Abstract
Deamination of bases is a form of DNA damage that occurs spontaneously via the hydrolysis and nitrosation of living cells, generating hypoxanthine from adenine. E. coli endonuclease V (eEndoV) cleaves hypoxanthine-containing double-stranded DNA, whereas human endonuclease V (hEndoV) cleaves hypoxanthine-containing RNA; however, hEndoV in vivo function remains unclear. To date, hEndoV has only been examined using hypoxanthine, because it binds closely to the base located at the cleavage site. Here, we examined whether hEndoV cleaves other lesions (e.g., AP site, 6-methyladenine, xanthine) to reveal its function and whether 2′-nucleoside modification affects its cleavage activity. We observed that hEndoV is hypoxanthine-specific; its activity was the highest with 2′-OH modification in ribose. The cleavage activity of hEndoV was compared based on its base sequence. We observed that it has specificity for adenine located on the 3′-end of hypoxanthine at the cleavage site, both before and after cleavage. These data suggest that hEndoV recognizes and cleaves the inosine generated on the poly A tail to maintain RNA quality. Our results provide mechanistic insight into the role of hEndoV in vivo.
Keywords: Deamination, Endonuclease V, Inosine, DNA repair, RNA editing, Poly-A tail
Subject terms: Biochemistry, Enzymes, RNA, Molecular biology, DNA damage and repair, RNA metabolism
Introduction
DNA damage can be caused by spontaneous deamination of bases through hydrolysis and nitrosation, leading to the substitution of adenine, cytosine, and guanine bases with hypoxanthine, uracil, and xanthine, respectively1. The substitution of deoxyinosine, a nucleoside formed when hypoxanthine is attached to a deoxyribose ring, occurs predominantly via two mechanisms: (1) spontaneous hydrolysis or nitrosation within the cell and (2) the insertion of dITP (a deaminated product of dATP) by DNA polymerase during replication, both necessitating repair to safeguard the genome. Consequently, deoxyinosine pairs with cytosine during replication because of its structural similarity to guanine, resulting in an A: T to G: C transition mutation2,3.
Two DNA repair pathways remove this mutagenic deoxyinosine in cells4: (a) the base excision repair (BER) pathway initiated by alkylidene DNA glycosylase (AAG)5, and (b) the alternative excision repair (AER) pathway6, which produces a single nick on one side of the damaged base and removes deoxyinosine using the exonuclease activity of DNA polymerase7. The AER pathway, involving E. coli endonuclease V (eEndoV), is biochemically distinct from the major DNA repair pathways, such as nucleotide excision repair (NER) and BER8–11. eEndoV recognizes double-stranded and single-stranded DNAs containing deoxyinosine, cleaving the second and third phosphodiester bonds at the 3′-end of deoxyinosine. Subsequent 3′- to 5′-exonuclease activity removes deoxyinosine, completing the DNA repair in the eEndoV-dependent AER pathway. Since deoxyinosine is an error-prone lesion, mutations in EndoV result in a strong mutator phenotype in E. coli and S. pombe2,3,12. However, the role of AER pathway in mammals remains unclear.
Deamination reactions may occur in RNA. Inosine is a nucleoside formed when hypoxanthine is attached to a ribose ring. Three fundamental mechanisms contribute to inosine production in RNA4: (a) DNA, spontaneous, and incidental modifications of inosine through deamination via hydrolytic and nitrosative reactions1,13; and (b) inosine triphosphate (ITP) is incorporated into the mRNA by RNA polymerase during transcription. Although ITP is removed from the cell nucleotide pool by inosine triphosphatase (ITPase), the incorporation of ITP is caused by an imbalance in purine nucleotide metabolism14,15, and (c) adenosine is converted to inosine by the RNA-editing enzymes, adenosine deaminases acting on RNA (ADAR) and tRNA (ADAT)16–19. The ADAR family comprises three members: ADAR1, ADAR2, and ADAR3. ADAR1 and ADAR2 catalyze RNA editing, whereas ADAR3 does not. ADAT converts adenosine into inosine in tRNA. Inosine is present at the wobble position of the anticodons.
Quantitative analyses of E. coli and S. cerevisiae revealed that the inosines in the RNA pools were present at background levels of 11 and 42 per 106 nucleotides, respectively. These levels were higher than those of deoxyinosine in DNA (1.2 and 2.0 per 106 nucleotides, respectively)14,15. Therefore, several inosine-containing RNAs are presumed to be present in living mammalian cells.
Previously, we analyzed the function of human endonuclease V (hEndoV) and confirmed that it cleaves DNA-containing deoxyinosines20. Because the amino acid sequences of eEndoV and hEndoV share 37% identity, we assumed that hEndoV could repair deoxyinosine-containing DNA. We observed that this enzyme can cleave RNA-containing inosines. A comparison of the cleavage activity of hEndoV on DNA and RNA showed that it preferentially cleaves inosine-containing RNA20,21. Therefore, hEndoV is an enzyme that removes RNA containing inosine but does not function in DNA repair20. Similarly, Arabidopsis thaliana endonuclease V is a ribonuclease specific for inosine-containing RNA22.
EndoV structures are similar in bacteria and mammals23. hEndoV interacts with inosine before and after the base at the cleavage site. eEndoV and T. thermophilus EndoV (tthEndoV) recognize DNA containing apurinic/apyrimidinic (AP) sites, urea base mismatches, and xanthines8,24,25. Unlike hEndoV, eEndoV, and tthEndoV recognize sites other than inosine. In this study, we investigated the following three factors to analyze the hEndoV function: (a) whether hEndoV cleaves RNA-containing inosines, (b) whether hEndoV needs a 2′-OH substitution in ribose, and (c) whether hEndoV prefers specific sequences. In this study, an inosine 3′ endonuclease activity of hEndoV revealed that hEndoV can cleave poly-A tails containing inosine. Our results suggested that hEndoV is involved in RNA metabolism.
