Abstract
The Fanconi anemia (FA) repair pathway governs repair of highly genotoxic DNA interstrand crosslinks (ICLs) and relies on translesion synthesis (TLS). TLS is facilitated by REV1 or site-specific monoubiquitination of proliferating cell nuclear antigen (PCNA) (PCNA-Ub) at lysine 164 (K164). A PcnaK164R/K164R but not Rev1−/− mutation renders mammals hypersensitive to ICLs. Besides the FA pathway, alternative pathways have been associated with ICL repair (1, 2), though the decision making between those remains elusive. To study the dependence and relevance of PCNA-Ub in FA repair, we intercrossed PcnaK164R/+; Fancg−/+ mice. A combined mutation (PcnaK164R/K164R; Fancg−/−) was found embryonically lethal. RNA-seq of primary double-mutant (DM) mouse embryonic fibroblasts (MEFs) revealed elevated levels of replication stress-induced checkpoints. To exclude stress-induced confounders, we utilized a Trp53 knock-down to obtain a model to study ICL repair in depth. Regarding ICL-induced cell toxicity, cell cycle arrest, and replication fork progression, single-mutant and DM MEFs were found equally sensitive, establishing PCNA-Ub to be critical for FA-ICL repair. Immunoprecipitation and spectrometry-based analysis revealed an unknown role of PCNA-Ub in excluding mismatch recognition complex MSH2/MSH6 from being recruited to ICLs. In conclusion, our results uncovered a dual function of PCNA-Ub in ICL repair, i.e. exclude MSH2/MSH6 recruitment to channel the ICL toward canonical FA repair, in addition to its established role in coordinating TLS opposite the unhooked ICL.
Keywords: PCNA monoubiquitination, Fanconi anemia, interstrand crosslink, mismatch repair, replication stress
Significance Statement.
This study presents, for the first time, a comparative analysis of Fanconi anemia (FA) repair and DNA damage tolerance (DDT)-deficient models. Proliferating cell nuclear antigen (PCNA) monoubiquitination at lysine 164 (PCNA-Ub), which is at the heart of DDT, is essential for FA repair of interstrand crosslinks (ICLs) as its absence demonstrates phenotype similar to the FA model. Additionally, PCNA-Ub prevents an alternative pathway, mismatch repair from intervening in ICL repair. These insights indicate a novel dual role of PCNA-Ub and establish its prominence in ICL repair.
Introduction
Fanconi anemia (FA) is a rare genetic disorder characterized by aplastic anemia, developmental defects, and a high predisposition to cancer (3–5). FA is caused by a mutation in one of the 23 Fanconi complementation groups. The FA pathway is central to the repair of interstrand crosslinks (ICLs) (6). ICLs are highly toxic lesions that covalently link both strands of the DNA. Consequently, key biological processes such as replication and transcription are impeded (7, 8).
At the site of the ICL, eight FA proteins (FANCA, B, C, E, F, G, L, and M) form the FA core complex with FA-associated proteins (FAAP-20, FAAP-24, FAAP-100, MHF1, MHF2, and HES1) (3, 9). This complex encompasses the E3 ubiquitin ligase FANCL, which collaborates with the ubiquitin-conjugating enzyme UBE2T (10). The core complex monoubiquitinates FANCI (11) and FANCD2 proteins mediating the recruitment of endonucleases such as the XPF-ERCC1 (12) and SLX1-SLX4 complexes among others (13,14). These endonucleases cut the sugar phosphate backbone on both flanks of the ICL, resulting in a gap on one and an unhooked ICL on the other DNA strand. The FA pathway relies on the activity of TLS polymerases to mediate the replicative bypass of the noninstructive unhooked ICL lesion (15–17). Subsequently, the components of homologous recombination (HR) and nucleotide excision repair (NER) pathways are recruited to complete the repair process (16, 18).
Besides the FA pathway, alternative pathways such as the NEIL3 pathway (1) and mismatch repair pathway (MMR) (2), among others (19), have been implicated in ICL repair. These alternative pathways process ICLs independent of the canonical FA route and can vary depending on the cell cycle phase. While the former requires the glycosylase NEIL3 to cleave one of the two N-glycosyl bonds of the crosslink, the latter involves the MMR recognition complex MSH2–MSH6 that can sense helix distorting ICLs and recruit MLH1-PMS2 and EXO1 to the ICL to unhook the crosslink. Subsequently, a damage-tolerant DNA polymerase replicates over the unhooked, noninstructive ICL. Finally, NER completes the repair process by removing the remaining bulky adduct (2, 20, 21).
Regardless of the mode of the ICL unhooking, noninstructive lesions are central intermediates. Replicative bypass opposite these processed ICLs requires translesion synthesis (TLS) polymerases. TLS is a mode of DNA damage tolerance (DDT) that enables specialized TLS polymerases to continue replication in the presence of lesions that otherwise block the replicative polymerases POLδ or POLε (22). During standard TLS, stalling of replicative polymerases segregates the helicase from the polymerase, exposing single-stranded DNA (ssDNA). Replication protein A binds to ssDNA and recruits RAD6–RAD18, an E2–E3 ubiquitin conjugase–ligase complex. This complex monoubiquitinates proliferating cell nuclear antigen (PCNA) site-specifically at lysine 164 (K164) (23–27). Ubiquitinated PCNA (PCNA-Ub) facilitates the switching of the replicative polymerases with a specialized damage-tolerant TLS polymerase that continues replication opposite the persisting lesion (22, 28). Thereby, DDT prohibits prolonged replication fork stalling, fork collapse, and associated genome instability. However, ICL repair studied using plasmids in Xenopus extracts does not demonstrate extensive ssDNA formation as replisomes approach the ICL from both directions (13).
Model systems using nonmodifiable PCNA-K164R (lysine-to-arginine mutation) have been highly informative in understanding the role of PCNA-Ub and SUMO modifications in tolerating a plethora of replication-blocking impediments (23, 29–31). Data on whether which mode of TLS required for TLS polymerase recruitment and ICL repair are conflicting (13, 17, 30). While unbiased mass spec data revealed recruitment of RAD6/18 to chromatin (32), REV1, shown to be required for ICL repair (17, 33, 34), has recently been demonstrated to be dispensable to mammalian ICL repair (30). This determines that the mode of TLS primary to ICL repair is dictated by PCNA-Ub. Moreover, PCNA-K164R primary PreB cells (B-cell precursors), MEFs, or lymphomas and POLK-deficient lymphomas are sensitive to commonly used ICL-inducing cancer chemotherapeutics such as cisplatin (CDDP) (30, 35–37), while REV1-deficient counterparts are not (30). To determine the relevance of PCNA-Ub in FA-ICL repair in mammals, we requested a direct comparison between the two systems i.e. FA and PCNA-K164R. Moreover, PCNA-Ub has also been implied to recruit the Fanconi complexes to the sites of ICLs (38) and is responsible for POLK recruitment (30), suggesting a critical role for PCNA-Ub in the early steps of ICL repair. Altogether, these aspects necessitated a comprehensive systemic, cellular, and molecular analysis of PCNA-Ub in ICL repair.
To address these subjects, we intercrossed Fancg+/− mouse model (39) with the preestablished PCNAK164R/+ model (40, 41). A homozygous combination of both alterations [double mutants (DMs)] was found to be embryonically lethal, indicating that the lack of both DDT and FA is incompatible with mammalian life. In line, primary cell cultures could not be established from these mice. Inactivation of Trp53 rescued the proliferation defect of DM mouse embryonic fibroblasts (MEFs). Furthermore, upon exposure to ICL-inducing agents, PCNA-Ub-deficient cells and FANCG-deficient cells were found equally sensitive as measured by colony formation, EdU incorporation, and ssDNA formation. This further establishes PCNA-Ub as an essential regulator in the FA repair pathway in mammalian cells. Interestingly, in the absence of PCNA-Ub, the mismatch recognition complex MSH2–MSH6 was recruited to ICL site, indicating a novel function of PCNA-Ub in the decision making between canonical FA and noncanonical MMR-directed ICL repair. Our findings reveal a dual role of PCNA-Ub in ICL repair: excluding MMR from processing ICLs and facilitating TLS to resolve ICLs efficiently.
