Summary
HIV-1 hijacks host proteins involved in membrane trafficking, endocytosis, and autophagy that are critical for virus replication. Molecular details are lacking but are essential to inform on the development of alternative antiviral strategies. Despite their potential as clinical targets, only a few membrane trafficking proteins have been functionally characterized in HIV-1 replication. To further elucidate roles in HIV-1 replication, we performed a CRISPR-Cas9 screen on 140 membrane trafficking proteins. We identified phosphatidylinositol-binding clathrin assembly protein (PICALM) that influences not only infection dynamics but also CD4+ SupT1 biology. The knockout (KO) of PICALM inhibited viral entry. In CD4+ SupT1 T cells, KO cells exhibited defects in intracellular trafficking and increased abundance of intracellular Gag and significant alterations in autophagy, immune checkpoint PD-1 levels, and differentiation markers. Thus, PICALM modulates a variety of pathways that ultimately affect HIV-1 replication, underscoring the potential of PICALM as a future target to control HIV-1.
Subject areas: Human genetics, Virology, Cell biology
Graphical abstract

Highlights
-
•
PICALM knockout cells reveal a link between endocytosis and metabolic pathways
-
•
PICALM modulates HIV-1 entry, viral replication, and autophagy flux
-
•
PICALM knockout leads to PD-1 downregulation and modulates cell differentiation
-
•
The upregulation PICALM may represent a new avenue to restrict HIV-1 replication
Human genetics; Virology; Cell biology
Introduction
Human immunodeficiency virus type 1 (HIV-1) infects 1.5 million people per year and represents a prominent global threat despite progress made with prevention and combined antiretroviral drug therapies (UNAIDS). Further research toward the development of new strategies and therapeutic approaches remains a high priority. Considering that HIV-1 and other viruses are obligatory intracellular pathogens, host factors are usurped and are critical for all stages of their replication cycles. HIV-1 replication hijacks various components of membrane trafficking pathways, including several Rab proteins implicated in virion budding.1,2,3,4,5 Interplay of HIV-1 with autophagic and lysosomal trafficking pathways has also been demonstrated, with HIV-1 negatively modulating autophagic flux by counteracting trafficking and the acidification of late endosomes and lysosomes to block their fusion with autophagosomes.6 These findings underscore that HIV-1 has evolved to reprogram intracellular host membrane trafficking proteins to facilitate infection, but the complete network of host proteins and their mechanisms of action during HIV-1 replication are unknown. Elucidating host networks and performing functional characterization of key proteins will contribute to our understanding of HIV-1 replication dynamics and provide new antiviral therapeutic targets.
The recent development of the ingenious clustered regularly interspaced short palindromic repeats (CRISPR) system has revolutionized research.7,8,9 CRISPR-associated protein 9 (Cas9) screens are indeed more robust and specific than shRNA screens,10 and have recently been used to identify novel host factors required for viral replication.11,12,13,14 In this study, we applied a CRISPR-Cas9 screen targeting 140 membrane trafficking proteins to identify those that are essential for HIV-1 replication, and performed validation studies of several candidate proteins by generating stable knockout (KO) cell lines to elucidate their roles in HIV-1 replication.
Among other proteins of interest, the screen identified phosphatidylinositol-binding clathrin assembly protein (PICALM; CALM, clathrin assembly lymphoid myeloid leukemia protein) as pertinent for HIV-1 infection of CD4+ T cells. The PICALM gene encodes the PICALM protein, which is one of the three most abundant proteins of clathrin-coated vesicles,15 and critical in mediating clathrin-mediated endocytosis through their binding to clathrin-coated vesicles in order to regulate their size, maturation and endocytic rates at the plasma membrane.15,16,17 The interaction of complex mammalian membrane trafficking protein networks with numerous other diverging cellular pathways positions PICALM as an important regulator of macroautophagy. Decreased PICALM expression in neurodegenerative diseases is the most significant genetic risk factor for Alzheimer’s disease, where PICALM deregulates autophagic flux by increasing autophagosomes and an associated hindrance in autophagosome-lysosome fusion.18,19,20,21,22,23 PICALM is recognized for its involvement in immune functions, including the regulation of pro-inflammatory TLR4 signaling,24 and has been implicated in the replication of herpes simplex viruses and enteroviruses.25,26
In this work, we show that CRISPR-mediated PICALM KO leads to impaired HIV-1 entry in CD4+ SupT1 cells, providing evidence that entry of HIV-1 into T cells also occurs via clathrin-mediated endocytosis. PICALM KO CD4+ SupT1 T cells also showed an increased autophagic flux resulting in increased HIV-1 Gag expression, increased productive infection, and altered PD-1 expression that led to a highly active and proliferative state.
Results
CRISPR screen of membrane trafficking genes reveals HIV-1 modulators
CRISPR genetic screens have greatly improved discovery efforts aimed at uncovering essential host factors required for HIV-1 replication. Several studies have focused on identifying new host dependency factors (HDFs) that maintain viral latency,27,28,29,30 while others have identified factors involved in viral integration processes.31,32,33 However, a CRISPR screen specifically focused on identifying new HDFs involved in membrane trafficking pathway has yet to be reported.
To uncover membrane trafficking pathway HDFs that may serve critical roles in HIV-1 replication, we performed an arrayed KO screen on a library of 140 membrane trafficking genes in CD4+ HeLa-derived TZMbl-Cas9 cells (Table S1). The Protein Analysis Through Evolutionary Relationships (PANTHER) gene ontology (GO) tool was used to perform enrichment analysis of the gene set to classify them into subgroups with diverse molecular functionalities in membrane trafficking, where it mostly identified proteins with catalytic functions (40%; e.g., Rab GTPases), binding functions (39%; e.g., clathrin adaptors), and those involved in cytoskeletal maintenance and actin remodeling (10%) (Figure S5C).
The well-established TZM-bl cell line was chosen as the parental cell line for our functional CRISPR screen due to its amenability to transfection, permissibility to HIV-1 infection, and integration of the Tat-responsive LTR-driven luciferase cassette that permits quantitative measures of HIV-1 infectivity in relative light units (RLUs). TZM-bl cells were made to constitutively and stably express the Cas9 endonuclease using the lentiCas9-Blast lentiviral vector containing wild-type (WT) C-terminal FLAG-tagged Streptococcus pyogenes Cas9 protein and conferring blasticidin resistance (Figure S1A). Western blots from lentiCas9-Blast transduction titrations confirmed volumes providing optimum Cas9 expression (Figures S1B and S1C), and confirmed homogeneous population of Cas9-positive cells (99.6%) (Figure S1D), herein designated as the TZM-bl-Cas9 cell line used for the CRISPR screen.
To ensure that the Cas9 endonuclease in TZM-bl-Cas9 cells could effectively target and cleave the correct genomic targets, we designed a sgRNA (i.e., sgLTR362) targeting the highly conserved NF-κB binding site of the HIV-1 LTR in TZM-bl cells, which is required for provirus transcription and expression of the luciferase gene cassette. The sgLTR362 was cloned within the lentiGuide-Puro vector (Figure S2A), which was then transduced into TZM-bl-Cas9 cells which were then infected with NL4.3 virus. This resulted in 20-fold decrease (p < 0.0001) in luciferase expression by TZM-bl-Cas9 cells (Figures S2B and S2C), validating both the presence and functionality of the Cas9 protein in TZM-bl-Cas9 cells.
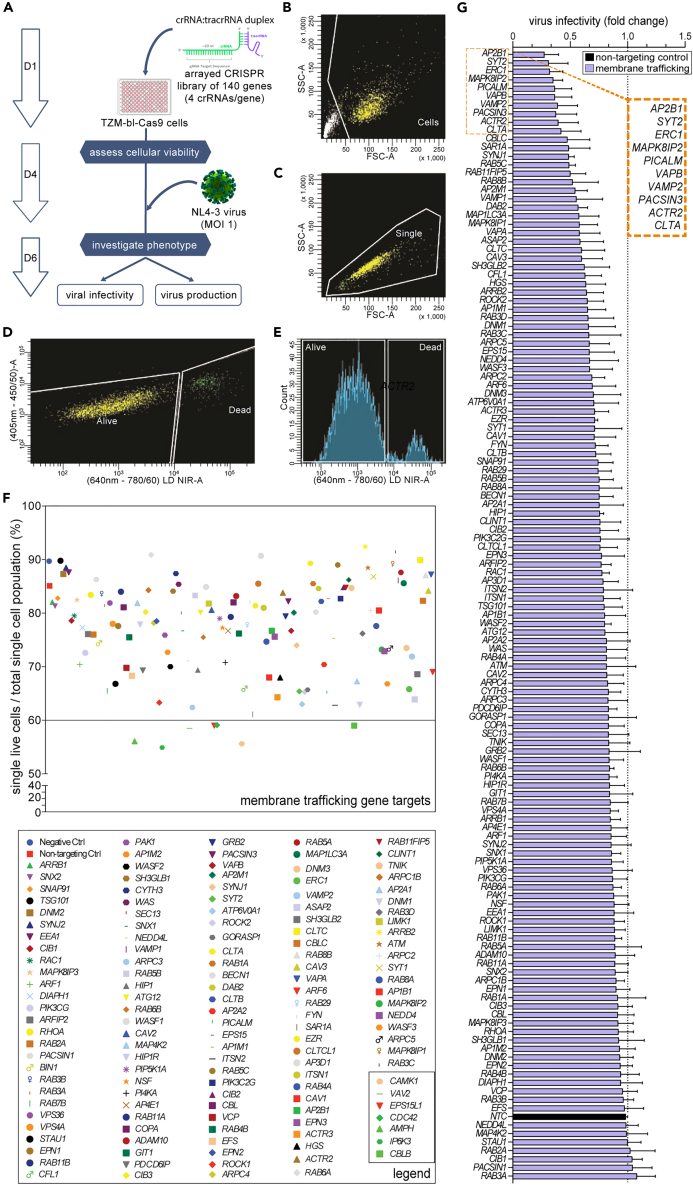
We performed three independent arrayed CRISPR screens in the TZM-bl-Cas9 cells using the 140 membrane trafficking gene library to uncover genes that may be essential to HIV-1 replication (Figure 1A; Table S1). The crRNA library was transfected into the TZM-bl-Cas9 cells at a 1:1 ratio with the respective tracrRNA, and a non-targeting crRNA was used as a negative control. TZM-bl-Cas9 cells were assessed for viability four days post crRNA transfection using flow cytometry, with 70% of targets displaying ≥75% cellular viability following gene perturbation (Figures 1B–1F) relative to ∼90% for non-transfected and non-targeting controls crRNA transfected cells. The seven gene targets identified as detrimental to cellular viability (i.e., <60%) were excluded from subsequent experiments (Figure 1F).
Figure 1.
Viability and viral infectivity of 140 membrane trafficking CRISPR screen KO cells
(A) Schematic representation of the arrayed CRISPR screen protocol, where negative control uses a non-targeting control crRNA (NTC). The 140 gene crRNA library was transfected into TZM-bl-Cas9 cells at a 1:1 ratio with the respective tracrRNA.
(B–E) Flow cytometry was used to assess cellular viability of the CRISPR genetically edited cells three days following transfection, with gating strategy involving (B), forward and side scatter (FSC-A vs. SSC-A) to identify cells, followed by (C), forward area versus height (FSC-A vs. FSC-H) to differentiate singlets from doublets, then (D), a live/dead amine-binding kit, also demonstrated here via histogram function (E).
(F) Cellular viability of individual CRISPR-edited gene screen KOs is represented as a percentage of single live cells to total single cell population (top), with gene legend (bottom), with KOs having lower than 60% cellular viability boxed at bottom right corner.
(G) Representative bar graph demonstrating average fold-change in viral infectivity from three independent CRISPR screen knockout replicate experiments infected with NL4.3 as measured by TZM-bl luciferase assay, with dotted line added as cut-off value relative to effect of non-targeting control (NTC, black bar). The error bars including that of the NTC represent standard deviation (±SD) calculated from the inter-assay variance.
CRISPR-Cas9 gene edited cells were infected with purified HIV-1 NL4.3 virus (MOI 1) followed by measures of cell viability, viral infectivity and HIV-1 production 48 h later from cell lysates and supernatants using the TZM-bl luciferase and p24 antigen enzyme-linked immunosorbent assays, respectively (Table S2). To ensure that the luciferase assay was compliant with the TZM-bl-Cas9 cells and with the sgRNAs, we compared measures with WT TZM-bl cells, non-transfected, mock transfected (i.e., transfection reagent only) and non-targeting controls and found no differences (Figures S3A and S3B). Satisfied with these controls, three independent sgRNA screens identified 10 genes that, when knocked out, resulted in a greater than 2-fold decrease in infectivity compared to non-targeting controls (Figures 1G and S3C). A 2-fold threshold was chosen as a criterion to determine biological relevance in the context of HIV-1 infectivity as it is often used to represent a meaningful change in gene expression analysis.34,35 These genes were AP2B1 (0.28-fold), SYT2 (0.32-fold), ERC1 (0.33-fold), MAPK8IP2 (0.36-fold), PICALM (0.37-fold), VAPB (0.37-fold), VAMP2 (0.38-fold), PACSIN3 (0.38-fold), ACTR2 (0.39-fold) and CLTA (0.42-fold), suggesting that these ten genes are essential for the early stages of HIV-1 replication.
