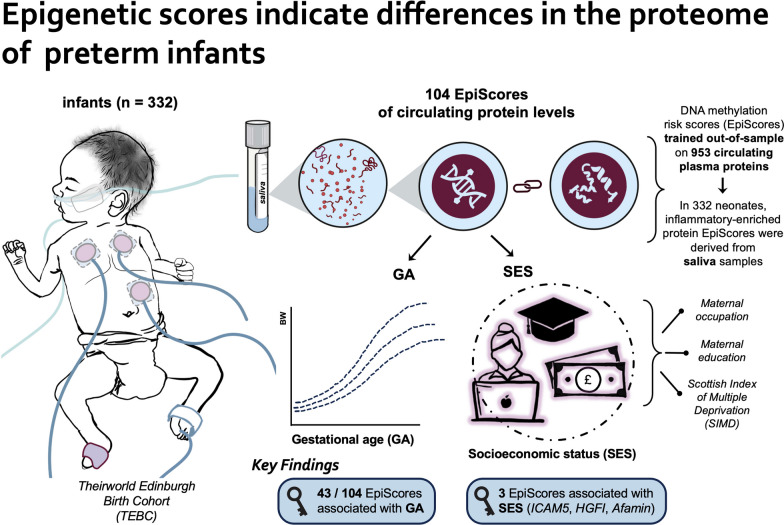
Abstract
Background
Epigenetic scores (EpiScores), reflecting DNA methylation (DNAm)-based surrogates for complex traits, have been developed for multiple circulating proteins. EpiScores for pro-inflammatory proteins, such as C-reactive protein (DNAm CRP), are associated with brain health and cognition in adults and with inflammatory comorbidities of preterm birth in neonates. Social disadvantage can become embedded in child development through inflammation, and deprivation is overrepresented in preterm infants. We tested the hypotheses that preterm birth and socioeconomic status (SES) are associated with alterations in a set of EpiScores enriched for inflammation-associated proteins.
Results
In total, 104 protein EpiScores were derived from saliva samples of 332 neonates born at gestational age (GA) 22.14 to 42.14 weeks. Saliva sampling was between 36.57 and 47.14 weeks. Forty-three (41%) EpiScores were associated with low GA at birth (standardised estimates |0.14 to 0.88|, Bonferroni-adjusted p-value < 8.3 × 10−3). These included EpiScores for chemokines, growth factors, proteins involved in neurogenesis and vascular development, cell membrane proteins and receptors, and other immune proteins. Three EpiScores were associated with SES, or the interaction between birth GA and SES: afamin, intercellular adhesion molecule 5, and hepatocyte growth factor-like protein (standardised estimates |0.06 to 0.13|, Bonferroni-adjusted p-value < 8.3 × 10−3). In a preterm subgroup (n = 217, median [range] GA 29.29 weeks [22.14 to 33.0 weeks]), SES–EpiScore associations did not remain statistically significant after adjustment for sepsis, bronchopulmonary dysplasia, necrotising enterocolitis, and histological chorioamnionitis.
Conclusions
Low birth GA is substantially associated with a set of EpiScores. The set was enriched for inflammatory proteins, providing new insights into immune dysregulation in preterm infants. SES had fewer associations with EpiScores; these tended to have small effect sizes and were not statistically significant after adjusting for inflammatory comorbidities. This suggests that inflammation is unlikely to be the primary axis through which SES becomes embedded in the development of preterm infants in the neonatal period.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-024-01701-2.
Keywords: Epigenetic, Inflammation, Neonatal, Preterm birth, Socioeconomic status
Background
Preterm birth (delivery <37 weeks' gestation) affects around 10% of births worldwide and is closely associated with increased likelihood of cerebral palsy, neurocognitive impairment, behavioural, social and communication difficulties, and mental and cardiometabolic health diagnoses across the life course [1–5]. These adverse outcomes can be explained, in part, by deleterious effects of early exposure to extrauterine life on brain and cardiac development, and they are often accompanied by changes in blood proteins, including those reflecting the perinatal innate and adaptive immune response [6–8].
Socioeconomic status (SES) is also associated with the adverse neurodevelopmental and health outcomes listed above [9–12], and social deprivation is consistently overrepresented among preterm children and their families [13, 14]. In a meta-analysis of 43 studies (n = 111,156 individuals), low SES associated with increased inflammatory markers of disease risk (C-reactive protein [CRP] and interleukin-6 [IL6]), which suggests that pro-inflammatory pathways may be important mechanisms for translating social inequalities into health disparities [15]. However, only four studies included participants under 10 years of age, leaving uncertainty about SES-inflammation correlations in early life [16–19].
Although protein levels are commonly used as biomarkers of exposure and disease risk, their use is limited because they are often phasic in the systemic circulation, rely on venepuncture, and may not capture baseline status or chronicity. For example, inflammation is often measured using acute-phase inflammatory proteins such as CRP [20, 21], but it is not always reliable [22], particularly in neonates, and a single-time-point measure may not reflect baseline inflammation or capture chronic inflammation [23]. These challenges have been addressed by the development of DNA methylation (DNAm) markers of protein expression (EpiScores), which are derived from a linear weighted sum of DNAm sites that are correlated with protein levels. Several EpiScores are associated with magnetic resonance imaging (MRI) measures of brain health, cognition, child mental health, stroke, ischaemic heart disease, Alzheimer’s disease, and lung cancer [24–31]. In neonates, DNAm CRP is associated with birth gestational age (GA), perinatal inflammatory processes, and MRI features of encephalopathy of prematurity [32]. Childhood SES is associated with differential DNAm in inflammation-related genes [33, 34] and at CpG sites that correlate with an inflammation index [35]. Adult SES and social mobility are associated with variations in DNAm in inflammation-related genes [33, 36]. Importantly, SES-related DNAm variations are associated with differences in gene expression so may have functional consequences [33, 36].
Several maternal factors are associated with DNAm in term infants sampled soon after birth, including maternal smoking [37], diabetes [38, 39], obesity [40, 41], and mode of delivery [42, 43]. It is unknown whether these associations apply in preterm infants, who have a curtailed in utero exposure, and are sampled after prolonged exposure to neonatal intensive care, which is known to have widespread effects on the methylome [44].
We investigated relationships between preterm birth, SES, and 104 EpiScores enriched for inflammation-related proteins [26–28, 31, 45]. We tested the following hypotheses: First, low GA is associated with differences in EpiScores; and second, SES is correlated with EpiScores, and interacts with birth GA, but the relationship is attenuated by inflammatory disease burden in preterm infants.
Methods
Participants
Participants were preterm infants (born 33-weeks' gestation) and term-born infants born at the Royal Infirmary of Edinburgh, UK. These infants were recruited to a longitudinal cohort study designed to investigate the effect of preterm birth on brain development and outcomes with multimodal data collection [46]. Infants were recruited between February 2012 and December 2021.
Exclusion criteria were congenital malformation, chromosomal abnormality, congenital infection, cystic periventricular leukomalacia, haemorrhagic parenchymal infarction, and post-haemorrhagic ventricular dilatation. These criteria mean the cohort is representative of the majority of survivors of modern intensive care practices [46].