Results
A combined DDT and FA defect is embryonically and synthetically lethal
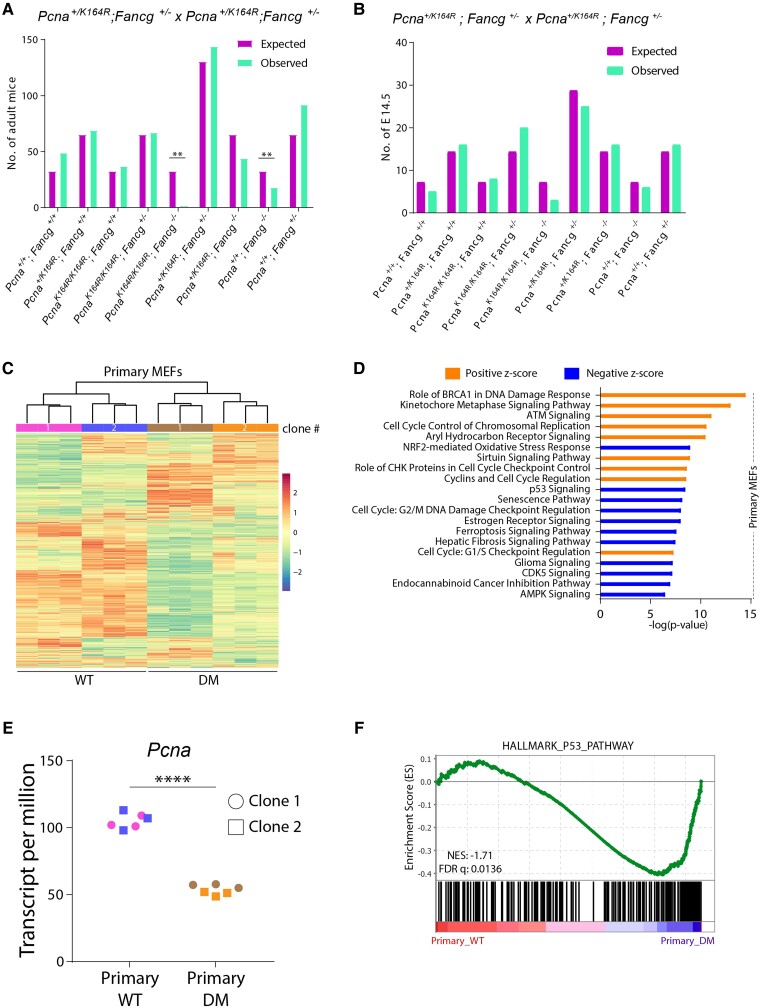
To determine the relevance of the PCNA-Ub-facilitated DDT in the FA pathway, Fancg+/−; Pcna+/KR mice (KR indicates a targeted lysine 164 to arginine mutation in the Pcna locus) were intercrossed as a homozygous mutation rendered the mice infertile. Breeding data revealed that wild type (WT) (Pcna+/+; Fancg+/+) and KR (PcnaKR/KR; Fancg+/+) mice were born at expected Mendelian frequencies (Figure 1A). In contrast, single-mutant FGko (Pcna+/+; Fancg−/−) and DM (PcnaKR/KR; Fancg−/−) mice were born sub-Mendelian. Of note, while the Fancg deficiency had an intermediate effect on the litter size (observed/expected: 18/33), absence of both PcnaK164 and Fancg was very severe (3/33). Apparently, a homozygous DM is embryonically lethal.
Fig. 1.
A combined DDT and FA defect increases cell cycle stress and cell death in pMEFs. A) Observed and expected numbers of offspring obtained from Pcna+/KR; Fancg+/− intercrosses. P-values were calculated using Pearson's Chi-square test, **P < 0.01. B) Observed and expected numbers of E14.5 embryos obtained from timed mating of PCNA+/KR; FG+/− intercrosses. C) Heatmap showing unsupervised clustering of all genes expressed in two independent clones of WT and DM pMEFs as determined by RNA sequencing. Each MEF clone was sequenced in three technical replicates. D) RNA-seq data from WT and DM pMEFs were analyzed using IPA (right-tailed Fisher's exact test; FDR < 0.05). Graph displays top 15 canonical pathways that have a positive or negative Z-score. The Z-score indicates predicted pathway activation or inhibition. E) Graph displays Pcna TPM counts from clones shown in C. P-values were calculated using one-way ANOVA. ****P < 0.0001. F) GSEA comparing transcriptomes of WT and DM pMEFs using the hallmark p53 gene set. Normalized enrichment score (NES) and FDR are indicated.
To understand this embryonically lethal interaction in the DM, timed mating was conducted to obtain 14.5-day-old embryos (E14.5), which were genotyped subsequently (Figure 1B). In contrast to the sub-Mendelian frequency of FGko mice, FGko embryos were found at expected Mendelian ratio. However, DM embryos were still obtained at sub-Mendelian frequency. This analysis demonstrated that the sub-Mendelian frequency of adult FGko mice is not attributed to an early but rather late embryonic defect. In contrast, the adverse impact of a combinatorial DM mutation on embryonic fitness initiates prior to E14.5.
To understand the impact of a combined DDT and FA deficiency in relation to embryonic lethality, we investigated the impact of these DDR defects by establishing primary cell cultures from E14.5. Despite several attempts, we selectively failed to establish primary PreB cell cultures from the E14.5 fetal livers of DM embryos. This apparent synthetically lethal interaction between both pathways further highlights the relevance of the DDT and FA pathways in warranting cellular fitness and survival. Like PreB cell cultures, DM primary MEFs (pMEFs) failed to grow beyond the first two passages, limiting their experimental usage. To understand the selective disadvantage of the DM background on fitness of primary cells, we generated RNA-seq data from two independent WT and DM pMEFs. Fancg transcript per million (TPM) count across the DM clones validated the genotypic background of the clones (Supplementary Figure S1A). Unsupervised clustering grouped the independent pMEF clones according to their genotype (Figure 1C), which led to us to pool the RNA-seq data for further downstream analyses.
Ingenuity pathway analysis (IPA) identified the top 15 majorly affected pathways (Figure 1D). The majority of these pathways were involved in the DNA damage repair (DDR) network and cell cycle regulation. The latter involved transcriptomic changes in key cell cycle checkpoints activated in pMEFs, implying severe defects in cell cycle progression in DM pMEFs. Cell cycle impairment was further confirmed by comparison of the Pcna TPM counts, a well-documented S-phase marker, between WT and DM. As expected, Pcna transcripts were reduced about two-fold in DM (Figure 1E), suggesting that the reduced proliferation of DM pMEFs relates to Trp53-based checkpoint induction, as confirmed by other well-known cell cycle markers (Supplementary Figure S1B).
To further elaborate the pathways affecting the cellular fitness of DM pMEFs, enrichment of specific “HALLMARK” gene sets obtained from the MSigDB server was determined by gene set enrichment analysis (GSEA) (Supplementary Figure S1C). In line with the IPA, GSEA also showed significant enrichment of the p53 pathway in the DM pMEFs (Figure 1F). As the guardian of the genome, Trp53 maintains a delicate balance between DNA repair and cell death (42–44). The lack of critical DDR elements in DM pMEFs increases replicative stress and DNA damage, which results in Trp53 pathway activation and eventually cell death.
In summary, lack of PCNA-Ub and FANCG is embryonically and synthetically lethal. RNA-seq from pMEFs indicated that a combined defect induces DNA damage-based Trp53 activation, contributing to the early demise of embryos, PreB cells, and pMEFs.
Trp53kd rescues DM-induced genotoxic stress
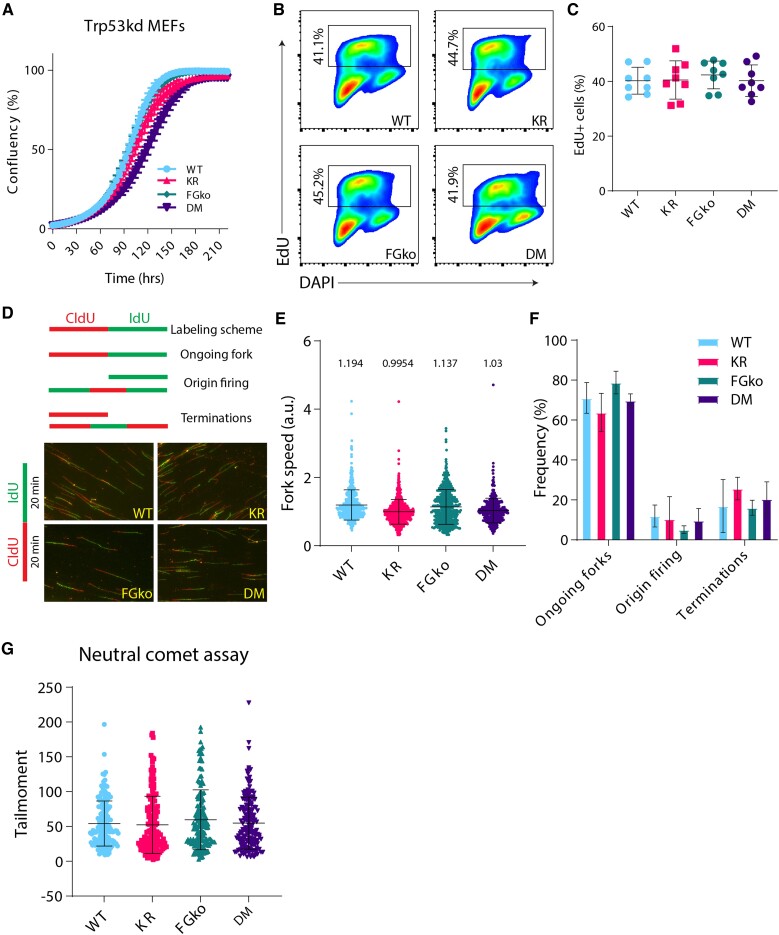
Given these observations, we reasoned that Trp53 knock-down would rescue the synthetic lethality of the DM pMEFs. Indeed, as determined by revised cluster analysis of RNA-seq data comparing Trp53kd WT and DM MEFs, a Trp53kd of DM pMEFs (Supplementary Figure S1D) restored Trp53-related phenotypes between WT and DM (Supplementary Figure S1E). Additionally, the Pcna TPM counts were comparable between Trp53kd WT and DM MEFs (Supplementary Figure S1F). In line, IPA of the immortalized MEFs also no longer displayed any differential DDR or cell cycle-associated pathways observed for the pMEFs (Supplementary Figure S1G). To determine the dynamic effect of the double mutation on cell proliferation in the absence of Trp53, growth kinetics of all Trp53kd immortalized MEFs, i.e. WT, KR, FGko, and DM, were recorded using the IncuCyte Live cell imaging system (Figure 2A). Immortalized MEFs of all four genotypes grew at comparable rates, indicating that in the absence of p53, the status of PCNA-Ub or/and FANCG does not influence proliferation kinetics of MEFs under normal growth conditions. These insights further highlighted the impact of stress-induced activation of p53 in DM pMEFs.
Fig. 2.
