Abstract
Background
Immune checkpoint blockade targeting the adaptive immune system has revolutionized the treatment of cancer. Despite impressive clinical benefits observed, patient subgroups remain non-responsive underscoring the necessity for combinational therapies harnessing additional immune cells. Natural killer (NK) cells are emerging tools for cancer therapy. However, only subpopulations of NK cells that are differentially controlled by inhibitory receptors exert reactivity against particular cancer types. How to leverage the complete anti-tumor potential of all NK cell subsets without favoring the emergence of NK cell-resistant tumor cells remains unresolved.
Methods
We performed a genome-wide CRISPR/Cas9 knockout resistance screen in melanoma cells in co-cultures with human primary NK cells. We comprehensively evaluated factors regulating tumor resistance and susceptibility by focusing on NK cell subsets in an allogenic setting. Moreover, we tested therapeutic blocking antibodies currently used in clinical trials.
Results
Melanoma cells deficient in antigen-presenting or the IFNγ-signaling pathways were depleted in remaining NK cell-co-cultured melanoma cells and displayed enhanced sensitivity to NK cells. Treatment with IFNγ induced potent resistance of melanoma cells to resting, IL-2-cultured and ADCC-activated NK cells that depended on B2M required for the expression of both classical and non-classical MHC-I. IFNγ-induced expression of HLA-E mediated the resistance of melanoma cells to the NKG2A+ KIR− and partially to the NKG2A+ KIR+ NK cell subset. The expression of classical MHC-I by itself was sufficient for the inhibition of the NKG2A− KIR+, but not the NKG2A+ KIR+ NK cell subset. Treatment of NK cells with monalizumab, an NKG2A blocking mAb, enhanced the reactivity of a corresponding subset of NK cells. The combination of monalizumab with lirilumab, blocking KIR2 receptors, together with DX9, blocking KIR3DL1, was required to restore cytotoxicity of all NK cell subsets against IFNγ-induced resistant tumor cells in melanoma and tumors of different origins.
Conclusion
Our data reveal that in the context of NK cells, IFNγ induces the resistance of tumor cells by the upregulation of classical and non-classical MHC-I. Moreover, we reveal insights into NK cell subset reactivity and propose a therapeutic strategy involving combinational monalizumab/lirilumab/DX9 treatment to fully restore the antitumor response across NK cell subsets.
Keywords: Natural killer - NK, Immune Checkpoint Inhibitor, Immunotherapy
WHAT IS ALREADY KNOWN ON THIS TOPIC.
Immune checkpoint blockade by monoclonal antibodies has revolutionized the treatment of patients with cancer. Natural killer (NK) cells, known to eliminate tumor cells, especially in the early stage of tumor development, are innate immune effector cells divided into distinct subsets on the basis of their receptor expression. Tumor resistance to immunotherapy remains a major challenge in the clinics.
WHAT THIS STUDY ADDS
Our study highlights that IFNγ shuts off the cytotoxicity of distinct NK cell subsets characterized by KIR and/or NKG2A receptors by induction of classical and non-classical MHC class I molecules in a time-dependent manner. We provide detailed insight into the regulation of distinct NK cell subsets in allogeneic settings and show that the therapeutic blocking mAbs monalizumab and lirilumab do not act uniformly on all NK cells but act on specific NK cell subsets. Our results reveal a minimal effect of KIR2DL1/L2/L3 or KIR3DL1 blockade on restoring the function of the NKG2A+ KIR+ NK cell subset unless a simultaneous NKG2A blockade was used.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our study provides compelling evidence supporting the pursuit of combinational therapy involving monalizumab and lirilumab while also advocating for the development and clinical testing of KIR3DL1 blockade for additional effectiveness. Since therapeutic strategies leading to NK cell activation such as ADCC-mediating mAbs, NK engagers or application of cytokines such as IL-12 induce IFNγ production with many beneficial effects on adaptive immunity, our results support concomitant NKG2A/KIR blockade to unleash the NK cells’ full antitumor potential. The results from our experiments performed in allogeneic settings might be highly relevant for allogenic NK cell products used for adoptive transfers in the clinics.
Introduction
Natural killer (NK) cells are cytotoxic innate lymphocytes that play a crucial role in eliminating tumor or virus-infected cells. Their activity is tightly regulated by multiple germ-line encoded activating and inhibitory receptors.1 The differential expression of several receptors, including CD56, CD16, TIGIT, NKG2A, NKG2C, or Killer-cell immunoglobulin-like receptors (KIRs), separates NK cells into subsets, whose reactivity depends on the receptor expression patterns.2–4 The ligands for these receptors, which can trigger or inhibit NK cell responses, are expressed on tumor cells or upregulated in the context of cellular stress. NK cells not only kill but also produce cytokines in response to tumor targets, such as interferon-γ (IFNγ), that can further affect both innate and adaptive immunity.5 Thus, encounter of NK cells with target cells can lead not only to direct cellular cytotoxicity but also to the release of soluble factors, which can exert subsequent and long lasting effects on immune responses. The mechanisms of how the antitumor reactivity of distinct subsets of NK cells is regulated at early and later time points of NK/tumor cell encounters are still incompletely understood.
Despite effective antitumor immune reactivity, tumor cells employ strategies to avoid and escape from innate and adaptive immune responses. Tumor cells often develop resistance to standard treatments, such as chemotherapy, radiotherapy or targeted immune therapy.6 Immune checkpoint blockade (ICB) was shown to enhance the immune system’s ability to recognize and eliminate tumor cells by neutralizing signals that cancer cells use to avoid and escape from the immune system. ICB therapy showed impressive clinical benefits in both prolonging patients’ survival or even reaching tumor remission.7 8 However, subsets of patients remain unresponsive to ICB therapy, develop resistance and relapse. Currently, antibodies blocking CTLA-4, PD-1/PD-L1 and LAG-3 are FDA-approved ICB treatment options for some cancer types, with many ongoing clinical trials involving other targets, such as TIGIT, CD112R, NKG2A, TIM-3.8–13 Since most current ICB therapies target the adaptive immune system, there is an emerging need in discovering novel immunomodulatory tools and developing ICBs unleashing innate immunity.7 14
Here, we used melanoma cells that express multiple ligands involved in NK cell recognition as a model for discovering innate immune checkpoints that impair the antitumor cytotoxic activity of NK cells.15 We performed a genome-wide (GW) CRISPR-Cas9 knockout (KO) resistance screen, a powerful tool to detect single factors capable to shift the balance towards increased susceptibility or resistance of tumor cells to immune cell attack. Recently published screens identified multiple ligands, whose genetic targeting led to improved T, CAR-T and NK cell function against leukemia and different solid tumors.16–27 However, so far, no GW CRISPR/Cas9 KO screen addressed key resistance mechanisms of melanoma cells to human primary NK cells. Here, we reveal that the resistance of melanoma cells to NK cell killing depended on NK cell-produced IFNγ. IFNγ acted as NK cell shut-off mechanism by inducing the expression of HLA-E, HLA-B and HLA-C molecules that differentially inhibited distinct NK cell subsets defined by the expression of NKG2A and KIRs. We further used therapeutic mAbs currently tested in clinical settings that block these checkpoints and confirmed our results not only in melanoma cells but also in other tumor entities. Our insights into the role of IFNγ on unveiling checkpoints regulating the activation of individual NK cell subsets will contribute to leverage the full antitumor potential of all NK cells.
Results
IFNγ induces melanoma cell resistance to NK cells dependent on the expression of B2M
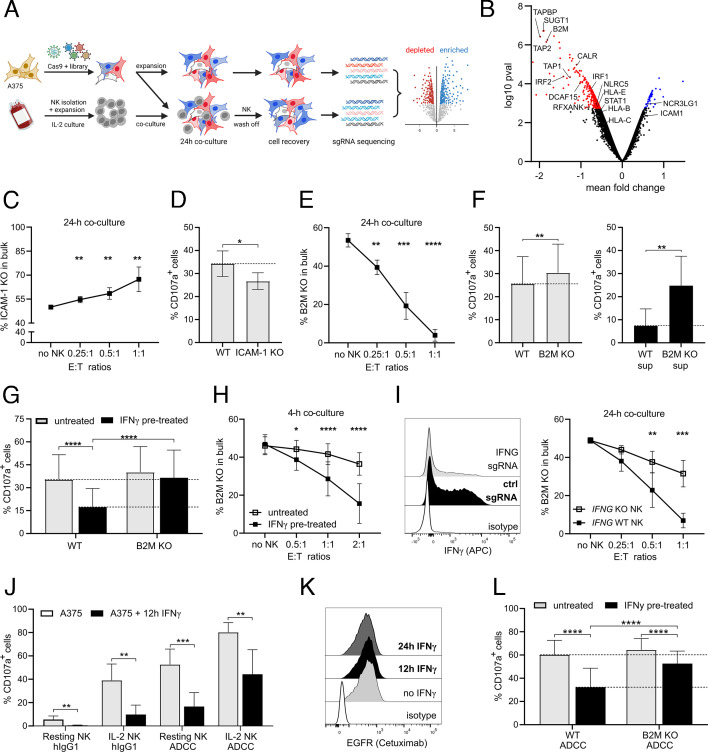
To identify mechanisms involved in the resistance of melanoma cells to NK cell cytotoxicity, we performed a GW CRISPR/Cas9 KO screen on the A375 melanoma cell line co-cultured with IL-2-cultured primary human NK cells. We co-cultured NK cells with sgRNA library-transduced melanoma cells for 24 hours. Subsequently, we separated melanoma cells that had survived co-culture with NK cells, and determined the changes in sgRNA library representation in comparison to melanoma cells cultured alone (figure 1A). Genes, whose targeting affected cell proliferation and survival independent of the presence of NK cells, were filtered out and excluded. Cells bearing sgRNAs targeting TAPBP, TAP2, B2M, TAP1, CALR genes were under-represented indicating the antigen-presenting machinery as one major contributor to the resistance of melanoma cells to NK cell cytotoxicity (figure 1B). In addition, sgRNAs targeting genes involved in transcriptional regulation of MHC-I, such as SUGT1, RFXANK, DCAF15, and NLR5, as well as genes from the IFNγ signaling pathway, such as IRF1, IRF2, and STAT1 were present at comparatively low abundancy.24 28–30 Melanoma cells deficient in ICAM-1, a gene encoding an adhesion molecule, were enriched after co-culture with NK cells (figure 1B). Next, we set up a co-culture assay mimicking the 24 hours of screen co-culture. We mixed equal numbers of candidate gene-deficient (KO) cells with control wild-type (WT) melanoma cells and co-cultured them with NK cells at different effector to target ratios (E:T). After the co-culture, the ratio between WT and KO cells was determined to assess the increase in resistance or sensitivity of KO cells in comparison to WT cells (online supplemental figure 1A). ICAM-1 KO melanoma cells displayed increased resistance after 24 hours co-culture with NK cells, which aligned with decreased degranulation of NK cells observed during 4 hours of co-culture (figure 1C,D, online