Final participants included were 217 preterm infants (born 33-weeks' gestation) and 115 term-born infants, with median birth GA of 29.29 and 39.71 weeks, respectively. Their demographic characteristics are shown in Table 1. The three SES measures (Scottish Index of Multiple Deprivation (SIMD 2016) [47], maternal education, and maternal occupation) differed between the preterm and term groups (Cohen’s d effect sizes 0.52–0.68). Ethnicity did not differ between groups and is representative of the Edinburgh area [48].
Table 1.
Participant characteristics
| Demographic measure | Preterm (n = 217) | Term (n = 115) | |
|---|---|---|---|
| Sex | Male | 114/217 (52.5%) | 64/115 (55.7%) |
| Female | 103/217 (47.5%) | 51/115 (44.3%) | |
| Birth GA (weeks)—median (range) | 29.29 (22.14–33.0) | 39.71 (37.0–42.14) | |
| Birthweight (g)—median (range) | 1200 (370–2510) | 3450 (2346–4670) | |
| Birthweight z-score—median (range) | 0.10 (-3.13–2.07) | 0.43 (-2.3–2.96) | |
| SIMD rank—median (range)A | 3720 (6–6966) | 5344 (267–6967) | |
| Maternal ethnicity | African | 1/217 (0.5%) | 0/115 (0%) |
| Bangladeshi | 0/217 (0%) | 1/115 (0.9%) | |
| Caribbean | 0/217 (0%) | 0/115 (0%) | |
| Chinese | 0/217 (0%) | 1/115 (0.9%) | |
| Indian | 3/217 (2.4%) | 1/115 (0.9%) | |
| Pakistani | 4/214 (1.8%) | 1/115 (0.9%) | |
| White | 195/217 (89.9%) | 106/115 (92.2%) | |
| White/Asian | 2/217 (0.9%) | 0/115 (0%) | |
| White/Black African | 0/217 (0%) | 1/115 (0.9%) | |
| White/Black Caribbean | 1/217 (0.5%) | 1/115 (0.9%) | |
| Other AsianB | 1/217 (0.5%) | 2/115 (1.7%) | |
| Other ethnic groupC | 6/217 (2.8%) | 0/115 (0%) | |
| Other mixed ethnic backgroundD | 4/217 (1.8%) | 1/115 (0.9%) | |
| Maternal education | None | 7/208 (3.4%) | 0/115 (0%) |
| Basic high school qualification (1–4) | 5/208 (2.4%) | 2/115 (1.7%) | |
| Basic high school qualification (≥5) | 8/208 (3.8%) | 1/115 (0.9%) | |
| Advanced high school qualification | 32/208 (15.4%) | 3/115 (2.6%) | |
| College qualification | 46/208 (22.1%) | 8/115 (7.0%) | |
| University undergraduate | 61/208 (29.3%) | 50/115 (43.5%) | |
| University postgraduate | 49/208 (23.6%) | 51/115 (44.3%) | |
| Maternal occupation | Unemployed | 12/213 (5.6%) | 1/115 (0.9%) |
| Homemaker | 10/213 (4.7%) | 1/115 (0.9%) | |
| Still in full time education | 9/213 (4.2%) | 1/115 (0.9%) | |
| Sheltered employment | 1/213 (0.5%) | 0/115 (0%) | |
| Unskilled | 11/213 (5.2%) | 3/115 (2.6%) | |
| Partly skilled | 7/213 (3.3%) | 2/115 (1.7%) | |
| Manual skilled | 25/213 (11.7%) | 9/115 (7.8%) | |
| Non-manual skilled | 44/213 (20.7%) | 11/115 (9.6%) | |
| Professional | 94/213 (44.1%) | 87/115 (75.7%) | |
| Sample GA (weeks)—median (range) | 40.57 (36.57–45.86) | 42.0 (39.86–47.14) | |
| Batch | 1 | 92/217 (42.4%) | 46/115 (40.0%) |
| 2 | 62/217 (28.6%) | 56/115 (48.7%) | |
| 3 | 32/217 (14.7%) | 1/115 (0.9%) | |
| 4 | 31/217 (14.3%) | 12/115 (10.4%) | |
| Maternal smoking | 41/214 (19.2%) | 3/115 (2.6%) | |
| Maternal diabetes | 12/217 (5.5%) | 6/115 (5.2%) | |
| Maternal obesity | 44/212 (20.8%) | 19/114 (16.7%) | |
| Mode of delivery | Vaginal delivery | 72/217 (33.2%) | 43/115 (37.4%) |
| Instrumental delivery | 4/217 (1.8%) | 17/115 (14.8%) | |
| Caesarean delivery | 141/217 (65.0%) | 55/115 (47.8%) | |
A Preterm n = 216, term n = 115, B “Other Asian” includes Sri Lankan (n = 1), Malay (n = 1), C “Other ethnic group” includes Arab (n = 1), Iraqi (n = 1), white Bulgarian (n = 1), Fijian (n = 1), Japanese (n = 1), Hong Kong (n = 1), D “Other mixed ethnic background” includes Pakistani/Scottish (n = 2), British/Arab (n = 1), Sri Lankan/Black (n = 1), Sri Lankan/Indian (n = 1)
GA Gestational age, SIMD Scottish index of multiple deprivation
DNA methylation
Saliva samples for DNAm were collected at term equivalent age using Oragene OG-575 Assisted Collection kits (DNA Genotek, ON, Canada), and DNA was extracted using prepIT.L2P reagent (DNA Genotek, ON, Canada). Saliva sampling was used due to accessibility and the non-invasiveness of the method; DNAm patterns measured via saliva samples correlate with brain and other tissue DNAm patterns [49, 50]. We chose to sample at the term equivalent gestation time point to include the allostatic load of both prenatal and early postnatal exposures.
DNA was bisulphite converted and methylation levels were measured using Illumina HumanMethylationEPIC BeadChip (Illumina, San Diego, CA, USA) at the Edinburgh Clinical Research Facility (Edinburgh, UK). The arrays were imaged on the Illumina iScan or HiScan platform, and genotypes were called automatically using GenomeStudio Analysis software version 2011.1 (Illumina). DNAm was processed in four batches.
Raw intensity (.idat) files were read into the R environment using minfi. wateRmelon and minfi were used for preprocessing, quality control, and normalisation [51]. The pfilter function in wateRmelon was used to exclude samples with 1% of sites with a detection p-value > 0.05, sites with beadcount < 3 in 5% of samples, and sites with 1% of samples with detection p-value > 0.05. Cross-hybridising probes, probes targeting single-nucleotide polymorphisms with overall minor allele frequency ≥ 0.05, and control probes were also removed. Samples were removed if there was a mismatch between predicted sex (minfi) and recorded sex (n = 3), or if samples did not meet preprocessing quality control criteria (n = 29). Data were danet normalised, which includes background correction and dye bias correction [51]. Saliva contains different cells types, including buccal epithelial cells. Epithelial cell proportions were estimated with epigenetic dissection of intra-sample heterogeneity with the reduced partial correlation method implemented in the R package EpiDISH [52]. Probes located on sex chromosomes were removed before analysis. The cohort includes twins (n = 32); these were randomly removed leaving one participant per twin pair. This left a final sample size of n = 332.