Trp53kd KR; FGko MEFs have a normal proliferation and cell cycle profile. A) Trp53kd cell lines were seeded at 250 cells/well in a 96-well plate, and cell confluency was measured every 4 hours using IncuCyte live cell imaging system from two independent experiments. Dots at each timepoint indicate mean, and bars represent SD. B) Representative flow cytometry plots showing gating strategy to identify and compare EdU-positive population in Trp53kd MEFs cultured under standard conditions. Cells in S-phase were revealed by EdU incorporation for 20 minutes before harvesting and fixed in 70% ethanol. In addition, the DNA stain DAPI was used as a cell cycle marker to distinguish G1 from G2/M cells. C) Quantification of EdU-positive cells from two independent experiments. Each dot indicates a sample, and bars represent mean ± SD. D) Schematic representation of different replication fork structures (45) identified after pulse labeling with CldU (red) and IdU (green) (upper panel). Representative images of DNA fibers from Trp53kd MEFs (lower panel). Cells were pulse-labeled with CldU and IdU for 20 minutes each. Ongoing forks were used to calculate fork speed (kb/min). E) Replication fork speeds from Trp53kd MEFs cultured under standard conditions. Each dot represents an ongoing fork. At least 350 track lengths of ongoing forks were measured (from three independent experiments) with ImageJ. Bars represent mean ± SD. F) Quantification of ongoing forks, origin firing, and terminations as a frequency of all replication fork structures identified in fiber images used in E. G) Quantification of tail moments from Trp53kd MEFs as determined by neutral comet assay (technical replicates = 50, n = 3). Bars represent mean ± SD. Tail moments were obtained using CASP software.
To detect any potential cellular alteration in cell cycle progression, cells were pulse-labeled with thymidine analogue EdU for 20 minutes and analyzed by flow cytometry (Figure 2B). Complementing the results from the IncuCyte assay, the frequency of EdU-labeled cells was comparable across all the genotypes. Combined with the growth kinetics data, these independent approaches portray that Trp53kd normalizes the cell growth in our MEFs irrespective of their DDR defect (Figure 2C).
Involvement of both PCNA-Ub and FA in overcoming impediments during DNA replication led us to study the impact of the mutations on a single-molecule level using the DNA fiber assay (45). To determine the replication fork speed, we analyzed the length of DNA fibers that were prepared from cells pulsed for 20 minutes of CldU followed by 20 minutes of IdU (Figure 2D). The replication fork speed quantified from ongoing forks was comparable across all genotypes (Figure 2E). Additionally, the frequency of ongoing forks, origin firing, and replication terminations, as defined in (45), also displayed no significant differences (Figure 2F). Altogether, these analyses revealed that lack of DDT causes a slight but insignificant reduction in fork speed, probably related to endogenous lesions, but overall does not affect replication fidelity.
Prolonged replication stress results in fork collapse and is associated with the formation of recombinogenic double-stranded breaks (DSBs) (46). To address whether replication stress induced by endogenous impediments leads to an increase in DSBs, we performed neutral comet assays on unchallenged MEFs. The analyses of the tail moments as readouts for DSBs did not display a significant increase in DSBs in single-mutant or DM MEFs in comparison with WT. Based on this assay, a K164R mutation, with or without FANCG, does not generate more DSBs in response to endogenous replication impediments (Figure 2G).
Taken together, a knock-down of Trp53 in DM MEFs rescued the synthetic lethality observed in primary DM MEFs. Apart from a restored proliferative capacity in Trp53kd DM MEFs, these immortalized cells closely phenocopied those of KR MEFs regarding fork speed, origin firing and DSB formation. This finding provided an ideal system to address the relation and relevance of both PCNA-Ub and FA in the context of crosslink repair.
PCNA-Ub is a central player in crosslink repair
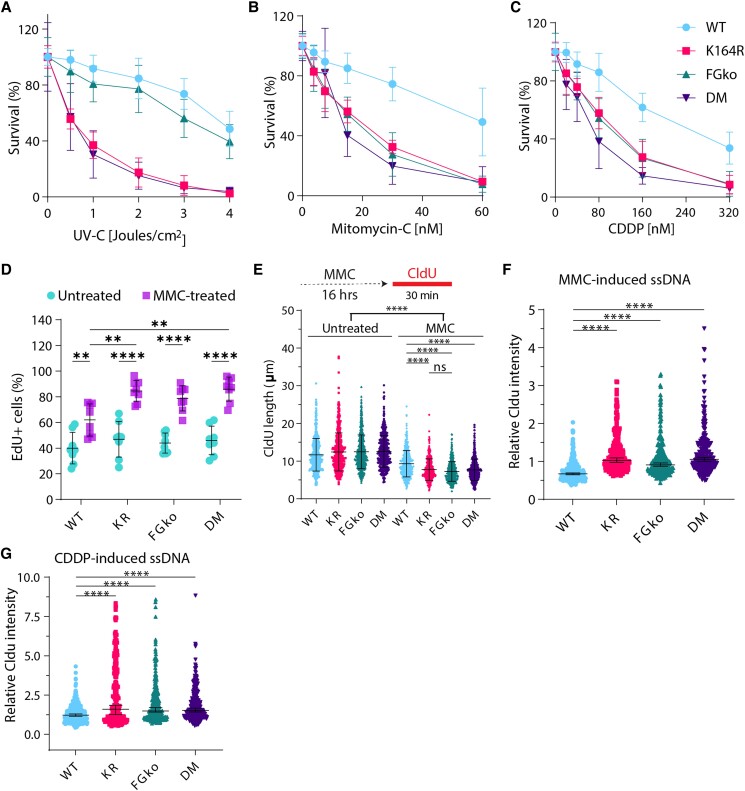
To determine the effect of the combined lack of PCNA-Ub and FANCG on cell proliferation in the presence of ICLs, IncuCyte assays were performed. Upon treatment with lower doses of MMC or CDDP, the single mutants as well as the DM were hypersensitive compared to WT MEFs (Supplementary Figure S2A). Interestingly, the DM was as sensitive and displayed a similar growth profile to the single mutants, demonstrating epistasis between PCNA-Ub and FANCG in the presence of ICLs. This was also observed in the mutant cells treated with higher doses of the compounds.
However, as IncuCyte assays do not account for clonal variability, drug-resistant clones may outgrow and occupy enter surface of the wells. A more robust method to assess the impact of ICLs on the cells is to assess the single-cell survival. This assay determines the potential of single cells to establish a colony in the presence of exogenous insults, such as UV-C, CDDP, and mitomycin C (MMC). Survival plots revealed that KR and DM MEFs were hypersensitive to UV-C-induced lesions (Figure 3A). This agreed with our previous work, highlighting the relevance of PCNA-Ub in tolerating UV-C-induced lesions (35, 48) and re-confirmed the irrelevance of FANCG in repair of these lesions. Treatment with CDDP and MMC provided several key observations (Figure 3B and C). First, as expected FGko MEFs were sensitive to both crosslinking agents. Second, KR MEFs were found equally sensitive as FGko MEFs, indicating the dependence of FA-ICL repair on PCNA-Ub. Third, DM MEFs were as sensitive as KR and FGko MEFs, i.e. PCNA-Ub is as relevant as a key component of the FA pathway, demonstrating an epistatic relation between the FA pathway and the RAD6/18 pathway.
Fig. 3.
PCNA-Ub is a central player in crosslink repair. A–C) Survival of WT, KR, FGko, and DM Trp53kd MEFs obtained by colony formation assay. Cells were seeded at increasing densities in 10-cm dishes, treated with increasing doses of A) UV-C, B) CDDP, and C) MMC and fixed, stained, and counted on day 7 after seeding. Colonies were counted using a colony counter from three independent experiments. For each dose, graph displays mean ± SD. D) Percentage of EdU-positive cells in untreated or MMC-treated MEFs. Each dot indicates a sample, and bars represent mean ± SD. Cells were left untreated or treated with 1.5 µM MMC for 16 hours followed by EdU labeling for 20 minutes before being harvested and fixed in 70% ethanol. In addition, the DNA stain DAPI was used as a cell cycle marker to distinguish G1 from G2/M cells. E) Length of CldU-labeled DNA fibers. Cells were left untreated or treated with 1.5 µM MMC for 16 hours followed by CldU labeling for 30 minutes before being harvested. At least 400 fibers were measured. Bars represent mean ± SD. P-values were calculated using one-way ANOVA. **P < 0.01, ****P < 0.0001. F), G) Quantification of CldU-based ssDNA based on protocol from Koundrioukoff et al. (47). Cells were grown in the presence of CldU for 24 hours before treatment or not with F) 1.5 µM MMC or G) 8 µM CDDP for 16 hours prior to fixation. CldU was immuno-detected without DNA denaturation, which only permits visualization of single-stranded regions. Nuclei were counterstained with DAPI. Bars represent median ± 95% CI.
To address the effect of crosslinks on the cell cycle progression, EdU incorporation assays were performed using MEFs that either were mock- or MMC-treated for 16 hours prior to EdU labeling. As expected, the frequency of EdU + replicating cells increased upon MMC exposure (Figure 3D, Supplementary Figure S2B). Interestingly, the mutants had a higher proportion of EdU + cells as compared to WT. Therefore, the lack of PCNA-Ub and/or FANCG, both of which participate in ICL repair, delays S/G2-phase.
The EdU incorporation assay demonstrated that induction of crosslinks arrested mutant MEFs to a similar extent but did not provide molecular insights regarding DNA replication. To address the impact of crosslinks at the single-molecule level, DNA fiber assays were performed on cells exposed to a high dose of MMC and subsequently labeled with CldU. Analyzing the length of CldU tracks under MMC-challenged conditions revealed a reduced CldU track length as compared to unchallenged controls (Figure 3E). Apparently, MMC-induced lesions hinder replication across all conditions. The hindrance was further augmented in KR, FGko, and DM cell lines. Apparently, the absence of PCNA-Ub had a similar effect as the lack of FANCG, caused by unresolved crosslinks that impact replication fork dynamics.