To supplement viral infectivity data, virus production from CRISPR screen KO cells was also quantified using a p24 ELISA assay on cell-free supernatants. We found that numerous CRISPR screen KOs significantly affected virus production compared to the non-targeting control (Figures S4A–S4C). Interestingly, the VPS36 (EAP45) gene KO consistently exhibited >2-fold decrease in virus production (Figure S4C), implicating key functions in the later stages of HIV-1 replication, confirming our earlier work.36
CRISPR screen hits regulate clathrin-mediated endocytosis and autophagy
The ten genes identified by the KO screen as HDFs were analyzed using the Search Tool for the Retrieval of Interacting Genes (STRING) and Cytoscape software, permitting visualization of protein-protein interaction networks. The overwhelming majority of these (8/10) are well-known cofactors of clathrin-mediated endocytosis (Figure S5A), highlighting the importance of this endocytic pathway during HIV-1 replication which we speculate facilitates viral entry (Figure S5B). While CLTA (i.e., clathrin light chain A) represents another gene involved in clathrin-mediated endocytosis, and its KO also showed an effect on HIV-1 infectivity (Figure 1G), it was not pursued further since CLTA has been previously characterized for the early stages of HIV-1 replication,37 but homozygous CLTA KO mice are frequently non-viable.38
Unlike many other enveloped RNA viruses, HIV-1 entry has been shown to be pH-independent, rather relying on direct hemi-fusion with the plasma membrane to access the cytoplasm.39 Nonetheless, several lines of evidence support that in certain cell types, HIV-1 may also require endocytic support for cellular entry. Indeed, HIV-1 engages in vesicular-mediated endocytosis in macrophages40,41 and in T cells.42,43,44 The precise mechanisms by which HIV-1 is endocytosed remain controversial as it is still unclear which host factors are involved in this process. Our KO screen results broaden our knowledge of how clathrin cofactors may contribute to this mechanism, revealing a cluster of ten genes that may jointly contribute to clathrin-mediated endocytosis (Figure S5B). Our results imply that HIV-1 may depend on the coordinated actions of all these gene products, such that the depletion of any one of these may significantly interfere with this entire pathway. There is however accumulating evidence that some of these proteins also mediate non-endocytic effects that can regulate cellular functions linked to membrane biology, including autophagy.45 It has been observed that the inhibition of clathrin-mediated internalization affects the formation of mature autophagosomes since the plasma membrane directly contributes to the formation of early VAMP2-positive autophagosome precursors.46 In addition, it is well established that as one of the three main proteins of mammalian endocytic clathrin coated vesicles (CCVs) (i.e., clathrin, AP2, and PICALM), PICALM modulates the autophagic degradation of the Tau protein causatively linked to Alzheimer’s disease.19,21 Indeed, the downregulation of PICALM has been observed to affect autophagosome degradation resulting in a larger number of autophagosomes as represented by increased LC3B-II expression.18
PICALM intersects with several stages of HIV-1 replication
To validate and explore the effects of the CRISPR KO screen hits in the context of HIV-1 replication, we genetically altered the gene hits in a CD4+ SupT1 T lymphoblastic cell line, a more biologically relevant model. SupT1 T cells resemble T cells phenotypically,47 display similar mechanisms of cell-free HIV-1 entry to that found for primary CD4+ T cells,48 and are widely used to model HIV-1 latency and HTLV-1 infection.49,50,51 Since CRISPR-Cas9-mediated KO of AP2B1, ERC1, SYT2, MAPK8IP2, ACTR2 and VAPB resulted in non-viable cells, these proteins were not pursued for further study. Selective KO of VAMP2, PACSIN3 and PICALM did result in viable cells, with protein expression levels confirmed by western blotting (Figure S5D). The VAMP2 and PACSIN3 proteins were represented by single bands in western blots, while WT SupT1 cells provided three detectable PICALM protein bands. Indeed, while four PICALM isoforms have been identified (i.e., isoforms 1–4: NCBI RefSeq: NP_009097.2, NP_001008660.1, NP_001193875.1 and NP_001193876.1, respectively),23 we were only able to detect two of these in SupT1 cells (Figure S5D), with the top, 71 kDa band representing isoform 1, and the lower, 66 kDa band representing isoform 2. PICALM isoforms were successfully knocked out by the CRISPR sgRNAs (Figure S5D).
The VAMP2, PACSIN3 and PICALM proteins are known to play roles in membrane trafficking19,52,53,54 and autophagy,18 the latter involved in eliminating molecules and subcellular elements via lysosome-mediated degradation to promote cellular homeostasis and survival.55 We next examined whether modulation of autophagy pathways contributed to observed reduction in HIV-1 replication. At the initial steps of autophagosome formation, the microtubule-associated protein 1 light chain 3 (MAP1LC3; LC3B-I) is cleaved and lipidated to generate LC3B-II,56 which initiates autophagy with autophagosome formation. Autophagosome maturation involves their transformation into degradative autolysosomes through their fusion with lysosomal-associated membrane protein 1 (LAMP1; CD107a) positive lysosomes.57 Although prior studies have shown elevated expression of LC3B-II in HIV-1-infected primary CD4+ T cells,58,59 it has also been observed that HIV-1 inhibits autophagosome maturation, thereby impeding the targeting and degradation of viral proteins.60,61,62
To assess the impacts of these KOs on virus expression, we performed immunofluorescence (IF) for HIV-1 Gag and fluorescence in situ hybridization to visualize the full-length vRNA63 in WT, VAMP2, PACSIN3 and PICALM KO HIV-1 infected cells. Additionally, we performed IF for LC3B-II and LAMP1 expression to examine their effects on autophagy. An elevation of LC3B-II would indicate increased autophagosome formation, while heightened colocalization with LAMP1 would suggest enhanced autophagosome-lysosome fusion or autophagy. Visualization of only LC3B-II using an antibody that also recognizes cytosolic LC3B-I was made possible by treating cells with saponin to deplete the latter, leaving behind only membrane-bound LC3B-II.64 Here, we observed that while all three KOs had effects on the expression of endosomal proteins (e.g., increased LAMP1 and LAMP1-LC3B-II colocalization by PACSIN3 KO), the PICALM KO cells displayed the most interesting phenotype. While there was no difference in vRNA expression, PICALM KO cells led to a striking increase in both LC3B-II expression (p < 0.0001) and Gag expression (p < 0.0001) and increased LC3B-II-Gag colocalization, in addition to reduced colocalization of LAMP1 and LC3B-II (Figures 2A–2C). Together, these data suggest that PICALM does not affect viral transcription but may play a role in regulating autophagy during other stages of HIV-1 infection.
Figure 2.
PICALM KO increases expression and colocalization of virus and autophagy proteins, can produces more virus, but have diminished infection rates
(A and B) KO cell lines were infected with WT NL4-3 provirus (MOI = 5; n = 3) for 48 h. Cells were treated with 0.1% saponin to deplete LC3B-I from cells. Cells were collected for FISH analysis for the HIV-1 vRNA and immunofluorescence for HIV-1 Gag protein and host proteins LC3B-II and LAMP1.
(C) Corresponding boxplots demonstrate that PACSIN3 KO caused increased LAMP1 and its colocalization with LC3B-II. LC3B-II colocalization with the vRNA is also increased in PACSIN3 KO. PICALM KO caused increased LC3B-II expression, increased Gag expression, along with increased LC3B-II colocalization with Gag. VAMP2 KO caused increased LAMP1, and increased colocalization between LC3-II and Gag, LC3B-II and the vRNA, and Gag and the vRNA. Note: LAMP1ՈLC3B-II colocalization in the PICALM KO is particularly low and is thus practically juxtaposing the x axis (2.33 ± SD 2.06 MFI).
(D) Lysates were collected and immunoblotted for Gag p55 and p24.
(E) Bar graph from data in (D).
In parallel, culture supernatant was collected and (F) virus were determined by ELISA p24. One-Way ANOVA; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Error bars from bar graphs indicate ±SD. MFI, mean fluorescence intensity; %, percent; rel., relative; UI, uninfected; Inf., infected. Ո, colocalization (i.e., intersection); KO, knock out; μm, micrometer.
To confirm observations that PICALM KO increased Gag expression in T cells, we performed western blot analysis of cell lysates to demonstrate significant increase in Gag expression (p < 0.01) (Figures 2D and 2E). This was further confirmed by increased p24 release into the culture supernatant, indicative of increase viral production (Figure 2F; Figure S6C). Therefore, from these experiments, it was determined that PICALM KO cells had the most striking effects on both HIV-1 infection and replication kinetics and was further pursued for the remainder of this study.
PICALM KO supports enhanced HIV-1 replication by usurping the autophagy machinery
We initially identified PICALM as a protein important for the infection of cells by HIV-1 (Figure 1G), and further observed that its absence perturbed autophagy regulation (Figures 2A–2C), which resulted to the increased HIV-1 Gag expression in SupT1 cells (Figures 2D–2F). We next hypothesized that SupT1 PICALM KO cells modulated virus uptake by inhibiting the cell membrane fusion of HIV-1 during infection. To test this, we performed the β-lactamase (BlaM) activity assay,65 and show that PICALM KO cells displayed reduced viral fusion efficiencies compared to WT cells (Figures 3A and 3B; Figures S6A and S6B). HIV-1 fusion/internalization is preceded by the critical step of HIV-1 envelope glycoprotein (Env) binding to the CD4 T cell receptor and co-receptors, CXCR4 or CCR5.66 Since the CD4 glycoprotein is constantly internalized via clathrin-mediated endocytosis and interaction with the complex adaptor protein 2 (AP2; known to interact with PICALM),67 and that silencing of the clathrin heavy chain can inhibit CXCR4 internalization,68 we decided to use flow cytometry to examine cell surface CD4 and CXCR4 expression. Despite the expected downregulation in CD4 expression by HIV-1 infection that prevents superinfection and promotes virus budding,69 which served as an internal control for infection in these experiments (Figures 3C and 3D), we observed no perturbation on the cell surface expression of these markers in PICALM KO cells compared to WT cells (Figures 3C–3E). These results suggest that PICALM does not affect the internalization of these receptors and supports a role for PICALM/clathrin-mediated endocytosis in productive HIV-1 entry, as shown previously in investigations demonstrating that HIV-1 co-opts clathrin adaptors to mediate entry into cells through CD4/co-receptor-mediated endocytosis.42,70,71
Figure 3.
KOs alter virus entry and the expression of HIV-1 Gag and autophagy proteins LAMP1 and LC3B-II
(A and B) WT and PICALM KO cell lines were infected with WT NL4-3 or -heat-inactivated virus containing BlaM-Vpr (MOI = 1) for 2 h and virus fusion was evaluated by the expression of cleaved CCF2 (acquired in Pacific blue channel, LSR Fortessa BD biosciences). (A) Representative dot plots and (B) bar graphs of % of cells + for cleaved CCF2 (BlaM +).
(C and D) WT and PICALM KO cells were infected with NL4.3 for 48 h and CD4-BV421 expression were analyzed by Flow cytometry.
(E) Data representative of uninfected WT and PICALM KO stained for CXCR4 BB700. WT and PICALM KO were infected with NL4.3 for 48 h (MOI 5, n = 3), and 2 h before harvesting, treated with BafA1 (100 nM), then collected for IF, coverslips were treated with 0.1% Saponin to deplete free cytoplasmic LC3BI, then stained for Gag (AF488), LAMP1 (AF647), LC3B (AF594) and DAPI.
(F and G) Representative images and plotted data showing (H) LAMP1, (I) LC3B-II expression, and (J) LAMP1 and LC3B-II colocalization. Relative values (%) were calculated by setting WT values to 100%. μm, micrometer; KO, knock out; ∩, colocalization (intersection); UI, uninfected; Inf., infected. Ո, colocalization (i.e., intersection). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; 1-way ANOVA, Tukey post-test.
We previously observed that PICALM KO cells displayed increased LC3B-II expression and reduced LC3B-II colocalization with LAMP1 (Figures 2A–2C). Colocalization of LC3B-II and LAMP1 suggests autophagosome-lysosome fusion typically leading to the degradation of autophagic targets, therefore, we speculated that PICALM KO might have a direct effect on autophagy, and consequently on HIV-1 infection. In order to confirm this hypothesis, we used Bafilomycin A1 (Baf; Baf-A1), which is commonly used to evaluate autophagy flux. Baf-A1 inhibits the H+ vATPase to blunt autophagosome-lysosome acidification and thus degradation of its contents, which allows a more clear visualization of autophagosome delivery to lysosomes and quantification of autophagic flux.72 Therefore, an increase in colocalization of LC3B-II and LAMP1 upon Baf-A1 treatment suggests increased autophagy flux.