EpiScore calculation
The 104 protein EpiScores included 100 EpiScores from Gadd et al. [26], a study enriched for inflammatory-related proteins, excluding those where the required CpGs were not available, owing to differences in assay platform CpG coverage relative to Gadd et al. [26]. For duplicate proteins, those developed using Olink platform-identified proteins (antibody-based assays) were prioritised over those from SOMAscan platforms (aptamer-based assays), due to specificity and reproducibility, and the variable correlation between the two methods [53–56] (for details see Supplementary eMethods, Additional File 1). In addition, we included EpiScores for IL6 [28], growth and differentiation factor 15 (GDF15) and N-terminal-pro B-type natriuretic peptide (NTproBNP) [45], and CRP. The CRP EpiScore used was Barker et al.’s seven-CpG variation of Ligthart et al.’s CRP EpiScore [27, 31], as this is known to correlate with birth GA, perinatal pro-inflammatory exposures, and neonatal brain development [32].
For each individual, EpiScores were obtained by multiplying the methylation proportion at a given CpG by the effect size from previous studies. This was performed using the MethylDetectR platform [57] for those inflammatory proteins currently included and using R for those not currently included (CRP, GDF15, IL6, NTproBNP). All CpG sites and coefficients required to calculate the 104 EpiScores are in Supplementary Table 1 (Additional File 2).
Statistics
The predictor variables were SES and birth GA. SES was operationalised in three ways: neighbourhood-level SES using the Scottish Index of Multiple Deprivation (SIMD) [47], and two measures of family-level SES, which were maternal education (highest educational qualification) and maternal occupation (current or most recent occupation). For further details, see Supplementary eMethods, Additional File 1. Birth GA was a continuous variable to maximise statistical power [58, 59]. We adjusted for GA at saliva sampling, DNAm batch, infant sex, and birthweight z-score.
All statistical analyses were performed in R (version 4.3.1) and were preregistered [60].
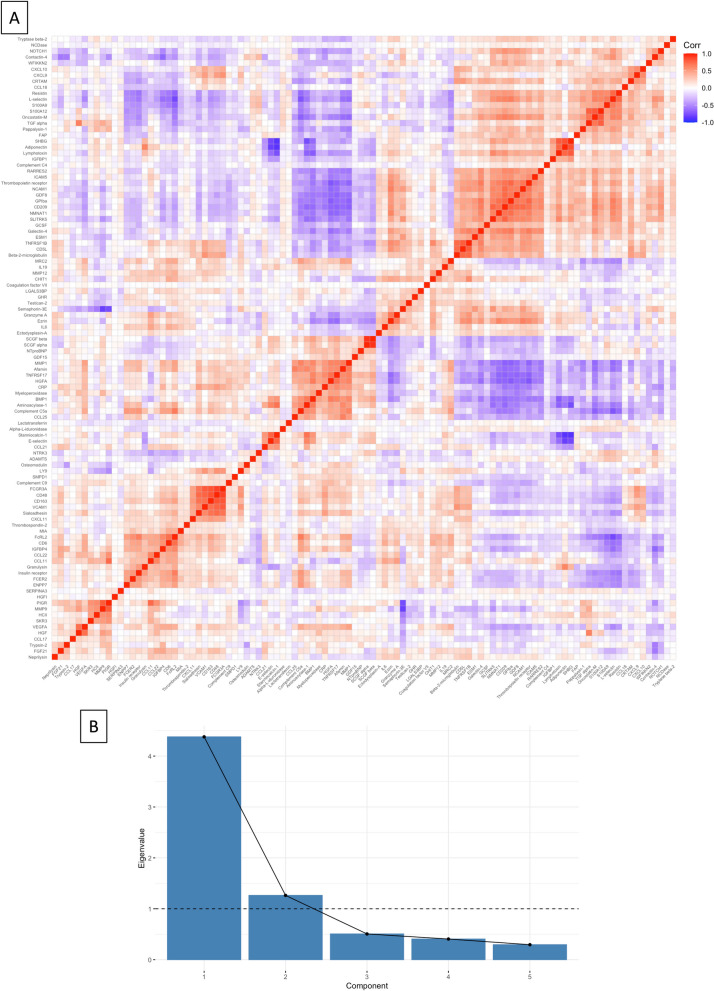
Principal component analysis (PCA) was used to determine the significance threshold for controlling type 1 error in analyses of multiple EpiScores [61]. We began with a correlation analysis, which showed correlation coefficients between EpiScores of |0.01 to 0.93| (Fig. 1A). To determine the number of statistical “families” among the 104 EpiScores, PCA was performed. This yielded two principal components with eigenvalues > 1, our pre-specified threshold, which explained 59.5 and 17.2% of variance, respectively (Fig. 1B). Standardised component loadings are provided in Supplementary Table 2 (Additional File 1). In all subsequent analyses, we corrected for multiple comparisons across EpiScores and SES measures using a Bonferroni-adjusted p-value threshold of 8.3 × 10−3. This is 0.05/(2 × 3), with two reflecting the two principal components for EpiScores and three reflecting the number of SES measures used.
Fig. 1.
Determining significance threshold. Principal component analysis was used to determine the adjusted statistical significance threshold, given multiple statistical comparisons. A A correlation matrix of 104 EpiScores, showing correlation coefficient as red for positive and blue negative associations when significant (p < 0.05). B A scree plot of principal components, with the eigenvalues for each component. Standardised component loadings for principal components one and two are provided in Supplementary Table 3 (Additional File 1)
We constructed general linear regression models for each EpiScore as outcome measure to assess associations between GA, each of the three SES measures (separate models for each of SIMD, maternal education, and maternal occupation), and the product interaction term SES*birth GA (removing the term if not significant), and adjusting for GA at sampling, sex, and batch.
For the preterm subgroup, we additionally adjusted for perinatal inflammatory exposures known to be associated with the CRP EpiScore as, to our knowledge, this is the only DNAm proxy of an inflammatory protein that has been studied in this context [32]. These were histological chorioamnionitis (HCA), sepsis, bronchopulmonary dysplasia (BPD), and necrotising enterocolitis (NEC). Maternal smoking and preeclampsia were not associated with DNAm CRP, so are not included as covariates [32, 44]. For definitions see Supplementary eMethods (Additional File 1) and for frequencies see Supplementary Table 3 (Additional File 1).
For EpiScores with significant associations with GA or SES, we performed a post hoc sensitivity analysis, adjusting for maternal factors that have been associated with neonatal methylome in term infants in prior research: maternal smoking, diabetes, obesity, and mode of delivery. A change of standardised by 20% or change of p-value to 0.05 was considered significant, and we report adjusted R2 values of each model. For definitions, see Supplementary eMethods (Additional File 1), and for frequencies, see Table 1.