To determine whether the damage-induced replication stress favors the formation of ssDNA, cells were labeled with CldU for 24 hours before subjecting them to MMC or CDDP for 16 hours. Cells were immunostained for CldU without denaturation, and nuclear CldU intensity was measured as a representation of ssDNA (Supplementary Figure S2C). Fold changes of MMC- and CDDP-treated WT MEFs in comparison with untreated condition showed no increase in CldU intensity (Figure 3F and G), indicating that ssDNA did not accumulate, as crosslinks were resolved efficiently. On the other hand, CldU intensity significantly increased in the mutant cells, implying that exogenous crosslinks were not resolved efficiently in the absence of either PCNA-Ub or FANCG resulting in ssDNA, potentially due to the halted replication process (Figure 3D, Supplementary Figure S2B).
Together, these observations determine that PCNA-Ub and FANCG are epistatic, i.e. function in the same pathway, regarding ICL repair in Trp53kd MEFS. Additionally, these results reflect the significance of the posttranslational modification of PCNA at K164 in FA-mediated repair to resolve ICLs effectively, prevent ssDNA formation, and warrant genome stability. Ultimately, these insights cement PCNA-Ub as a core member of the FA repair pathway.
PCNA-Ub facilitates FA repair and prohibits recruitment of MMR
Having demonstrated that PCNA-Ub plays a central role in FA-mediated crosslink repair, we next determined the impact of a PcnaK164R mutation on the repli/repair-osome composition around FANCG in the presence of endogenous or MMC-induced crosslinks.
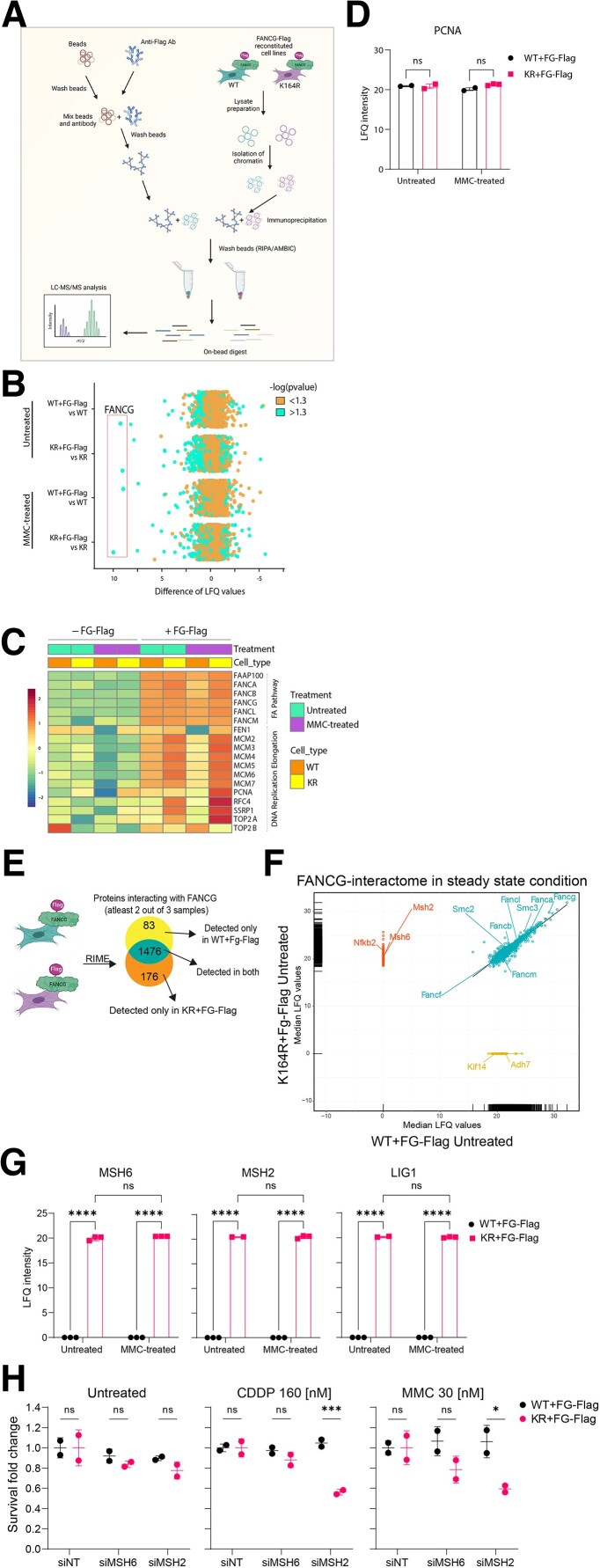
Due to lack of a functional anti-FANCG antibody, we utilized a flagged FANCG overexpression system (39). Potential alterations in the FANCG interactome were addressed by rapid immunoprecipitation mass spectrometry of endogenous protein (RIME) (49) (Figure 4A). Experiments were performed on WT and KR MEFs reconstituted with or without FG-Flag and were either untreated or treated with MMC for 16 hours before crosslinking with formaldehyde. Immunoprecipitation was performed on the chromatin extracts using an anti-Flag antibody. FANCG was significantly enriched in all reconstituted cell lines in comparison with nonreconstituted ones. This was observed under both untreated and treated conditions, highlighting the specificity of the pull down (Figure 4B). Differences in label-free quantification (LFQ) values were calculated for each protein and a -log (P-value) > 1.3 (P < 0.05) was considered significant. First, we determined the relative enrichment of FANCG under each condition. As the FANCG levels were comparable across the distinct conditions, we then analyzed for known FANCG interactors. Among others, members of FA pathway and “DNA replication elongation” clusters, as annotated by Räschle et al (50), were enriched in the reconstituted cell lines. These insights further validated the quality of the RIME approach (Figure 4C). Interestingly, when comparing the amount of co-precipitated PCNA, the presence or absence of lysine 164-specific ubiquitination appeared to be irrelevant (Figure 4D). This suggests that the interaction of PCNA with chromatin is not affected by the PcnaK164R mutation.
Fig. 4.
PCNA-Ub facilitates FA repair and prohibits recruitment of MMR. A) Schematic displaying the steps involved in RIME. Figure was made with BioRender.com. B) Scatterplot depicting enrichment of WT + FG-Flag and KR + FG-Flag RIME experiments over their respective nonreconstituted controls under both untreated and MMC-treated conditions. FANCG is highly enriched (LFQ > I7.5I) under all FG-Flag-containing conditions. n = 3. Cells were either left untreated or treated for 16 hours with 1.5 µM MMC. C) Heatmap depicting mean LFQ intensity in nonreconstituted and FG-Flag-reconstituted WT and KR cell lines for proteins detected from the “Fanconi anemia pathway” and “replication elongation” clusters as defined by Räschle et al. (32). D) Graph displaying raw LFQ intensities of PCNA in untreated and MMC-treated WT + FG-Flag and KR + FG-Flag RIMEs. Bars represent mean ± SD. P-values were calculated using two-way ANOVA. E) Schematic displaying the steps involved in analysis of the RIME data. F) Regression plot displaying median LFQ values for proteins detected in two out of three samples in the untreated WT + FG-Flag and KR + FG-Flag RIMEs. Upon IP on FG-Flag, proteins were enriched in KR only (upper left cluster), WT only (lower right cluster), or both (diagonal cluster) RIMEs. G) Graph displaying raw LFQ intensities of MSH2, MSH6, and LIG1 in untreated and MMC-treated WT + FG-Flag and KR + FG-Flag RIMEs. Bars represent mean ± SD. P-values were calculated using two-way ANOVA. *P < 0.05, **P < 0.01, ****P < 0.0001. H) Cells were treated with siRNAs against no target (NT), MSH2, and MSH6 for 6 hours. 48 hours after transfection, cells were plated and subsequently treated with CDDP or MMC or left untreated for 5 days. Crystal violet absorbance was used to determine survival, and fold change was plotted for siMSH2- and siMSH6-treated WT and KR MEFs where data were normalized to the respective siNT controls for all conditions.
The differential interactome of FANCG under the untreated condition was determined by searching for proteins that were enriched specifically in the KR + FG-Flag MEFs and not in the WT + FG-Flag MEFs (Figure 4E). We found member of the FA core complex and other replication-associated proteins to be equally enriched in both WT + FG-Flag and KR + FG-Flag RIMEs (Figure 4F, green cluster). 176 specific proteins were identified and fed into the STRING database for potential network/pathway enrichment (Figure 4F, orange cluster). The KEGG pathway analysis revealed the MMR to be the sole pathway enriched with an FDR of 0.00065. Upon looking at the raw LFQ values of individual replicates, MSH6, MSH2, and LIG1 were significantly enriched in the KR + FG-Flag IP under untreated as well as MMC-treated conditions (Figure 4G), indicating that in the absence of PCNA-Ub the mismatch recognition complex MSH2/MSH6 is preferentially recruited to sites of the FA core complex of endogenous/exogenous crosslinks.
To determine whether the recruitment of MSH2–MSH6 to the ICL site is advantageous to cell survival or not, we utilized a smartpool of siRNAs against MSH2 and MSH6 to check for cell survival in the presence/absence of exogenous DNA crosslinking agents. Cells were either untreated or treated with moderate doses of CDDP and MMC for 5 days 48 hours post-siRNA transfection, fixed and stained with crystal violet. Upon data normalization to the nontargeting siRNA (siNT), MSH2- and MSH6-deficient KR cells showed mild but nonsignificant reduction in survival under the untreated condition (Figure 4H). In contrast, when challenged with CDDP- or MMC-induced crosslinks, the presence MSH2 provided a significant survival advantage (Figure 4H). The trend seen for the MSH6 knock-down condition likely attributes to the fact that MSH2/MSH3 can compete with MSH2/MSH6 complexes in the detection of ICLs.
The insights generated by this unbiased RIME approach put forward a model, where PCNA-Ub exerts two critical functions during ICL repair: (i) facilitate TLS across the unhooked ICL to sustain canonical FA repair and (ii) subsequently prevent MSH2/MSH6 from binding to the ICL site and permitting MMR to resolve the ICL in an FA-independent manner.