Results
hEndoV is an inosine-specific ribonuclease
EndoV homologs have highly conserved amino acid sequences and structures. eEndoV and tthEndoV cleave DNA containing AP sites, urea base mismatches, or xanthines (Fig. S1A; Table 1). To investigate whether hEndoV could cleave lesions other than deoxyinosine, we tested the cleavage activity of hEndoV on substrates (Fig. S1A), containing deoxyinosine (Fig. S1B, lanes 1–4), and AP sites (Fig. S1B, lanes 5–8), and 6-methyladenine (Fig. S1B, lanes 9–12), and xanthine produced by the deamination of guanine (Fig. S1B, lanes 13–16). Once the oligonucleotides containing deoxyinosine were cleaved by hEndoV (Fig. S1B, lanes 1–4, Fig. S1C), nuclease activity of hEndoV was not observed (Fig. S1B, lanes 5–16, Fig. S1C).
Table 1.
DNA and RNA oligos for human EndoV nuclease activity assay (Related to Figs. 1–5 and Figures S1–S3).
| Nuclease activity assay of human EndoV for inosine | |||||||
|---|---|---|---|---|---|---|---|
| Name | Sequence | ||||||
| ssDNA containing deoxyinosine | 5'-CTGTATGAT | G | dI | rA | G | A | TGCTGAC-3' |
| ssDNA containing AP site | 5'-CTGTATGAT | G | AP | rA | G | A | TGCTGAC-3' |
| ssDNA containing 6-methyladenine | 5'-CTGTATGAT | G | Me-A | rA | G | A | TGCTGAC -3' |
| ssDNA containing xanthine | 5'-CTGTATGAT | G | X | rA | G | A | TGCTGAC-3' |
| Nuclease activity assay of human EndoV for ribo-adenine | |||||||
|---|---|---|---|---|---|---|---|
| Name | Sequence | ||||||
| dI next to ribo-A | 5'-CTGTATGAT | G | dI | rA | G | A | TGCTGAC -3' |
| dI next to deoxy-A | 5'-CTGTATGAT | G | dI | dA | G | A | TGCTGAC -3' |
| dI next to 2'-O-Me-A | 5'-CTGTATGAT | G | dI | O-Me-A | G | A | TGCTGAC -3' |
| dI next to 2'-F-A | 5'-CTGTATGAT | G | dI | F-A | G | A | TGCTGAC -3' |
| Nuclease activity assay of human EndoV for nucleotide located -1 of cleavage site | |||||||
|---|---|---|---|---|---|---|---|
| Name | Sequence | ||||||
| dI next to ribo-A | 5'-CTGTATGAT | G | dI | rA | G | A | TGCTGAC -3' |
| dI next to ribo-C | 5'-CTGTATGAT | G | dI | rC | G | A | TGCTGAC -3' |
| dI next to ribo-G | 5'-CTGTATGAT | G | dI | rG | G | A | TGCTGAC -3' |
| dI next to riboU | 5'-CTGTATGAT | G | dI | rU | G | A | TGCTGAC -3' |
| Nuclease activity assay of human EndoV for nucleotide located + 1 cleavage site | |||||||
|---|---|---|---|---|---|---|---|
| Name | Sequence | ||||||
| dI two next to ribo-A | 5'-CTGTATGAT | G | dI | rA | rA | A | TGCTGAC-3' |
| dI two next to ribo-C | 5'-CTGTATGAT | G | dI | rA | rC | A | TGCTGAC-3' |
| dI two next to ribo-G | 5'-CTGTATGAT | G | dI | rA | rG | A | TGCTGAC-3' |
| dI two next to ribo-U | 5'-CTGTATGAT | G | dI | rA | rU | A | TGCTGAC-3' |
| Nuclease activity assay of human EndoV for nucleotide located + 2 of cleavage site | |||||||
|---|---|---|---|---|---|---|---|
| Name | Sequence | ||||||
| dI third next to ribo-A | 5'-CTGTATGAT | G | dI | rA | rA | rA | TGCTGAC-3' |
| dI third next to ribo-C | 5'-CTGTATGAT | G | dI | rA | rA | rC | TGCTGAC-3' |
| dI third next to ribo-G | 5'-CTGTATGAT | G | dI | rA | rA | rG | TGCTGAC-3' |
| dI third next to ribo-U | 5'-CTGTATGAT | G | dI | rA | rA | rU | TGCTGAC-3' |
| Nuclease activity assay of human EndoV for nucleotide located -3 of cleavage site | |||||||
|---|---|---|---|---|---|---|---|
| Name | Sequence | ||||||
| dI ahead of ribo-A | 5'-CTGTATGAT | rA | dI | A | G | A | TGCTGAC-3' |
| dI ahead of ribo-C | 5'-CTGTATGAT | rC | dI | A | G | A | TGCTGAC-3' |
| dI ahead of ribo-G | 5'-CTGTATGAT | rG | dI | A | G | A | TGCTGAC-3' |
| dI ahead of ribo-U | 5'-CTGTATGAT | rU | dI | A | G | A | TGCTGAC-3' |
| Nuclease activity assay of human EndoV for ribo-adenine | |||||||
|---|---|---|---|---|---|---|---|
| Name | Sequence | ||||||
| dl next to rA dA dA | 5'-CTGTATGAT | G | dI | rA | dA | dA | TGCTGAC-3' |
| dl next to rA rA dA | 5'-CTGTATGAT | G | dI | rA | rA | dA | TGCTGAC-3' |
| dl next to rA dA dA | 5'-CTGTATGAT | G | dI | rA | rA | rA | TGCTGAC-3' |
| Nuclease activity assay of human EndoV for poly-A | |||||||
|---|---|---|---|---|---|---|---|
| Name | Sequence | ||||||
| Poly-A | 5'-rArArArArArArArArArArArArArArArArArArArArA- 3' | ||||||
| i-PolyA | 5'-rArArArArArArArArArArIrArArArArArArArArArA-3' | ||||||
Effect of hEndoV cleavage activity on qualified 2′ of nucleoside
eEndoV cleaves ssDNA-containing deoxyinosines more specifically than ssRNA-containing deoxyinosines. On the contrary, hEndoV preferred ssRNA containing inosine over ssDNA containing deoxyinosine and a hydroxy substitution in ribose located at 3′ to inosine20. Thus, we assumed that hydroxy in the 2′ position in nucleoside affects the cleavage activity of hEndoV compared to DNA and RNA.