Results
Associations between gestational age and EpiScores
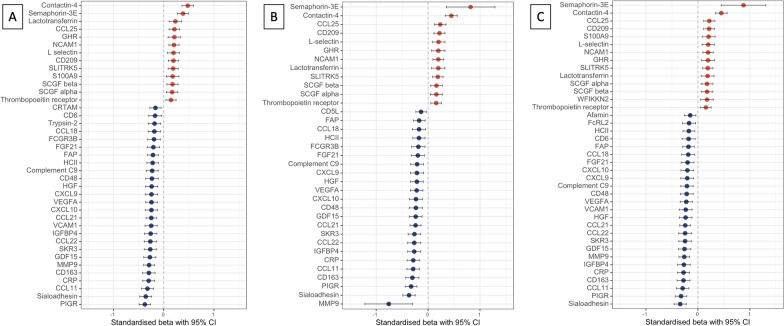
Gestational age associated with 43 of the 104 EpiScores after adjustment for SIMD, maternal education, or maternal occupation (Fig. 2A–C). The proteins represented by the 43 EpiScores are listed in Table 2 categorised by functional annotation adapted from the STRING database [62], and their broader roles in immune processes and inflammation, and the pathogenesis of neonatal diseases, where known, are described in Supplementary Table 4 (Additional File 1).
Fig. 2.
EpiScores associated with gestational age in regression models adjusted for socioeconomic status. EpiScores associated with gestational age in regression models with A Scottish Index of Multiple Deprivation, B maternal education, and C maternal occupation. A (n = 331) shows 39 associations, B (n = 323) shows 35 associations, and C shows (n = 328) shows 39 associations. Points and bars represent standardised beta and 95% confidence intervals, with red indicating positive and blue negative associations. Covariates included in all models: age at sample, birthweight z-score, sex, and methylation processing batch. Bonferroni-adjusted p-value < 8.3 × 10−3 CCL11 C-C chemokine 11, CCL18 C-C chemokine 18, CCL21 C-C chemokine 21, CCL22 C-C chemokine 22, CCL25 C-C chemokine 25, CD5L CD5 antigen-like protein, CD6 T-cell differentiation antigen, CD163 scavenger receptor cysteine-rich type 1 protein M130, CI confidence interval, CRP C-reactive protein, CRTAM cytotoxic and regulatory T-cell molecule, CXCL9 C-X-C motif chemokine 9, CXCL10 C-X-C motif chemokine 10, FAP fibroblast activation protein alpha, FCGR3B low-affinity immunoglobulin gamma Fc region receptor III-B, FcRL2 Fc receptor-like protein 2, FGF21 Fibroblast growth factor 21, GDF15 growth/differentiation factor 15, GHR growth hormone receptor, HCII heparin cofactor II, HGF hepatocyte growth factor alpha chain, IGFBP4 insulin-like growth factor-binding protein 4, MMP9 matrix metalloproteinase-9, NCAM1 neural cell adhesion molecule 1, PIGR polymeric immunoglobulin receptor, SCGF stem cell growth factor, SIMD Scottish Index of Multiple Deprivation, SKR3 serine/threonine protein kinase receptor R3, SLITRK5 SLIT and NTRK-like protein 5, VCAM1 vascular cell adhesion protein 1, VEGFA vascular endothelial growth factor A, WFIKKN2 WAP Kazal immunoglobulin Kunitz and NTR domain-containing protein 2
Table 2.
Protein EpiScores associated with birth gestational age
| Chemokines | Growth factors | Neurogenesis | Vascular development | Cell membrane proteins/receptors | Other immune response |
|---|---|---|---|---|---|
| CCL11 | FGF21 | NCAM1 | SKR3 | Afamin | Complement C9 |
| CCL18 | GDF15 | Semaphorin 3E | VCAM1 | CD5L | CRP |
| CCL21 | GHR | SLITRK5 | VEGFA | CD6 | CRTAM |
| CCL22 | HGF | CD48 | FAP | ||
| CCL25 | IGFBP4 | CD163 | HCII | ||
| CXCL9 | SCGF alpha | CD209 | L-selectin | ||
| CXCL10 | SCGF beta | Contactin-4 | Lactotransferrin | ||
| WFIKKN2 | FcRL2 | MMP9 | |||
| FcGR3B | S100A9 | ||||
| PIGR | Sialoadhesin | ||||
| Thrombopoeitin receptor | Trypsin-2 |
Forty-three EpiScores associated with birth gestational age in regression models adjusted for socioeconomic status. Roles adapted from the STRING database [62]. See Supplementary Table 4, Additional File 1, for further details of the functional roles of each protein, including roles in immunity and inflammation, and in preterm infants specifically
CCL11 C-C chemokine 11, CCL18 C-C chemokine 18, CCL21 C-C chemokine 21, CCL22 C-C chemokine 22, CCL25 C-C chemokine 25, CD5L CD5 antigen-like protein, CD6 T-cell differentiation antigen, CD163 Scavenger receptor cysteine-rich type 1 protein M130, CRP C-reactive protein, CRTAM cytotoxic and regulatory T-cell molecule, CXCL9 C-X-C motif chemokine 9, CXCL10 C-X-C motif chemokine 10, FAP fibroblast activation protein alpha, FCGR3B low-affinity immunoglobulin gamma Fc region receptor III-B, FcRL2 Fc receptor-like protein 2, FGF21 fibroblast growth factor 21, GDF15 growth/differentiation factor 15, GHR growth hormone receptor, HCII heparin cofactor II, HGF hepatocyte growth factor alpha chain, IGFBP4 insulin-like growth factor-binding protein 4, MMP9 matrix metalloproteinase-9, NCAM1 neural cell adhesion molecule 1, PIGR polymeric immunoglobulin receptor, SCGF stem cell growth factor, SIMD Scottish Index of Multiple Deprivation, SKR3 serine/threonine protein kinase receptor R3, SLITRK5 SLIT and NTRK-like protein 5, VCAM1 vascular cell adhesion protein 1, VEGFA vascular endothelial growth factor A, WFIKKN2 WAP Kazal immunoglobulin Kunitz and NTR domain-containing protein 2
Twenty-nine (67%) EpiScores negatively associated with birth GA (standardised estimates |0.14–0.76|, adjusted p-value < 8.3 × 10−3), and 14 (33%) EpiScores positively associated with birth GA (standardised estimates 0.14–0.88, adjusted p-value < 8.3 × 10−3).
Thirty-three EpiScores associated with low GA irrespective of SES measure used in the model. The results for all 104 EpiScores are provided in Supplementary Figs. 1–9 (Additional File 1).
Associations between SES, EpiScores, and the effect of inflammatory comorbidities of preterm birth.
Three out of 104 EpiScores associated with SES measures or the interaction between SES and GA (Fig. 3). There was a small effect size association between higher afamin EpiScore and higher maternal occupation (standardised = 0.06, 95% confidence interval (CI) 0.02–0.11, p = 0.0082), and DNAm afamin associated with the birth GA*maternal education interaction term such that afamin positively correlated with birth GA among babies with mothers without university education, and negatively correlated with birth GA among babies with mothers with university education (undergraduate or postgraduate) (standardised = , 95% CI − 0.20 to − 0.04, p = 0.0041, Supplementary Fig. 10, Additional File 1).