Discussion
PCNA ubiquitination is an integral step in FA-ICL repair
Repair of highly genotoxic ICLs depends on DDT system as an essential intermediate step in accomplishing TLS across the unhooked ICL (51). Despite previous insights (24, 26, 52), the actual relevance of PCNA-Ub as a contributing factor in FA-mediated ICL repair remained to be fully determined on the basis of direct comparisons of PcnaK164R and Fancg−/− mice as well as intercrosses thereof.
We now demonstrate for the first time that a combined DM is embryonically lethal. This embryonic lethality coincides with a synthetic lethality, as demonstrated by the selective failure in establishing primary PreB and MEF cell cultures from DM embryos. This finding likely relates to the fact that PCNA-K164 modification, in addition to the ICLs, controls the tolerance of a plethora of DNA lesions, as a part of DDT pathway, in addition to the ICLs. This effect is similar to the embryonic lethality observed upon removing FANCG and FANCD2 simultaneously, where FANCD2 has additional roles associated with DNA double-stranded ends and Holliday junctions (53). At the systemic level, this highlights a nonepistatic relation of the two pathways with specific independent functions in genome maintenance.
Trp53 dictates the cell fate of our DM cells (Figure 1). Interestingly, Garaycoechea et al. (54) observed that the embryonic lethality in Aldh2−/−Fancd2−/− mice could be rescued by crossing with Trp53−/− mice. The hematopoietic stem cell containing Lineage(-)Sca-1(+)c-Kit(+) (LSK) population returned to WT status in Trp53−/−Aldh2−/−Fancd2−/− mice. Similarly, a Trp53kd in our DM MEFs restored them to WT status (Figure 1). Therefore, in the presence of Trp53, the lack of both DDT and FA pathways raises the DNA damage response to a p53-dictated threshold that is incompatible with mammalian life, potentially delineating the nonepistatic relation between the two pathways observed in our mice and embryos.
In contrast, in the absence of Trp53, DM cells are rendered more tolerant to endogenous stressors (Figure 2), which enabled a detailed study of the relationship between the two pathways regarding repair of ICLs introduced by exogenous compounds such as CDDP or MMC. In the context of these compounds, single-mutant and DM cells are comparable in terms of growth kinetics, colony-forming ability, cell arrest in S-phase, nucleotide incorporation, and ssDNA formation (Figure 3, Supplementary Figure S2), revealing an epistatic relationship between PCNA-Ub and FANCG.
Based on these data, we conclude that Trp53kd provided us the opportunity to measure specific cell behavior in the presence of abundant ICLs imposed by exogenous compounds. Following this approach, we conclude that PCNA-Ub and FANCG act epistatic in regulating ICL repair.
PCNA-Ub determines the choice between FA-ICL repair and MMR-ICL repair
The multifaceted interactome of PCNA comprises a number of repair components including FA repair and MMR elements (6, 26, 38, 55–58). In addition, experiments in Xenopus egg extracts show that FA proteins are recruited to the chromatin in a strictly replication-initiation-dependent manner that increases upon crosslink induction (59). Using FANCG-dependent RIME, we here determined the dependency of FA assembly on PCNA-Ub as well as other potential replisome changes induced by the K164R mutation in the presence of endogenous or MMC-induced crosslinks. Interestingly, MMR components MSH2 and MSH6 were only detected in the KR + FG-Flag RIMEs, indicating that the recruitment of this mismatch recognition complex to the ICL repair site is prohibited by PCNA-Ub. We here provide first evidence that the exclusion of MMR components during FA repair relies on site-specific PCNA ubiquitination (Figure 5).
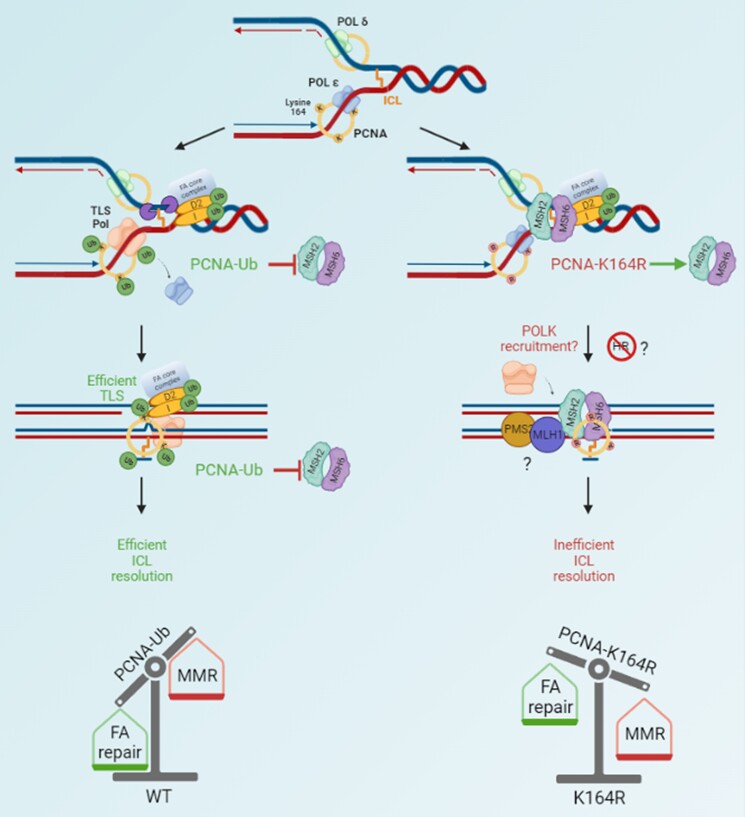
Fig. 5.
Dual role of PCNA-Ub in facilitating FA-mediated ICL repair. ICL repair initiates with the assembly of the FA core complex and ubiquitination of downstream components I-D2, which results in unhooking of the lesion via various endonucleases. When PCNA can be monoubiquitinated at K164 (left), TLS polymerases are actively recruited that can replicate across the unhooked lesion and simultaneously prevent MMR members MSH2-MSH6 from being recruited. This facilitates efficient TLS and downstream repair, resulting in a resolved ICL. In the absence of PCNA-Ub (right), MSH2–MSH6 complex is able to intervene and compete with FA repair. Additionally, inefficient TLS but prevention/delay of HR-repair results in inefficiently resolved ICLs, which amplifies upon MMC exposure. Therefore, PCNA-Ub acts as a molecular balance, that in its ubiquitinated form warrants FA repair and excludes MMR activities. Figure was made with BioRender.com.
The interaction of PCNA with MMR is well established (60–62). Also, MMR has been implicated in ICL repair, in both FA-dependent (63–65) and independent contexts (2, 66–72). Furthermore, deficiency of different MMR components can have an effect on FA cancer cell lines, suggesting that MMR provides an alternative mode to process ICLs. Thus, ICL repair by noncanonical MMR can be initiated but likely comes at the expense of efficiency. Likewise, TLS can also be performed upon encountering lesions in the absence of PCNA-Ub, albeit inefficient (73). Ubiquitination has an impact on protein function, and the presence of the Ub moiety on PCNA influences its interaction with other proteins (74–76). Hence, we speculate that in the absence of mono/poly-Ub-driven steric hindrance, MMR competes with FA repair to process the ICL.
Collectively, our findings delineate a dual role of PCNA-Ub in FA pathway-mediated crosslink repair: Facilitate TLS and exclude MMR activities to warrant effective FA-repair. Future studies will have to uncover if the entire MMR pathway is recruited or only specific components, and if so, which ones. Furthermore, the involvement of HR and NER factors in resolving the ICLs and the impact of this alternative pathway on genomic integrity also needs to be determined. In turn, this may provide novel insights that potentially can be exploited to improve efficacy of ICL-based chemotherapy.
Materials and methods
Generation of primary cell lines and cell culture
The generation of Pcna+/K164R(KR) and Fancg−/− mice has been described elsewhere (39, 40, 77). Timed matings of PcnaKR/+ Fancg+/− females were set up with corresponding males to obtain embryos. On day 14.5 of gestation, embryos were isolated to prepare primary MEFs. From fetal liver, PreB cells were generated by culturing on irradiated ST2 feeder cells in complete Iscove Modified Dulbecco Medium (IMDM) medium (Iscoves, supplemented with 8% fetal calf serum, 50 μM 2-mercapthoethanol, penicillin/streptomycin) supplemented with IL-7. pMEFs (2, from independent embryos per genotype) were isolated using trypsin and cell strainers and cultured under low (3%) oxygen condition, with 5% CO2 at 37°C. To immortalize MEFs, pMEFs were transduced with a lentivirus encoding a p53-specific shRNA. The immortalized Trp53kd MEFs were grown in complete IMDM medium under normal oxygen levels with 5% CO2 at 37°C. All animal experiments were approved by an independent animal ethics committee of the Netherlands Cancer Institute (Amsterdam, The Netherlands) (DEC number 14053) and executed according to national and European guidelines.
IncuCyte proliferation assay
The IncuCyte ZOOM instrument (Essen Bioscience) live cell imaging system was used to monitor cell growth. 250 cells were plated in a 96 Greiner micro clear black well plate and imaged every 4 hours. The default software parameters for a 96-well plate with a 10 × objective were used for imaging. The IncuCyte software was used to calculate mean confluence from four nonoverlapping bright-phase images of each well.
EdU incorporation assay
MEFs cultured in the presence or absence of 1.5 µM MMC (concentrations) were labeled with 5-ethynyl-2´-deoxyuridine (EdU) (10 µM) for 20 minutes, fixed in 70% EtOH, and stored at 4°C. On the day of analysis, cells were permeabilized using PBS/0.2% Triton X-100 and a Click-It reaction was performed for 1 hour. Cells were stained with DAPI (Sigma), and data acquisition was performed on a BD LSR FortessaTM. Data analysis (cell cycle) was performed using the FlowJo software (Tree Star, Ashland, OR, USA).