supplemental figure 1B,C). Melanoma cells deficient in NCRL3LG1 (B7H6), a ligand for the activating receptor NKp30, were also enriched after NK cell co-culture. Accordingly, blocking of NKp30 during co-culture with melanoma cells resulted in reduced degranulation of NK cells (online supplemental figure 1D). In contrast, when we targeted the B2M gene (B2M KO), which resulted in abolished expression of MHC-I, the resistance of melanoma cells after 24 hours of co-culture with NK cells was greatly reduced, particularly at higher E:T ratios resulting in overall higher killing of melanoma cells (figure 1E, online supplemental figure 1E). The loss of B2M in melanoma cells increased the degranulation of NK cells; however, not to an extent that aligns with high sensitivity of B2M KO melanoma cells to NK cell killing observed during 24 hours of co-culture (figure 1F). Thus, we decided to mimic the cytokine environment present during the 24 hours of co-culture by pretreating melanoma cells with supernatant collected from 24 hours of NK cell/melanoma cell co-cultures. NK cells displayed more than a threefold increase in degranulation against supernatant-treated B2M KO cells in comparison to WT cells (figure 1F). After tumor cell encounter, besides cytotoxicity, NK cells also produce the proinflammatory cytokine IFNγ (online supplemental figure 1F), which increases the expression of MHC-I.31 Next, we pretreated WT and B2M KO cells with recombinant IFNγ and observed reduced degranulation of NK cells against WT cells (figure 1G, online supplemental figure 1G). The reduction in the degranulation of NK cells after IFNγ pretreatment of melanoma cells was not mediated by soluble factors and was absent upon deletion of B2M from melanoma cells (figure 1G, online supplemental figure 1H). Accordingly, in a short 4 hours of co-culture, B2M KO cells were as sensitive as WT to NK cell-mediated killing, since the ratio between WT and B2M KO cells did not dramatically change as observed during 24 hours of co-culture (figure 1H). However, overnight (o/n) IFNγ pretreatment of a mixture of WT/B2M KO cells followed by a short 4 hours of co-culture with NK cells resulted in a higher representation of WT cells in the cells that had resisted NK cell co-culture indicating an enhanced resistance of WT cells and an increased sensitivity of B2M KO cells to NK cell killing (figure 1H). These changes in ratios between WT versus B2M KO cells were due to NK cell-mediated killing and not caused by reduced or increased melanoma cell proliferation by soluble factors present in the co-culture supernatant such as IFNγ or TNFα (online supplemental figure 1I). To exclude a cell line-specific effect of IFNγ, we used other MHC-I-expressing (MHC-I+) and MHC-I-deficient (MHC-I−) melanoma cell lines (online supplemental figure 1J). IFNγ pretreatment of melanoma cells did not cause a reduction in the degranulation of NK cells against MHC-I− cell lines but resulted in a reduced response against MHC-I+ cell lines (online supplemental figure 1K). Next, we generated IFNG-deficient primary NK cells (IFNG KO), using CRISPR/Cas9 ribonucleoprotein (RNP) technology, to prevent induction of melanoma cells resistance to NK cell killing (figure 1I). Upon reduction of IFNγ availability during 24 hours of co-culture with mixed WT/B2M KO cells, a lower proportion of WT cells and a higher amount of B2M KO melanoma cell resisted co-culture with IFNG KO NK cells compared to co-culture with WT NK cells (figure 1I). To address the effect of IFNγ in a more physiological context, we tested responses of freshly isolated (resting) NK cells. As with IL-2-cultured cells, we observed impaired degranulation of resting NK cells against IFNγ-pretreated melanoma cells in the absence and the presence of an antibody-dependent cellular cytotoxicity (ADCC)-mediating mAb (figure 1J). This impairment in ADCC-mediated degranulation of NK cells was not caused by reduced binding of the ADCC-mediating antibody, cetuximab, to IFNγ-pretreated melanoma cells (figure 1K). Moreover, B2M KO melanoma cells failed to efficiently inhibit ADCC after IFNγ pretreatment and induced minor inhibition of NK cells (figure 1L). Besides upregulation of MHC-I, IFNγ treatment is reported to reduce the expression of MICA/B, ligands for the activating NKG2D receptor.32 33 We confirmed the involvement of NKG2D in the degranulation of NK cells against A375 cells (online supplemental figure 1L); however, despite the reduction in the expression of MICA/B, we observed only a minor reduction in NKG2D-Fc binding to melanoma cells after IFNγ treatment (online supplemental figure 1M). Taken together, our data confirm that ICAM-1 and B7H6 on melanoma cells mediate NK cell activation while the expression of MHC-I is essential to induce the inhibition of NK cells. Moreover, we show that pretreatment of melanoma cells with IFNγ resulted in reduced NK cell activation, which depended on the ability of melanoma cells to express MHC-I. Accordingly, in melanoma/NK cell co-culture targeting IFNG in NK cells or B2M in melanoma cells avoided NK cell inhibition. Therefore, the IFNγ pathway acts as NK cells shut-off mechanism by inducing the resistance of melanoma cells to NK cells, which is dependent on the expression of B2M/MHC-I.
Figure 1.
IFNγ-mediated MHC-I-dependent resistance of A375 cells to NK cell killing. (A) Set-up of a GW CRISPR/Cas9 NK cell/melanoma screen co-culture. Created with BioRender.com (B) Volcano plot showing depleted and enriched sgRNAs by fold change mean versus p-value. Significant sgRNAs below 0.05 false discovery rate are marked as red (sensitive) and blue (resistant). (C) Increase in ICAM-1 KO cell population after 24 hours of co-culture with 2 days IL-2-cultured NK cells. WT and KO cells were mixed at a 1:1 ratio and co-cultured with NK cells at different effector to target (E:T) ratios (n=6). (D) Degranulation of NK cells after 4 hours co-culture with WT or ICAM-1 KO A375 cells (n=6). (E) Reduction in B2M KO cell population after 24 hours of co-culture of NK cells with mixed WT:B2M KO cells (1:1) at different E:T ratios (n=9). (F) Degranulation of NK cells after 4 hours of co-culture with B2M KO and WT A375 cells either untreated (left) or pretreated o/n with supernatant collected from 24 hours of NK/A375 cell co-culture (right) (n=6). (G) Degranulation of NK cells after 4 hours of co-culture with WT or B2M KO A375 cells pretreated or not with recombinant IFNγ for 12 hours (n=17). (H) Reduction in B2M KO cell population after 4 hours of co-culture of NK cells with mixed WT:B2M KO cells (1:1) at different E:T ratios. A375 cells were pretreated or not with IFNγ o/n before the co-culture (n=6). (I) Representative histograms showing the expression of intracellular IFNγ after 4 hours of stimulation of WT NK or IFNG sgRNA electroporated primary NK cells with IL-12/18 (left). Reduction in B2M KO cell population after 24 hours of co-culture of mixed WT:B2M KO cells (1:1) at different E:T ratios with WT or IFNG KO NK cells (right) (n=3). (J) Degranulation of freshly isolated (resting) NK cells and 2 days IL-2-cultured NK cells after 4 hours of co-culture with A375 cells pretreated or not with IFNγ 12 hours before the co-culture in the presence of hIgG1 or ADCC-inducing cetuximab (5 µg/mL) (n=6). (K) Histogram showing binding of cetuximab to EGFR on A375 cells pretreated or not with IFNγ for 12 hours. (L) Degranulation of 2 days IL-2-cultured NK cells after 4 hours of co-culture with WT or B2M KO A375 cells pretreated or not with IFNγ 12 hours before the co-culture in the presence of ADCC-inducing cetuximab mAb (n=6). Statistical analysis was performed by one-way ANOVA (multiple comparisons) test (C, E, G, L); two-way ANOVA (multiple comparisons) test (H, I) and two-tailed Student’s t-test (D, F, J), *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ANOVA, analysis of variance; KO, knockout; NK, natural killer; WT, wild-type.
jitc-2024-009410supp001.pdf (968.8KB, pdf)
IFNγ-induced expression of HLA-E protects melanoma cells from NKG2A+ NK cells
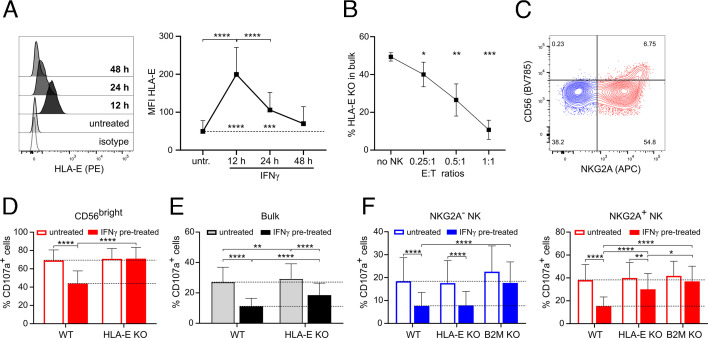
Since IFNγ-mediated tumor resistance to NK cells depended on the expression of B2M in melanoma cells, we dissected the role of individual MHC-I molecules and their effect on NK cell activity. In our screen, among all MHC-I molecules, only the cells with targeted deletion of the non-classical MHC-I molecule HLA-E were significantly depleted after co-culture with NK cells. The surface expression of HLA-E, absent on untreated A375 cells, was transiently upregulated by IFNγ, peaking at 12 hours post-IFNγ treatment (figure 2A, online supplemental figure 2A). Next, we generated melanoma cells deficient in the HLA-E gene (HLA-E KO) (online supplemental figure 2B). In a 24-hour co-culture of mixed WT/HLA-E KO cells with NK cells, HLA-E KO cells showed reduced resistance to NK cells in comparison to WT cells (figure 2B). HLA-E is a ligand for the activating NKG2C and the inhibitory NKG2A receptor.34 While the expression of NKG2C was detected only on a minor fraction of NK cells, the expression of NKG2A consistently defined two NK cell subsets (figure 2C, online supplemental figure 2C). The degranulation of CD56bright NK cells, which display high expression of NKG2A, was reduced after co-culture with IFNγ-pretreated WT cells, but not HLA-E KO cells (figure 2D). The overall degranulation of NK cells displayed only partial inhibition after co-culture with IFNγ-pretreated HLA-E KO cells, since NKG2A+ NK cells were no longer inhibited by HLA-E (figure 2E,F). In contrast, NKG2A− NK cells remained inhibited after the IFNγ pretreatment of melanoma cells. The inhibition of both NKG2A+ and NKG2A− NK cell subsets after IFNγ pretreatment of melanoma cells was absent during co-culture with B2M KO cells (figure 2F). Similar results were obtained with SKMel-37 melanoma cells (online supplemental figure 2D-F). Here, we show that the deletion of HLA-E or B2M in IFNγ-pretreated melanoma cells abolished their resistance to NK cells. HLA-E that was induced by IFNγ inhibited the responses of CD56bright and NKG2A+ NK cells while the classical MHC-I mediated the inhibition of NKG2A− NK cells after IFNγ treatment of melanoma cells.
Figure 2.