Fig. 3.
EpiScores associated with socioeconomic status or an interaction between socioeconomic status and birth gestational age. EpiScores associated with socioeconomic status (Scottish Index of Multiple Deprivation, maternal education, or maternal occupation), or with an interaction between socioeconomic status and birth gestational age. Regression models with gestational age at birth, gestational age at sample, birthweight z-score, sex, and methylation processing batch. Sample sizes: for the Scottish Index of Multiple Deprivation n = 331, for maternal education n = 323, and for maternal occupation n = 328. 3/104 EpiScores were significant (Bonferroni-adjusted p-value < 8.3 × 10−3). Points and bars represent standardised beta and 95% confidence intervals, with red indicating positive and blue negative associations. CI Confidence interval, GA Gestational age, HGFI Hepatocyte growth factor-like protein alpha chain, ICAM5 Intercellular adhesion molecule 5, SIMD Scottish Index of Multiple Deprivation
Higher intercellular adhesion molecule 5 (ICAM5) EpiScore associated with higher SIMD (standardised = 0.13, 95% CI 0.03–0.23, p = 0.0079). Hepatocyte growth factor-like protein (HGFI) associated with the birth GA*maternal occupation interaction term (standardised = −0.10, 95% CI − 0.16– − 0.04, p = 0.0021, Supplementary Fig. 11, Additional File 1), such that HGFI EpiScore positively correlated with birth GA among babies with mothers who were unemployed, homemakers, in full time education, or in unskilled or manual occupations, but negatively correlated with birth GA among babies with mothers in partly skilled, non-manual skilled or professional occupations.
In the planned analysis of preterm-born babies only, when controlling for inflammatory exposures (sepsis, HCA, NEC, and BPD), a greater proportion of R2 was explained (adjusted R2 = 0.031–0.262 in unadjusted models, adjusted R2 = 0.047–0.392 in adjusted models), but the EpiScores no longer met our statistical threshold (p-values > 0.045 with adjusted p-value threshold < 8.3 × 10−3, see Supplementary Table 5, Additional File 1).
Sensitivity analyses
There were few changes to the significant associations between 43 EpiScores and GA, when models were adjusted for maternal smoking, diabetes, obesity, and mode of delivery (see Supplementary Table 6, Additional file 1). There were minor changes in proportion of R2 explained (change in adjusted R2 =|0.0001–0.051|) and change in standardised was by 0–12%. All adjusted models retained the threshold of p < 0.05, although 4/43 (6.2%) EpiScores no longer met the adjusted p-value threshold < 8.3 × 10−3.
For associations between EpiScores and SES or an interaction between SES and birth GA, there was change in adjusted R2 = |0.002–0.004| and in standardised by 6–11% (see Supplementary Table 6, Additional file 1). All adjusted models retained p < 0.05, although 3/4 models no longer met the adjusted p-value threshold < 8.3 × 10−3.
Discussion
In this study, we identified several associations between a set of EpiScores enriched for inflammatory proteins and low GA at birth. Few EpiScores associated with SES within the whole sample and these associations were partially attenuated in preterm infants who experienced inflammatory comorbidities. This is the first study to assess the impact of preterm birth and social status using epigenetic signatures designed to reflect the circulating proteome.
Associations between birth GA and EpiScores
43 EpiScores associated with preterm birth when sampled at term equivalent age. The EpiScores reflect chemokines, growth factors, proteins required for neurogenesis and vascular development, cell membrane proteins and receptors, and immune response proteins (Table 2). As well as having specific immunoregulatory roles, in the neonatal setting or relevant animal models, these proteins are associated with several comorbidities and developmental consequences of preterm birth. These include lung development and disease such as BPD [63–75], in utero and postnatal growth failure [76–79], HCA [7, 80, 81], patent ductus arteriosus [82–85], retinopathy of prematurity [86–90], NEC [91–93], hyperglycaemia [94], sepsis [93, 95–98], brain injury [32, 99–102], and neurodevelopmental outcomes [103–107].
Associations between SES measures and EpiScores
SES appears to play a much smaller role in the patterning of EpiScores compared to birth GA. SES measures, or interactions between birth GA and SES measures, were associated with only three of the 104 EpiScores studied: afamin, ICAM5, and HGFI. Afamin and ICAM5 positively associated with maternal occupation and SIMD, respectively. Afamin and HGFI both associated with the interaction term between SES and birth GA. Afamin is involved in vitamin E transport [108], ICAM5 has a role in microglial regulation [109], and HGFI is a macrophage-stimulating protein [110]. The relationships between these proteins and SES have not previously been investigated, although afamin is associated with the development of metabolic syndrome [111], which varies with SES [112].
Among preterm infants, no SES-EpiScore associations survived adjustment for inflammatory exposures, which suggests that the weak effects of SES on the neonatal proteome that we observed in a small number of EpiScores are at least partially accounted for by inflammatory pathologies in early life. Taken together, the results suggest immune dysregulation, proxied by EpiScores, may not be the primary axis through which SES becomes embedded in the development of preterm infants during neonatal intensive care.
SES has been consistently associated with inflammation in adulthood, including in relation to childhood deprivation [15, 113], but less is known about the relationship between SES and inflammation in the neonatal period. A longitudinal study by Leviton et al. [114], with five sampling time points during the first month of life after preterm birth, showed that maternal eligibility for Medicaid associated with levels of 14 inflammatory proteins (IL6R, TNFR1, TNFR2, IL8, ICAM1, VCAM1, TSH, EPO, bFGF, IGF1, VEGF, PIGF, Ang-1, Ang-2). However, only three were significant at more than one time point during the month after preterm birth (TSH, bFGF, Ang-1), and none was associated at all five measurement time points. These studies, taken together with our results, suggest that the impact of SES on immune regulation is relatively modest and inconsistent in the newborn period but accrues through to adulthood. Further research is required to understand how and when SES becomes embedded in child development and whether early life events such as preterm birth modify that process; EpiScores could be a powerful tool for investigating the temporal dynamics of social determinants of child health.
Sensitivity analyses
Post hoc analyses showed potential small effect of maternal variables on the neonatal methylome in this cohort enriched for preterm birth sampled at term equivalent age. This is consistent with previous findings of no association between smoking or preeclampsia and DNAm within this cohort [32]. This may be due to the reduced in utero exposure to maternal factors for preterm infants, or that the samples were taken at term equivalent age, so neonatal unit exposures may outweigh maternal factors.
Strengths and limitations
Strengths of this study include the large sample of term and preterm neonates; to the best of our knowledge, this is the first examination of multiple DNA methylation-based estimators of circulating proteins in a neonatal sample. We derived EpiScores from minimally invasive sampling (buccal cells from saliva) which overcomes the ethical challenge of venepuncture for research in children. The EpiScores, serving as proxies of inflammatory proteins and sampled at term equivalent age in preterm infants, were selected for their potential to capture chronic, cumulative inflammation associated with preterm birth and neonatal intensive care exposures [23–25]. We adjusted for variables associated with DNAm, and additionally for inflammatory exposures to increase the clinical validity of our results.