Fiber analysis
Cells were pulse-labeled with 25 µM CldU followed by 250 µM IdU for 20 minutes each. After labeling, cells were trypsinized and lysed in a spreading buffer (200 mM Tris–HCl pH 7.4, 50 mM ethylenediaminetetraacetic acid (EDTA), and 0.5% sodium dodecyl sulfate (SDS) before spreading on a microscope slide (Menzel-Gläser, Superfrost). Slides were fixed in methanol/acetic acid 3:1. Before immunodetection, slides were treated with 2.5 M HCl for 1 hour and 15 minutes. To detect CldU- and IdU-labeled tracts, slides were incubated for 1 hour with rat anti-BrdU (Clone BU1/75, Novus Biologicals; 1:500) and mouse anti-BrdU (clone B44, Becton Dickinson; 1:750), respectively. Subsequently, slides were fixed with 4% paraformaldehyde for 10 minutes and incubated with Alexa 488-labeled goat antimouse and Alexa 555-labeled goat antirat (Molecular probes; 1:500) for 1 hour and 30 minutes. Pictures were taken with a Zeiss AxioObserver Z1 inverted microscope using a 63 × lens equipped with a cooled Hamamatsu ORCA AG Black and White CCD camera, and track lengths and fork structures were analyzed with ImageJ software. Replication track lengths were calculated using the conversion factor 1 µm = 2.59 kb (78).
Neutral comet assay
Neutral comet assays were performed as described by Olive et al (79). Pictures of individual cells were taken with a Zeiss AxioObserver Z1 inverted microscope equipped with a cooled Hamamatsu ORCA AG Black and White CCD camera and analyzed with CASP software (http://www.casp.of.pl), where head center threshold was set at 0.8.
Immunoblotting
MEFs were harvested and subcellular fractionation kit for cultured cells (Thermofisher) was used according to the manufacturer's protocol. Nuclear lysates were sonicated for 15 minutes using a BioRuptor (30 seconds on, 30 seconds off, maximum power, at 4°C). Samples were spun at 20,000 g for 10 minutes, and the protein concentration was measured via the bicinchoinic acid (BCA) assay. Samples were run on a 3–8 Tris–acetate gel at 150 V for 3 hours. The gels were transferred using a wet transfer system. After staining with Ponceau-S, samples were blocked for 1 hour using PBS-T containing 5% skim milk powder, followed by incubation with primary antibody against FANCD2 and VINCULIN overnight at 4°C on a roller bank in PBS-T 1% milk. The next day, blots were washed four times with PBS-T for 5 minutes, followed by a 1-hour incubation with the secondary antibodies at room temperature (RT). The membrane was washed three times with PBS-T for 5 minutes, after which the membrane was imaged on an Odyssey scanner (LiCor).
siRNA transfection
MEFs were grown in 6-cm dishes and transfected with SMARTpool siRNAs against murine Msh2 and Msh6 (Horizon-Discovery) using RNAImax according to the manufacturer's specifications. Nontargeting control SMARTpool siRNAs were used as control.
Colony formation assay
MEFs were seeded in 10-cm dishes at various seeding densities in complete medium. One day later, the medium was removed and replaced with a complete medium containing the indicated concentrations of respective drugs. For UV-C, cells were treated with varying doses using the UV-MAT irradiation controller (Opsystec Dr. Grebel). After 6 days, medium was aspirated and the cells were washed with PBS and fixed in 5 ml 3:1 v/v methanol/acetic acid for 1 hour. Following the fixation, colonies were stained by adding 3 ml of 0.3% Coomassie brilliant blue (Merck) solution prepared in H2O. After 1.5 hours, the staining solution was removed, and the dishes were washed with H2O and dried overnight. Colonies were counted using the Colcount (Oxford Optronix), and each condition was corrected for the number of seeded cells before being normalized to the untreated condition. Data points represent the mean survival relative to the untreated control cells.
ssDNA-based immunofluorescence
Immunofluorescence was performed as described previously (20). Cells were grown on coverslips in 10 µM CldU for 24 hours before the treatment with CDDP or MMC for 16 hours. After treatment, cells were washed with PBS and preextracted with PBS/0.5% Triton X-100) on ice for 1 minute. Cells were then fixed using 4% formaldehyde for 15 minutes at RT. Fixed cells were then incubated with primary Abs against CldU at 37°C for 90 minutes. Cells were washed and incubated with secondary Abs alongside DAPI for 1 hour at RT. After washing, coverslips were mounted onto glass slides using Aqua Poly/Mount. Pictures were taken with a Zeiss AxioObserver Z1 inverted microscope using a 63 × lens equipped with a cooled Hamamatsu ORCA AG Black and White CCD camera. Nuclear intensities were measured using a macro designed with ImageJ software.
Rapid immunoprecipitation mass spectrometry of endogenous protein
Cells reconstituted with flagged FANCG cDNA constructs were used. Cells were left untreated or treated with 1.5 µM MMC for 16 hours. Cells were fixed, lysed, and sonicated as previously described (49). The nuclear lysate was incubated with 100 µl magnetic beads (Dynabeads, Thermo Fisher Scientific) prebound with 10 µg anti-Flag antibody (Clone M2, Merck).
Tryptic digestion of bead-bound proteins was performed as described previously (80). With the exception that LC–MS/MS analysis of the tryptic digests was performed on a Q Exactive HF-X hybrid quadrupole-Orbitrap mass spectrometer equipped with an EASY-NLC 1200 system (Thermo Fisher Scientific). The samples were eluted from the analytical column in a 90-minute linear gradient, containing a 74-minute linear increase from 7 to 29% solvent B, followed by a 16-minute wash at 100% solvent B.
Raw data were analyzed by MaxQuant (version 1.6.17.0) (81) using standard settings. MS/MS data were searched against the Mus Musculus Swissprot database (17,027 entries, release 2020_02) complemented with a list of common contaminants and concatenated with the reversed version of all sequences. The maximum allowed mass tolerance was 4.5 ppm in the main search and 20 ppm for fragment ion masses. False discovery rates (FDRs) for peptide and protein identification were set to 1%. Trypsin/P was chosen as cleavage specificity allowing two missed cleavages. Carbamidomethylation (C) was set as a fixed modification, while oxidation (M) and deamidation (NQ) were used as variable modifications. LFQ intensities were log2-transformed in Perseus (version 1.6.14.0) (82), and the proteins were filtered for at least two out of three valid values in one condition. Missing values were replaced by imputation based on the standard settings of Perseus, i.e. a normal distribution using a width of 0.3 and a downshift of 1.8. Differentially expressed proteins were determined using a t test (threshold: P ≤ 0.05 and [x/y] ≥ 1.5 | [x/y] ≤ −1.5). The raw data were submitted to ProteomeXchange via the PRIDE database with the dataset identifier PXD035337. Data corresponding to RIME experiments were plotted using the ggplot2 package in R.
RNA sequencing
RNA-seq sample preparation
MEFs were resuspended in TRIzol (Ambion Life Technologies), and total RNA was extracted according to the manufacturer's protocol. Quality and quantity of the total RNA were assessed by the 2100 Bioanalyzer using a Nano chip (Agilent). Only RNA samples having an RNA integrity number > 8 were subjected to library generation.
RNA-seq library preparation
Strand-specific cDNA libraries were generated using the TruSeq Stranded mRNA sample preparation kit (Illumina) according to the manufacturer's protocol. The libraries were analyzed for size and quantity of cDNAs on a 2100 Bioanalyzer using a DNA 7500 chip (Agilent), diluted, and pooled in multiplex sequencing pools. The libraries were sequenced as 65 base single reads on a HiSeq2500 (Illumina).
RNA-seq preprocessing
Strand-specific RNA reads (11–33 million reads per sample), 65-bp single end, were aligned against the mouse reference genome (Ensembl build 38) using Tophat (version 2.1, bowtie version 1.1). Tophat was supplied with a file in a Gene Transfer Format (GTF, Ensembl version 77) and with the following parameters: “--prefilter-multihits –no-coverage-search –bowtie1 –library-type fr-firststrand”. To count the number of reads per gene, a custom script which is based on the same ideas as HTSeq count was used. A list of the total number of uniquely mapped reads for each gene that is present in the provided GTF file was generated.
RNA-seq analysis
Differential gene expression analysis was performed in R language (version 4.0.2) using the edgeR package (3.30.3) with default arguments. Genes that have no expression across all samples within the dataset were removed, and the analysis was restricted to those genes which have at least two counts per million value in all the samples in a specific contrast, to exclude very low expressing genes. Differential expression analysis was performed on only relevant samples with the design set to either genotypes or Trp53 status of the cell type. Genes were considered to be differentially expressed when the FDR was below 0.05 after the Benjamini–Hochberg multiple testing correction. After differential analysis, expression of genes was plotted by using the ggplot2 package (3.3.3). Read counts were corrected for gene length based on the longest transcript of the gene followed by normalization for the library size and shown as transcript per million (TPM). TPMs were used for showing the expression of a set of genes across different conditions by using pheatmap package (1.0.12) with default arguments, and the genes were clustered based on Z-score.
Ingenuity pathway analysis
Lists of differentially expressed genes (FDR < 0.05) between different conditions were submitted to IPA with the log fold change (logFC) threshold set at 0.1, to identify the potential pathways that are related to the transcriptional changes.
Gene set enrichment analysis
GSEA was performed using GSEA software (v. 4.0.3) (83, 84) on RNA-seq data of WT/DM pMEFs, on genesets obtained from mSigDB Hallmark gene sets (Liberzon et al., 2015). Permutations for each gene set were conducted 1,000 times to obtain an empirical null distribution.