Thus, to evaluate further the role of PICALM in autophagy flux, we again infected WT and PICALM KO cells with NL4-3 virus and treated cells with BafA1 for 2 h prior to fixation and LC3B-II and LAMP1 staining for IF analyses. While a reduction in colocalization between LC3B-II and LAMP1 was observed in control cells, (p < 0.0005) (Figures 3F–3I), Baf-A1 treatment revealed increased colocalization in PICALM-infected cells relative to WT cells. This result suggests that there are more autophagosomes being delivered to lysosomes in cells lacking PICALM, and may lower LAMP1 levels due to accelerated degradation of lysosomal contents. In summary, these data suggests that PICALM KO cells exhibit increased autophagy flux relative to WT cells and an increased colocalization of Gag and LC3B-II and increased overall Gag expression (Figure 2). These data provide a mechanistic link between viral replication and autophagy pathways in CD4+T cells.
PICALM KO induces an activated phenotype deficient in immune checkpoint PD-1
In culturing and expanding PICALM KO cells, we consistently observed that these cells appeared larger and proliferated more rapidly than the WT and the other KO cells. To validate these observations, we used flow cytometry to analyze the expression of proliferation marker, Ki67, which was found to be increased in PICALM KO cells (Figure 4A; Figure S6D). Since Ki67 is often used as an activation marker,73 we confirmed these findings using additional activation markers.74 Indeed, early activation marker CD69 was also increased in PICALM KO cells independent of HIV-1 infection (Figure 4B). PD-1 is also another molecule that is rapidly co-expressed and upregulated in conjunction with CD69,75 but surprisingly it was found to be markedly decreased in both HIV-1 infected and uninfected PICALM KO cells (Figure 4C), and validated by a different anti-PD-1 antibody (Figure 4D). As activated CD4+ T cells secrete cytokines such interferon-gamma (IFN-γ) involved in the PD-1/PD(L)-1 axis, we found increased IFN-γ expression in PICALM KO cells relative to WT cells (Figure 4E), a finding that is supported by reports of opposing PD-1 and IFN-γ expression in CD4+ T cells.76,77
Figure 4.
PICALM KO causes increased cell proliferation and deregulates activation and PD-1 pathways
(A) SupT1 PICALM KO T cells were transfected with a ‘rescue’ plasmid to recover PICALM expression. SupT1 WT, PICALM KO T cells and KO PICALM expressing T cells were evaluated by flow cytometry for expression of Ki67, demonstrating that PICALM cells have increased propensity to proliferate.
(B and C) WT and PICALM KO T cells were infected with WT NL4-3 virus (MOI = 5) for 48 h. Cells were analyzed for cell surface expression of PD-1 and CD69 expression flow cytometry, demonstrating that PICALM KO reduces expression of PD-1, and increases CD69, independently of virus infection.
(D) Flow cytometry was used to assess cell surface expression of PD-1 of PICALM KO cells using two different antibodies, validating that PICALM KO decreases PD-1 expression.
(E) Flow cytometry was used to measure expression of INF-γ of PICALM KO, demonstrating that PICALM KO increases INF-γ expression.
(F–I) WT and PICALM KO T cells were infected with WT NL4-3 virus (MOI = 5) for 48 h. Cells were analyzed for both cell surface expression and overall cellular expression of numerous immunological biomarkers using a cell permeabilization staining flow cytometry protocol. Bar graphs demonstrate that PICALM modulated expression of both PD-1 (F), CD25 (G), and TLR4 (H) is cell surface dependent. (I) CD4 was also analyzed by this assay, demonstrating that both cell surface and whole cell CD4 are downregulated by HIV-1 infection, but that CD4 levels are unaffected by PICALM KO in either case. (A) Data represents the percentage (%) of cells positive for Ki67. (B-I) Data were calculated from MFI (mean fluorescence intensity) of respective marker and relative % is calculated by setting wildtype uninfected (WT UI) values to 100%. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; 1-way ANOVA, Tukey post-test.
Next, because of PICALM’s roles in endosomal pathways, we were curious to determine if the decreased levels of PD-1 were due to defects in endosome recycling in PICALM KO cells. We thus analyzed the distribution of cell surface and intracellular PD-1 and other key molecules by flow cytometry. Indeed, while PD-1 expression is decreased on PICALM KO cell surface, its expression levels are relatively unchanged within whole cells (Figure 4F). While CD4+CD25+ regulatory T cells (Tregs) are a population known to retain intracellular PD-1, we could rule this out from observations that PICALM KO cells had significantly diminished CD25 expression, despite an expected increase in cell surface expression of this molecule as a result of HIV-1 infection78 (Figure 4G).
To reexamine the possibility that the PICALM KO modulates cell surface PD-1 through a defect in endocytosis, we examined the TLR4 receptor which is a known substrate for clathrin-mediated endocytosis,79 and whose internalization and signaling can be abolished by direct inhibition of clathrin-mediated pathways.80 Indeed, we observed that TLR4 is maintained at the cell surface in PICALM KO cells and is upregulated by HIV-1 infection as previously demonstrated81 (Figure 4H), whereas CD4 is unaffected by the PICALM KO (Figure 4I). These results suggest that PICALM KO cells may have impeded endocytic pathways maintaining a functional, activated, and proliferating phenotype.
PICALM KO cells exhibit increased productive HIV-1 infection and contrasting differentiation markers
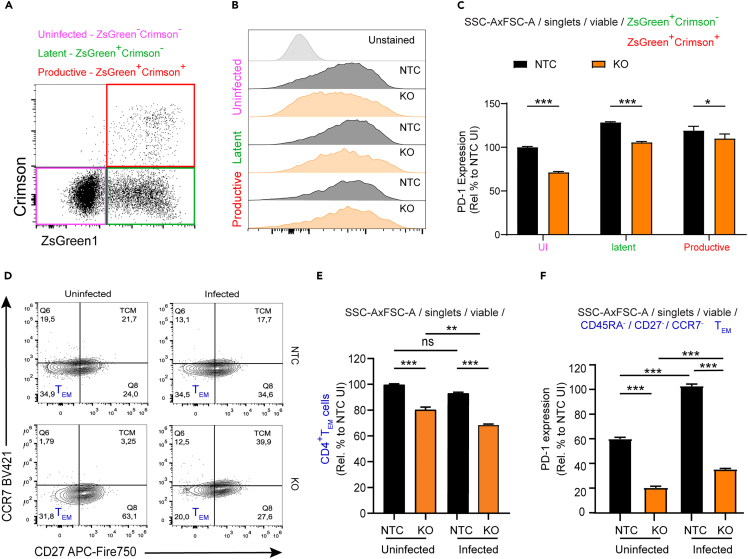
We observed that PICALM KO led to increased Gag expression and virus production (Figures 2 and 3), while these cells also exhibited increased activation and proliferation markers. Considering that the PD-1 pathway promotes control of chronic HIV-1 infection,82,83 we queried whether decreased levels of PD-1 in PICALM KO cells would also be correlated with an increase of productive infection promoting Gag expression and virus production. For this determination, we used a full-length dual fluorescent HIV-1 reporter system capable of quantifying both productively and latently infected cells longitudinally. This reporter contains a Nef-Crimson fusion protein under the control of the HIV-1 LTR promoter, and ZsGreen1 under the control of the mammalian EF1α promoter (called HIV Nef-CRIMZs; Figure 5A). To validate the ability of the HIV Nef-CRIMZs to identify productively and latently infected cells, primary CD4+ T cells were isolated, activated via TCR triggering, expanded in culture for seven days, and then infected with HIV Nef-CRIMZs at an MOI of 0.1 in 3D collagen gels.84 These cells were analyzed five days later using flow cytometry, providing evidence of clearly distinguishable populations of both productively infected cells (Nef-CrimsonhiZsGreen1hi) or latently infected cells (Nef-CrimsonnegZsGreen1hi). Importantly, both Env and Nef proteins of this construct are functional, as extensive CD4 downregulation is observed in productively infected (p24hi), but not in latently infected (p24neg), T cells (Figure 5B).
Figure 5.
PICALM KO increases productive infection
(A) Schematic of the HIV Nef-CRIMZs reporter construct used to assess latency, containing the WT NL4-3 HIV-1 genome, with HIV-1 Nef fused in frame with E2-Crimson for the productive infection readout, followed by the ZsGreen reporter under the control of the EF1α promoter for a latent infection readout. Human CD4+ T cells were isolated from PBMC, activated with Dynabeads CD3/CD28 antibody and infected with HIV Nef_CRIMZs with MOI 0.1 for 5 days.
(B) Representative dot plots of flow cytometry analyses demonstrating that the co-expression of Nef-Crimson and ZsGreen quantifies productive infection, with concomitant CD4 downregulation and increased Gag expression, as compared to latent infection by only ZsGreen1 expression were CD4 is increased in expression and Gag is decreased in expression.
(C) Flow cytometry gating strategy for infected SupT1 cells. Gating shows strategy in unstained samples. All SupT1 cells were selected and individualized by crossing the singlets gate, viable cells were gated using fixable viability staining (FVS)-510 and TEM-like cells were visualized using (CDR45RA−) and (CCR7−CD27−) gates. Productively (ZsGreen+Nef-Crimson+) and latently (ZsGreen+Nef-Crimson-) infected populations were identified following gating of viable cells.
(D and E) SupT1 NTC and PICALM KO cells were infected with HIV Nef-CRIMZs (MOI 1), Crimson and ZSGreen1 expression were evaluated 48 h post-infection by flow cytometry, (D) Corresponding dot plots of data in (E) demonstrating that PICALM KO induces an increase in productive infection. (E) Quantification of latent and productively-infected cells obtained by gating in ZsGreen+Crimson− and ZsGreen+Crimson+, respectively. Data shown is mean ± SEM. Relative % is calculated by setting uninfected non-targeting control (UI NTC) values to 100%, ∗∗∗p < 0.001; unpaired two-tailed t-test; %, percent; UI, uninfected; Inf., infected.
Additionally, to confirm that the downregulation on PD-1 expression in the PICALM KO cells was not an off-target effect of CRISPR/Cas9, the following experiments were performed comparing PICALM KO cells against a non-targeting control (NTC) stable cell line, created using only the LentiCRISPRv2 empty vector and following the gating strategy denoted in Figure 5C. Indeed, infecting cells with the HIV-1 dual reporter provided evidence that PICALM KO cells showed a higher proportion of productively infected cells (Crimson+ZsGreen+) 48 h post infection compared to the NTC cells (Figures 5D and 5E). As the dual reporter virus allows us to analyze latently and productively infected cells (Figure 6A), PD-1 expression was evaluated in both populations. Interestingly, we observed that PD-1 expression was not downregulated in productively infected cells to the same extent as those in uninfected and latently infected cells (Figures 6B and 6C). These results support the notion that the PD-1 pathway is important to regulate HIV-1 expression and underscore a possible role of PICALM in this pathway.
Figure 6.
PICALM KO cells exhibit low PD-1 levels and a modulated differentiation into TEM-like cells
SupT1 NTC and PICALM KO cells were infected with HIV Nef-CRIMZs virus and were harvested 48 h post-infection.
(A) Dot plot representation of analyzed populations in (B and C). (B) Representative histograms of PD-1 expression in uninfected, latently- and productively-infected cells. (C) Bar graph quantification of data shown in (B) demonstrating that PD-1 levels in PICALM KO cells are modulated and are differently expressed in uninfected, productive and latently infected cells.
(D and E) CD45RA, CCR7 and CD27 were measured to determine that PICALM KO cells show a decreased TEM-like phenotype. Gating strategy was performed as mentioned in Figure 5C, following the gating of viable cells, cells were gated on CD45Ra−, then on CCR7−CD27− as shown in panel (D). (E) Graphical representation of data in (D) illustrating the frequency of TEM-like cells.
(F) Quantification of PD-1 expression (MFI) in TEM-like cells, showing that PICALM KO leads to lower levels of PD-1. Relative values (%) were calculated by setting NTC values to 100%. Graphs show mean ± SEM. ANOVA one-way, with Tukey post-test. ∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001.
Since the PD-1 pathway may also play a role in the differentiation and function of memory CD4+ T cells,85 we next assessed expression of common cell surface CD45RA, CD27 and CCR7 markers. PICALM KO cells lose their CD45RA−CD27−CCR7- TEM-like phenotype (Figures 6D and 6E) and displayed a marked downregulation of PD-1 expression compared to NTC cells (Figure 6F). These results provide evidence that the downregulation of PD-1 in PICALM KO cells may also have a role in cell differentiation.
Discussion
To successfully infect and replicate in target cells, HIV-1 must hijack host cellular proteins from several different cellular pathways including the endocytic and autophagy/lysosomal degradation pathway.60,61,62,86,87 However, the precise mechanisms remain unclear. In this study, we demonstrate that PICALM, a component of both clathrin-mediated endocytosis and autophagy pathways18 plays an important role in HIV-1 entry into CD4+ T cells, and that it is also an important protein in the host response to HIV-1 infection. The absence of PICALM also leads to the accumulation and release of HIV-1 viral Gag protein by infected cells and most astoundingly, it decreases the expression of the immune checkpoint PD-1, eliciting a highly active state and possibly disturbing CD4+ T cell differentiation.