To confirm whether substitution at 2′ of nucleoside affects the cleavage activity of hEndoV, we tested the nuclease activity of hEndoV on single-strand oligonucleotides containing ribo (hydroxy), deoxy, methoxy, and fluorine substitutions (Fig. S2A; Table 1). We observed that hEndoV preferred hydroxy substitution at 2′ in nucleoside (Fig. S2B, lanes 1–4, Fig. S2C) compared to deoxy, methoxy, and fluorine substitutions (Fig. S2B, lanes 5–16, Fig. S2C). In other words, this protein shows high RNA cleavage activity. Our results suggest that substituting nucleoside at 2′ plays a key role in the cleavage activity of hEndoV.
The cleavage activity of hEndoV depends on the nucleotide sequence
Previous studies on the crystal structure of EndoV and a comparison of the alignment of the amino acid sequences of EndoV homologs revealed the following characteristic similarities between hEndoV and eEndoV: identity = 74/199 (37%), positivity = 108/199 (54%), and gap = 19/199 (10%)20, demonstrating that these proteins are highly conserved in bacteria and humans. Notably, electrophoretic mobility shift assays revealed that hEndoV has affinity for both RNA- and DNA-containing inosines. This enzyme also has an affinity for dsRNA containing no inosine, indicating that the inosine in RNA is dispensable for RNA binding of hEndoV20.
However, our structural analyses revealed that EndoV binds closely to the 1 and + 1 bases of the cleavage site23. Therefore, we investigated whether the base sequence of the deoxyinosine affected the cleavage activity of hEndoV.
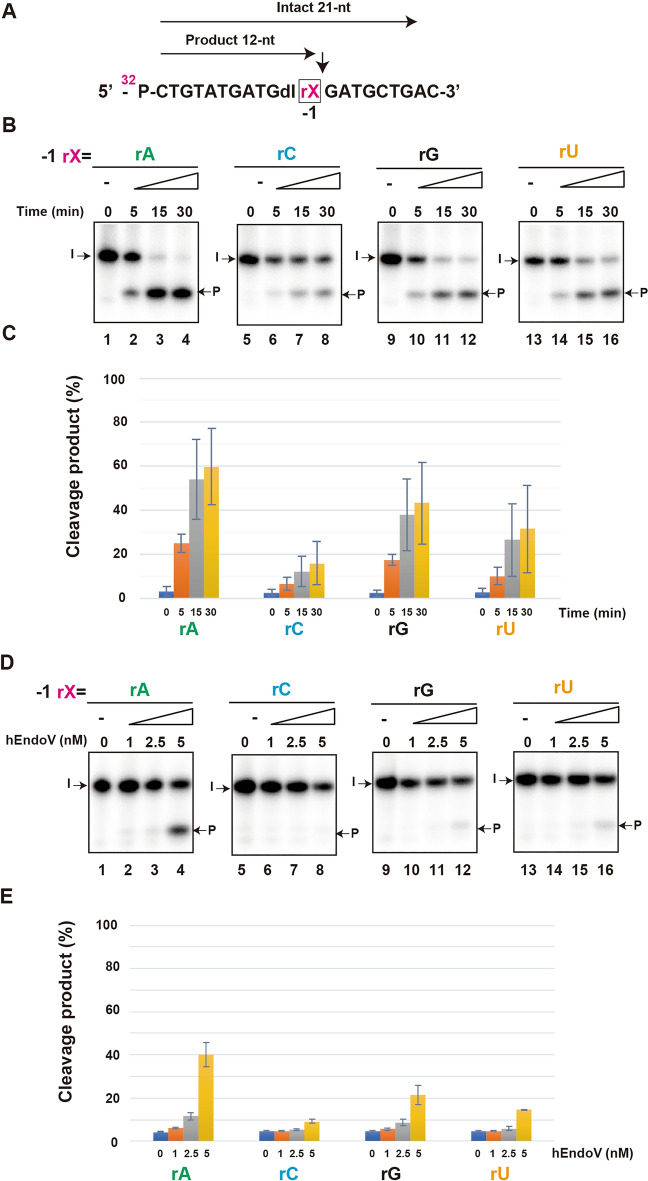
First, we compared the cleavage activity by changing the base located next to the 3′ of deoxyinosine, replacing the –1 located base of the cleavage site with adenosine (rA), cytidine (rC), guanosine (rG), and uridine (rU) (Fig. 1A; Table 1). For statistical analysis, time-course and concentration experiments were performed thrice (Fig. 1B,D), and the cleavage products were quantified (Fig. 1C,E). We observed that the cleavage activity of hEndoV was the highest with the adenine base in a time- and concentration-dependent manner (Fig. 1B,D, lanes 1–4, Fig. 1C,E).