IFNγ-induced expression of HLA-E on A375 cells inhibits NKG2A+ NK cells. (A) Expression of HLA-E on A375 cells treated or not with IFNγ for 12, 24 and 48 hours (n=6). (B) Reduction in HLA-E KO cell population after 24 hours of co-culture of NK cells with mixed WT:HLA-E KO cells (1:1) at different E:T ratios (n=6). (C) Representative dot plot showing the expression of NKG2A and CD56 on NK cells. (D, E) Degranulation of CD56bright (D) and bulk NK cells (E) after 4 hours of co-culture with HLA-E KO or WT A375 cells. A375 cells were pretreated or not o/n with IFNγ (n=11). (F) Degranulation of NKG2A− and NKG2A+ NK cells after 4 hours of co-culture with WT, HLA-E KO and B2M KO A375 cells pretreated or not with IFNγ 12 hours before the co-culture (n=6–9). Statistical analysis was performed by one-way ANOVA (multiple comparisons) test (A, B, D, E, F), *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ANOVA, analysis of variance; E:T, effector to target; KO, knockout; NK, natural killer; WT, wild-type.
IFNγ-induced expression of classical MHC-I on melanoma cells inhibits the degranulation of NKG2A− NK cells
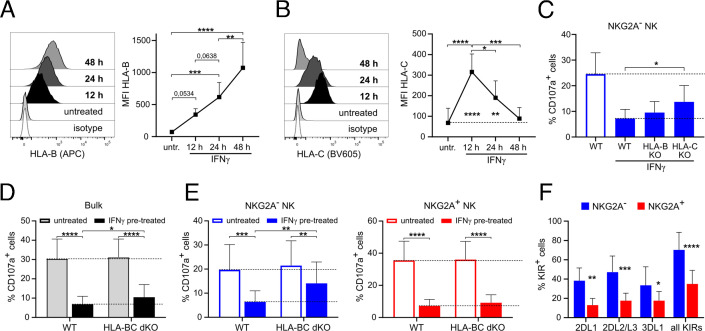
Next, we explored the role of the individual classical MHC-I molecules in regulating melanoma cell susceptibility to NK cells. A375 cells expressed HLA-A at a steady state while HLA-B and HLA-C were only expressed upon IFNγ treatment (figure 3A,B, online supplemental figure 3A). The expression of HLA-A and HLA-B increased with the duration of the treatment while the expression of HLA-C was transient peaking at 12 hours post-IFNγ treatment (figure 3A,B, online supplemental figure 3A). To generate HLA-ABC KO melanoma cells, we transfected melanoma cells with sgRNA targeting classical MHC-I genes, which resulted in impaired expression of HLA-A, HLA-B and HLA-C (online supplemental figure 3B). Upon co-culture with NK cells, IFNγ-pretreated HLA-ABC KO melanoma cells failed to inhibit the degranulation of NKG2A− NK cells (online supplemental figure 3C). However, the KO of classical MHC-I also led to impaired upregulation of HLA-E, which relies on signal peptides derived from classical MHC-I for its expression on cell surface35 36 (online supplemental figure 3B). Therefore, HLA-ABC KO cells also failed to inhibit the degranulation of NKG2A+ NK cells. Next, we focused on individual HLA-B and HLA-C molecules, as their expression was dependent on the presence of IFNγ, and generated HLA-B KO and HLA-C KO cells (online supplemental figure 3D). We observed that the degranulation of NKG2A− NK cells was higher after co-culture with IFNγ-pretreated HLA-B KO and HLA-C KO cells in comparison to WT melanoma cells (figure 3C). Next, we generated HLA-BC double KO (dKO) melanoma cells that kept the expression of HLA-A to provide signal peptides for the expression of HLA-E (online supplemental figure 3E). Simultaneous KO of HLA-B and HLA-C only slightly increased the degranulation of bulk NK cells against IFNγ-pretreated melanoma cells; however, it specifically increased the degranulation of the NKG2A− NK cells but not NKG2A+ NK cells (figure 3D,E). Accordingly, the expression of ILT2 (LIR-1a, CD85j) and KIRs, inhibitory receptors engaged by classical MHC-I molecules, was higher on NKG2A- NK cells (figure 3F, online supplemental figure 3F,G). Of note, the blockade of ILT2 failed to restore the degranulation of NKG2A− NK cells against IFNγ-pretreated melanoma cells (online supplemental figure 3H). Taken together, these data show that NKG2A− NK cells are inhibited by IFNγ-induced classical MHC-I molecules on melanoma cells in an ILT2-independent manner.
Figure 3.
Inhibition of NKG2A− NK cells by IFNγ-pretreated A375 cells deficient in classical MHC-I molecules. (A, B) Expression of HLA-B and HLA-C on A375 cells treated or not with IFNγ for 12, 24 and 48 hours (n=6). (C) Degranulation of NKG2A− NK after 4 hours of co-culture with WT, HLA-B KO and HLA-C KO A375 cells pretreated or not with IFNγ 12 hours before the co-culture (n=6). (D) Degranulation of bulk NK cells and (E) NKG2A− and NKG2A+ NK cells after 4 hours of co-culture with WT and HLA-BC dKO A375 cells pretreated or not with IFNγ 12 hours before the co-culture. (n=6). (F) Expression of KIRs on 2 days IL-2-cultured NKG2A− or NKG2A+ NK cell subsets (n=5–9). Statistical analysis was performed by one-way ANOVA (multiple comparisons) test (A–E) and two-tailed Student’s t-test (F), *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ANOVA, analysis of variance; KO, knockout; NK, natural killer; WT, wild-type.
NK cell subsets defined by the expression of NKG2A and KIRs are differentially inhibited by HLA-E and classical MHC-I
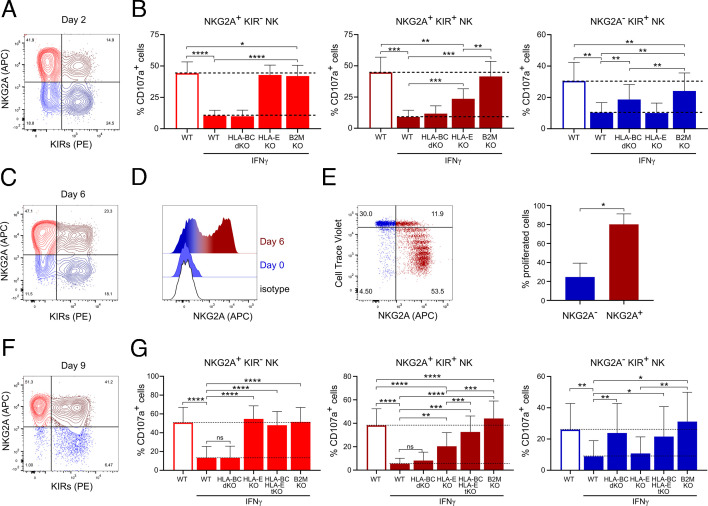
Next, we set out to dissect the reactivity of individual NK cell subsets defined by the expression of NKG2A and/or KIRs towards IFNγ-pretreated melanoma cells. Co-staining of KIRs and NKG2A revealed different NK cell subsets, which all showed impaired degranulation in co-cultures with IFNγ-pretreated WT melanoma cells (figure 4A,B, online supplemental figure 4A). In co-culture with HLA-E KO cells, the degranulation of NKG2A+ KIR− NK cells was not inhibited after IFNγ pretreatment (figure 4B). NKG2A+ NK cells that co-expressed KIRs still showed partially reduced degranulation after co-culture with HLA-E KO cells, but no inhibition of degranulation on co-culture with B2M KO melanoma cells. The degranulation of NKG2A− KIR+ NK cells displayed only partial inhibition after co-culture with HLA-BC dKO as well as B2M KO cells in comparison to co-culture with IFNγ-pretreated WT melanoma cells. However, no reduction in the inhibition of KIRs+ NK cells that also expressed NKG2A was observed during co-culture with IFNγ-pretreated HLA-BC dKO cells (figure 4B). So far, in our experiments, we used NK cells that were cultured in the presence of IL-2 for 2 days. The expression of KIRs on NK cells either freshly isolated or cultured in the presence of IL-2 for 2 days was lower on NKG2A+ NK cells compared with NKG2A− NK cells (figures 3F and 4A). However, in concordance with previously published data,37 after 9 days of culture, the NKG2A/KIRs subset composition changed toward >90% of NK cells being NKG2A+, comprising both KIR− and KIR+ NK cells (figure 4A,C and F). We confirmed that these changes were not due to a preferential proliferation of the NKG2A+ NK cell subset, but rather a result of proliferation of both KIR− and KIR+ NKG2A− NK cells that upregulated NKG2A during expansion (figure 4D,E, online supplemental figure 4B). The upregulation of NKG2A by NKG2A− KIR+ NK cells resulted in masking the effect of HLA-B and HLA-C loss in melanoma cells on the degranulation of NK cells (figure 4B,G). To exclude the role of HLA-E during the co-culture of NKG2A+ KIR+ NK cells with HLA-BC dKO cells, we knocked out HLA-B and HLA-C in HLA-E KO cells (HLA-BCE triple KO (tKO)). The degranulation of NKG2A+ KIR+ NK cells against IFNγ-pretreated HLA-BCE tKO cells was higher in comparison to co-culture with HLA-E KO cells, suggesting the major role of HLA-E in mediating the inhibition of NKG2A+ KIR+ NK cells (figure 4G). These data show differential regulation of individual NKG2A/KIRs NK cell subsets, where the function of NKG2A+ KIR− NK cells is inhibited by HLA-E while the function of NKG2A− KIR+ NK cells is inhibited by classical MHC-I molecules. Moreover, the combination of both HLA-E and classical MHC-I molecules on melanoma cells inhibits the function of the NKG2A+ KIR+ NK cell subset. Taken together, NKG2A+ KIR+ NK cells originate from NKG2A− KIR+ NK cells by upregulating NKG2A on proliferating NK cells as more potent inhibitory receptor than KIRs in our allogenic settings.
Figure 4.
Different inhibition of NKG2A/KIRs NK cell subsets by IFNγ-pretreated A375 cells. (A) Representative dot plot showing the expression of NKG2A and KIRs on NK cells cultured for 2 days in IL-2-containing media. (B) Degranulation of 2 days IL-2-cultured NKG2A/KIRs NK cell subsets after 4 hours of co-culture with WT, HLA-E KO, HLA-BC dKO and B2M KO A375 cells pretreated or not with IFNγ 12 hours before the co-culture (n=7). (C) Expression of NKG2A and KIRs on NK cells cultured for 6 days in IL-2-containing media. (D) Expression of NKG2A on isolated NKG2A- NK cells at days 0 and 6 of culture in IL-2-containing media. (E) Expression of NKG2A and the reduction in Cell Trace Violet (CTV) staining on isolated NKG2A- NK cells cultured for 6 days in IL-2-containing media. NKG2A− NK cells were isolated by depleting of NKG2A+ NK cells using NKG2A magnetic beads, stained with CTV and analyzed after 6 days of culture. NK cell were gated as NKG2A− and NKG2A+ and the percentage of NK cells that have proliferated was quantified (n=3). (F) Representative dot plot showing the expression of NKG2A and KIRs on NK cells cultured for 9 days in IL-2-containing media. (G) Degranulation of 9 days IL-2-cultured NKG2A/KIR NK cell subsets after 4 hours of co-culture with WT, HLA-E KO, HLA-B/C dKO, HLA-BCE tKO and B2M KO A375 cells pretreated or not with IFNγ 12 hours before the co-culture (n=8). Statistical analysis was performed by one-way ANOVA (multiple comparisons) test (B, G) and two-tailed Student’s t-test (E), *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ANOVA, analysis of variance; KO, knockout; NK, natural killer; WT, wild-type.