Statistical analysis
To assess the statistical significance of our data, appropriate tests were performed using GraphPad Prism.
Supplementary Material
Acknowledgments
The authors would like to thank the biotechnical staff of the NKI-AVL for support, especially Anita Pfauth, Frank van Diepen, and Martijn van Baalen, for their expertise and help with flow cytometry experiments and cell sorting. The authors moreover thank the Animal Pathology Facility for providing H&E stains of mouse organs. The authors thank Dr. Ron van Kerkhoven, Marja Nieuwland, and Roel Kluin for their help with the RNA sequencing. The authors finally thank Prof. Dr. Titia Sixma for critical reading of the manuscript.
Contributor Information
Ronak Shah, Department of Tumor Biology and Immunology, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Muhammad Assad Aslam, Department of Tumor Biology and Immunology, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands; Department/Institute of Molecular Biology and Biotechnology, Bahauddin Zakariya University, Bosan Road, 60800 Multan, Pakistan.
Aldo Spanjaard, Department of Tumor Biology and Immunology, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Daniel de Groot, Department of Tumor Biology and Immunology, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Lisa M Zürcher, Department of Tumor Biology and Immunology, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Maarten Altelaar, Proteomics Facility, The Netherlands Cancer Institute, Plesmanlaan 121, 1066CX Amsterdam, The Netherlands; Biomolecular Mass Spectrometry and Proteomics, Bijvoet Center for Biomolecular Research, Utrecht Institute for Pharmaceutical Sciences, Utrecht University and Netherlands Proteomics Centre, Utrecht, Padualaan 8, 3584 CH Utrecht, The Netherlands.
Liesbeth Hoekman, Proteomics Facility, The Netherlands Cancer Institute, Plesmanlaan 121, 1066CX Amsterdam, The Netherlands.
Colin E J Pritchard, Mouse Clinic for Cancer and Aging Transgenic Facility, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Bas Pilzecker, Department of Tumor Biology and Immunology, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Paul C M van den Berk, Department of Tumor Biology and Immunology, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Heinz Jacobs, Department of Tumor Biology and Immunology, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
This work was supported and made possible by two grants from the Dutch Cancer Society: KWF NKI-2017-10032 and NKI-2017-10796 to H.J. This research was supported by an institutional grant of the Dutch Cancer Society and of the Dutch Ministry of Health, Welfare and Sport.
Author Contributions
R.S. and H.J. designed research; R.S. and L.M.Z. performed research; M.A.A. analyzed RNA-seq data and prepared scripts for various figures generated using R tool; A.S. and D.dG. contributed analytically and critical reading of the manuscript; M.A. and L.H. prepared proteomics samples and provided preliminary data; C.E.J.P. and B.P. designed and generated the Fancg-ko mouse; P.C.M.vdB. maintained the mouse matings and assisted in cell line isolation; and R.S. and H.J. wrote the paper.
Data Availability
The RNA-seq datasets reported in this article have been deposited at the National Center for Biotechnology Information under the accession number GSE165989, which can be accessed with token number elipwiiszlsdtqz. The RIME dataset has been deposited to the ProteomeXchange via the PRIDE database under the dataset identifier PXD035337. The reviewer account details for this dataset are as follows: username: reviewer_pxd035337@ebi.ac.uk and password: ey92LRuB.
References
- 1. Semlow DR, Zhang J, Budzowska M, Drohat AC, Walter JC. 2016. Replication-Dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell. 167(2):498–511.e14. 10.1016/j.cell.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kato N, et al. 2017. Sensing and processing of DNA interstrand crosslinks by the mismatch repair pathway. Cell Rep. 21(5):1375–1385. 10.1016/j.celrep.2017.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nalepa G, Clapp DW. 2018. Fanconi anaemia and cancer: an intricate relationship. Nat Rev Cancer. 18(3):168–185. 10.1038/nrc.2017.116 [DOI] [PubMed] [Google Scholar]
- 4. Che R, Zhang J, Nepal M, Han B, Fei P. 2018. Multifaceted Fanconi anemia signaling. Trends Genet. 34(3):171–183. 10.1016/j.tig.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ceccaldi R, Sarangi P, D’Andrea AD. 2016. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 17(6):337–349. 10.1038/nrm.2016.48 [DOI] [PubMed] [Google Scholar]
- 6. Kim H, D’Andrea AD. 2012. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 26(13):1393–1408. 10.1101/gad.195248.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lopez-Martinez D, Liang C-C, Cohn MA. 2016. Cellular response to DNA interstrand crosslinks: the Fanconi anemia pathway. Cell Mol Life Sci. 73(16):3097–3114. 10.1007/s00018-016-2218-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Noll DM, Mason TM, Miller PS. 2006. Formation and repair of interstrand cross-links in DNA. Chem Rev. 106(2):277–301. 10.1021/cr040478b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deans AJ, West SC. 2011. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 11(7):467–480. 10.1038/nrc3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rodríguez A, D’Andrea A. 2017. Fanconi anemia pathway. Curr Biol. 27(18):R986–R988. 10.1016/j.cub.2017.07.043 [DOI] [PubMed] [Google Scholar]
- 11. Smogorzewska A, et al. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 129(2):289–301. 10.1016/j.cell.2007.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klein Douwel D, et al. 2014. XPF-ERCC1 Acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 54(3):460–471. 10.1016/j.molcel.2014.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Knipscheer P, et al. 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 326(5960):1698–1701. 10.1126/science.1182372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamamoto KN, et al. 2011. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci. 108(16):6492–6496. 10.1073/pnas.1018487108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ho TV, Schärer OD. 2010. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 51(6):552–566. 10.1002/em.20573 [DOI] [PubMed] [Google Scholar]
- 16. Kottemann MC, Smogorzewska A. 2013. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 493(7432):356–363. 10.1038/nature11863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Budzowska M, Graham TG, Sobeck A, Waga S, Walter JC. 2015. Regulation of the Rev1–Pol ζ Complex during bypass of a DNA interstrand cross-link. EMBO J. 34(14):1971–1985. 10.15252/embj.201490878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Silva IU, McHugh PJ, Clingen PH, Hartley JA. 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 20(21):7980–7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hodskinson MR, et al. 2020. Alcohol-Derived DNA crosslinks are repaired by two distinct mechanisms. Nature. 579(7800):603–608. 10.1038/s41586-020-2059-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hopfner K-P, Tainer JA. 2000. DNA mismatch repair: the hands of a genome guardian. Structure. 8(12):R237–R241. 10.1016/S0969-2126(00)00545-1 [DOI] [PubMed] [Google Scholar]
- 21. Peña-Diaz J, Jiricny J. 2012. Mammalian mismatch repair: error-free or error-prone? Trends Biochem Sci. 37(5):206–214. 10.1016/j.tibs.2012.03.001 [DOI] [PubMed] [Google Scholar]
- 22. Friedberg EC, Lehmann AR, Fuchs RPP. 2005. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 18(5):499–505. 10.1016/j.molcel.2005.03.032 [DOI] [PubMed] [Google Scholar]
- 23. Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. 2002. RAD6 -Dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 419(6903):135–141. 10.1038/nature00991 [DOI] [PubMed] [Google Scholar]
- 24. McIntyre J, Woodgate R. 2015. Regulation of translesion DNA synthesis: posttranslational modification of lysine residues in key proteins. DNA Repair (Amst). 29:166–179. 10.1016/j.dnarep.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cipolla L, Maffia A, Bertoletti F, Sabbioneda S. 2016. The regulation of DNA damage tolerance by ubiquitin and ubiquitin-like modifiers. Front Genet. 7:105. 10.3389/fgene.2016.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moldovan G-L, Pfander B, Jentsch S. 2007. PCNA, the maestro of the replication fork. Cell. 129(4):665–679. 10.1016/j.cell.2007.05.003 [DOI] [PubMed] [Google Scholar]
- 27. Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. 2008. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 29(5):625–636. 10.1016/j.molcel.2007.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sale JE. 2013. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol. 5(3):a012708. 10.1101/cshperspect.a012708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arakawa H, et al. 2006. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 4(11):e366. 10.1371/journal.pbio.0040366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spanjaard A, et al. 2022. Division of labor within the DNA damage tolerance system reveals non-epistatic and clinically actionable targets for precision cancer medicine. Nucleic Acids Res. 50(13):7420–7435. 10.1093/nar/gkac545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stelter P, Ulrich HD. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 425(6954):188–191. 10.1038/nature01965 [DOI] [PubMed] [Google Scholar]