Figure 1.
hEndoV prefers adenine base located at the –1 position site. (A) 32P-labeled substrates containing rX (rA, rC, rG, or rU) at the –1 position are indicated. Arrows indicate the position of cleavage. Intact 21-nt; Intact oligonucleotide. Product 12-nt; cleavage products. (B) The 32P-labeled substrate containing rA (lanes 1–4), rC (lanes 5–8), rG (lanes 9–12), and rU (lanes 13–16) were incubated with 5 nM hEndoV at 37 ℃ for indicated time. The cleavage products were analyzed by denaturing 12.5% urea gel electrophoresis. The arrows mark the positions of intact oligonucleotide (I), and product (P) after hEndoV endonuclease action. (C) Graphs showing the yield of cleavage products obtained from (B) hEndoV cleavage activity. (D) The 32P-labeled substrate containing rA (lanes 1–4), rC (lanes 5–8), rG (lanes 9–12), and rU (lanes 13–16) were incubated with indicated concentrations of hEndoV for 30 min. (E) Graphs showing the yield of cleavage products obtained from (D) hEndoV cleavage activity. Abbreviations: hEndoV, human endonuclease V; P, phosphorus; rA, riboadenine; rC, ribocytosine; rG, riboguanine; rU, ribouracil. Representatives of three independent experiments are shown.
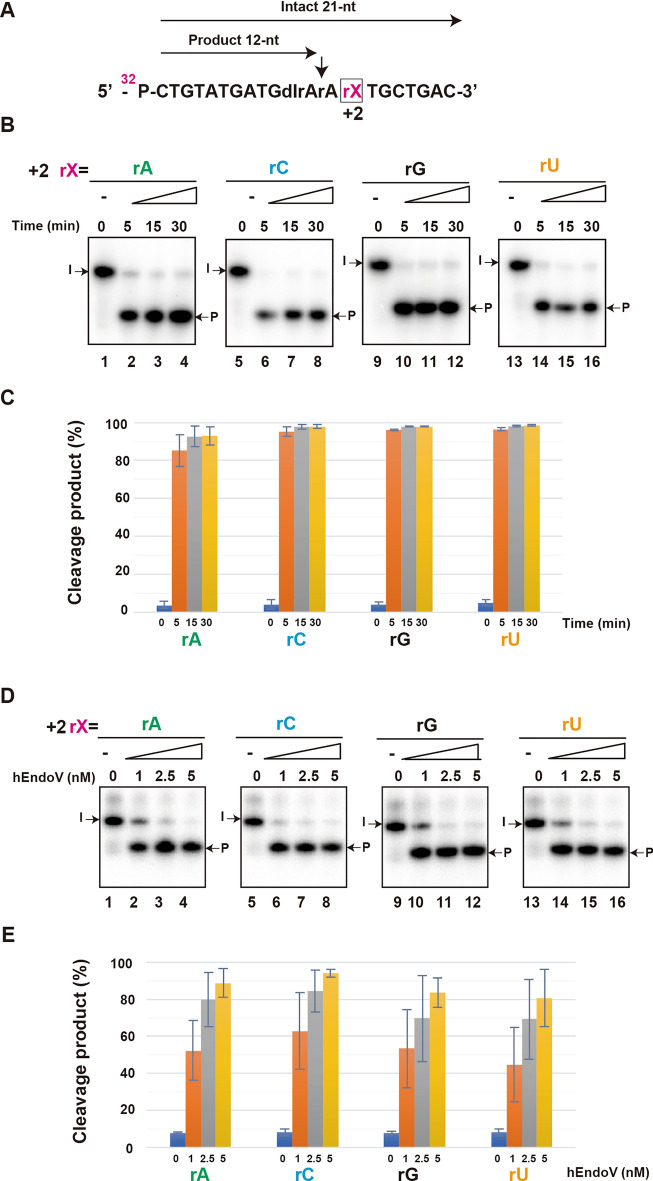
Next, we changed the second base located at 3′ of inosine, replacing the + 1 located base of the cleavage site with rA, rC, rG, and rU to compare the cleavage activity of hEndoV (Fig. 2A; Table 1). The cleavage activity of hEndoV with rA and rG was higher than that with rC and rU in a time- and concentration-dependent manner (Fig. 2B,E, lanes 1–4 and lanes 9–12, Fig. 2C,E). These results suggest that the –1 and + 1 bases, to which hEndoV binds, are important for its cleavage activity and that hEndoV has specificity for rA.
Figure 2.
hEndoV prefers adenine base located at the + 1 position site. (A) 32P-labeled substrates containing rX (rA, rC, rG, or rU) at the + 1 position. Arrows indicate cleavage positions. Intact 21-nt; Intact oligonucleotide. Product 12-nt; cleavage products. (B) The 32P-labeled substrate containing rA (lanes 1–4), rC (lanes 5–8), rG (lanes 9–12), and rU (lanes 13–16) were incubated with 5 nM hEndoV at 37 ℃ for indicated time. The cleavage products were analyzed by denaturing 12.5% urea gel electrophoresis. Arrows mark the positions of the intact oligonucleotide (I) and the product (P) after hEndoV endonuclease activity. (C) Graphs showing the yield of the cleavage products obtained from (B) hEndoV cleavage. (D) 32P-labeled substrates containing rA (lanes 1–4), rC (lanes 5–8), rG (lanes 9–12), and rU (lanes 13–16) were incubated with the indicated concentrations of hEndoV for 30 min. (E) Graphs showing the yield of cleavage products obtained from (D) hEndoV cleavage. Abbreviations: hEndoV, human endonuclease V; P, phosphorus; rA, riboadenine; rC, ribocytosine; rG, riboguanine; rU, ribouracil Representatives of three independent experiments are shown.