Blockade of NKG2A and KIRs restores NK cell cytotoxicity against IFNγ-pretreated tumor cells
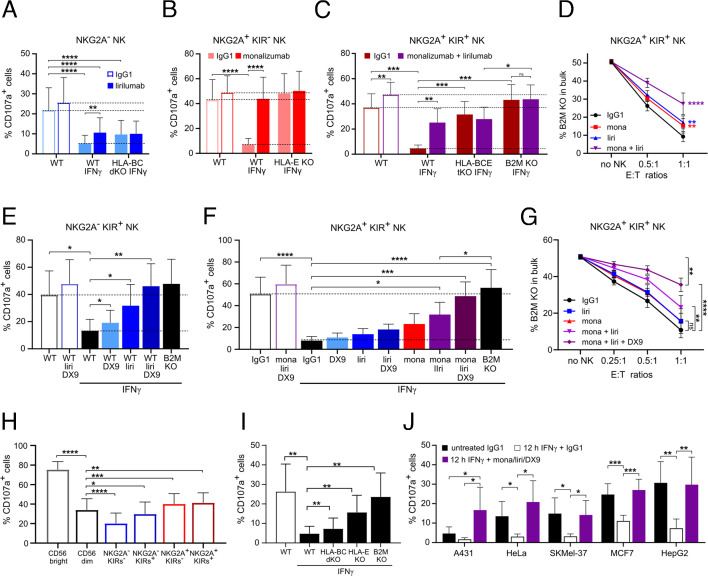
To overcome the inhibitory effect of melanoma-expressed classical MHC-I, we used lirilumab, a monoclonal antibody (mAb) blocking KIR2DL1, KIR2DL2, and KIR2DL3. Lirilumab treatment of NK cells during the co-culture with IFNγ-pretreated WT cells resulted in increased degranulation of NKG2A− NK cells comparable to their response against HLA-BC dKO cells (figure 5A, online supplemental figure 5A). Treatment with monalizumab, a mAb blocking NKG2A, increased the degranulation of NKG2A+ KIR− NK cells against IFNγ-pretreated WT cells, comparable to the response observed against HLA-E KO cells (figure 5B, online supplemental figure 5B). Combined monalizumab and lirilumab treatment of NKG2A+ KIR+ NK cells induced a response comparable to the degranulation measured against HLA-BCE tKO cells (figure 5C). Single mAb treatment of the NKG2A+ KIR+ NK cell subset failed to abolish the resistance of WT melanoma cells, but the treatment with a combination of monalizumab with lirilumab mAbs highly reduced the resistance of WT cells and the preferential killing of B2M KO melanoma cells by NKG2A+ KIR+ NK cells during 24 hours of co-culture (figure 5D). However, monalizumab with lirilumab treatment was not able to fully abolish the resistance of WT melanoma cells to NK cell killing to the same extent as deletion of the B2M gene. Since the lirilumab mAb does not block KIR3DL1, we used DX9, a mAb blocking KIR3DL1 (online supplemental figure 5A). DX9 treatment alone partially increased the degranulation of the NKG2A− KIR+ NK cell subset and in combination with lirilumab completely restored the degranulation of NKG2A− KIR+ NK cells against IFNγ-pretreated WT melanoma cells (figure 5E). Similar to treatment with lirilumab mAb, no effect of KIR3DL1 blockade alone on the degranulation of NKG2A+ KIR+ NK cells against IFNγ-pretreated WT melanoma cells was observed (figure 5F). The combination of DX9 mAb together with monalizumab and lirilumab treatment almost fully restored the degranulation of NKG2A+ KIR+ NK cells against IFNγ-pretreated melanoma cells (figure 5F). The addition of DX9 to monalizumab and lirilumab treatment reduced the resistance of WT melanoma cells and the preferential killing of B2M KO melanoma cells by NKG2A+ KIR+ NK cells in comparison to combined monalizumab and lirilumab treatment (figure 5G, online supplemental figure 5C). Next, we addressed the contribution of different NK cell subsets. CD56bright NK cells that express NKG2A and NKG2A+ CD56dim NK cells displayed the highest degranulation against melanoma cells (figure 5H). Accordingly to the high activity of NKG2A+ NK cell subsets, the loss of HLA-E on melanoma cells had the highest effect on the overall degranulation of NK cells against IFNγ-pretreated melanoma cells while the loss of classical MHC-I showed only a minor effect on the degranulation of NK cells (figure 5I). To exclude a mechanism only relevant for melanoma cells, we used other MHC-I+ tumor entities, such as the A431 epidermoid carcinoma, SKMel-37 melanoma, HeLa cervical cancer, HepG2 hepatoblastoma, MCF7 breast cancer and K562 lymphoblast cell line (online supplemental figure 5D). In concordance with data obtained with melanoma cells, IFNγ pretreatment of all tumor cell lines tested caused a reduction in the degranulation of NK cells. The presence of monalizumab, lirilumab and DX9 mAbs during co-culture was sufficient to fully restore the degranulation of NK cells against IFNγ-pretreated tumor cells (figure 5J, online supplemental figure 5E and 6). Together, these data demonstrate that the use of therapeutic mAbs blocking the HLA-E/NKG2A and classical MHC-I/KIRs axes restored NK cell cytotoxicity against IFNγ-induced resistant MHC-I+ tumor cells.
Figure 5.
Effect of monalizumab, lirilumab and DX9 mAb treatment on the degranulation of NK cell subsets against tumor cells. (A) Degranulation of NKG2A− NK cells after 4 hours of co-culture with untreated or 12 hours IFNγ-pretreated WT or HLA-BC dKO A375 cells in the presence of 5 µg/mL IgG1 or lirilumab (liri) mAb (n=6). (B) Degranulation of NKG2A+ KIR− NK cells after 4 hours of co-culture with untreated or 12 hours IFNγ-pretreated WT or HLA-E KO A375 cells in the presence of 5 µg/mL IgG1 or monalizumab (mona) mAb (n=6). (C) Degranulation of expanded FACS-sorted NKG2A+ KIRs+ NK cells after 4 hours of co-culture with untreated or 12 hours IFNγ-pretreated WT and HLA KO A375 cells in the presence of 5 µg/mL IgG1 or combined monalizumab+lirilumab mAbs (n=9). (D) Reduction in B2M KO cell population after 24 hours of co-culture of NK cells with mixed WT:B2M KO cells (1:1) at different E:T ratios in the presence of 5 µg/mL of indicated mAbs. Statistical significance is displayed for E:T ratio 1:1 (n=6). (E) Degranulation of NKG2A− KIR+ NK cells after 4 hours of co-culture with untreated or 12 hours IFNγ-pretreated WT and B2M KO A375 cells in the presence of indicated mAbs (n=6). (F) Degranulation of NKG2A+ KIR+ NK cells after 4 hours of co-culture with untreated or 12 hours IFNγ-pretreated WT and B2M KO A375 cells in the presence of indicated mAbs (n=5). (G) Reduction in B2M KO cell population after 24 hours of co-culture of NKG2A+ KIRs+ NK cells with mixed WT/B2M KO (1:1) cells in the presence of indicated mAbs. Statistical significance is displayed for E:T ratio 1:1 (n=6). (H) Degranulation of CD56bright, CD56dim and CD56dim NKG2A/KIRs NK cell subsets after 4 hours of co-culture with A375 cells (n=15). (I) Degranulation of 2 days IL-2-cultured NK cells after 4 hours of co-culture with untreated or 12 hours IFNγ-pretreated WT and HLA KO A375 cells (n=8). (J) Degranulation of 9 days IL-2-cultured NK cells after 4 hours of co-culture with MHC-I+ tumor cell lines pretreated or not with IFNγ 12 hours before the co-culture in the presence of indicated mAbs (n=6). Statistical analysis was performed by two-way ANOVA (multiple comparisons) test (A–D, G) and one-way ANOVA (multiple comparisons) test (E, F, H–J), *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ANOVA, analysis of variance; E:T, effector to target; KO, knockout; NK, natural killer; WT, wild-type.
Discussion
In this study, we harnessed the power of a GW CRISPR/Cas9 KO screen as a tool to uncover molecules involved in the resistance of melanoma cells to the cytotoxicity of primary human NK cells. Our results highlight that antigen-presenting machinery and IFNγ signaling pathways represent a dominant inhibitory axis protecting tumor cells against NK cell cytotoxicity. Here, we dissect the relative contribution of different MHC class I molecules and their impact on distinct NK cell subsets.