An interesting finding is that PICALM regulates HIV-1 entry into CD4+ T cells. As observed in the BlaM assays (Figures 3A and 3B), PICALM KO cells are resistant to viral entry, and that this was independent of CD4 or CXCR4 expression (Figures 3C and 3D). Altogether, these results provide new mechanistic insights into the alternative cell entry models of HIV-1, and supports earlier reports that demonstrate that HIV-1 entry into CD4+ T cells can occur via clathrin-mediated endocytosis.66,71
Infection of PICALM KO SupT1 cells displayed an accumulation of LC3B-II puncta, and despite showing diminished colocalization with LAMP1 positive organelles, the Baf-A1 treatment showed that PICALM KO cells have higher autophagic flux (Figures 3E–3I). Indeed, it was previously observed that PICALM regulates autophagosome-lysosome fusion via the soluble NSF attachment protein receptor (SNARE) Vesicle Associated Membrane Protein 8 (VAMP8).18,19,22 Additionally, we have observed that infected PICALM KO cells exhibit increased Gag expression and virus release (Figures 2A–2F). These observations first appeared counterintuitive i.e., that PICALM KO cells exhibited both higher Gag expression and increased autophagic flux. However, although high levels of colocalization of LC3B-II and LAMP1 were observed in PICALM KO cells, endosomal acidification and autophagy can still be inhibited due to the presence of Nef.88,89
Although a previous report showed that the autophagy machinery contributes to the expression of Gag in macrophages,90 autophagy has yet to be linked to Gag expression in CD4+ T cells. In our work, we observed increased colocalization of LC3B-II and Gag in the absence of PICALM (Figures 2A–2C and 3F–3I), supporting the possibility that autophagy could be exploited to favor Gag expression in PICALM KO CD4+ T cells. While lysosomal degradation induces the accumulation of Gag in endosomes and increased virus production,91 the absence of PICALM may nevertheless allow HIV-1 to use autophagosomal membranes as scaffolds for virion assembly, rather than being degraded by them. Together, our observations indicate that PICALM likely counteracts HIV-1-mediated hijacking of the autophagy machinery to increase viral load in CD4+ T cells.
We have also demonstrated that PICALM KO cells have deregulated PD-1 expression and possess an augmented proliferative and activated phenotype (Figures 4, 5, and 6). High PD-1 expression is generally associated with T cell exhaustion during cancer and chronic viral infections, while a blockade increases T cell activation and proliferation. Therefore, inhibitors of this immune checkpoint have been proven to help in both viral control and the treatment of more than 20 types of cancer.92,93,94,95 Recent studies have however demonstrated that the PD-1 pathway also plays a fundamental role in shaping effector and memory T cell responses.96,97
Our work shows that PICALM KO cells have low PD-1 levels, and elevated proliferation marker Ki67, correlating with increased activation and enhanced HIV-1 production as reflected in earlier observations.98,99 A negative relationship between PD-1 and Ki67 has also been observed for HIV-1 specific IFN-γ–producing CD4+ T cells100 (Figure 4A), and increased activation is also consistent with elevated TLR4 levels in our PICALM KO cells (Figure 4H). Evidence shows that TLR4 promotes proliferation and survival of CD4+ T cells, but it is also correlated to central nervous system inflammation.101 PICALM KO-mediated effects could be due to its involvement in both endocytosis and autophagy,102 or to decreased PD-1 expression.92 This observation leads to important considerations since the timing of a PD-1 blockade will determine memory T cell responses during chronic viral infection.97,103 While it is not known how PD-1 is recycled to the plasma membrane, its internalization and degradation can be blocked by the inhibition of clathrin-mediated endocytosis.104 Palmitoylation of PD-1 also promotes localization to recycling endosomes and prevents lysosome-dependent degradation.105 Our observations that PICALM KO reduces cell surface PD-1 but not whole cell levels of PD-1 (Figures 4C and 4F) suggest that PICALM may be involved in the presentation and/or recycling of PD-1 from the cell surface.19,106 In support of this, it has been suggested that PD-1 is recycled through direct interaction with Rab11,105 and that PICALM also guides intracellular trafficking of amyloid-β to Rab11+ recycling endosomes.19
PICALM KO cells led to consistently increased productive HIV-1 infection (Figures 5D and 5E) when assessed in an HIV-1 latency reporter system, and consistent with our ELISA results (Figure 2F; Figure S6C). Although surface PD-1 expression levels were low in uninfected and latent populations, productively infected cells displayed increased levels of PD-1 (Figures 6A–6C). This finding is in line with earlier reports demonstrating that HIV-1 infection is associated with increased expression of PD-1.83,107 Our findings suggest that PICALM restricts the PD-1 expression but it does not completely impair the PD-1 axis and that the functional PICALM pathway may intersect with HIV-1, thwarting PD-1 overexpression.
Finally, PICALM KO cells exhibited diminished CD45RA−CD27−CCR7- expression 48 h post-infection compared to control cells (Figures 6D and 6E), indicating reduction in the TEM-like population. However, we would need to conduct further analyses to determine the precise phenotype of the PICALM KO cells, since cell differentiation involves various factors such as cytokines, transcription factors, and metabolic programs.108 Moreover, we observed that PICALM KO TEM-like cells exhibited markedly lowered PD-1 expression compared to NTC cells (Figure 6F). Since PD-1 signaling has a significant effect in modifying the transcriptome and function of T cell subsets,85,109 we believe that the change in differentiation markers could possibly be related to the change in PD-1 expression. Nevertheless, since the PD-1 axis also regulates activation and function of T follicular helper cells (Tfh) and polarized T helper 17 cells (Th17) cells,110 it would be interesting to determine whether these cells are affected by PD-1 downregulation in the context of PICALM KO cells. The secretion of IFN-γ is a key signature of Tfh and Th17 cells,111 which is consistent with our observation in PICALM KO cells (Figure 4E). Both Tfh and Th17 are more susceptible to HIV-1 infection, potentially explaining why there is increased viral production in PICALM KO cells.
In summary, the endocytosis-related protein PICALM has pleiotropic effects on T cell biology and HIV-1 replication. The study of PICALM KO cells has revealed functional links between endocytosis and other pathways, including pro-inflammatory responses and autophagy. Our results highlight that the components of clathrin-mediated endocytosis also have non-endocytotic functions that influence HIV-1 replication. Finally, our data suggest that the targeting of PICALM affects CD4+ T cell health, increasing their proliferation, preserving their activation status and possibly influencing cell differentiation while reducing PD-1 expression. Considering that the SupT1 is a CD4+/CXCR4+/CCR5- cell line is derived from a T cell lymphoblastic lymphoma112 which likely differs from primary T cells found in vivo, it will be interesting to knock-out PICALM in primary CD4+ T cells to confirm the observations made in this study. Further investigations on the precise mechanisms are warranted, as this host protein may represent an interesting target that can benefit HIV-1 and cancer patients. The anti-malaria drug artesunate has recently been shown to prevent amyloid-β pathology by upregulating PICALM at the blood-brain barrier.113 Artesunate reverses immunosuppression,114 possesses anti-viral activities against human cytomegalovirus, herpes viruses, hepatitis B and C viruses, and inhibits the replication of M- and T-tropic HIV-1.115,116 Considering that artesunate increases PICALM expression and inhibits virus replication, it still remains unclear if PICALM is the target of this drug. Further investigation into the mechanistic target of artesunate and its relation to PICALM functions may identify their utility as an antiviral therapy for HIV-1 infection.
Limitations of the study
The results of this study are largely derived from experimentation with the cell line, SupT1 that may limit interpretation in physiological conditions. Furthermore, limitations might include the role of PICALM in HIV-1 infected primary CD4+ T cells as well as the impact on various primary HIV-1 isolates and transmitted founder viruses, that were not examined in the current manuscript.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-p24 | NIH AIDS Reference and Reagent Program | Cat# ARP-6521 |
| Sheep anti-p17 polyclonal serum | NIH AIDS Reference and Reagent Program | Cat# 6123 |
| Sheep anti-Digoxigenin-AP, Fab fragments | Sigma-Aldrich | Cat# 11093274910, RRID:AB_2734716 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor® 594 | Thermo Fisher Scientific | Cat# A-21207, RRID:AB_141637 |
| Donkey anti-Sheep IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor® 488 | Thermo Fisher Scientific | Cat# A-11015, RRID:AB_2534082 |
| Donkey anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor® 647 | Thermo Fisher Scientific | Cat# A-31571, RRID: AB_162542 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed, Alexa Fluor® 647 | Thermo Fisher Scientific | Cat# A-31573, RRID:AB_2536183 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed, Alexa Fluor® 594 | Thermo Fisher Scientific | Cat# A-21203, RRID:AB_141633 |
| goat anti-mouse IgG (H + L) Polyclonal Antibody (HRP (Horseradish Peroxidase)) | Rockland Immunochemicals | Cat# ROCK610-1319, RRID:AB_219659 |
| goat anti-rabbit IgG (H + L) Polyclonal Antibody (HRP (Horseradish Peroxidase)) | Rockland Immunochemicals | Cat# ROCK611-1322, RRID:AB_219723 |
| APC-H7 Mouse Anti-Human CD27 | BD Biosciences | Cat# 560222 RRID:AB_1645474 |
| BUV737 Mouse Anti-Human CD45RA | BD Biosciences | Cat# 612846 RRID:AB_2738810 |
| APC Mouse Anti-Human CCR7 (CD197) | BD Biosciences | Cat# 566762 RRID:AB_2869854 |
| BV421 Mouse Anti-Human CD4 | BD Biosciences | Cat# 566907 RRID:AB_2739448 |
| BB700 Mouse Anti-Human CD279 (PD-1) | BD Biosciences | Cat# 566460 RRID:AB_2744348 |
| BUV395 Mouse Anti-Ki67 | BD Biosciences | Cat# 564071 |
| AF647 Mouse Anti-human CD279 (PD-1) | BD Biosciences | Cat# 560838 |
| CD184 BB700 anti-human (CXCR4) | BD Biosciences | Cat# 566553 |
| CD69 Monoclonal Antibody (H1.2F3), PE | Invitrogen | Cat# 12-0691-82 RRID: AB_465732 |
| Ki67 | BD Biosciences | Cat# 564071 RRID: AB_2738577 |
| TLR4 | BD Biosciences | Cat# 564215 RRID: AB_2738674 |
| BUV395 Mouse Anti-Human IFN-γ | BD Biosciences | Cat# 563563 RRID:AB_2738277 |
| Recombinant Anti-PICALM antibody | Abcam | Cat# ab172962 |
| Recombinant Anti-VAMP2 antibody | Abcam | Cat# ab181869 |
| PACSIN3 Polyclonal Antibody | Thermo Fisher Scientific | Cat# PA5-83690 RRID:AB_2790843 |
| LC3B Antibody | Cell Signaling | Cat# 2775 |
| Anti-LAMP1 antibody | Abcam | Cat# ab25630 |
| Bacterial and virus strains | ||
| HIV-1 (NL4-3) | NIH AIDS Reference and Reagent Program | Cat# 114 |
| HIV-1 Nef_CRIMZs | This study | |
| Biological samples | ||
| Peripheral blood mononuclear cells – Montreal | Réseau SIDA MI - FRQS | N/A |
| Peripheral blood mononuclear cells - Manitoba | NetCad (Canadian Blood Service) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | Thermo Fisher Scientific | Cat# 11965-118 |
| RPMI 1640 | Invitrogen | Cat# 11875-119 |
| Opti-MEM | Thermo Fisher Scientific | Cat# 31985070 |
| FBS | Wisent | Cat# 080-150 |
| Penicillin-streptomycin | Wisent | Cat# 450-201-EL |
| JetPrime (PolyPlus) | VWR | Cat# CA89129-924 |
| D-PBS, 1X | Wisent | Cat# 311-425-CL |
| EDTA-free Protease inhibitor tablets | Roche | Cat# 11873580001 |
| Bradford protein assay | BioRad | Cat# 500-0006 |
| Pierce ECL Plus western blotting Substrate | Thermo Fisher Scientific | Cat# 32132 |
| DIG RNA Labeling Kit | Invitrogen | Cat# 18033-019 |
| DharmaFECT 1 Transfection Reagent | Dharmacon™ | Cat# T-2001-02 |
| Puromycin | Invivogen | Cat# ant-pr-1 |
| Poly-L-lysine solution | Sigma-Aldrich | Cat# 25988-63-0 |
| Fixable Viability Stain 510 | BD Biosciences | Cat# 564406 |
| Dynabeads™ Human T-Activator CD3/CD28 for T cell Expansion and Activation | Thermo Fisher Scientific | Cat# 11131D |
| Critical commercial assays | ||
| Human Edit-RTM - Membrane Trafficking | Dharmacon™ | Cat# GC-005505-01 |
| LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit, for 633 or 635 nm excitation | Thermo Fisher Scientific | Cat# L10119 |
| LiveBLAzer™ FRET-B/G Loading Kit with CCF2-AM | Thermo Fisher Scientific | Cat #K1032 |
| Experimental models: Cell lines | ||
| HeLa (ATCC® CCL-2) | American Type Culture Collection | N/A |
| HEK293T/17 (ATCC® CRL-11268) | American Type Culture Collection | N/A |
| TZM-bl | NIH AIDS Reference and Reagent Program | Cat# 8129 |
| SUP-T1 (ATCC® CRL-1942) | American Type Culture Collection | N/A |
| MAGI.CCR5 | NIH AIDS Reagent Program | Cat# 3522 |
| Recombinant DNA | ||
| lentiCRISPRv2 | Sanjana et al., 2014117 | Addgene Plasmid #52961 |
| pCMV-VSV-G | Stewart et al., 2003118 | Addgene Plasmid #8454 |
| psPAX2 | Didier Trono Lab (unpublished) | Addgene Plasmid #12260 |
| pNL4-3 | NIH AIDS Reference and Reagent Program | Cat# 114 |
| pCMV4-BlaM-Vpr | Cavrois et al., 200266 | Addgene Plasmid #21950 |
| Hi.fate | Ratnapriya et al., 2021119 | |
| CALM-pmCherryN1 | Taylor et al., 201181 | Addgene Plasmid #27691 |
| Oligonucleotides | ||
| 5′-TGC ACG CGT GGA GGG GGC GGT ATG GAT AGC ACT GAG AAC G-3 | Forward PCR primer | This work |
| 5′-GCT ACC CGG GTC AGG GCA AGG CGG AGC CGG AGG CG-3′ | Reverse PCR primer | This work |
| Restriction Enzymes | ||
| XmaI | NEB | Cat# R0180S |
| Mlul | NEB | Cat# R0198S |
| Software and algorithms | ||
| Imaris | Bitplane/Andor | https://imaris.oxinst.com/packages |
| Volocity 6.3 | Perkin Elmer | https://www.perkinelmer.com |
| GraphPad v9 Prism | https://www.graphpad.com/scientific-software/prism | |
| FlowJo | FlowJo | https://www.flowjo.com/solutions/flowjo |
| BD FACSDiva™ Software | BD Biosciences | https://www.bdbiosciences.com/en-ca/products/software/instrument-software/bd-facsdiva-software |
| Cytoscape | Cytoscape | https://cytoscape.org/ |
| Search Tool for the Retrieval of Interacting Genes (STRING) | STRING | https://string-db.org/ |
| Other | ||
| Leica DM16000B laser confocal microscope | Leica | N/A |
| WaveFX spinning disk confocal head | Quorum Technologies | N/A |
| Hamamatsu EM-CCD digital camera Hamamatsu | N/A | |
| LSM800 Confocal Microscope with Airyscan | Zeiss | N/A |
| DeNovix DS-11 FX Spectrophotometer/Fluorometer | DeNovix | N/A |
| 4-chamber Lab-Tek®II Chambered #1.5 German Coverglass System | VWR | Cat# CA62406-165 |
| 12-well plates | DIAMED | Cat# DIATE610-3167 |
| 25 × 75 mm × 1 mm thick glass slides | VWR | Cat# 16004-300 |
| 18 mm ø No. 1 cover glasses | VWR | Cat# 28143-310 |
| BD LSRFortessa™ Cell Analyzer | BD Biosciences | N/A |
| ID7000 Spectral Cell Analyzer | Sony Biotechnology | N/A |
| Anti-Mouse Ig, κ/Negative Control Compensation Particles Set | BD Biosciences | Cat# 552843 |
| UltraComp eBeadsTM Compensation Beads | Thermo Fisher Scientific | Cat# 01-3333-41 |
Resources availability
Lead contact
Further information and request for reagents should be directed to and will be fulfilled by the lead contact, Andrew J. Mouland (andrew.mouland@mcgill.ca).