Finally, we changed the third base located at 3′ of inosine (+ 2 located base of cleavage site), and the base located at 5′ to inosine (–3 located base of cleavage site), replacing them with rA, rC, rG, and rU to compare the cleavage activity of hEndoV (Fig. 3A and Fig. S3A; Table 1).
Figure 3.
hEndoV prefers adenine base located at the + 2 position site. (A) 32P-labeled substrates containing rX (rA, rC, rG, or rU) at the + 2 position. Arrows indicate cleavage positions. Intact 21-nt; Intact oligonucleotide. Product 12-nt; cleavage products. (B) The 32P-labeled substrate containing rA (lanes 1–4), rC (lanes 5–8), rG (lanes 9–12), and rU (lanes 13–16) were incubated with 5 nM hEndoV at 37 ℃ for indicated time. The cleavage products were analyzed by denaturing 12.5% urea gel electrophoresis. Arrows mark the positions of the intact oligonucleotide (I) and the product (P) after hEndoV endonuclease activity. (C) Graphs showing the yield of the cleavage products obtained from (B) hEndoV cleavage. (D) 32P-labeled substrates containing rA (lanes 1–4), rC (lanes 5–8), rG (lanes 9–12), and rU (lanes 13–16) were incubated with the indicated concentrations of hEndoV for 30 min. (E) Graphs showing the yield of cleavage products obtained from (D) hEndoV cleavage. Abbreviations: hEndoV, human endonuclease V; P, phosphorus; rA, riboadenine; rC, ribocytosine; rG, riboguanine; rU, ribouracil Representatives of three independent experiments are shown.
However, we observed no difference in the cleavage activity of hEndoV, regardless of the type of replacement at the + 2 located base (Fig. 3B–E) in a time- and concentration-dependent manner or at the 3 located base (Fig. S3), in a time-dependent manner. Based on these results, we suggest that the cleavage activity of hEndoV is specific to the adenine base located at the cleavage site, implying that hEndoV prefers the IAA base sequence.
hEndoV prefers a hydroxy substitution in adenosine at + 1 position
A previous study showed that hEndoV recognizes both ssDNA and dsDNA containing deoxyinosine, and ssRNA and dsRNA containing inosine. Additionally, we confirmed that this protein exhibits nuclease activity against RNA. Only slight cleavage activity was observed for ssDNA, albeit only slightly26. Therefore, we compared the cleavage activity of hEndoV for 2′-OH substituted ribose by changing the – 1, + 1, and + 2 located base sequences to deoxyadenosine (A) and adenosine (rA) to confirm whether hEndoV requires a hydroxy substitution at 2′ of ribose and an adenine base (Fig. 4A; Table 1). Cleavage activity was slightly lower for the rAAA sequence and approximately the same for the rArAA and rArArA sequences. These results suggest that the OH group in adenosine at the + 1 position contributes to activity (Fig. 4B,C). These results confirmed that hEndoV prefers the IAA sequence (Fig. 4D).
Figure 4.
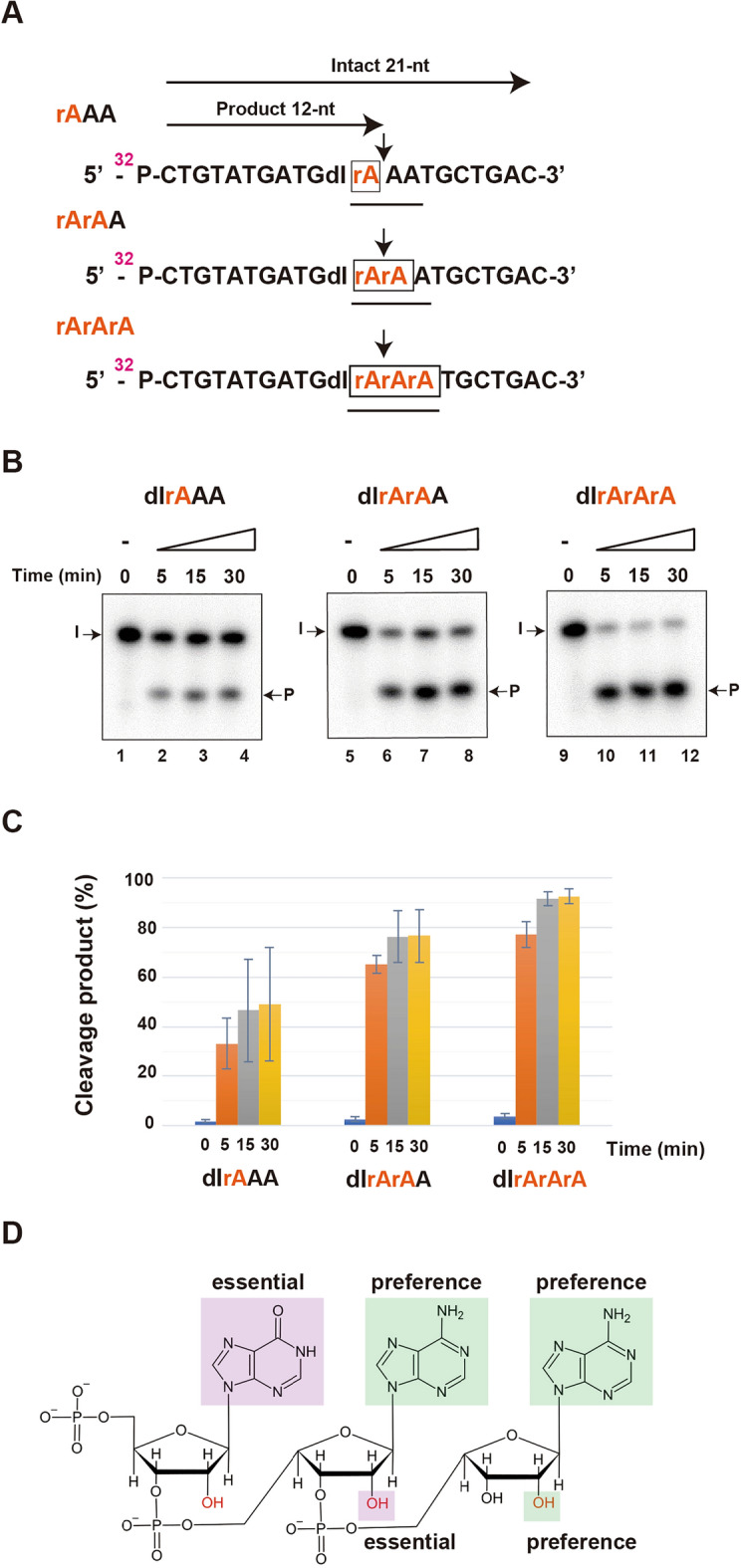