Despite the well-known role of MHC-I that was also identified as a resistance mechanism to NK cells in previous screens with other tumor entities,18–21 24–27 we observed a relatively minor effect of MHC-I on the resistance of melanoma cells to allogenic NK cells in a short (4 hours) co-culture. In contrast, o/n pretreatment of tumor cells with IFNγ greatly increased the MHC-I-mediated inhibition of not only IL-2-cultured NK cells, but also of freshly isolated, resting NK cells in context of ADCC. Correspondingly, the inhibitory effects of MHC-I were revealed in a longer (24 hours) co-cultures, where NK cell-derived IFNγ-induced resistance of melanoma cells to NK cells, which was completely dependent on the expression of MHC-I on melanoma cells. The discrepancies between short (4 hours) and longer (24 hours) co-cultures and the IFNγ-dependent inhibitory effect of MHC-I align with previously published in vivo data, where NK cells were able to control and eliminate lung-disseminated melanoma cells only within 24 hours on tumor cell arrival.38 Within the first 4 hours 50% of NK-tumor cell interactions resulted in tumor cell death while after 24 hours approximately 100% of interactions resulted in tumor cell survival and formation of macro metastatic nodules, which can be explained by IFNγ-induced expression of MHC-I on tumor cells. A previous study based on a GW screen with K562 leukemia cells and NK cells activated by autologous feeder cells, IL-2 and phytohemagglutinin focused on HLA-E mediating the resistance of K562 cells to NK cell cytotoxicity as their main hit.18 This expansion protocol leads to high proliferation presumably skewing most NK cells to an NKG2A+ phenotype. Since in our study we validated hits with freshly isolated, ADCC-activated and IL-2-cultured NK cells, we were able to dissect the relative impact of classical and non-classical MHC class I molecules on distinct NKG2A and/or KIR expressing NK cell subsets in our setting. Additionally, other significantly enriched or depleted candidates from our screen align with other screens performed using NK cells and other tumor entities suggesting a shared mechanism across multiple tumor types.18–20 24–27
In our setting, which involves MHC-I+ melanoma cells, IFNγ induced the expression of HLA-E, HLA-B and HLA-C, which are, unlike HLA-A, not expressed at a steady state. Their induced expression potently inhibited NK cell function and protected melanoma cells from NK cell-mediated attack. However, these signals do not act uniformly on all NK cells, but specifically on individual NK cell subsets. HLA-E is the ligand for the inhibitory NKG2A receptor expressed and frequently upregulated on NK and T cell subsets within TME across many human cancers and associated with poor outcome and reduced NK and T cell cytotoxicity.39–46 Classical MHC-I molecules are major ligands for KIRs that define educated NK cells responsible for the protection of cells that express MHC-I alleles native to the host system.47 Accordingly, we stratified NK cells into subsets based on the expression of NKG2A and KIRs. We observed that melanoma cell-expressed HLA-E fully inhibited the NKG2A+ KIR- NK cell subset while only partially the NKG2A+ KIR+ NK cell subset. The loss of HLA-B and HLA-C in melanoma cells specifically increased the degranulation of the NKG2A− KIR+ NK cell subset but had no effect on the NKG2A+ KIR+ NK cell subset. Moreover, we showed that NKG2A+ KIR+ NK cells can originate from NKG2A− KIR+ NK cells by upregulating NKG2A during proliferation as a more potent inhibitory receptor than KIRs in our setting. We confirmed our results with therapeutic mAbs currently used in clinical trials. Monalizumab, a NKG2A blocking mAb that promotes antitumor function of both T and NK cells, shows promising patient responses in clinical trials, especially if combined with other therapeutic agents.13 39 48–53 Treatment of NK cells with monalizumab reproduced the effect of HLA-E loss in melanoma cells on the degranulation of NKG2A+ NK cells. To target KIR+ NK cells, we used lirilumab, a KIR2D blocking mAb used in clinical trials with so far limited efficacy on patient survival.54–56 Lirilumab treatment had a comparable effect on the function of NKG2A/KIRs NK cell subsets as the depletion of HLA-B and HLA-C in melanoma cells and confirmed the involvement of KIRs in mediating the inhibition of NK cells in allogeneic settings. Combined use of lirilumab together with KIR3DL1 blockade by DX9 mAb57 fully restored the cytotoxicity of the NKG2A− KIR+ NK, but not NKG2A+ KIR+ NK cell subset which remained inhibited by engagement of NKG2A after co-culture with IFNγ-pretreated melanoma cells. The fact that the effect of lirilumab treatment on restoring the function of the NKG2A+ KIR+ NK cell subset depended on simultaneous use of monalizumab opens up a potential explanation for the limited benefits of lirilumab mAb observed in the clinical trials. The presence of KIR3DL1 and NKG2A inhibitory signaling might have masked the potential of lirilumab in restoring the NK cell function against IFNγ-induced resistant tumor cells in cancer patients. We confirmed that combined monalizumab, lirilumab and DX9 treatment restored the degranulation of NKG2A+ KIR+ NK cells and abolished IFNγ-induced resistance of not just melanoma cells but also other tumor entities to the cytotoxicity of distinct NK cell subsets. This interplay of NKG2A, KIR3DL1 and KIR2s inhibiting function of NK cell subsets underscores the importance of a combined treatment in contrast to a single treatment. Besides KIRs and NKG2A, ILT2, binding to classical MHC-I and HLA-G, might also affect the resistance of tumor cells. In our experimental set-up where HLA-G was not expressed, we did not observe an involvement of ILT2 in mediating the inhibition of NK cells after IFNγ pretreatment of melanoma cells. In addition to increased inhibition of NK cells, a reduction in the expression of NKG2D activating ligands was also observed after IFNγ pretreatment in concordance with literature.32 33 However, the frequencies of expression of all NKG2D-ligands detected by the NKG2D-Fc were only marginally changed on treatment with IFNγ presumably due to other NKG2D-ligands (ULBPs) expressed on the tumor cells. Thus, depending on the levels of different NKG2D-ligands expressed by the target cells, IFNγ treatment might also impair NKG2D-mediated cytotoxicity. The relative contribution of inhibitory pathways might not only be affected by the type of NK effector cells studied, but also by the ligand expression on tumor cells. Thus, it is intriguing that we confirmed the main resistance pathways, antigen presentation and IFNγ signaling, across many NK donors and tumor entities in concordance with other published screens highlighting their general relevance.
Mutations in the antigen-presenting and IFNγ signaling pathway are frequently found in human tumors.58–62 However, patient data obtained from already established tumors reflect the tumor escape from the adaptive immune system and not the initial tumor status in the context of innate immune surveillance. In this context, an in vivo GW CRISPR/Cas9 KO screen in mouse tumor models in the presence of an intact immune system, revealed the pivotal role of MHC-I and IFNγ as tumor resistance mechanisms.26 Moreover, our model of IFNγ-induced tumor resistance in MHC-I+ tumors aligns with an in vivo study by Kelly et al where NK cells were able to reject only MHC-I- tumor while the rejection of MHC-I+ tumor cells was mediated by effector T cells.63 IFNγ pretreatment of melanoma cells prior to injection enhanced their tumorigenicity and increased the formation of lung metastasis in vivo further supporting increased resistance of melanoma cells to initial NK cell killing.64 The inhibitory effect of MHC-I and IFNγ highlighted a crucial role of the NK cell response over T cells in the early phase of tumor cell elimination. Additional studies underlined the importance of NK cells during the early phase of tumor development and in the elimination of circulating metastases while the adaptive immune system, mostly cytotoxic T cells, was crucial to control already established tumor.38 65–68 The induction of melanoma cell resistance by NK cell-produced IFNγ was previously observed by Balsamo et al, who correlated this finding with the induction of MHC-I without investigating the mechanistic underpinnings.69 70 In our study, we comprehensively validated the effect of IFNγ-induced classical MHC-I and HLA-E in mediating the inhibition of distinct NK cell subsets in our allogenic settings. Our data support a model in which, NK cells encountering tumor cells display similar immediate cytotoxicity regardless of the capability of tumor cells to express MHC-I. However, at later time points, NK cell activation accompanied by high IFNγ production leads to an induced expression of HLA-E and classical MHC-I greatly reducing NK cell cytotoxicity of multiple distinct NK cell subsets by inhibitory NKG2A and KIR receptors. Therefore, IFNγ, one of the main effector molecules of NK cells, functions as a self-regulating NK cell shut-off mechanism by inducing the resistance of tumor cells. This mechanism could potentially be one of the reasons why NK cells often fail to control already established tumors, where IFNγ is frequently present. Our study investigates the resistance of tumor cells to NK cells in the absence of additional factors and cells in the tumor microenvironment. We are aware that several factors including TGF-β and suppressive cells like Treg and MDSC will further impact NK cell-mediated tumor killing and the resistance of tumor cells to NK cell-based therapies in vivo. Since also in vivo screens identified IFNγ signaling and antigen presentation as main resistance pathways to NK cell mediated killing,26 we are confident that our results, although obtained in in vitro co-cultures, might be relevant in more complex in vivo settings.
The opposing nature of IFNγ and the classical MHC-I on NK and T cell recognition of tumor cells allows T cells to continue tumor surveillance initiated by NK cells that become inhibited due to the upregulation of MHC-I. Since IFNγ is crucial in further tumor surveillance, neutralizing IFNγ or targeting IFNGR as a potential therapeutic use is not favorable as IFNγ itself was also shown to have antiproliferative effect and in some cases inducing direct apoptosis of tumor cells.71–73 Thus, instead of IFNγ neutralization, a direct blockade of NKG2A and KIRs that restores both NK and a subset of NKG2A+ T cells while keeping the beneficial effect of IFNγ on tumor surveillance, can be a highly attractive approach to tackle IFNγ-mediated tumor resistance. Monalizumab is currently tested as a combination therapy with other therapeutic agents such as durvalumab (anti-PD-L1), trastuzumab (anti-HER2) or cetuximab (anti-EGFR).49 51–53 74 Thus, the synergistic action of monalizumab with lirilumab/DX9 could also complement other immune checkpoint antibodies and effectively avoid and combat tumor cell resistance by simultaneously unleashing innate and adaptive immunity. Combination of multiple checkpoint blockade antibodies in therapy could lower the risk of development of treatment resistant tumors, but might increase the risk of side effects, however, both monalizumab and lirilumab showed relatively low toxicity in initial clinical trials.48 51 56 Regardless, careful dose adjustment or personalized treatment strategy is necessary to avoid toxicity connected to using multiple antibodies. To date, KIR3DL1 blocking antibodies have not entered clinical trials, to complement lirilumab or other ICB therapy options. Knowing the regulation of individual NK cell subsets could also highly benefit personalized medicine by targeting a specific NK cell subsets present within tumor.
In addition to the application of ICB in cancer patients, there is now emerging evidence of clinical benefits of adoptive transfer of NK cells or NK cells harnessed by chimeric antigen receptors (CARs) as off the shelf cell products.75 For these cell products, that undergo high expansion resulting in the upregulation of NKG2A, the additional use of combined monalizumab, lirilumab and KIR3DL1 blockade mAbs treatment could neutralize the resistance of tumor cells and increase the antitumor efficacy of NK cells products. Moreover, engineering NK cells by genetic depletion of NKG2A and KIRs might render them unresponsive to IFNγ-mediated tumor cell resistance mechanisms, potentially improving current cancer therapies in the clinics.
Materials and methods
Cells and functional antibodies
A375 (ATCC; Cat# CRL-1619, RRID:CVCL_0037), A431 (ATCC; Cat# CRL-1555), HeLa (ATCC; Cat# CRM-CCL-2, CVCL_0030), K562 (ATCC; Cat# CCL-243, RRID:CVCL_0004), SKMel-28 (ATCC; Cat# HTB-72, RRID:CVCL_0526), MaMel-86b (RRID:CVCL_A221)68 and UKRV-Mel02 (RRID:CVCL_A700)69 cell lines were cultured in RPMI 1640 media supplemented with 10% FCS, 2 mM L-glutamine, and 1% Penicillin/Streptomycin. HEK293T (ATCC; Cat# CRL-3216, RRID:CVCL_0045), HepG2 (ATCC; Cat# HB-8065, RRID:CVCL_0027), MCF7 (ATCC; Cat# HTB-22, RRID:CVCL_0031) and SKMel-37 (Sigma-Aldrich; Cat# SCC262, RRID:CVCL_3878) were cultured in DMEM media supplemented with 10% FCS, 2 mM L-glutamine, and 1% Penicillin/Streptomycin. Primary NK cells were cultured in SCGM (CellGenix, Cat# 20 802–0500) or in NK MACS media (Miltenyi Biotec, Cat# 130-114-429), both supplemented with 10% human AB serum (PAN-Biotech Cat# P30-2901), 1% Penicillin/Streptavidin, 2 nm L-Glutamine and 400 U/mL of IL-2 (TECINTM (Teceleukin) provided by the National Cancer Institute). Antibodies used for blocking NK cell receptors were: ILT2 (BioLegend Cat# 333702, RRID:AB_1089089), ILT2 (Thermo Fisher Scientific Cat# 16-5129-82, RRID:AB_10669632) NKG2D (BioLegend Cat# 320814, RRID:AB_2561488), NKp30 (BioLegend Cat# 325224, RRID:AB_2814183), DX9/KIR3DL1 (Miltenyi Biotec Cat# 130-092-555, RRID:AB_871611), lirilumab (MedChemExpress Cat# HY-P99208) and monalizumab (MedChemExpress Cat# HY-P99032). Antibodies used for ADCC experiments were: Cetuximab (InvivoGen Cat# hegfr-mab1, RRID:AB_3064809) and human IgG1 (BioLegend Cat# 403501 RRID:AB_2927629).