Materials availability
Requests for materials and reagents should be addressed to the lead contact (andrew.mouland@mcgill.ca).
Data and code availability
This paper does not report original code.
Experimental models
Experimental model and study participant details
HEK293T and TZM-bl cells were grown in Dulbecco’s Modified Eagle’s medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 1% penicillin and streptomycin (Invitrogen). TZM-bl cells (NIH AIDS Reagent Program, Cat #8129) are a HeLa-derived cell line expressing CD4, CCR5, and CXCR4, and have a luciferase gene cassette driven by the HIV-1 LTR. Human T cell Lymphoma SupT1 cells (SupT1 cells) (NIH AIDS Reagent Program, Cat# ARP-100) are derived from human T cell lymphoblast and express low levels of CCR5 and high levels of CXCR4 co-receptors,120 and are grown in Roswell Park Memorial Institute (RPMI) (Life Technologies) medium supplemented with L-glutamine (Sigma-Aldrich), 10% FBS and 1% penicillin and streptomycin. The knockouts were kept with RPMI + Puromycin (2 μg/mL). All cell lines were maintained at 37°C and 5% CO2. Human CD4+ T cell cells were isolated from the PBMC fraction of healthy donors obtained from NetCAD (Canadian Blood Services), expanded in vitro using Dynabeads coated with anti-human CD3/CD28 antibody (1:1, bead:cell ratio, Life Technologies Cat #11131D) in RPMI1640 supplemented with 10% FBS (VWR Seradigm Cat #1500-500), 2 mM GlutaMAX (Gibco Cat #3050-061), 1 mM sodium pyruvate (Corning Cat #25-000-CI) and 10 mM HEPES (Sigma-Aldrich Cat #H4034). All work with human blood has been approved by the University of Manitoba Biomedical Research Ethics Board.
Method details
Generation of Cas9-expressing TZM-bl cell line
We used a lentiviral vector to deliver the Cas9 nuclease to the TZM-bl cells. For that, we co-transfected HEK293T cells with the plasmids LentiCas9-Blast (Addgene #52962), Indiana vesiculovirus G protein (VSV-G)121 (Addgene #14888) and psPAX2 (Addgene #12260) using PEI (Polysciences). Viral supernatants were collected at 48 h post-transfection and used to transduce TZM-bl with polybrene (Sigma-Aldrich). After 48 h, cells were subjected to the blasticidin selection. Different volumes of lentivirus were tested to determine the best transduction efficiency, and we determined, by western blotting and Flow cytometry, that 300 μL was the optimum volume (viral titration was not done) to provide a homogeneous population of Cas9-positive cells (99.6%) with low cellular toxicity.
Arrayed screen of membrane trafficking library and viral infectivity assay
The crRNA library consists of a panel of 140 human membrane trafficking genes (4 crRNAs per gene and 1 non-targeting control (NTC) crRNA) in an arrayed 96-well plate format for one gene-per-well analysis (Dharmacon Edit-R crRNA Library, Cat #GC-005505-01). Each crRNA and the associated tracrRNA were stored in 10 mM Tris-HCl buffer pH 7.4 (Dharmacon) at −20°C. TZM-bl-Cas9 cells were platted in 96 well plates (5×103 cells/well in serum-free RPMI). The following day, cells were transfected with 20 μL of transfection mix, containing 25 nM crRNA and tracrRNA (1:1), DharmaFECT 1 transfection reagent (Dharmacon) diluted in serum-free RPMI and then, incubated for 24 h at 37°C with 5% CO2. After 24 h, the serum-free RPMI was changed to complete media.
Viral infectivity was evaluated by infecting the transfected-TZM-bl -cas9 cells, after crRNA/tracrRNA transfection described above, with NL4.3 (MOI 1) and evaluating the luciferase production. Briefly, cells were lysed, frozen at −80°C for 1 h, and then mixed with 1X Luciferase substrate (1M KPO4, 1m MgSO4, 1M DTT, 100 mM ATP, 10× Luciferin) and activity was measured in relative light units (RLUs) using MicroBeta TriLux luminescence counter (PerkinElmer).
Cellular viability determination
Cellular viability was determined at 72 h post-transfection of the crRNA library using the LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit as recommended by the manufacturer (ThermoFisher Scientific, Cat #L10119). Following the viability staining, cells were removed with Trypsin and fixed with PFA 4% for 15 min at room temperature. Cells were stored in PBS at 4°C until analysis. Results and cell population statistics were acquired on a BD LSRFortessa cell analyzer (BD Biosciences).
CRISPR editing of SupT1 cells
The gRNAs were cloned into the lentiCRISRPRv2 vector (Addgene plasmid #52961), which expresses a mammalian codon-optimized S. pyogenes Cas9 and confers resistance to puromycin. HEK293T cells were co-transfected with LentiCRISPRv2,118 VSV-G, and psPAX2 using JetPRIME (Polyplus), as recommended by the manufacturer. Viral supernatants were harvested and filtered at 48 h post-transfection and used to infect SupT1 cells by spinoculation (1800 rpm, 45 min, at room temperature). The empty LentiCRISPRv2 vector was used to generate the non-targeting control (NTC) SUPT1 cell line. At 48 h post-infection, cells were expanded from a single-cell culture and positively selected with 2 μg/mL puromycin (Invitrogen) to generate monoclonal cell lines.
Western immunoblotting
Cells lysates were collected in NP40 lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40), protein concentration was quantified by Bradford assay (Bio-Rad). Equal amounts of protein (20 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (Bio-Rad). Membranes were blocked (5% milk TBST), incubated with primary antibody overnight at 4°C, and 1 h with secondary HRP α-mouse or Rabbit (Rockland Immunochemicals), then developed using Western Lightning Plus-ECL (PerkinElmer) and signal was acquired using ChemiDoc (BioRad). Signal intensities were analyzed by densitometry using ImageJ software (NIH, Bethseda, USA).
HIV-1 NL4.3 and HIV Nef-CRIMZs viral stocks and infection
NL4.3 and HIV Nef-CRIMZs virus particles were produced by transfecting HEK293T cells with the HIV-1 NL4.3 or HIV Nef-CRIMZs + VSV-G (to increase infectivity) provirus-encoding plasmid pNL4.3 using 1 μL/μg JetPrime. Supernatants were collected 48 h post-transfection, filtered through a 0.45 μm pore-sized filter (Pall Corporation). Virus was concentrated by ultra-centrifugation (20,000 rpm for 1 h at 4°C) and stored at −80°C. The viral titer was quantified using the X-gal staining assay in TZM-bl indicator cells as described previously in the literature (67) and used to determine the volume for the indicated multiplicity of infection.4
SupT1 cells expressing CXCR4 were infected with NL4.3, an X4- (CXCR4) tropic virus,117 or HIV Nef_CRIMZs by spinoculation (1,800 rpm for 45 min at room temperature), and incubated for extra 2.5 h and, then replenished with complete media. Cell and supernatants were collected at the indicated time points.
HIV-1 viral fusion assay
HIV-1 particles containing β-lactamase–Vpr (BlaM-Vpr) chimera proteins were produced by co-transfecting HEK293T cells with HIV-1 NL4.3 provirus-encoding plasmid pNL4.3, and pCMV-BlaM-Vpr DNA65 using PEI (Polysciences). Virus was collected after 48 h and concentrated as described before. SupT1 cell lines were spinoculated with HIV-1 particles containing BlaM-Vpr. After a 2-h incubation at 37°C, the cells were washed with CO2-independent medium (Invitrogen) and incubated with 1X loading solution (CCF2/AM substrate, Invitrogen), for 1 h protected from light. After washing, cells were fixed with PFA 1% for 30 min. The levels of CCF2/AM and its cleaved products were measured by flow cytometry.
Antibodies and fluorescent probes
Primary antibodies used were as follows: mouse anti-p24 (IF, 1:250; western blotting, 1:10,000; NIH AIDS ARRP); sheep anti-p17 polyclonal serum (IF, 1:250; NIH AIDS ARRP), LC3B (IF, 1:250; Cell signaling technology, cat# 2775S), mouse anti-LAMP1 (IF, 1: 50; Abcam cat# ab25630), sheep anti-Digoxigenin-AP, Fab fragments (IF, 1:250; Roche #11093274910). For IF, secondary antibodies used were as follows: Donkey anti-Sheep IgG (H + L) Cross-Adsorbed, Alexa Fluor 488 (1:500; Invitrogen-Thermo Fisher Scientific #A-11015); Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed, Alexa Fluor 594 (1:500; Invitrogen-Thermo Fisher Scientific #A-21207); Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed, Alexa Fluor 647 (1:500; Invitrogen-Thermo Fisher Scientific #A-31571); Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed, Alexa Fluor 647 (1:500; Invitrogen-Thermo Fisher Scientific #A-31573); Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed, Alexa Fluor 594 (1:500; Invitrogen-Thermo Fisher Scientific #A-21203). For western blotting, secondary antibodies used were goat anti-mouse or goat anti-rabbit IgG polyclonal antibodies conjugated to horseradish peroxidase (Rockland Immunochemicals).
Immunofluorescence
Immunofluorescence (IF) analyses were performed exactly as described previously (68). Briefly, for suspension cells, sterile 18 mm ø No. 1 cover glasses (VWR) were treated with 0.1% poly-L-lysine solution (Sigma) overnight at 4°C. Cover glasses were dropped into wells, and 4x105 WT or KOs cell lines were allowed to settle onto these for 1 h at 37°C prior to fixing cells onto cover glasses. For fixing cells onto cover glasses, cells were washed once in D-PBS (Wisent) and fixed with 4% paraformaldehyde for 15 min. Fixed cells were then washed with D-PBS, quenched in 0.1 M glycine for 10 min, washed with D-PBS, permeabilized in 0.2% Triton X-100 for 5 min and washed twice with D-PBS. Cells were briefly washed in PBS before being blocked in 1 X blocking solution.122 Primary antibodies were applied for 1 h at 37°C, and then washed for 10 min in PBS followed by secondary antibodies for 1 h. Cells were washed for 20 min in PBS before being mounted on glass slides using ProLong Gold Antifade Reagent with DAPI (Life Technologies). Negative isotype-matched antibodies were used to control staining specificity.