hEndoV prefers a hydroxy (–OH) modification of adenosine. (A) 32P-labeled substrates containing deoxyinosine and a series of adenosine modifications are shown. Intact 21-nt; Intact oligonucleotide. Product 12-nt; cleavage products. (B) 32P-labeled substrate containing deoxyinosine and rAAA (lanes 1–4), rArAA (lanes 5–8) and rArArA (lanes 9–12) were incubated with 5 nM hEndoV at 37 ℃ for 0, 5, 15 and 30 min. (C) The cleavage products were analyzed by denaturing 12.5% urea gel electrophorese. (D) Preference of chemical structure in hEndoV cleavage activity. Abbreviations: hEndoV, human endonuclease V; P, phosphorus; rA, ribo-adenine; dI, deoxyinosine; A, deoxy-adenine Representatives of three independent experiments are shown.
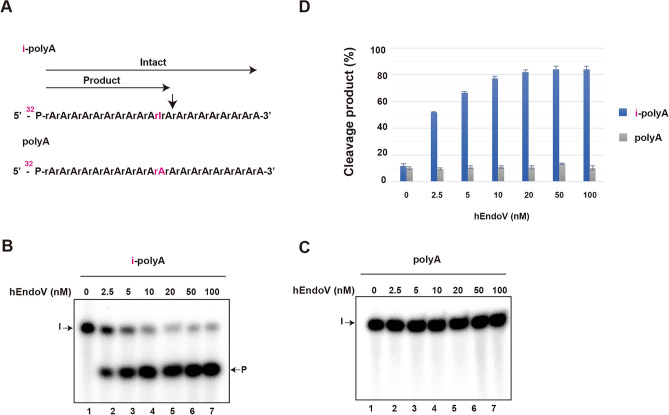
hEndoV can cleave poly-A tail containing inosine
As our data indicate that hEndoV prefers ssRNAs containing inosine and adenine, which are located at the cleavage site before and after cleavage, we hypothesized that hEndoV recognizes and cleaves the poly (A) tail of RNA, including inosine. To confirm this, we examined whether hEndoV cleaves the poly A tail containing inosine using 21-mer-labeled poly A tail substrates and poly A tail containing inosine (i-poly A tail substrate) (Fig. 5A; Table 1). As expected, the results showed that hEndoV cleaved the i-poly-A tail in a concentration-dependent manner, but not the poly-A tail substrates (Fig. 5B,C,D).
Figure 5.
hEndoV can cleave poly-A tail containing inosine. (A) 32P-labeled inosine-containing poly-A (i-poly-A) or poly-A substrates are shown. 32P-labeled (B) i-poly-A substrates and (C) poly-A were incubated with indicated concentrations of hEndoV for 30 min. (D) The cleavage products were analyzed by denaturing 12.5% urea gel electrophorese. Abbreviations: hEndoV, human endonuclease V; P, phosphorus; rA, ribo-adenine; rI, inosine; i-polyA, inosine-containing poly-A; polyA, poly-A Representatives of three independent experiments are shown.
Discussion
In the present study, we analyzed the cleavage activity of hEndoV. Although hEndoV is structurally similar to eEndoV, which recognizes AP sites and mismatched bases, it shows high specificity only for inosines8,24,25. We examined whether hEndoV can cleave substrates other than RNA and observed that the cleavage activity was higher in the presence of a 2′ modification in ribose sugar. The cleavage activity of hEndoV was compared based on alterations in the base sequences. hEndoV showed the highest preference for adenine residues and its cleavage activity was highest when the bases before and after the cleavage site were adenine residues. In addition, cleavage activity was higher when the bases before and after the cleavage site contained ribose rather than deoxyribose, indicating that hEndoV prefers RNA to DNA. Taken together, we showed that hEndoV has endonuclease activity, which specifically cleaves single-stranded RNA containing inosine; the cleavage activity tends to be higher when the base on the 3′ end of inosine is adenine.