Isolation of NK cells
Healthy donors’ blood buffy coats were provided by DRK-Blutspendedienst Baden-Württemberg-Hessen (Mannheim, Germany). Peripheral blood mononuclear cells (PBMCs) were enriched using Pancoll (PAN Biotech, Cat# P04-60500) density centrifugation. NK cells were isolated from PBMC by negative selection using the Human NK Cell Isolation Kit, according to manufacturer’s protocol, yielding ~95% purity (Miltenyi Biotec, Cat# 130-092-657). NKG2A− NK cells were further selected using biotinylated NKG2A antibody (Miltenyi Biotec Cat# 130-113-564, RRID:AB_2783968) and anti-Biotin MicroBeads (Miltenyi Biotec Cat# 130-090-485, RRID:AB_244365). Isolated NK cells were cultured at a concentration of 2×106 cells/mL for 2 days in media containing 400 U/mL of IL-2, if not indicated otherwise.
Lentiviral library production
Heidelberg (HD) CRISPR library sub-library A was synthesized and cloned into the HD_CRISPRv1 sgRNA-puromycin expression vector, as previously described.70 Lentivirus was produced in HEK293T cells transfected with psPAX2 G (Addgene, Cat# 12260, RRID:Addgene_12260 and pMD2.G (Addgene, Cat# 12259, RRID:Addgene_12259) packaging plasmids using Lipofectamine 2000 (ThermoFisher Cat# 11668019). Virus-containing supernatant was filtered through a 0.45 low protein-binding membrane (Milipore Sterilflip HV/PVDF), concentrated by the Amicon 100 kDa Ultra Centrifugal Filter Units, and stored at −80°C.
Cas9 overexpression in melanoma cell lines
Lentiviral vectors encoding Cas9-EGFP (Addgene, Cat# 63592, RRID: Addgene_63592) or Cas9-Blasticidin (Addgene, Cat# 52962, RRID:Addgene_52962) were used to transduce melanoma cells at low MOI. GFPhigh-expressing melanoma cells were sorted 2 days after transduction by BD FACSAria Fusion. Melanoma cells transduced with Cas9-Blasticidin vector were cultured 7 days in Blasticidin (ThermoFisher, Cat# A1113903)-containing media (10 µg/mL), to select Cas9+ cells.
GW CRISPR/Cas9 KO screen
The Cas9-expressing A375 cell line was transduced with HD CRISPR library sub-library A68 MOI of 0.3 or lower to ensure that the large majority of cells receive only one sgRNA for gene editing. On average 500 cells per sgRNA were infected to achieve enough coverage for subsequent statistical analysis. Two days postinfection, transduced cells were selected using 2.5 µg/mL of Puromycin (SigmaAldrich, Cat.; P9620)—ontaining media (2.5 µg/mL). Selected melanoma cells were cultured for 24 hours in RPMI media with different amounts of NK cells expanded for 7 days in complete SCGM media with 400 U/mL of IL-2, to determine the ratio for the screen reaching 20%, 50% or 80% melanoma killing. For the screen co-culture, NK cells and melanoma cells were co-cultured at selected E:T ratios (0.5:1 for 25% killing, 1:1 for 50% killing, 1.5:1 for 75% killing) for 24 hours. Each condition consisted of 50 million melanoma cells to ensure that each sgRNA is represented by at least 500 cells. After 24 hours, NK cells were removed by washing with PBS. The adherent melanoma cells were detached, washed, and seeded in fresh media for 1 day to recover. Afterwards, cells were used for DNA isolation. Genomic DNA was isolated using QIAGEN Blood & Cell Culture DNA Maxi Kit (10) (QIAGEN, Cat# 13362) according to the manufacturer’s protocol. DNA concentrations in all subsequent steps were determined using TECAN Spark or Qubit dsDNA HS and BR Assay Kits (ThermoFisher, Cat# Q32851, Q32850). The sgRNA cassette was amplified using 100 ug of genomic DNA and indexed with unique primers for each sample. Quality and quantity of the amplified PCR products were determined by Agilent 2100 Bioanalyzer. DNA concentrations from all experimental conditions were adjusted and pooled at a 1:1 ratio. Sequencing was performed with Illumina NextSeq 500/550 High Output Kit v2.5 (75 Cycles) to read the 20-nt sgRNA sequence and quantify the number of copies. Absolute sgRNA read-counts were collected and demultiplexed using MAGeCK package.76 To process the raw data, read counts were first normalized and log-transformed. Fold changes between conditions were determined by subtracting the log-normalized read-counts of the control samples from the corresponding treated sample. The significance was calculated by a Wilcoxon rank sum test on the average sgRNA fold change of a gene and a defined set of the non-targeting controls. Replicates were collapsed by arithmetic mean for each gene and each sgRNA. P values were corrected for multiple testing by Benjamini Hochberg correction. A false discovery rate cut-off of 0.05 was applied for hit selection. The hit-list was further filtered by eliminating A375 “core-essential genes”, defined as genes which, when targeted by targeted by sgRNAs, altered cell viability and/or proliferation.
Generation of melanoma KO cell lines
To target candidate genes of interest, individual sgRNAs were designed using the https://design.synthego.com and cloned into the BfuA1-cleaved HDCRISPRv1 sgRNA-puromycin expression vector, as previously described.77 SgRNA-expressing vectors were transfected into the Cas9-expressing melanoma cell lines using jetOPTIMUS (Polyplus, Cat# 117-15). After 24 hours, cells were treated with Puromycin (SigmaAldrich, Cat.; P9620) containing media (2.5 µg/mL) for 2 days. Expanded cells deficient in expression of protein encoded by targeted genes were sorted using BD FACSAria Fusion cell sorter. Following sgRNA were used (5’ to 3’): CCTGCCTGGGAACAACCGGA (ICAM-1), CAGTAAGTCAACTTCAATGT (B2M), GTGAATCTGCGGACGCTGCG (HLA-E), GATCTGAGCCGCCGTGTCCG (HLA-C). GCTGTCGAACCTCACGAACT (HLA-B), GGGTCCGGAGTATTGGGACG (HLA-A, -B, -C).
Generation of IFNγ-deficient NK cells
NK cells were transfected by electroporation (Neon Transfection System device, Thermo Fisher) with Cas9 2NLS protein (Synthego) and control (IDT, Cat# 072544) or the crRNAs targeting the IFNG gene (5’ CTTCTTTTACATATGGGTCC 3’), with electroporation parameters as described.78 Electroporated cells were identified by flow cytometry using an Atto-550-coupled tracrRNA (IDT, Cat# 1075928). After transfection, NK cells were kept in antibiotic-free NK cell medium for 24 hours, followed by sorting Atto-550+cells. Sorted NK cells were kept for 48 hours in NK cell medium before further experimental applications.
Flow cytometry
Cells were stained with Zombie Aqua (Biolgened, Cat# 423102) together with fluorochrome-conjugated mAbs for 30 min at RT in the dark. Cells were washed and either analyzed directly on LSRFortessa X-20, or fixed with eBioscience Foxp3 (ThermoFisher, Cat# 00-5523-00) fixation buffer for 1 hour. For intracellular staining, cells were permeabilized, stained with fluorochrome-conjugated mAbs for 30 min at RT, washed, and analyzed by flow cytometry. For sorting, cells were stained with mAbs for 15 min at 4°C in the dark. Cells were washed with PBS and diluted in PBS/EDTA (2 mM) to a concentration of 1–2×106 cells/mL. For analysis of proliferation, freshly isolated NK cells were stained with CellTrace Violet dye (Thermofisher, Cat# C34557), cultured in media containing 400 U/mL of IL-2 and analyzed on day 6 and day 9. Following antibodies were used for flow cytometry: CD107a (BioLegend Cat# 328606, RRID:AB_1186036), CD3 (BioLegend Cat# 317332, RRID:AB_2561943), CD45 (BioLegend Cat# 304022, RRID:AB_493655), CD56 (BioLegend Cat# 362550, RRID:AB_2566059), KIR2DL1 (Biolegend, Cat# 374904, RRID:AB_2832736), KIR2DL2/L3 (BioLegend Cat# 312606, RRID:AB_2130554), KIR2DL4 (BioLegend Cat# 347006, RRID:AB_2130692), KIR2DL5 (BioLegend Cat# 341304, RRID:AB_2130701), KIR3DL1 (BioLegend Cat# 312708, RRID:AB_2249498), KIR3DL2 (R and D Systems Cat# FAB2878P, RRID:AB_2687490), KIR3DL3 (R and D Systems Cat# FAB8919P), NKG2A (Miltenyi Biotec Cat# 130-113-563, RRID:AB_2726170) and NKG2C (Miltenyi Biotec Cat# 130-117-398, RRID:AB_2727933). Antibodies used for tumor cell lines were B7H6 (RND, Cat# FAB7144P), HLA-A1 (Antibodies-online, Cat# ABIN786685), HLA-A2 (BioLegend Cat# 343308, RRID:AB_2561567), HLA-ABC (BioLegend Cat# 311410, RRID:AB_314879), HLA-Bw4 (Miltenyi Biotec, Cat# 130-132-419), HLA-C (BioLegend Cat# 373309, RRID:AB_2894583), HLA-E (BioLegend Cat# 342604, RRID:AB_1659249), HLA-F (BioLegend Cat# 373203, RRID:AB_2650871), HLA-G (BioLegend Cat# 335905, RRID:AB_1227710), ICAM-1 (BioLegend Cat# 322708, RRID:AB_535980). Biotin conjugated mAb were further stained with fluorochrome conjugated Streptavidin (Biolegend, Cat# 405229). NKG2D ligands were stained using NKG2D-Fc fusion chimera protein (RND, Cat# 1299-NK-050).
NK cell degranulation assay
Melanoma cells were pretreated or not with IFNγ (500–1000 U/mL) for o/n or 12 hours before co-culture with NK cells at 1:1 ratio for 4 hours in the presence of a fluorochome-labeled anti-CD107a mAb and protein transport inhibitor GolgiStop (BD, Cat# BDB554724). When indicated, mAb blocking receptor-ligand interaction, were added to the co-culture at a final concentration 5–10 ug/mL. To test potential soluble mediators of NK cell inhibition, melanoma were cells pretreated or not with IFNγ, washed after 24 hours or 48 hours and cultured fresh media for additional 4 hours. Supernatant was collected and used for NK cell co-culture with WT melanoma cells. After 4 hours, cells were harvested, stained and analyzed by flow cytometry.
