides: correlation with cervical pathology. Cancer Res. 1996;56:3967–3974. [PubMed] [Google Scholar]
- 32.Zinkernagel R M, Doherty P C. MHC-restricted cytotoxic T-cells: studies on the biological role of polymorphic major transplantation antigens determining T-cell restriction-specificity, function, and responsiveness. Adv Immunol. 1979;27:51–177. doi: 10.1016/s0065-2776(08)60262-x. [DOI] [PubMed] [Google Scholar]
- 33.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1992;1288:55–78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]