Intracellular functions of hEndoV
Studies have shown that eEndoV efficiently cleaves DNA-containing deoxyinosine and acts as a DNA repair enzyme in the AER pathway9,10,27. In contrast, hEndoV is less active on deoxyinosine-containing DNA and preferentially cleaves inosine-containing RNA. It has been reported that hEndoV is localized in the cytoplasm20. Furthermore, human alkyladenine DNA glycosylase (hAAG) in the BER pathway is a DNA glycosylase responsible for recognizing deoxyinosine28,29. Therefore, hEndoV is believed to remove RNAs with inosine rather than repair deoxyinosine-damaged DNAs. Our data suggests that hEndoV preferred inosine-containing RNA substrates with the 5′-IAA-3′ sequence and could cleave an inosine-containing poly-A RNA substrate. Therefore, we hypothesized that hEndoV functions against the inosine generated in the poly A tail of RNA and removes untranslatable mature mRNAs containing inosine due to RNA damage.
The poly A tail is a significant modification in RNA processing and is involved in RNA stability and translation efficiency30,31. Polyadenylation involves the addition of up to 250 adenosine residues by poly A polymerase. The 5′-capping structure in mature mRNA is directly bound by the translation factors with the poly-A binding protein (PABP), thus forming a complex that circularizes the mRNA32,33. When inosine is introduced into the poly A tail, it is recognized as guanine, and PABP barely binds to the poly A tail, forming an unstable mRNA ring.
We believe that inosine is generated in the poly (A) tail through three main mechanisms. First is incorporating ITP by poly-A polymerase, which recognizes the 2- carbon of ATP and uses only ATP as a substrate. This enzyme cannot incorporate guanosine triphosphate (GTP) in which the 2′- carbon is present in an amino group14,18. However, it may be able to incorporate ITP, the deaminated product of ATP, because 2′- carbon in ITP has the same structure as that in ATP.
The second method is RNA editing using ADARs16–18. These enzymes convert adenosine residues into inosines in double-stranded RNA but not in single-stranded RNA. Because poly (A) tails are single-stranded RNA, ADARs may barely produce inosine.
The third is spontaneous deamination by hydrolysis or nitrosation reactions1,13. Chemical reactions are generally reliable in living cells. As these deamination reactions occur automatically in a time-dependent manner, the adenosine in the poly A tail of the mRNA may be partially converted to inosine. We believe that hEndoV contributes to the removal of mature mRNA by recognizing and cleaving the inosine generated on the poly A tails for RNA quality control.
Materials and methods
Proteins
The hEndoV protein was expressed in E. coli Rosetta 2 (Novagen) using pGEX-hEndoV and purified using DEAE Sepharose FF, Glutathione Sepharose 4 Fast Flow, and HiTrap Heparin HP columns (GE Healthcare) following the manufacturer's instructions. The proteins from the HiTrap Heparin HP column were eluted using a solution containing 300 mM KCl, 20 mM Tris–HCl buffer (pH 8.0), 10% glycerol, 0.1 mM ethylenediaminetetraacetic acid, and 1 mM dithiothreitol. Protein concentrations were measured using the Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, CA, USA). hEndoV was observed as a single band on SDS-PAGE with Coomassie blue staining, according to a previous report20. The T4 polynucleotide kinase was obtained from New England Biolabs (Ipswich, MA, USA).
DNA and RNA oligonucleotides
The DNA and RNA oligonucleotides used in this study are listed in Table 1. These oligonucleotides were synthesized at FASMAC (Kanagawa, Japan) and purified using high-performance liquid chromatography (HPLC). For the nuclease assay, oligonucleotides were phosphorylated at 5′ position using (γ-32P)-ATP and T4 polynucleotide kinase. The unincorporated nucleotides were removed using a MicroSpin G-25 column (Cytiva, Marlborough, MA, USA). Because the nuclease activity of hEndoV is unchanged between inosine and deoxyinosine and requires a second ribose at 3′ to the lesion20, here we used a DNA substrate containing the second ribose at 3′ to the deoxyinosine (Table 1).
hEndoV nuclease assays
Reaction mixtures (5 μL) containing 0.2 nM of 32P-labelled oligonucleotides and the indicated amount of hEndoV were added in a reaction buffer containing 50 mM potassium acetate, 20 mM Tris–acetate, 10 mM magnesium acetate, and 1 mM dithiothreitol (DTT) (pH 7.9). The mixtures were incubated at 37 ℃ for the indicated time or concentration and then added to sequencing stop buffer (5 μL) containing 98% formamide, 20 mM ethylenediaminetetraacetic acid (EDTA), 0.5% bromophenol blue, and 0.5% xylene cyanol to terminate the reaction. The reaction products were separated on 12.5% denaturing urea gels. The dried gel was analyzed using a Fuji FLA-7000 phosphorimeter (Fujifilm, Tokyo, Japan). The products were quantified using the Fuji MultiGauge Software (Fujifilm, Tokyo, Japan). Photostimulated luminescence (PSL) was used for product counting. Cleavage products (%) = (products / (products + intact oligonucleotides)) × 100. The same experiment was performed thrice for statistical analysis. The error bars represent the standard error of the mean (SEM).
Supplementary Information
Acknowledgements
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan [Grant-in-Aid for Scientific Research (B) 26650006] and the Central Research Institute of Fukuoka University [GR2310]. We thank Editage (www.editage.com) for providing editing support for this manuscript.
Author contributions
S. I. and I. K. conceived the project; K. M., J. K., A. Y., A. M., N. S., and I. K. conducted the experiments; K. M., J. K., A. Y., A. N. S., M., S. I., and I. K. interpreted the data; and K. M., S. I., and I. K. wrote the manuscript. All the authors discussed the results and commented on the manuscript.