Confluency and WT/KO ratio co-culture assay
WT and KO melanoma cells were cultured at similar density, then harvested and mixed at a 1:1 ratio. NK cells were added to mixed WT/KO cells at the E:T ratios of 0.25-4:1 and co-cultured for 24 hours. The medium with NK cells and detached melanoma cells was carefully removed. Each well was gently washed and filled with warm media. The cell confluency was measured using a TECAN Spark plate reader. To determine the postculture WT/KO ratio, melanoma cells were harvested, stained and analyzed by flow cytometry. For determining the ration of WT/ HLA-E KO cells, melanoma cells were pretreated with IFNγ for 12 hours at 37°C prior to the analysis.
Statistical analysis
Data were analyzed by using R Studio V.4 (RRID:SCR_001905), GraphPad Prism V.8 (RRID:SCR_002798), MS Excel 2016 (RRID:SCR_016137) and FlowJo V.10 (RRID:SCR_008520). Data are presented as mean with SD.
Acknowledgments
We thank the FlowCore Mannheim facility of the Medical Faculty Mannheim at Heidelberg University, especially Stefanie Uhlig, for her assistance with cell sorting. We thank the next-generation sequencing (NGS) core facility of the Medical Faculty Mannheim at Heidelberg University and of German Cancer Research Center (DKFZ). We thank the State of Baden-Württemberg Foundation special program ‘Angioformatics Single Cell Platform’. We thank Jens Pahl for initial project supervision. We thank Ana Stojanovic and Jennifer Wischhusen for manuscript proofreading.
Footnotes
Contributors: TH designed and performed experiments, analyzed data and wrote the paper. IG-L generated IFNγ KO primary NK cells and helped with NK cell isolations. SWN analyzed CRISPR/Cas9 screen data and provided statistical support. FH demultiplexed and analyzed raw CRISPR/Cas9 screen data. MB provided GW CRISPR/ Cas9 library and sgRNA vector plasmids. AC designed and supervised the study, acquired funding and co-wrote the manuscript. AC is responsible for the overall content as the guarantor.
Funding: The project was supported by a network grant of the European Commission (H2020-MSCA-MC-ITN-765104-MATURE-NK), and the German Research Foundation: SFB1366 (Project number 394046768-SFB 1366; C02 to AC), SPP 1937 (CE 140/2-2 to AC), TRR179 (TP07 to AC), SFB-TRR156 (B10N to AC), and RTG2727 – 445549683 (B1.2 to AC) and RTG 2099 (Project number: 259332240 - RTG2099; P9 to AC) and ExU 6.1.11 (to AC) and by the German Cancer Aid translational oncology program “NK fit against AML” (74114180) (to AC).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplemental information. The data that support this study are available from the corresponding author on reasonable request. The CRISPR/Cas9 KO screen candidate data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through accession number GSE261626 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE261626).
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Written informed consent from the blood donors was obtained and ethical approval 87/04 was granted by the Ethik Kommission II of the Medical Faculty Mannheim (Mannheim, Germany).
References
- 1. Lanier LL. NK cell recognition. Annu Rev Immunol 2005;23:225–74. 10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed] [Google Scholar]
- 2. Esen F, Deniz G, Aktas ECP-1. CTLA-4, LAG-3, and TIGIT: the roles of immune Checkpoint receptors on the regulation of human NK cell phenotype and functions. Immunol Lett 2021;240:15–23. 10.1016/j.imlet.2021.09.009 [DOI] [PubMed] [Google Scholar]
- 3. Björkström NK, Riese P, Heuts F, et al. Expression patterns of Nkg2A, KIR, and Cd57 define a process of Cd56Dim NK-cell differentiation Uncoupled from NK-cell education. Blood 2010;116:3853–64. 10.1182/blood-2010-04-281675 [DOI] [PubMed] [Google Scholar]
- 4. Lopez-Vergès S, Milush JM, Schwartz BS, et al. Expansion of a unique Cd57. Proc Natl Acad Sci U S A 2011;108:14725–32. 10.1073/pnas.1110900108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alspach E, Lussier DM, Schreiber RD. Interferon γ and its important roles in promoting and inhibiting spontaneous and therapeutic cancer immunity. Cold Spring Harb Perspect Biol 2019;11:a028480. 10.1101/cshperspect.a028480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi Z-D, Pang K, Wu Z-X, et al. Tumor cell plasticity in targeted therapy-induced resistance: mechanisms and new strategies. Sig Transduct Target Ther 2023;8:1–21. 10.1038/s41392-023-01383-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou X, Ni Y, Liang X, et al. Mechanisms of tumor resistance to immune Checkpoint blockade and combination strategies to overcome resistance. Front Immunol 2022;13:915094. 10.3389/fimmu.2022.915094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vaddepally RK, Kharel P, Pandey R, et al. Review of indications of FDA-approved immune Checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers (Basel) 2020;12:738. 10.3390/cancers12030738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tawbi HA, Schadendorf D, Lipson EJ, et al. Relatlimab and Nivolumab versus Nivolumab in untreated advanced Melanoma. N Engl J Med 2022;386:24–34. 10.1056/NEJMoa2109970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Hall T, André P, Horowitz A, et al. Monalizumab: inhibiting the novel immune Checkpoint Nkg2A. J Immunother Cancer 2019;7:263. 10.1186/s40425-019-0761-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai L, Li Y, Tan J, et al. Targeting LAG-3, TIM-3, and TIGIT for cancer Immunotherapy. J Hematol Oncol 2023;16:101. 10.1186/s13045-023-01499-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zeng T, Cao Y, Jin T, et al. The Cd112R/Cd112 axis: a breakthrough in cancer Immunotherapy. J Exp Clin Cancer Res 2021;40:285. 10.1186/s13046-021-02053-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Home | ClinicalTrials.gov, Available: https://clinicaltrials.gov/
- 14. Sun Q, Hong Z, Zhang C, et al. Immune Checkpoint therapy for solid tumours: clinical dilemmas and future trends. Sig Transduct Target Ther 2023;8:1–26. 10.1038/s41392-023-01522-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morgado S, Sanchez-Correa B, Casado JG, et al. NK cell recognition and killing of Melanoma cells is controlled by multiple activating receptor-ligand interactions. J Innate Immun 2011;3:365–73. 10.1159/000328505 [DOI] [PubMed] [Google Scholar]
- 16. Shifrut E, Carnevale J, Tobin V, et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 2018;175:1958–71. 10.1016/j.cell.2018.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang D, Prager BC, Gimple RC, et al. CRISPR screening of CAR T cells and cancer stem cells reveals critical dependencies for cell-based therapies. Cancer Discov 2021;11:1192–211. 10.1158/2159-8290.CD-20-1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhuang X, Veltri DP, Long EO. Genome-wide CRISPR screen reveals cancer cell resistance to NK cells induced by NK-derived IFN-γ. Front Immunol 2019;10:2879. 10.3389/fimmu.2019.02879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheffer M, Lowry E, Beelen N, et al. Genome-scale screens identify factors regulating tumor cell responses to natural killer cells. Nat Genet 2021;53:1196–206. 10.1038/s41588-021-00889-w [DOI] [PubMed] [Google Scholar]
- 20. Chiba M, Shimono J, Ishio T, et al. Genome-wide CRISPR screens identify Cd48 defining susceptibility to NK cytotoxicity in peripheral T-cell Lymphomas. Blood 2022;140:1951–63. 10.1182/blood.2022015646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dufva O, Koski J, Maliniemi P, et al. Integrated drug profiling and CRISPR screening identify essential pathways for CAR T-cell cytotoxicity. Blood 2020;135:597–609. 10.1182/blood.2019002121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freeman AJ, Vervoort SJ, Ramsbottom KM, et al. Natural killer cells suppress T cell-associated tumor immune evasion. Cell Rep 2019;28:2784–94. 10.1016/j.celrep.2019.08.017 [DOI] [PubMed] [Google Scholar]
- 23. Bernareggi D, Xie Q, Prager BC, et al. Chmp2A regulates tumor sensitivity to natural killer cell-mediated cytotoxicity. Nat Commun 2022;13:1899. 10.1038/s41467-022-29469-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pech MF, Fong LE, Villalta JE, et al. Systematic identification of cancer cell Vulnerabilities to natural killer cell-mediated immune surveillance. Elife 2019;8:e47362. 10.7554/eLife.47362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patel SJ, Sanjana NE, Kishton RJ, et al. Identification of essential genes for cancer Immunotherapy. Nature 2017;548:537–42. 10.1038/nature23477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dubrot J, Du PP, Lane-Reticker SK, et al. In vivo CRISPR screens reveal the landscape of immune evasion pathways across cancer. Nat Immunol 2022;23:1495–506. 10.1038/s41590-022-01315-x [DOI] [PubMed] [Google Scholar]
- 27. Dufva O, Gandolfi S, Huuhtanen J, et al. Single-cell functional Genomics reveals determinants of sensitivity and resistance to natural killer cells in blood cancers. Immunity 2023;56:2816–35. 10.1016/j.immuni.2023.11.008 [DOI] [PubMed] [Google Scholar]
- 28. Meissner TB, Liu Y-J, Lee K-H, et al. Nlrc5 cooperates with the RFX transcription factor complex to induce MHC class I gene expression. J Immunol 2012;188:4951–8. 10.4049/jimmunol.1103160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Durand B, Sperisen P, Emery P, et al. RFXAP, a novel subunit of the RFX DNA binding complex is Mutated in MHC class II deficiency. EMBO J 1997;16:1045–55. 10.1093/emboj/16.5.1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dersh D, Phelan JD, Gumina ME, et al. Genome-wide screens identify Lineage- and tumor-specific genes Modulating MHC-I- and MHC-II-restricted Immunosurveillance of human Lymphomas. Immunity 2021;54:116–31:S1074-7613(20)30467-2. 10.1016/j.immuni.2020.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou F. Molecular mechanisms of IFN-γ to up-regulate MHC class I antigen processing and presentation. Int Rev Immunol 2009;28:239–60. 10.1080/08830180902978120 [DOI] [PubMed] [Google Scholar]
- 32. Schwinn N, Vokhminova D, Sucker A, et al. Interferon-gamma down-regulates Nkg2D ligand expression and impairs the Nkg2D-mediated cytolysis of MHC class I-deficient Melanoma by natural killer cells. Int J Cancer 2009;124:1594–604. 10.1002/ijc.24098 [DOI] [PubMed] [Google Scholar]
- 33. Salih HR, Rammensee H-G, Steinle A. Cutting edge: down-regulation of MICA on human tumors by proteolytic shedding. J Immunol 2002;169:4098–102. 10.4049/jimmunol.169.8.4098 [DOI] [PubMed] [Google Scholar]
- 34. Braud VM, Allan DS, O’Callaghan CA, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A. Nature 1998;391:795–9. 10.1038/35869 [DOI] [PubMed] [Google Scholar]