Microscopy and imaging analyses
Laser confocal microscopy was performed using a Leica DM16000B microscope equipped with a WaveFX spinning disk confocal head (Quorum Technologies) and HCX PL APO/40X, Oil/0.75–1.25 NA CS and HCX PL APO/63X, Oil/0.60–1.40 NA BL objectives, and images were acquired with a Hamamatsu EM-charge coupled device digital camera. Images were recorded from laser-scanned cell layers with a thickness of 1 μm and were digitized at a resolution of 1024 × 1024 pixels. For multi-color image capture, Alexa Fluor 647, -594, 488, conjugated secondary antibody emissions were sequentially captured with 665–715, 570–620, and 500–550, bandpass filters, followed by 435–485 nm [for 4′,6-diamidino-2-phenylindole (DAPI) staining] followed by differential image capture image capture. Raw.liff files were exported by the Volocity software (PerkinElmer) for import into Imaris software v. 9.6.0 (Bitplane/Oxford Instruments) used for generation of colocalization channels, and.xlsx file exports of quantitative measurements of mean signal intensity values used for downstream data harmonizing and statistical analyses using Excel (Microsoft) and GraphPad v9 (Prism), respectively.
Flow cytometry staining and acquisition
Cells were collected at the indicated time points. Prior to staining, cells were incubated in Fc blocker (BD Biosciences, catalog #564220) and Fixed Viability Stain 510 (BD Biosciences, catalog #564406) or Zombie NIR (Biolegend, #423106) for 15 min at room temperature, followed by incubation with antibodies (Table S2) for 30 min at room temperature. Cells were then fixed with 1% paraformaldehyde (PFA) for 30 min. For the intracellular staining, cells were permeabilized and fixed with CytoFix/CytoPerm kit (BD Bioscience) prior to staining with intracellular markers and after extracellular staining. For the analysis of cell surface vs. whole cell, the extracellular staining in the whole cell group was performed after the permeabilization. An average of 100,000 cells were analyzed using a 4-laser LSR Fortessa (BD Biosciences) or using, where indicated, the Spectral Cytometer Sony ID7000 at the Flow Cytometry Core Facility, Lady Davis Research Institute. Single stain controls for each fluorochrome were prepared using Compensation Beads (BD Biosciences, catalog #552843) or UltraComp eBeads (ThermoFisher). Data was analyzed using FlowJo V10 (Tree Star).
Construction, validation, and usage of the HIV Nef-CRIMZs reporter
The E2-Crimson-EF1a-ZsGreen DNA insert was amplified from the Hi.Fate latency plasmid123 using primers 5′-TGC ACG CGT GGA GGG GGC GGT ATG GAT AGC ACT GAG AAC G-3′ and 5′-GCT ACC CGG GTC AGG GCA AGG CGG AGC CGG AGG CG-3’ by PCR, and inserted into the R5-tropic ‘HIV-GFP’ proviral vector124 using unique restriction enzyme sites XmaI 73R0180S and MluI 73R0198S. A flexible 4 amino acid linker DNA was added between Nef and E2-Crimson. The resulting plasmid, termed ‘HIV Nef-CRIMZs’, was sequenced on both strands before transfection into HEK293T cells. Viral supernatant was collected and concentrated by ultracentrifugation, as previously described.124 Viral stocks were titrated using MAGI.CCR5 and expressed as blue forming units (b.f.u.) per mL.
Quantification and statistical analysis
All experiments were performed in triplicate with similar results, unless otherwise indicated in figure legends as n = #. Two independent observers validated phenotypes resulting from all experimental conditions tested. Cellular imaging statistics reported for mean fluorescence (MFI) are from observation of average of n = 130 cells per condition tested. Statistical analysis and plotted graphs were performed and generated using GraphPad Prism v.8.0.1, and one-way ANOVA (with Tukey’s multiple-comparisons test) with 95% CI was used for multiple comparisons, and an unpaired 2-tailed Student’s t test with 95% CI was used to compare two groups. p-values <0.05 were considered statistically significant. Box-plot center lines indicate median, box limits are interquartile range, and whisker are minimum to maximum. Error bars on bar graphs represent ±SEM or SD, as indicated in figure legends.
Acknowledgments
We are grateful for detailed discussions, generous offers of reagents and advice from Gregory Melikian. We thank Christian Young and Mathew Duguay from the Lady Davis Institute Core Facility for technical support, Qinghua Pan for advice and the Blam-Vpr plasmid, Rémi Fromentin, and Nicolas Chomont for providing antibodies and advice, Petronela Ancuta for critical reading of the manuscript and Sergio P. Alpuche-Lazcano for technical discussions. We thank Gabriel Guajardo Contreras for artwork and blood donors and NetCAD (Canadian Blood Services) for providing PBMCs. P.G. was supported by a scholarship from the Mexican Instituto de Financiamiento e Información para la Educación (EDUCAFIN). R.E.C. is a recipient of doctoral scholarships from the Fonds de recherche du Québec and the McGill Center for Viral Diseases. This work was funded by the Canadian Institutes of Health Research HB2-164064 and PJT-481126 to T.M., PJT-166048 to C.L., and grant FRN-162447 to A.J.M.
Author contributions
A.J.M. conceived of the project; P.G, A.L.A., A.M., and K.D. conceived of aspects of the project; K.D. performed the initial CRISPR-Cas9 screen and analyzed data; P.G. generated CRISPR stable cell lines and performed validation experiments; A.L.A performed validation cell sorting, flow cytometry analyses and analyzed data; A.M. carried out imaging experiments and analyzed data; R.E.C. analyzed flow cytometry data and interpreted results; M.N. performed and contributed expertise to infection experiments; X.L., O.E.A., and T.T.M generated Nef_CRIMZs vector, methodology as a latency model cell system, validated its use and provided expertise; P.G., A.L.A., A.M., K.D., and A.J.M. contributed to the drafting of the manuscript with editing by all authors; P.G., A.L.A., R.E.C., and A.J.M. performed all revisions suggested by the reviewers though experimentation and revisions to the manuscript; T.T.M., C.L., and A.J.M. supervised and secured infrastructure and funding.
Declaration of interests
The authors declare no competing interests.
Published: May 27, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110131.
Supplemental information
References
- 1.Qi M., Williams J.A., Chu H., Chen X., Wang J.-J., Ding L., Akhirome E., Wen X., Lapierre L.A., Goldenring J.R., Spearman P. Rab11-FIP1C and Rab14 Direct Plasma Membrane Sorting and Particle Incorporation of the HIV-1 Envelope Glycoprotein Complex. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirschman J., Qi M., Ding L., Hammonds J., Dienger-Stambaugh K., Wang J.-J., Lapierre L.A., Goldenring J.R., Spearman P. HIV-1 Envelope Glycoprotein Trafficking through the Endosomal Recycling Compartment Is Required for Particle Incorporation. J. Virol. 2018;92 doi: 10.1128/JVI.01893-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray J.L., Mavrakis M., McDonald N.J., Yilla M., Sheng J., Bellini W.J., Zhao L., Le Doux J.M., Shaw M.W., Luo C.-C., et al. Rab9 GTPase Is Required for Replication of Human Immunodeficiency Virus Type 1, Filoviruses, and Measles Virus. J. Virol. 2005;79:11742–11751. doi: 10.1128/JVI.79.18.11742-11751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerber P.P., Cabrini M., Jancic C., Paoletti L., Banchio C., von Bilderling C., Sigaut L., Pietrasanta L.I., Duette G., Freed E.O., et al. Rab27a controls HIV-1 assembly by regulating plasma membrane levels of phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 2015;209:435–452. doi: 10.1083/jcb.201409082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caillet M., Janvier K., Pelchen–Matthews A., Delcroix-Genête D., Camus G., Marsh M., Berlioz-Torrent C. Rab7A Is Required for Efficient Production of Infectious HIV-1. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cinti A., Le Sage V., Milev M.P., Valiente-Echeverría F., Crossie C., Miron M.-J., Panté N., Olivier M., Mouland A.J. HIV-1 enhances mTORC1 activity and repositions lysosomes to the periphery by co-opting Rag GTPases. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-05410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanna R.E., Doench J.G. Design and analysis of CRISPR–Cas experiments. Nat. Biotechnol. 2020;38:813–823. doi: 10.1038/s41587-020-0490-7. [DOI] [PubMed] [Google Scholar]
- 8.Puschnik A.S., Majzoub K., Ooi Y.S., Carette J.E. A CRISPR toolbox to study virus–host interactions. Nat. Rev. Microbiol. 2017;15:351–364. doi: 10.1038/nrmicro.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickar-Oliver A., Gersbach C.A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magro G., Calistri A., Parolin C. Targeting and Understanding HIV Latency: The CRISPR System against the Provirus. Pathogens. 2021;10:1257. doi: 10.3390/pathogens10101257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shue B., Chiramel A.I., Cerikan B., To T.-H., Frölich S., Pederson S.M., Kirby E.N., Eyre N.S., Bartenschlager R.F.W., Best S.M., Beard M.R. Genome-wide CRISPR screen identifies RACK1 as a critical host factor for flavivirus replication. J. Virol. 2021;95 doi: 10.1128/JVI.00596-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baggen J., Persoons L., Vanstreels E., Jansen S., Van Looveren D., Boeckx B., Geudens V., De Man J., Jochmans D., Wauters J., et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 2021;53:435–444. doi: 10.1038/s41588-021-00805-2. [DOI] [PubMed] [Google Scholar]
- 13.Marceau C.D., Puschnik A.S., Majzoub K., Ooi Y.S., Brewer S.M., Fuchs G., Swaminathan K., Mata M.A., Elias J.E., Sarnow P., Carette J.E. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savidis G., McDougall W.M., Meraner P., Perreira J.M., Portmann J.M., Trincucci G., John S.P., Aker A.M., Renzette N., Robbins D.R., et al. Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Rep. 2016;16:232–246. doi: 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 15.Miller S.E., Mathiasen S., Bright N.A., Pierre F., Kelly B.T., Kladt N., Schauss A., Merrifield C.J., Stamou D., Höning S., Owen D.J. CALM Regulates Clathrin-Coated Vesicle Size and Maturation by Directly Sensing and Driving Membrane Curvature. Dev. Cell. 2015;33:163–175. doi: 10.1016/j.devcel.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scotland P.B., Heath J.L., Conway A.E., Porter N.B., Armstrong M.B., Walker J.A., Klebig M.L., Lavau C.P., Wechsler D.S. The PICALM protein plays a key role in iron homeostasis and cell proliferation. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaksonen M., Roux A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018;19:313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 18.Moreau K., Fleming A., Imarisio S., Lopez Ramirez A., Mercer J.L., Jimenez-Sanchez M., Bento C.F., Puri C., Zavodszky E., Siddiqi F., et al. PICALM modulates autophagy activity and tau accumulation. Nat. Commun. 2014;5:4998. doi: 10.1038/ncomms5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Z., Sagare A.P., Ma Q., Halliday M.R., Kong P., Kisler K., Winkler E.A., Ramanathan A., Kanekiyo T., Bu G., et al. Central role for PICALM in amyloid-β blood-brain barrier transcytosis and clearance. Nat. Neurosci. 2015;18:978–987. doi: 10.1038/nn.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chae C.W., Lee H.J., Choi G.E., Jung Y.H., Kim J.S., Lim J.R., Kim S.Y., Hwang I.K., Seong J.K., Han H.J. High glucose-mediated PICALM and mTORC1 modulate processing of amyloid precursor protein via endosomal abnormalities. Br. J. Pharmacol. 2020;177:3828–3847. doi: 10.1111/bph.15131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narayan P., Sienski G., Bonner J.M., Lin Y.-T., Seo J., Baru V., Haque A., Milo B., Akay L.A., Graziosi A., et al. PICALM Rescues Endocytic Defects Caused by the Alzheimer’s Disease Risk Factor APOE4. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ando K., Tomimura K., Sazdovitch V., Suain V., Yilmaz Z., Authelet M., Ndjim M., Vergara C., Belkouch M., Potier M.-C., et al. Level of PICALM, a key component of clathrin-mediated endocytosis, is correlated with levels of phosphotau and autophagy-related proteins and is associated with tau inclusions in AD, PSP and Pick disease. Neurobiol. Dis. 2016;94:32–43. doi: 10.1016/j.nbd.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Thomas R.S., Henson A., Gerrish A., Jones L., Williams J., Kidd E.J. Decreasing the expression of PICALM reduces endocytosis and the activity of β-secretase: implications for Alzheimer’s disease. BMC Neurosci. 2016;17:50. doi: 10.1186/s12868-016-0288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mertins P., Przybylski D., Yosef N., Qiao J., Clauser K., Raychowdhury R., Eisenhaure T.M., Maritzen T., Haucke V., Satoh T., et al. An Integrative Framework Reveals Signaling-to-Transcription Events in Toll-like Receptor Signaling. Cell Rep. 2017;19:2853–2866. doi: 10.1016/j.celrep.