Data availability
All main data of the study appear in the submitted article. Supplementary data are available online.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-65814-7.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Schouten KA, Weiss B. Endonuclease V protects Escherichia coli against specific mutations caused by nitrous acid. Mutat. Res. 1999;435:245–254. doi: 10.1016/S0921-8777(99)00049-X. [DOI] [PubMed] [Google Scholar]
- 3.Weiss B. Endonuclease V of Escherichia coli prevents mutations from nitrosative deamination during nitrate/nitrite respiration. Mutat. Res. 2001;461:301–309. doi: 10.1016/S0921-8777(00)00062-8. [DOI] [PubMed] [Google Scholar]
- 4.Kuraoka I. Diversity of endonuclease V: From DNA repair to RNA editing. Biomolecules. 2015;5:2194–2206. doi: 10.3390/biom5042194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc. Natl Acad. Sci. U.S.A. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasui A. Alternative excision repair pathways. Cold Spring Harb. Perspect. Biol. 2013;5:a012617. doi: 10.1101/cshperspect.a012617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CC, et al. Endonuclease V-mediated deoxyinosine excision repair in vitro. DNA Repair. (Amst.) 2010;9:1073–1079. doi: 10.1016/j.dnarep.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Yao M, Hatahet Z, Melamede RJ, Kow YW. Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′ endonuclease, from Escherichia coli. J. Biol. Chem. 1994;269:16260–16268. doi: 10.1016/S0021-9258(17)34002-4. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, He B, Qing H, Kow YW. A deoxyinosine specific endonuclease from hyperthermophile, Archaeoglobus fulgidus: A homolog of Escherichia coli endonuclease V. Mutat. Res. 2000;461:169–177. doi: 10.1016/S0921-8777(00)00054-9. [DOI] [PubMed] [Google Scholar]
- 10.Moe A, et al. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucl. Acids Res. 2003;31:3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demple B, Linn S. On the recognition and cleavage mechanism of Escherichia coli endodeoxyribonuclease V, a possible DNA repair enzyme. J. Biol. Chem. 1982;257:2848–2855. doi: 10.1016/S0021-9258(19)81041-4. [DOI] [PubMed] [Google Scholar]
- 12.Dalhus B, Alseth I, Bjørås M. Structural basis for incision at deaminated adenines in DNA and RNA by endonuclease V. Prog. Biophys. Mol. Biol. 2015;117:134–142. doi: 10.1016/j.pbiomolbio.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog. Nucl. Acid Res. Mol. Biol. 1979;22:135–192. doi: 10.1016/S0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- 14.Pang B, et al. Defects in purine nucleotide metabolism lead to substantial incorporation of xanthine and hypoxanthine into DNA and RNA. Proc. Natl Acad. Sci. U.S.A. 2012;109:2319–2324. doi: 10.1073/pnas.1118455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol. 2010;6:733–740. doi: 10.1038/nchembio.434. [DOI] [PubMed] [Google Scholar]
- 16.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grice LF, Degnan BM. The origin of the ADAR gene family and animal RNA editing. BMC Evol. Biol. 2015;15:4. doi: 10.1186/s12862-015-0279-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melcher T, et al. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 19.Lai F, Chen CX, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol. Cell. Biol. 1997;17:2413–2424. doi: 10.1128/MCB.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita Y, et al. Human endonuclease v is a ribonuclease specific for inosine-containing RNA. Nat. Commun. 2013;4:2273. doi: 10.1038/ncomms3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vik ES, et al. Endonuclease v cleaves at inosines in RNA. Nat. Commun. 2013;4:2271. doi: 10.1038/ncomms3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endo M, Kim JI, Shioi NA, Iwai S, Kuraoka I. Arabidopsis thaliana endonuclease V is a ribonuclease specific for inosine-containing single-stranded RNA. Open Biol. 2021;11:210148. doi: 10.1098/rsob.210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Samara NL, Kuraoka I, Yang W. Evolution of inosine-specific endonuclease V from bacterial DNase to eukaryotic RNase. Mol. Cell. 2019;76:44–56.e3. doi: 10.1016/j.molcel.2019.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao M, Kow YW. Strand-specific cleavage of mismatch-containing DNA by deoxyinosine 3′-endonuclease from Escherichia coli. J. Biol. Chem. 1994;269:31390–31396. doi: 10.1016/S0021-9258(18)31706-X. [DOI] [PubMed] [Google Scholar]
- 25.Yao M, Kow YW. Further characterization of Escherichia coli endonuclease V. Mechanism of recognition for deoxyinosine, deoxyuridine, and base mismatches in DNA. J. Biol. Chem. 1997;272:30774–30779. doi: 10.1074/jbc.272.49.30774. [DOI] [PubMed] [Google Scholar]
- 26.Morita Y, Iwai S, Kuraoka I. A method for detecting genetic toxicity using the rna synthesis response to dna damage. J. Toxicol. Sci. 2011;36:515–521. doi: 10.2131/jts.36.515. [DOI] [PubMed] [Google Scholar]
- 27.Lee CC, et al. The excision of 3′ penultimate errors by DNA polymerase I and its role in endonuclease V-mediated DNA repair. DNA Repair. (Amst.) 2013;12:899–911. doi: 10.1016/j.dnarep.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012583–a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace SS. Base excision repair: A critical player in many games. DNA Repair. (Amst.) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 31.Tarun SZ, Sachs AB. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 32.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/S1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 33.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All main data of the study appear in the submitted article. Supplementary data are available online.