- 35. Lee N, Goodlett DR, Ishitani A, et al. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol 1998;160:4951–60. [PubMed] [Google Scholar]
- 36. Braud V, Jones EY, McMichael A. The human major Histocompatibility complex class IB molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur J Immunol 1997;27:1164–9. 10.1002/eji.1830270517 [DOI] [PubMed] [Google Scholar]
- 37. Pfefferle A, Jacobs B, Netskar H, et al. Intra-lineage plasticity and functional Reprogramming maintain natural killer cell repertoire diversity. Cell Rep 2019;29:2284–94. 10.1016/j.celrep.2019.10.058 [DOI] [PubMed] [Google Scholar]
- 38. Ichise H, Tsukamoto S, Hirashima T, et al. Functional visualization of NK cell-mediated killing of metastatic single tumor cells. Elife 2022;11:e76269. 10.7554/eLife.76269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. André P, Denis C, Soulas C, et al. Anti-Nkg2A mAb is a Checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell 2018;175:1731–43. 10.1016/j.cell.2018.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mamessier E, Sylvain A, Thibult M-L, et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest 2011;121:3609–22. 10.1172/JCI45816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sheu B-C, Chiou S-H, Lin H-H, et al. Up-regulation of inhibitory natural killer receptors Cd94/Nkg2A with suppressed intracellular Perforin expression of tumor-infiltrating Cd8+ T lymphocytes in human Cervical carcinoma. Cancer Res 2005;65:2921–9. 10.1158/0008-5472.CAN-04-2108 [DOI] [PubMed] [Google Scholar]
- 42. Katou F, Ohtani H, Watanabe Y, et al. Differing phenotypes between intraepithelial and Stromal lymphocytes in early-stage tongue cancer. Cancer Res 2007;67:11195–201. 10.1158/0008-5472.CAN-07-2637 [DOI] [PubMed] [Google Scholar]
- 43. Sun C, Xu J, Huang Q, et al. High NKG2A expression contributes to NK cell exhaustion and predicts a poor prognosis of patients with liver cancer. Oncoimmunology 2017;6:e1264562. 10.1080/2162402X.2016.1264562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee N, Llano M, Carretero M, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA 1998;95:5199–204. 10.1073/pnas.95.9.5199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Platonova S, Cherfils-Vicini J, Damotte D, et al. Profound coordinated alterations of Intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res 2011;71:5412–22. 10.1158/0008-5472.CAN-10-4179 [DOI] [PubMed] [Google Scholar]
- 46. Borst L, Sluijter M, Sturm G, et al. NKG2A is a late immune Checkpoint on CD8 T cells and marks repeated stimulation and cell division. Int J Cancer 2022;150:688–704. 10.1002/ijc.33859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boudreau JE, Hsu KC. Natural killer cell education in human health and disease. Curr Opin Immunol 2018;50:102–11. 10.1016/j.coi.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fayette J, Lefebvre G, Posner MR, et al. Results of a phase II study evaluating Monalizumab in combination with Cetuximab in previously treated recurrent or metastatic squamous cell carcinoma of the head and neck (R/M SCCHN). Annals of Oncology 2018;29:viii374. 10.1093/annonc/mdy287.005 [DOI] [Google Scholar]
- 49. Colevas DA, Misiukiewicz K, Pearson AT, et al. 123Mo Monalizumab, Cetuximab and Durvalumab in first-line treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (R/M SCCHN): A phase II trial. Annals of Oncology 2021;32:S1432. 10.1016/j.annonc.2021.10.142 [DOI] [Google Scholar]
- 50. Galot R, Le Tourneau C, Saada-Bouzid E, et al. A phase II study of Monalizumab in patients with recurrent/metastatic squamous cell carcinoma of the head and neck: the I1 cohort of the EORTC-HNCG-1559 UPSTREAM trial. Eur J Cancer 2021;158:17–26. 10.1016/j.ejca.2021.09.003 [DOI] [PubMed] [Google Scholar]
- 51. Patel SP, Alonso-Gordoa T, Banerjee S, et al. Phase 1/2 study of Monalizumab plus Durvalumab in patients with advanced solid tumors. J Immunother Cancer 2024;12:e007340. 10.1136/jitc-2023-007340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Herbst RS, Majem M, Barlesi F, et al. COAST: an open-label, phase II, multidrug platform study of Durvalumab alone or in combination with Oleclumab or Monalizumab in patients with Unresectable, stage III non–small-cell lung cancer. J Clin Oncol 2022;40:3383–93. 10.1200/JCO.22.00227 [DOI] [PubMed] [Google Scholar]
- 53. Cohen RB, Lefebvre G, Posner MR, et al. 1134P - Monalizumab in combination with Cetuximab in patients (Pts) with recurrent or metastatic (R/M) head and neck cancer (SCCHN) previously treated or not with PD-(L)1 inhibitors (IO): 1-year survival data. Annals of Oncology 2019;30:v460. 10.1093/annonc/mdz252.026 [DOI] [Google Scholar]
- 54. Hanna GJ, O’Neill A, Shin K-Y, et al. Neoadjuvant and adjuvant Nivolumab and Lirilumab in patients with recurrent, Resectable squamous cell carcinoma of the head and neck. Clin Cancer Res 2022;28:468–78. 10.1158/1078-0432.CCR-21-2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vey N, Dumas P-Y, Recher C, et al. Randomized phase 2 trial of Lirilumab (anti-KIR Monoclonal antibody, mAb) as maintenance treatment in elderly patients (Pts) with acute myeloid leukemia (AML): results of the Effikir trial. Blood 2017;130:889. 10.1182/blood.V130.Suppl_1.889.889 [DOI] [Google Scholar]
- 56. Vey N, Karlin L, Sadot-Lebouvier S, et al. A phase 1 study of Lirilumab (antibody against killer immunoglobulin-like receptor antibody Kir2D; Iph2102) in patients with solid tumors and hematologic malignancies. Oncotarget 2018;9:17675–88. 10.18632/oncotarget.24832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Litwin V, Gumperz J, Parham P, et al. Nkb1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med 1994;180:537–43. 10.1084/jem.180.2.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations associated with acquired resistance to PD-1 blockade in Melanoma. N Engl J Med 2016;375:819–29. 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gao J, Shi LZ, Zhao H, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 2016;167:397–404. 10.1016/j.cell.2016.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sucker A, Zhao F, Real B, et al. Genetic evolution of T-cell resistance in the course of Melanoma progression. Clin Cancer Res 2014;20:6593–604. 10.1158/1078-0432.CCR-14-0567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shin DS, Zaretsky JM, Escuin-Ordinas H, et al. Primary resistance to PD-1 blockade mediated by Jak1/2 mutations. Cancer Discov 2017;7:188–201. 10.1158/2159-8290.CD-16-1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Paschen A, Méndez RM, Jimenez P, et al. Complete loss of HLA class I antigen expression on Melanoma cells: A result of successive mutational events. Int J Cancer 2003;103:759–67. 10.1002/ijc.10906 [DOI] [PubMed] [Google Scholar]
- 63. Kelly JM, Darcy PK, Markby JL, et al. Induction of tumor-specific T cell memory by NK cell–mediated tumor rejection. Nat Immunol 2002;3:83–90. 10.1038/ni746 [DOI] [PubMed] [Google Scholar]
- 64. Zhou B, Basu J, Kazmi HR, et al. Interferon-gamma signaling promotes Melanoma progression and metastasis. Oncogene 2023;42:351–63. 10.1038/s41388-022-02561-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Koebel CM, Vermi W, Swann JB, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature 2007;450:903–7. 10.1038/nature06309 [DOI] [PubMed] [Google Scholar]
- 66. Lo HC, Xu Z, Kim IS, et al. Resistance to natural killer cell Immunosurveillance confers a selective advantage to polyclonal metastasis. Nat Cancer 2020;1:709–22. 10.1038/s43018-020-0068-9 [DOI] [PubMed] [Google Scholar]
- 67. Hanna N, Fidler IJ. Role of natural killer cells in the destruction of circulating tumor emboli. J Natl Cancer Inst 1980;65:801–9. 10.1093/jnci/65.4.801 [DOI] [PubMed] [Google Scholar]
- 68. Barlozzari T, Reynolds CW, Herberman RB. In vivo role of natural killer cells: involvement of large granular lymphocytes in the clearance of tumor cells in anti-Asialo Gm1-treated rats. J Immunol 1983;131:1024–7. [PubMed] [Google Scholar]
- 69. Balsamo M, Vermi W, Parodi M, et al. Melanoma cells become resistant to NK-cell-mediated killing when exposed to NK-cell numbers compatible with NK-cell infiltration in the tumor. Eur J Immunol 2012;42:1833–42. 10.1002/eji.201142179 [DOI] [PubMed] [Google Scholar]
- 70. Balsamo M, Pietra G, Vermi W, et al. Melanoma Immunoediting by NK cells. Oncoimmunology 2012;1:1607–9. 10.4161/onci.21456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kaplan DH, Shankaran V, Dighe AS, et al. Demonstration of an interferon γ-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA 1998;95:7556–61. 10.1073/pnas.95.13.7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kortylewski M, Komyod W, Kauffmann M-E, et al. Interferon-Γ-mediated growth regulation of Melanoma cells: involvement of Stat1-dependent and Stat1-independent signals. J Invest Dermatol 2004;122:414–22. 10.1046/j.0022-202X.2004.22237.x [DOI] [PubMed] [Google Scholar]
- 73. Kotredes KP, Gamero AM. Interferons as Inducers of apoptosis in malignant cells. Journal of Interferon & Cytokine Research 2013;33:162–70. 10.1089/jir.2012.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Geurts VCM, Voorwerk L, Balduzzi S, et al. Unleashing NK- and CD8 T cells by combining Monalizumab and Trastuzumab for metastatic HER2-positive breast cancer: results of the MIMOSA trial. Breast 2023;70:76–81. 10.1016/j.breast.2023.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang L, Meng Y, Feng X, et al. CAR-NK cells for cancer Immunotherapy: from bench to bedside. Biomark Res 2022;10:12. 10.1186/s40364-022-00364-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li W, Xu H, Xiao T, et al. Mageck enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 2014;15:554. 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Henkel L, Rauscher B, Schmitt B, et al. Genome-scale CRISPR screening at high sensitivity with an empirically designed sgRNA library. BMC Biol 2020;18:174. 10.1186/s12915-020-00905-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Riggan L, Hildreth AD, Rolot M, et al. CRISPR-Cas9 Ribonucleoprotein-mediated Genomic editing in mature primary innate immune cells. Cell Rep 2020;31:107651. 10.1016/j.celrep.2020.107651 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2024-009410supp001.pdf (968.8KB, pdf)
Data Availability Statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplemental information. The data that support this study are available from the corresponding author on reasonable request. The CRISPR/Cas9 KO screen candidate data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through accession number GSE261626 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE261626).