The study has some limitations. The EpiScores used were developed in adult cohorts [26–28, 45] and have not been validated with neonatal protein levels. A validation study would be challenging because the phasic nature of circulating proteins and maturational variation in protein expression would require serial venepuncture, which presents ethical and practical barriers in preterm infants. Of note, we have previously established that neonatal DNAm CRP scores correlate with cumulative clinical inflammatory exposures, which is corroborative evidence that the score developed in adults is relevant in neonates [32]. The 104 EpiScores we tested explain 1–58% of variance of protein levels [26–28, 45]. However, even those that capture a relatively low proportion of the variance associate with incident diseases such as cardiovascular disease, type 2 diabetes, cognitive function and brain health [26, 115–117]. This magnitude of variance explained is also comparable to that achieved with polygenic risk scores, which have proved useful in risk stratification [118–120]. The EpiScores were also trained using blood samples [26–28, 45], whereas we have projected these scores into saliva samples. However, previous studies have successfully used similar cross-tissue techniques [32, 121, 122], and in neonates saliva provides a noninvasive and accessible sample method. Not all inflammatory-related proteins are represented, as we were limited by available EpiScores, so we may have underestimated the full complexity of the relationship between birth GA, SES, and inflammation. Longitudinal investigations are imperative for elucidating whether the DNA methylation signatures associated with gestational age identified in this study exert a causal influence on the inflammation-associated mechanisms in preterm birth. It remains crucial to discern whether these signatures represent a direct downstream consequence of GA itself or are induced by specific factors correlated with GA, yet not necessarily driven by chronic inflammation. Mendelian randomisation studies, integrating genomic and epigenomic determinants, are a promising methodological approach to disentangle the directionality of these intricate relationships. The sensitivity analyses suggested a potential weak effect of maternal exposures on associations between GA and EpiScores at term equivalent age. However, the prevalence of some of the maternal factors was low, for example 18/332 (5.4%) mothers had diabetes. Therefore, larger sample sizes enriched for the variable of interest, with longitudinal sampling from birth, would be required to investigate the relative contributions of antenatal/intrapartum versus postnatal events on the associations we observed.
The study population is comparable to other neonatal populations in high-income, majority white settings, but these results may not generalise to settings with different socioeconomic or ethnicity profiles. We studied several measures of SES but were not able to include all that could be relevant, such as household income.
Conclusion
We identified 43 EpiScores enriched for inflammatory proteins that associated with low birth GA. These 43 proteins offer novel insights into the physiological response to preterm birth and warrant further study to explore their role in the relationship between preterm birth, inflammation, and longer-term outcomes. We found only three EpiScores associated with SES in the neonatal period, none of which survived adjustment for perinatal pro-inflammatory exposures, suggesting that inflammation is unlikely to be the primary axis through which SES becomes embedded in the development of preterm infants in the neonatal period.
Supplementary Information
Acknowledgements
The authors are grateful to the families who consented to take part in the study.
Author contributions
KM was responsible for conceptualisation, methodology, formal analysis, writing (original draft), writing (review/editing), and visualization. ELSC and JB took part in formal analysis and writing (review/editing). KV participated in formal analysis, writing (review/editing), and visualization. RH and DG took part in methodology, formal analysis, and writing (review/editing). GS contributed to investigation and writing (review/editing). AJS was involved in methodology and writing (review/editing). AC participated in investigation, resources, data curation, and writing (review/editing). LM helped with investigation, resources, and writing (review/editing). HCW assisted with methodology and writing (review/editing). HR took part in conceptualisation, writing (original draft), writing (review/editing), and supervision. REM and SRC participated in conceptualisation, methodology, writing (original draft), writing (review/editing), and supervision. JPB was responsible for conceptualisation, methodology, writing (original draft), writing (review/editing), supervision, project administration, and funding acquisition.
Funding
This work was supported by Theirworld (www.theirworld.org) and a UKRI MRC Programme Grant MR/X003434/1. KM receives salary from NHS Scotland. GS is funded by an MRC Clinician Scientist Fellowship MR/X019535/1. SRC is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (221890/Z/20/Z).
Availability of data and materials
DNA methylation data are available to researchers subject to the terms of the Data Access Policy: https://www.ed.ac.uk/centre-reproductive-health/tebc/about-tebc/for-researchers/data-access-collaboration.
Declarations
Ethics approval and consent to participate
Ethical approval for the study was obtained from the National Research Ethics Service, South-East Scotland Research Ethics Committee (REC 11/55/0061, 13/SS/0143, 16/SS/0154), and NHS Lothian Research and Development (2016/0255). Written informed consent was obtained from parents.
Consent for publication
Not applicable.
Competing interests
LM has received speaker and consultancy fees from Illumina. REM is a scientific advisor to the Epigenetic Clock Development Foundation and to Optima Partners. All other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ohuma EO, Moller A-B, Bradley E, Chakwera S, Hussain-Alkhateeb L, Lewin A, et al. National, regional, and global estimates of preterm birth in 2020, with trends from 2010: a systematic analysis. Lancet. 2023;402:1261–1271. doi: 10.1016/S0140-6736(23)00878-4. [DOI] [PubMed] [Google Scholar]
- 2.Johnson S, Marlow N. Early and long-term outcome of infants born extremely preterm. Arch Dis Child. 2017;102:97–102. doi: 10.1136/archdischild-2015-309581. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal S, Rao SC, Bulsara MK, Patole SK. Prevalence of autism spectrum disorder in preterm infants: a meta-analysis. Pediatrics. 2018;142:1–14. doi: 10.1542/peds.2018-0134. [DOI] [PubMed] [Google Scholar]