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris S.A., Harris E.A. Molecular Mechanisms for Herpes Simplex Virus Type 1 Pathogenesis in Alzheimer's Disease. Front. Aging Neurosci. 2018;10:48. doi: 10.3389/fnagi.2018.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J., Gu J., Shen L., Fang D., Zou X., Cao Y., Wang S., Mao L. Exosomal MicroRNA-155 Inhibits Enterovirus A71 Infection by Targeting PICALM. Int. J. Biol. Sci. 2019;15:2925–2935. doi: 10.7150/ijbs.36388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Wu J., Chavez L., Hoh R., Deeks S.G., Pillai S.K., Zhou Q. Reiterative Enrichment and Authentication of CRISPRi Targets (REACT) identifies the proteasome as a key contributor to HIV-1 latency. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H., Kong W., Jean M., Fiches G., Zhou D., Hayashi T., Que J., Santoso N., Zhu J. A CRISPR/Cas9 screen identifies the histone demethylase MINA53 as a novel HIV-1 latency-promoting gene (LPG) Nucleic Acids Res. 2019;47:7333–7347. doi: 10.1093/nar/gkz493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rathore A., Iketani S., Wang P., Jia M., Sahi V., Ho D.D. CRISPR-based gene knockout screens reveal deubiquitinases involved in HIV-1 latency in two Jurkat cell models. Sci. Rep. 2020;10:5350. doi: 10.1038/s41598-020-62375-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X., Wang Y., Lu P., Shen Y., Zhao X., Zhu Y., Jiang Z., Yang H., Pan H., Zhao L., et al. PEBP1 suppresses HIV transcription and induces latency by inactivating MAPK/NF-κB signaling. EMBO Rep. 2020;21 doi: 10.15252/embr.201949305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park R.J., Wang T., Koundakjian D., Hultquist J.F., Lamothe-Molina P., Monel B., Schumann K., Yu H., Krupzcak K.M., Garcia-Beltran W., et al. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat. Genet. 2017;49:193–203. doi: 10.1038/ng.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hultquist J.F., Schumann K., Woo J.M., Manganaro L., McGregor M.J., Doudna J., Simon V., Krogan N.J., Marson A. A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells. Cell Rep. 2016;17:1438–1452. doi: 10.1016/j.celrep.2016.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z., Chen S., Jin X., Wang Q., Yang K., Li C., Xiao Q., Hou P., Liu S., Wu S., et al. Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4(+) T cells from HIV-1 infection. Cell Biosci. 2017;7:47. doi: 10.1186/s13578-017-0174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarthy D.J., Smyth G.K. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics. 2009;25:765–771. doi: 10.1093/bioinformatics/btp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao Y., Hsiao T.H., Suresh U., Chen H.I.H., Wu X., Wolf S.E., Chen Y. A novel significance score for gene selection and ranking. Bioinformatics. 2014;30:801–807. doi: 10.1093/bioinformatics/btr671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghoujal B., Milev M.P., Ajamian L., Abel K., Mouland A.J. ESCRT-II's involvement in HIV-1 genomic RNA trafficking and assembly. Biol. Cell. 2012;104:706–721. doi: 10.1111/boc.201200021. [DOI] [PubMed] [Google Scholar]
- 37.Jäger S., Cimermancic P., Gulbahce N., Johnson J.R., McGovern K.E., Clarke S.C., Shales M., Mercenne G., Pache L., Li K., et al. Global landscape of HIV–human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu S., Majeed S.R., Evans T.M., Camus M.D., Wong N.M.L., Schollmeier Y., Park M., Muppidi J.R., Reboldi A., Parham P., et al. Clathrin light chains' role in selective endocytosis influences antibody isotype switching. Proc. Natl. Acad. Sci. USA. 2016;113:9816–9821. doi: 10.1073/pnas.1611189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashkenazi A., Viard M., Wexler-Cohen Y., Blumenthal R., Shai Y. Viral envelope protein folding and membrane hemifusion are enhanced by the conserved loop region of HIV-1 gp41. FASEB J. 2011;25:2156–2166. doi: 10.1096/fj.10-175752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maréchal V., Prevost M.C., Petit C., Perret E., Heard J.M., Schwartz O. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J. Virol. 2001;75:11166–11177. doi: 10.1128/jvi.75.22.11166-11177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carter G.C., Bernstone L., Baskaran D., James W. HIV-1 infects macrophages by exploiting an endocytic route dependent on dynamin, Rac1 and Pak1. Virology. 2011;409:234–250. doi: 10.1016/j.virol.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 42.Pauza C.D., Price T.M. Human immunodeficiency virus infection of T cells and monocytes proceeds via receptor-mediated endocytosis. J. Cell Biol. 1988;107:959–968. doi: 10.1083/jcb.107.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de la Vega M., Marin M., Kondo N., Miyauchi K., Kim Y., Epand R.F., Epand R.M., Melikyan G.B. Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology. 2011;8:99. doi: 10.1186/1742-4690-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondo N., Marin M., Kim J.H., Desai T.M., Melikyan G.B. Distinct requirements for HIV-cell fusion and HIV-mediated cell-cell fusion. J. Biol. Chem. 2015;290:6558–6573. doi: 10.1074/jbc.M114.623181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galluzzi L., Green D.R. Autophagy-Independent Functions of the Autophagy Machinery. Cell. 2019;177:1682–1699. doi: 10.1016/j.cell.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozbor D., Burioni R., Ar-Rushdi A., Zmijewski C., Croce C.M. Expression of members of immunoglobulin gene family in somatic cell hybrids between human B and T cells. Proc. Natl. Acad. Sci. USA. 1987;84:4969–4973. doi: 10.1073/pnas.84.14.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herold N., Anders-Ößwein M., Glass B., Eckhardt M., Müller B., Kräusslich H.-G. HIV-1 Entry in SupT1-R5, CEM-ss, and Primary CD4+ T Cells Occurs at the Plasma Membrane and Does Not Require Endocytosis. J. Virol. 2014;88:13956–13970. doi: 10.1128/JVI.01543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salasc F., Gludish D.W., Jarvis I., Boliar S., Wills M.R., Russell D.G., Lever A.M.L., Mok H.-P. A novel, sensitive dual-indicator cell line for detection and quantification of inducible, replication-competent latent HIV-1 from reservoir cells. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-55596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeeninga R.E., Westerhout E.M., van Gerven M.L., Berkhout B. HIV-1 latency in actively dividing human T cell lines. Retrovirology. 2008;5:37. doi: 10.1186/1742-4690-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu M., Yang L., Zhang L., Liu B., Merling R., Xia Z., Giam C.Z. Human T-cell leukemia virus type 1 infection leads to arrest in the G1 phase of the cell cycle. J. Virol. 2008;82:8442–8455. doi: 10.1128/jvi.00091-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mercer J.L., Argus J.P., Crabtree D.M., Keenan M.M., Wilks M.Q., Chi J.-T.A., Bensinger S.J., Lavau C.P., Wechsler D.S. Modulation of PICALM Levels Perturbs Cellular Cholesterol Homeostasis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C., Tu J., Zhang S., Cai B., Liu Z., Hou S., Zhong Q., Hu X., Liu W., Li G., et al. Different regions of synaptic vesicle membrane regulate VAMP2 conformation for the SNARE assembly. Nat. Commun. 2020;11:1531. doi: 10.1038/s41467-020-15270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dumont V., Lehtonen S. PACSIN proteins in vivo: Roles in development and physiology. Acta Physiol. 2022;234 doi: 10.1111/apha.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aman Y., Schmauck-Medina T., Hansen M., Morimoto R.I., Simon A.K., Bjedov I., Palikaras K., Simonsen A., Johansen T., Tavernarakis N., et al. Autophagy in healthy aging and disease. Nat. Aging. 2021;1:634–650. doi: 10.1038/s43587-021-00098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nath S., Dancourt J., Shteyn V., Puente G., Fong W.M., Nag S., Bewersdorf J., Yamamoto A., Antonny B., Melia T.J. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 2014;16:415–424. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hilverling A., Szegö E.M., Dinter E., Cozma D., Saridaki T., Falkenburger B.H. Maturing Autophagosomes are Transported Towards the Cell Periphery. Cell. Mol. Neurobiol. 2022;42:155–171. doi: 10.1007/s10571-021-01116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nardacci R., Amendola A., Ciccosanti F., Corazzari M., Esposito V., Vlassi C., Taibi C., Fimia G.M., Del Nonno F., Ippolito G., et al. Autophagy plays an important role in the containment of HIV-1 in nonprogressor-infected patients. Autophagy. 2014;10:1167–1178. doi: 10.4161/auto.28678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laforge M., Limou S., Harper F., Casartelli N., Rodrigues V., Silvestre R., Haloui H., Zagury J.F., Senik A., Estaquier J. DRAM triggers lysosomal membrane permeabilization and cell death in CD4(+) T cells infected with HIV. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Killian M.S. Dual role of autophagy in HIV-1 replication and pathogenesis. AIDS Res. Ther. 2012;9:16. doi: 10.1186/1742-6405-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leymarie O., Lepont L., Berlioz-Torrent C. Canonical and Non-Canonical Autophagy in HIV-1 Replication Cycle. Viruses. 2017;9 doi: 10.3390/v9100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santerre M., Arjona S.P., Allen C.N., Callen S., Buch S., Sawaya B.E. HIV-1 Vpr protein impairs lysosome clearance causing SNCA/alpha-synuclein accumulation in neurons. Autophagy. 2021;17:1768–1782. doi: 10.1080/15548627.2021.1915641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vyboh K., Ajamian L., Mouland A.J. Detection of viral RNA by fluorescence in situ hybridization (FISH) J. Vis. Exp. 2012;e4002 doi: 10.3791/4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaminskyy V., Abdi A., Zhivotovsky B. A quantitative assay for the monitoring of autophagosome accumulation in different phases of the cell cycle. Autophagy. 2011;7:83–90. doi: 10.4161/auto.7.1.13893. [DOI] [PubMed] [Google Scholar]
- 65.Cavrois M., de Noronha C., Greene W.C. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 2002;20:1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- 66.Marin M., Kushnareva Y., Mason C.S., Chanda S.K., Melikyan G.B. HIV-1 Fusion with CD4+ T cells Is Promoted by Proteins Involved in Endocytosis and Intracellular Membrane Trafficking. Viruses. 2019;11 doi: 10.3390/v11020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian Y., Chang J.C., Fan E.Y., Flajolet M., Greengard P. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer's APP-CTF for terminal degradation via autophagy. Proc. Natl. Acad. Sci. USA. 2013;110:17071–17076. doi: 10.1073/pnas.1315110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeNies M.S., Rosselli-Murai L.K., Schnell S., Liu A.P. Clathrin Heavy Chain Knockdown Impacts CXCR4 Signaling and Post-translational Modification. Front. Cell Dev. Biol. 2019;7:77. doi: 10.3389/fcell.2019.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benson R.E., Sanfridson A., Ottinger J.S., Doyle C., Cullen B.R. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J. Exp. Med. 1993;177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daecke J., Fackler O.T., Dittmar M.T., Kräusslich H.G. Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J. Virol. 2005;79:1581–1594. doi: 10.1128/jvi.79.3.1581-1594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G.B. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., Abeliovich H., Abildgaard M.H., Abudu Y.P., Acevedo-Arozena A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy. autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Motamedi M., Xu L., Elahi S. Correlation of transferrin receptor (CD71) with Ki67 expression on stimulated human and mouse T cells: The kinetics of expression of T cell activation markers. J. Immunol. Methods. 2016;437:43–52. doi: 10.1016/j.jim.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 74.Moran A.E., Polesso F., Weinberg A.D. Immunotherapy Expands and Maintains the Function of High-Affinity Tumor-Infiltrating CD8 T Cells In Situ. J. Immunol. 2016;197:2509–2521. doi: 10.4049/jimmunol.1502659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahn E., Araki K., Hashimoto M., Li W., Riley J.L., Cheung J., Sharpe A.H., Freeman G.J., Irving B.A., Ahmed R. Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl. Acad. Sci. USA. 2018;115:4749–4754. doi: 10.1073/pnas.1718217115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sakai S., Kauffman K.D., Sallin M.A., Sharpe A.H., Young H.A., Ganusov V.V., Barber D.L. CD4 T Cell-Derived IFN-γ Plays a Minimal Role in Control of Pulmonary Mycobacterium tuberculosis Infection and Must Be Actively Repressed by PD-1 to Prevent Lethal Disease. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dong Y., Li X., Zhang L., Zhu Q., Chen C., Bao J., Chen Y. CD4(+) T cell exhaustion revealed by high PD-1 and LAG-3 expression and the loss of helper T cell function in chronic hepatitis B. BMC Immunol. 2019;20:27. doi: 10.1186/s12865-019-0309-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Biancotto A., Iglehart S.J., Vanpouille C., Condack C.E., Lisco A., Ruecker E., Hirsch I., Margolis L.B., Grivel J.C. HIV-1 induced activation of CD4+ T cells creates new targets for HIV-1 infection in human lymphoid tissue ex vivo. Blood. 2008;111:699–704. doi: 10.1182/blood-2007-05-088435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Husebye H., Halaas Ø., Stenmark H., Tunheim G., Sandanger Ø., Bogen B., Brech A., Latz E., Espevik T. Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 2006;25:683–692. doi: 10.1038/sj.emboj.7600991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pascual-Lucas M., Fernandez-Lizarbe S., Montesinos J., Guerri C. LPS or ethanol triggers clathrin- and rafts/caveolae-dependent endocytosis of TLR4 in cortical astrocytes. J. Neurochem. 2014;129:448–462. doi: 10.1111/jnc.12639. [DOI] [PubMed] [Google Scholar]