- 32. Räschle M, et al. 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 134(6):969–980. 10.1016/j.cell.2008.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bezalel-Buch R, Cheun YK, Roy U, Schärer OD, Burgers PM. 2020. Bypass of DNA interstrand crosslinks by a rev1-DNA polymerase ζ Complex. Nucleic Acids Res. 48(15):8461–8473. 10.1093/nar/gkaa580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hicks JK, et al. 2010. Differential roles for DNA polymerases eta, Zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol. 30 (5):1217–1230. 10.1128/MCB.00993-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wit N, et al. 2015. Roles of PCNA ubiquitination and TLS polymerases κ and η in the bypass of methyl methanesulfonate-induced DNA damage. Nucleic Acids Res. 43(1):282–294. 10.1093/nar/gku1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buoninfante OA, et al. 2018. Precision cancer therapy: profiting from tumor specific defects in the DNA damage tolerance system. Oncotarget. 9(27):18832–18843. 10.18632/oncotarget.24777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pilzecker B, et al. 2017. DNA damage tolerance in hematopoietic stem and progenitor cells in mice. Proc Natl Acad Sci U S A. 114(33):E6875–E6883. 10.1073/pnas.1706508114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geng L, Huntoon CJ, Karnitz LM. 2010. RAD18-Mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J Cell Biol. 191(2):249–257. 10.1083/jcb.201005101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shah R, et al. 2023. C57BL/6J Fancg-KO mouse model generated by CRISPR/cas9 partially captures the human phenotype. Int J Mol Sci. 24(13):11129. 10.3390/ijms241311129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Langerak P, Nygren AOH, Krijger PHL, van den Berk PCM, Jacobs H. 2007. A/T mutagenesis in hypermutated immunoglobulin genes strongly Depends on PCNAK164 modification. J Exp Med. 204(8):1989–1998. 10.1084/jem.20070902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langerak P, Krijger PHL, Heideman MR, van den Berk PCM, Jacobs H. 2008. Somatic hypermutation of immunoglobulin genes: lessons from proliferating cell nuclear antigenK164R mutant mice. Philos Trans R Soc B Biol Sci. 364(1517):621–629. 10.1098/rstb.2008.0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Menendez D, Inga A, Resnick MA. 2009. The expanding universe of P53 targets. Nat Rev Cancer. 9(10):724–737. 10.1038/nrc2730 [DOI] [PubMed] [Google Scholar]
- 43. Green DR, Kroemer G. 2009. Cytoplasmic functions of the tumour suppressor P53. Nature. 458(7242):1127–1130. 10.1038/nature07986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stiewe T. 2007. The P53 family in differentiation and tumorigenesis. Nat Rev Cancer. 7(3):165–167. 10.1038/nrc2072 [DOI] [PubMed] [Google Scholar]
- 45. Quinet A, Carvajal-Maldonado D, Lemacon D, Vindigni A. 2017. Chapter three—DNA fiber analysis: mind the gap! in methods in enzymology. In: Eichman BF, editor. DNA repair enzymes: cell, molecular, and chemical biology. Vol. 591. Cambridge (MA): Academic Press. p. 55–82. [DOI] [PubMed] [Google Scholar]
- 46. Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat. Cell Biol. 16(1):2–9. 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koundrioukoff S, et al. 2013. Stepwise activation of the ATR signaling pathway upon increasing replication stress impacts Fragile site integrity. PLOS Genet. 9(7):e1003643. 10.1371/journal.pgen.1003643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jansen JG, et al. 2014. Redundancy of mammalian Y family DNA polymerases in cellular responses to genomic DNA lesions induced by ultraviolet light. Nucleic Acids Res. 42(17):11071–11082. 10.1093/nar/gku779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mohammed H, et al. 2016. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 11(2):316–326. 10.1038/nprot.2016.020 [DOI] [PubMed] [Google Scholar]
- 50. Räschle M, et al. 2015. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science. 348(6234):1253671. 10.1126/science.1253671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pilzecker B, Buoninfante OA, Jacobs H. 2019. DNA damage tolerance in stem cells, ageing, mutagenesis, disease and cancer therapy. Nucleic Acids Res. 47(14):7163–7181. 10.1093/nar/gkz531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mailand N, Gibbs-Seymour I, Bekker-Jensen S. 2013. Regulation of PCNA–protein interactions for genome stability. Nat Rev Mol Cell Biol. 14(5):269–282. 10.1038/nrm3562 [DOI] [PubMed] [Google Scholar]
- 53. Reliene R, Yamamoto ML, Rao PN, Schiestl RH. 2010. Genomic instability in mice is greater in Fanconi anemia caused by deficiency of Fancd2 than Fancg. Cancer Res. 70(23):9703–9710. 10.1158/0008-5472.CAN-09-1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garaycoechea JI, et al. 2018. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature. 553(7687):171–177. 10.1038/nature25154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Howlett NG, Harney JA, Rego MA, Kolling FW, Glover TW. 2009. Functional interaction between the Fanconi anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif. J Biol Chem. 284(42):28935–28942. 10.1074/jbc.M109.016352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Paul Solomon Devakumar LJ, Gaubitz C, Lundblad V, Kelch BA, Kubota T. 2019. Effective mismatch repair Depends on timely control of PCNA retention on DNA by the Elg1 Complex. Nucleic Acids Res. 47(13):6826–6841. 10.1093/nar/gkz441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Umar A, et al. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 87(1):65–73. 10.1016/S0092-8674(00)81323-9 [DOI] [PubMed] [Google Scholar]
- 58. Kleczkowska HE, Marra G, Lettieri T, Jiricny J. 2001. HMSH3 and HMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 15(6):724–736. 10.1101/gad.191201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sobeck A, et al. 2006. Fanconi anemia proteins are required to prevent accumulation of replication-associated DNA double-strand breaks. Mol Cell Biol. 26(2):425–437. 10.1128/MCB.26.2.425-437.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lau PJ, Kolodner RD. 2003. Transfer of the MSH2.MSH6 complex from proliferating cell nuclear antigen to mispaired bases in DNA. J Biol Chem. 278 (1):14–17. 10.1074/jbc.C200627200 [DOI] [PubMed] [Google Scholar]
- 61. Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. 2000. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J Biol Chem. 275(47):36498–36501. 10.1074/jbc.C000513200 [DOI] [PubMed] [Google Scholar]
- 62. Flores-Rozas H, Clark D, Kolodner RD. 2000. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition Complex. Nat Genet. 26(3):375–378. 10.1038/81708 [DOI] [PubMed] [Google Scholar]
- 63. Peng M, Xie J, Ucher A, Stavnezer J, Cantor SB. 2014. Crosstalk between BRCA-Fanconi anemia and mismatch repair pathways prevents MSH2-dependent aberrant DNA damage responses. EMBO J. 33(15):1698–1712. 10.15252/embj.201387530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang M, et al. 2011. Human MutS and FANCM complexes function as redundant DNA damage sensors in the Fanconi anemia pathway. DNA Repair (Amst). 10(12):1203–1212. 10.1016/j.dnarep.2011.09.006 [DOI] [PubMed] [Google Scholar]
- 65. Williams SA, et al. 2011. Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Hum Mol Genet. 20(22):4395–4410. 10.1093/hmg/ddr366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu Qi, Christensen LA, Legerski RJ, Vasquez KM. 2005. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 6 (6):551–557. 10.1038/sj.embor.7400418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yasin S-L, Rainbow AJ. 2011. A combination of MSH2 DNA mismatch repair deficiency and expression of the SV40 large T antigen results in cisplatin resistance of mouse embryonic fibroblasts. Int J Oncol. 39(3):719–726. 10.3892/ijo.2011.1065 [DOI] [PubMed] [Google Scholar]
- 68. Sawant A, Kothandapani A, Zhitkovich A, Sobol RW, Patrick SM. 2015. Role of mismatch repair proteins in the processing of cisplatin interstrand cross-links. DNA Repair (Amst). 35:126–136. 10.1016/j.dnarep.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Papouli E, Cejka P, Jiricny J. 2004. Dependence of the cytotoxicity of DNA-damaging agents on the mismatch repair Status of human cells. Cancer Res. 64(10):3391–3394. 10.1158/0008-5472.CAN-04-0513 [DOI] [PubMed] [Google Scholar]
- 70. Takahashi M, Koi M, Balaguer F, Boland CR, Goel A. 2011. MSH3 mediates sensitization of colorectal cancer cells to cisplatin, oxaliplatin, and a poly(ADP-ribose) polymerase inhibitor. J Biol Chem. 286(14):12157–12165. 10.1074/jbc.M110.198804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhao J, Jain A, Iyer RR, Modrich PL, Vasquez KM. 2009. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res. 37(13):4420–4429. 10.1093/nar/gkp399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kothandapani A, Sawant A, Dangeti VSMN, Sobol RW, Patrick SM. 2013. Epistatic role of base excision repair and mismatch repair pathways in mediating cisplatin cytotoxicity. Nucleic Acids Res. 41(15):7332–7343. 10.1093/nar/gkt479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hendel A, et al. 2011. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 7(9):e1002262. 10.1371/journal.pgen.1002262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chernorudskiy AL, Shorina AS, Garcia A, Gainullin MR. 2006. Evaluation of direct effects of protein ubiquitylation using computational analysis. Biophysics (Oxf). 51(1):39–43. 10.1134/S0006350906070086 [DOI] [Google Scholar]
- 75. Salas-Lloret D, et al. 20 May 2024. BRCA1/BARD1 Ubiquitinates PCNA in Unperturbed Conditions to Promote Replication Fork Stability and Continuous DNA Synthesis. Nature Commun. 15:4292. 10.1038/s41467-024-48427-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ball KA, et al. 2016. Non-Degradative ubiquitination of protein kinases. PLoS Comput Biol. 12(6):e1004898. 10.1371/journal.pcbi.1004898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Langerak P, Nygren AOH, Schouten JP, Jacobs H. 2005. Rapid and quantitative detection of homologous and non-homologous recombination events using three oligonucleotide MLPA. Nucleic Acids Res. 33(22):e188. 10.1093/nar/gni187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jackson DA, Pombo A. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140(6):1285–1295. 10.1083/jcb.140.6.1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Olive PL, Banáth JP. 2006. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 1(1):23–29. 10.1038/nprot.2006.5 [DOI] [PubMed] [Google Scholar]
- 80. Stelloo S, et al. 2018. Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis. Oncogene. 37(3):313–322. 10.1038/onc.2017.330 [DOI] [PubMed] [Google Scholar]
- 81. Cox J, et al. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 13(9):2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tyanova S, et al. 2016. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat Methods. 13(9):731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 83. Subramanian A, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102(43):15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. 2007. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 23(23):3251–3253. 10.1093/bioinformatics/btm369 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq datasets reported in this article have been deposited at the National Center for Biotechnology Information under the accession number GSE165989, which can be accessed with token number elipwiiszlsdtqz. The RIME dataset has been deposited to the ProteomeXchange via the PRIDE database under the dataset identifier PXD035337. The reviewer account details for this dataset are as follows: username: reviewer_pxd035337@ebi.ac.uk and password: ey92LRuB.