- 4.Twilhaar ES, Wade RM, deKieviet JF, vanGoudoever JB, vanElburg RM, Oosterlaan J. Cognitive outcomes of children born extremely or very preterm since the 1990s and associated risk factors: a meta-analysis and meta-regression. Jama Pediatr. 2018;172:361. doi: 10.1001/jamapediatrics.2017.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump C. An overview of adult health outcomes after preterm birth. Early Hum Dev. 2020;150:105187. doi: 10.1016/j.earlhumdev.2020.105187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagberg H, Mallard C, Ferriero DM, Vannucci SJ, Levison SW, Vexler ZS, et al. The role of inflammation in perinatal brain injury. Nat Rev Neurol. 2015;11:192–208. doi: 10.1038/nrneurol.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan G, Galdi P, Blesa-Cábez M, Borbye-Lorenzen N, Stoye DQ, Lamb GJ, et al. Interleukin-8 dysregulation is implicated in brain dysmaturation following preterm birth. Brain Behav Immun. 2020;90:311–318. doi: 10.1016/j.bbi.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Raisi-Estabragh Z, Cooper J, Bethell MS, McCracken C, Lewandowski AJ, Leeson P, et al. Lower birth weight is linked to poorer cardiovascular health in middle-aged population-based adults. Heart. 2023;109:535–541. doi: 10.1136/heartjnl-2022-321733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farah MJ. The neuroscience of socioeconomic status: correlates, causes, and consequences. Neuron. 2017;96:56–71. doi: 10.1016/j.neuron.2017.08.034. [DOI] [PubMed] [Google Scholar]
- 10.Kivimäki M, Batty GD, Pentti J, Shipley MJ, Sipilä PN, Nyberg ST, et al. Association between socioeconomic status and the development of mental and physical health conditions in adulthood: a multi-cohort study. Lancet Public Heal. 2020;5:e140–e149. doi: 10.1016/S2468-2667(19)30248-8. [DOI] [PubMed] [Google Scholar]
- 11.Duncan GJ, Ziol-Guest KM, Kalil A. Early-childhood poverty and adult attainment, behavior, and health. Child Dev. 2010;81:306–325. doi: 10.1111/j.1467-8624.2009.01396.x. [DOI] [PubMed] [Google Scholar]
- 12.Pillas D, Marmot M, Naicker K, Goldblatt P, Morrison J, Pikhart H. Social inequalities in early childhood health and development: a European-wide systematic review. Pediatr Res. 2014;76:418–424. doi: 10.1038/pr.2014.122. [DOI] [PubMed] [Google Scholar]
- 13.Thomson K, Moffat M, Arisa O, Jesurasa A, Richmond C, Odeniyi A, et al. Socioeconomic inequalities and adverse pregnancy outcomes in the UK and Republic of Ireland: a systematic review and meta-analysis. BMJ Open. 2021;11:e042753. doi: 10.1136/bmjopen-2020-042753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz M, Goldblatt P, Morrison J, Kukla L, Švancara J, Riitta-Järvelin M, et al. Mother’s education and the risk of preterm and small for gestational age birth: a DRIVERS meta-analysis of 12 European cohorts. J Epidemiol Commun H. 2015;69:826–833. doi: 10.1136/jech-2014-205387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muscatell KA, Brosso SN, Humphreys KL. Socioeconomic status and inflammation: a meta-analysis. Mol Psychiatr. 2020;25:2189–2199. doi: 10.1038/s41380-018-0259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broyles ST, Staiano AE, Drazba KT, Gupta AK, Sothern M, Katzmarzyk PT. Elevated C-reactive protein in children from risky neighborhoods: evidence for a stress pathway linking neighborhoods and inflammation in children. PLoS ONE. 2012;7:e45419. doi: 10.1371/journal.pone.0045419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowd JB, Zajacova A, Aiello AE. Predictors of inflammation in U.S. children Aged 3–16 Years. Am J Prev Med. 2010;39:314–320. doi: 10.1016/j.amepre.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmeer KK, Yoon AJ. Home sweet home? Home physical environment and inflammation in children. Soc Sci Res. 2016;60:236–248. doi: 10.1016/j.ssresearch.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmeer KK, Yoon A. Socioeconomic status inequalities in low-grade inflammation during childhood. Arch Dis Child. 2016;101:1043. doi: 10.1136/archdischild-2016-310837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiesa C, Natale F, Pascone R, Osborn JF, Pacifico L, Bonci E, et al. C reactive protein and procalcitonin: reference intervals for preterm and term newborns during the early neonatal period. Clin Chim Acta. 2011;412:1053–1059. doi: 10.1016/j.cca.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Brown JVE, Meader N, Cleminson J, McGuire W. C-reactive protein for diagnosing late-onset infection in newborn infants. Cochrane Database Syst Rev. 2019;10:16–78. doi: 10.1002/14651858.CD012126.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bower JK, Lazo M, Juraschek SP, Selvin E. Within-Person variability in high-sensitivity C-reactive protein. Arch Intern Med. 2012;172:1519–1521. doi: 10.1001/archinternmed.2012.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dammann O, Leviton A. Intermittent or sustained systemic inflammation and the preterm brain. Pediatr Res. 2014;75:376–380. doi: 10.1038/pr.2013.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevenson AJ, McCartney DL, Harris SE, Taylor AM, Redmond P, Starr JM, et al. Trajectories of inflammatory biomarkers over the eighth decade and their associations with immune cell profiles and epigenetic ageing. Clin Epigenet. 2018;10:159. doi: 10.1186/s13148-018-0585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson AJ, McCartney DL, Hillary RF, Campbell A, Morris SW, Bermingham ML, et al. Characterisation of an inflammation-related epigenetic score and its association with cognitive ability. Clin Epigenetics. 2020;12:113. doi: 10.1186/s13148-020-00903-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gadd DA, Hillary RF, McCartney DL, Zaghlool SB, Stevenson AJ, Cheng Y, et al. Epigenetic scores for the circulating proteome as tools for disease prediction. Elife. 2022;11:e71802. doi: 10.7554/eLife.71802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ligthart S, Marzi C, Aslibekyan S, Mendelson MM, Conneely KN, Tanaka T, et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016;17:255. doi: 10.1186/s13059-016-1119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson AJ, Gadd DA, Hillary RF, McCartney DL, Campbell A, Walker RM, et al. Creating and validating a DNA methylation-based proxy for interleukin-6. J Gerontol Ser Biol Sci Med Sci. 2021;76:2284–2292. doi: 10.1093/gerona/glab046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conole ELS, Stevenson AJ, Maniega SM, Harris SE, Green C, del Hernández M, CV, , et al. DNA methylation and protein markers of chronic inflammation and their associations with brain and cognitive aging. Neurology. 2021;97:e2340–e2352. doi: 10.1212/WNL.0000000000012997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green C, Shen X, Stevenson AJ, Conole ELS, Harris MA, Barbu MC, et al. Structural brain correlates of serum and epigenetic markers of inflammation in major depressive disorder. Brain Behav Immun. 2021;92:39–48. doi: 10.1016/j.bbi.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barker ED, Cecil CAM, Walton E, Houtepen LC, O’Connor TG, Danese A, et al. Inflammation-related epigenetic risk and child and adolescent mental health: a prospective study from pregnancy to middle adolescence. Dev Psychopathol. 2018;30:1145–1156. doi: 10.1017/S0954579418000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conole ELS, Vaher K, Blesa-Cábez M, Sullivan G, Stevenson AJ, Hall J, et al. Immuno-epigenetic signature derived in saliva associates with the encephalopathy of prematurity and perinatal inflammatory disorders. Brain Behav Immun. 2023;110:322–338. doi: 10.1016/j.bbi.2023.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Needham BL, Smith JA, Zhao W, Wang X, Mukherjee B, Kardia SLR, et al. Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: the multi-ethnic study of atherosclerosis. Epigenetics. 2015;10:958–969. doi: 10.1080/15592294.2015.1085139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stringhini S, Polidoro S, Sacerdote C, Kelly RS, van Veldhoven K, Agnoli C, et al. Life-course socioeconomic status and DNA methylation of genes regulating inflammation. Int J Epidemiol. 2015;44:1320–1330. doi: 10.1093/ije/dyv060. [DOI] [PubMed] [Google Scholar]