- 81.Hernández J.C., Stevenson M., Latz E., Urcuqui-Inchima S. HIV type 1 infection up-regulates TLR2 and TLR4 expression and function in vivo and in vitro. AIDS Res. Hum. Retrovir. 2012;28:1313–1328. doi: 10.1089/aid.2011.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raju S., Verbaro D.J., Egawa T. PD-1 Signaling Promotes Control of Chronic Viral Infection by Restricting Type-I-Interferon-Mediated Tissue Damage. Cell Rep. 2019;29:2556–2564.e3. doi: 10.1016/j.celrep.2019.10.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Velu V., Shetty R.D., Larsson M., Shankar E.M. Role of PD-1 co-inhibitory pathway in HIV infection and potential therapeutic options. Retrovirology. 2015;12:14. doi: 10.1186/s12977-015-0144-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lopez P., Ajibola O., Pagliuzza A., Zayats R., Koh W.H., Herschhorn A., Chomont N., Murooka T.T. T cell migration potentiates HIV infection by enhancing viral fusion and integration. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110406. [DOI] [PubMed] [Google Scholar]
- 85.Lerrer S., Tocheva A.S., Bukhari S., Adam K., Mor A. PD-1-stimulated T cell subsets are transcriptionally and functionally distinct. iScience. 2021;24 doi: 10.1016/j.isci.2021.103020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Januário Y.C., daSilva L.L.P. Hijacking of endocytosis by HIV-1 Nef is becoming crystal clear. Nat. Struct. Mol. Biol. 2020;27:773–775. doi: 10.1038/s41594-020-0486-5. [DOI] [PubMed] [Google Scholar]
- 87.Pereira E.A., daSilva L.L.P. HIV-1 Nef: Taking Control of Protein Trafficking. Traffic. 2016;17:976–996. doi: 10.1111/tra.12412. [DOI] [PubMed] [Google Scholar]
- 88.Campbell G.R., Rawat P., Bruckman R.S., Spector S.A. Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Castro-Gonzalez S., Shi Y., Colomer-Lluch M., Song Y., Mowery K., Almodovar S., Bansal A., Kirchhoff F., Sparrer K., Liang C., Serra-Moreno R. HIV-1 Nef counteracts autophagy restriction by enhancing the association between BECN1 and its inhibitor BCL2 in a PRKN-dependent manner. Autophagy. 2021;17:553–577. doi: 10.1080/15548627.2020.1725401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kyei G.B., Dinkins C., Davis A.S., Roberts E., Singh S.B., Dong C., Wu L., Kominami E., Ueno T., Yamamoto A., et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Molle D., Segura-Morales C., Camus G., Berlioz-Torrent C., Kjems J., Basyuk E., Bertrand E. Endosomal Trafficking of HIV-1 Gag and Genomic RNAs Regulates Viral Egress. J. Biol. Chem. 2009;284:19727–19743. doi: 10.1074/jbc.M109.019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schönrich G., Raftery M.J. The PD-1/PD-L1 Axis and Virus Infections: A Delicate Balance. Front. Cell. Infect. Microbiol. 2019;9:207. doi: 10.3389/fcimb.2019.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Day C.L., Kaufmann D.E., Kiepiela P., Brown J.A., Moodley E.S., Reddy S., Mackey E.W., Miller J.D., Leslie A.J., DePierres C., et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 94.Balança C.-C., Salvioni A., Scarlata C.-M., Michelas M., Martinez-Gomez C., Gomez-Roca C., Sarradin V., Tosolini M., Valle C., Pont F., et al. PD-1 blockade restores helper activity of tumor-infiltrating, exhausted PD-1hiCD39+ CD4 T cells. JCI Insight. 2021;6 doi: 10.1172/jci.insight.142513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goods B.A., Hernandez A.L., Lowther D.E., Lucca L.E., Lerner B.A., Gunel M., Raddassi K., Coric V., Hafler D.A., Love J.C. Functional differences between PD-1+ and PD-1- CD4+ effector T cells in healthy donors and patients with glioblastoma multiforme. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y., Chung Y.R., Eitzinger S., Palacio N., Gregory S., Bhattacharyya M., Penaloza-MacMaster P. TLR4 signaling improves PD-1 blockade therapy during chronic viral infection. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pauken K.E., Godec J., Odorizzi P.M., Brown K.E., Yates K.B., Ngiow S.F., Burke K.P., Maleri S., Grande S.M., Francisco L.M., et al. The PD-1 Pathway Regulates Development and Function of Memory CD8+ T Cells following Respiratory Viral Infection. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Simon S., Labarriere N. PD-1 expression on tumor-specific T cells: Friend or foe for immunotherapy? OncoImmunology. 2017;7 doi: 10.1080/2162402x.2017.1364828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Macatangay B.J.C., Gandhi R.T., Jones R.B., McMahon D.K., Lalama C.M., Bosch R.J., Cyktor J.C., Thomas A.S., Borowski L., Riddler S.A., et al. T cells with high PD-1 expression are associated with lower HIV-specific immune responses despite long-term antiretroviral therapy. AIDS. 2020;34:15–24. doi: 10.1097/qad.0000000000002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Neff C.P., Chain J.L., MaWhinney S., Martin A.K., Linderman D.J., Flores S.C., Campbell T.B., Palmer B.E., Fontenot A.P. Lymphocytic alveolitis is associated with the accumulation of functionally impaired HIV-specific T cells in the lung of antiretroviral therapy-naive subjects. Am. J. Respir. Crit. Care Med. 2015;191:464–473. doi: 10.1164/rccm.201408-1521OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reynolds J.M., Martinez G.J., Chung Y., Dong C. Toll-like receptor 4 signaling in T cells promotes autoimmune inflammation. Proc. Natl. Acad. Sci. USA. 2012;109:13064–13069. doi: 10.1073/pnas.1120585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ando K., Nagaraj S., Küçükali F., de Fisenne M.A., Kosa A.C., Doeraene E., Lopez Gutierrez L., Brion J.P., Leroy K. PICALM and Alzheimer's Disease: An Update and Perspectives. Cells. 2022;11:3994. doi: 10.3390/cells11243994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang J.-Y., Zhang Z., Wang X., Fu J.-L., Yao J., Jiao Y., Chen L., Zhang H., Wei J., Jin L., et al. PD-1 up-regulation is correlated with HIV-specific memory CD8+ T-cell exhaustion in typical progressors but not in long-term nonprogressors. Blood. 2007;109:4671–4678. doi: 10.1182/blood-2006-09-044826. [DOI] [PubMed] [Google Scholar]
- 104.Saad E.B., Oroya A., Rudd C.E. Abstract 6528: Anti-PD-1 induces the endocytosis of the co-receptor from the surface of T-cells: Nivolumab is more effective than Pembrolizumab. Cancer Res. 2020;80:6528. doi: 10.1158/1538-7445.Am2020-6528. [DOI] [Google Scholar]
- 105.Yao H., Li C., He F., Song T., Brosseau J.P., Wang H., Lu H., Fang C., Shi H., Lan J., et al. A peptidic inhibitor for PD-1 palmitoylation targets its expression and functions. RSC Chem. Biol. 2021;2:192–205. doi: 10.1039/d0cb00157k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nelson A.R., Sagare A.P., Zlokovic B.V. In: Developing Therapeutics for Alzheimer's Disease. Wolfe M.S., editor. Academic Press; 2016. Chapter 9 - Blood–Brain Barrier Transport of Alzheimer’s Amyloid β-Peptide; pp. 251–270. [DOI] [Google Scholar]
- 107.Porichis F., Kaufmann D.E. Role of PD-1 in HIV pathogenesis and as target for therapy. Curr. HIV AIDS Rep. 2012;9:81–90. doi: 10.1007/s11904-011-0106-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun L., Su Y., Jiao A., Wang X., Zhang B. T cells in health and disease. Signal Transduct. Targeted Ther. 2023;8:235. doi: 10.1038/s41392-023-01471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim M.J., Ha S.J. Differential Role of PD-1 Expressed by Various Immune and Tumor Cells in the Tumor Immune Microenvironment: Expression, Function, Therapeutic Efficacy, and Resistance to Cancer Immunotherapy. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.767466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shi J., Hou S., Fang Q., Liu X., Liu X., Qi H. PD-1 Controls Follicular T Helper Cell Positioning and Function. Immunity. 2018;49:264–274.e4. doi: 10.1016/j.immuni.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee Y., Awasthi A., Yosef N., Quintana F.J., Xiao S., Peters A., Wu C., Kleinewietfeld M., Kunder S., Hafler D.A., et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yuan Y., Jacobs C., Garcia I.L., Pereira P.M., Lawrence S.P., Laine R.F., Marsh M., Henriques R. Single-molecule super-resolution imaging of T-cell plasma membrane CD4 redistribution upon HIV-1 binding. bioRxiv. 2021 doi: 10.1101/2021.01.05.425371. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kisler K., Sagare A.P., Lazic D., Bazzi S., Lawson E., Hsu C.J., Wang Y., Ramanathan A., Nelson A.R., Zhao Z., Zlokovic B.V. Anti-malaria drug artesunate prevents development of amyloid-β pathology in mice by upregulating PICALM at the blood-brain barrier. Mol. Neurodegener. 2023;18:7. doi: 10.1186/s13024-023-00597-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shang S., Wu J., Li X., Liu X., Li P., Zheng C., Wang Y., Liu S., Zheng J., Zhou H. Artesunate interacts with the vitamin D receptor to reverse sepsis-induced immunosuppression in a mouse model via enhancing autophagy. Br. J. Pharmacol. 2020;177:4147–4165. doi: 10.1111/bph.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Efferth T., Romero M.R., Wolf D.G., Stamminger T., Marin J.J.G., Marschall M. The antiviral activities of artemisinin and artesunate. Clin. Infect. Dis. 2008;47:804–811. doi: 10.1086/591195. [DOI] [PubMed] [Google Scholar]
- 116.Efferth T., Marschall M., Wang X., Huong S.M., Hauber I., Olbrich A., Kronschnabl M., Stamminger T., Huang E.S. Antiviral activity of artesunate towards wild-type, recombinant, and ganciclovir-resistant human cytomegaloviruses. J. Mol. Med. 2002;80:233–242. doi: 10.1007/s00109-001-0300-8. [DOI] [PubMed] [Google Scholar]
- 117.Berkowitz R.D., Beckerman K.P., Schall T.J., McCune J.M. CXCR4 and CCR5 expression delineates targets for HIV-1 disruption of T cell differentiation. J. Immunol. 1998;161:3702–3710. [PubMed] [Google Scholar]
- 118.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ratnapriya S., Harris M., Chov A., Herbert Z.T., Vrbanac V., Deruaz M., Achuthan V., Engelman A.N., Sodroski J., Herschhorn A. Intra- and extra-cellular environments contribute to the fate of HIV-1 infection. Cell Rep. 2021;36:109622. doi: 10.1016/j.celrep.2021.109622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Esté J.A., Cabrera C., Blanco J., Gutierrez A., Bridger G., Henson G., Clotet B., Schols D., De Clercq E. Shift of clinical human immunodeficiency virus type 1 isolates from X4 to R5 and prevention of emergence of the syncytium-inducing phenotype by blockade of CXCR4. J. Virol. 1999;73:5577–5585. doi: 10.1128/jvi.73.7.5577-5585.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S., Sabatini D.M., Chen I.S.Y., Hahn W.C., Sharp P.A., et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen H.-J., Anagnostou G., Chai A., Withers J., Morris A., Adhikaree J., Pennetta G., de Belleroche J.S. Characterization of the Properties of a Novel Mutation in VAPB in Familial Amyotrophic Lateral Sclerosis. J. Biol. Chem. 2010;285:40266–40281. doi: 10.1074/jbc.M110.161398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ratnapriya S., Perez-Greene E., Schifanella L., Herschhorn A. Adjuvant-mediated enhancement of the immune response to HIV vaccines. FEBS J. 2022;289:3317–3334. doi: 10.1111/febs.15814. [DOI] [PubMed] [Google Scholar]
- 124.Murooka T.T., Deruaz M., Marangoni F., Vrbanac V.D., Seung E., von Andrian U.H., Tager A.M., Luster A.D., Mempel T.R. HIV-infected T cells are migratory vehicles for viral dissemination. Nature. 2012;490:283–287. doi: 10.1038/nature11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report original code.