- 35.McDade TW, Ryan C, Jones MJ, MacIsaac JL, Morin AM, Meyer JM, et al. Social and physical environments early in development predict DNA methylation of inflammatory genes in young adulthood. Proc Nat Acad Sci. 2017;114:7611–7616. doi: 10.1073/pnas.1620661114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith JA, Zhao W, Wang X, Ratliff SM, Mukherjee B, Kardia SLR, et al. Neighborhood characteristics influence DNA methylation of genes involved in stress response and inflammation: the multi-ethnic study of atherosclerosis. Epigenetics. 2017;12:662–673. doi: 10.1080/15592294.2017.1341026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98:680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howe CG, Cox B, Fore R, Jungius J, Kvist T, Lent S, et al. Maternal gestational diabetes mellitus and newborn DNA methylation: findings from the pregnancy and childhood epigenetics consortium. Diabetes Care. 2019;43:98–105. doi: 10.2337/dc19-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tobi EW, Juvinao-Quintero DL, Ronkainen J, Ott R, Alfano R, Canouil M, et al. Maternal glycemic dysregulation during pregnancy and neonatal blood DNA methylation: meta-analyses of epigenome-wide association studies. Diabetes Care. 2022;45:614–623. doi: 10.2337/dc21-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasaki A, Murphy KE, Briollais L, McGowan PO, Matthews SG. DNA methylation profiles in the blood of newborn term infants born to mothers with obesity. PLoS ONE. 2022;17:e0267946. doi: 10.1371/journal.pone.0267946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Godfrey KM, Reynolds RM, Prescott SL, Nyirenda M, Jaddoe VWV, Eriksson JG, et al. Influence of maternal obesity on the long-term health of offspring. Lancet Diabetes Endocrinol. 2017;5:53–64. doi: 10.1016/S2213-8587(16)30107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krumpolec P, Kodada D, Hadžega D, Petrovič O, Babišová K, Dosedla E, et al. Changes in DNA methylation associated with a specific mode of delivery: a pilot study. Front Med. 2024;11:1291429. doi: 10.3389/fmed.2024.1291429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Q, Ming Y, Gan Y, Huang L, Zhao Y, Wang X, et al. The impact of cesarean delivery on infant DNA methylation. BMC Pregnancy Childbirth. 2021;21:265. doi: 10.1186/s12884-021-03748-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wheater ENW, Galdi P, McCartney DL, Blesa-Cábez M, Sullivan G, Stoye DQ, et al. DNA methylation in relation to gestational age and brain dysmaturation in preterm infants. Brain Commun. 2022;4(2):fcac056. doi: 10.1093/braincomms/fcac056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gadd DA, Smith HM, Mullin D, Chybowska O, Hillary RF, Kimenai DM, et al. DNAm scores for serum GDF15 and NT-proBNP levels associate with a range of traits affecting the body and brain. Medrxiv. 2023;43:1715. [Google Scholar]
- 46.Boardman JP, Hall J, Thrippleton MJ, Reynolds RM, Bogaert D, Davidson DJ, et al. Impact of preterm birth on brain development and long-term outcome: protocol for a cohort study in Scotland. BMJ Open. 2020;10:e035854. doi: 10.1136/bmjopen-2019-035854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scottish National Statistics. SIMD—Scottish index of multiple deprivation: SIMD16 technical notes. Edinburgh: Scottish National Statistics; 2016 pp 1–69
- 48.National Records of Scotland. Scotland’s Census [Internet]. 2020 [cited 2023 Nov 23]. Available from: https://www.scotlandscensus.gov.uk/search-the-census/
- 49.Lowe R, Gemma C, Beyan H, Hawa MI, Bazeos A, Leslie RD, et al. Buccals are likely to be a more informative surrogate tissue than blood for epigenome-wide association studies. Epigenetics. 2013;8:445–454. doi: 10.4161/epi.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braun PR, Han S, Hing B, Nagahama Y, Gaul LN, Heinzman JT, et al. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl Psychiat. 2019;9:47. doi: 10.1038/s41398-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pidsley R, Wong CCY, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genom. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng SC, Webster AP, Dong D, Feber A, Graham DG, Sullivan R, et al. A novel cell-type deconvolution algorithm reveals substantial contamination by immune cells in saliva, buccal and cervix. Epigenomics. 2018;10:925–940. doi: 10.2217/epi-2018-0037. [DOI] [PubMed] [Google Scholar]
- 53.Katz DH, Robbins JM, Deng S, Tahir UA, Bick AG, Pampana A, et al. Proteomic profiling platforms head to head: leveraging genetics and clinical traits to compare aptamer- and antibody-based methods. Sci Adv. 2022;8:eabm5164. doi: 10.1126/sciadv.abm5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haslam DE, Li J, Dillon ST, Gu X, Cao Y, Zeleznik OA, et al. Stability and reproducibility of proteomic profiles in epidemiological studies: comparing the Olink and SOMAscan platforms. Proteomics. 2022;22:e2100170. doi: 10.1002/pmic.202100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raffield LM, Dang H, Pratte KA, Jacobson S, Gillenwater LA, Ampleford E, et al. Comparison of proteomic assessment methods in multiple cohort studies. Proteomics. 2020;20:1900278. doi: 10.1002/pmic.201900278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joshi A, Mayr M. In aptamers they trust: the caveats of the SOMAscan biomarker discovery platform from SomaLogic. Circulation. 2018;138:2482–2485. doi: 10.1161/CIRCULATIONAHA.118.036823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hillary RF, Marioni RE. MethylDetectR: a software for methylation-based health profiling. Wellcome Open Res. 2021;5:283. doi: 10.12688/wellcomeopenres.16458.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Preacher KJ. Extreme groups designs. Encycl Clin Psychol. 2015;2015:1–4. [Google Scholar]
- 59.Fisher JE, Guha A, Heller W, Miller GA. Extreme-groups designs in studies of dimensional phenomena: advantages, caveats, and recommendations. J Abnorm Psychol. 2020;129:14–20. doi: 10.1037/abn0000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mckinnon K, Conole ELS, Vaher K, Binkowska J, Sullivan G, Hillary R, 2023. The relationship between socioeconomic status, preterm birth and systemic inflammation using DNA methylation proxies. OSF. [DOI]
- 61.Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol. 2010;34:100–105. doi: 10.1002/gepi.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022;51:D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kandasamy J, Roane C, Szalai A, Ambalavanan N. Serum eotaxin-1 is increased in extremely-low-birth-weight infants with bronchopulmonary dysplasia or death. Pediatr Res. 2015;78:498–504. doi: 10.1038/pr.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Almudares F, Hagan J, Chen X, Devaraj S, Moorthy B, Lingappan K. Growth and differentiation factor 15 (GDF15) levels predict adverse respiratory outcomes in premature neonates. Pediatr Pulmonol. 2023;58:271–278. doi: 10.1002/ppul.26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Al-Mudares F, Reddick S, Ren J, Venkatesh A, Zhao C, Lingappan K. Role of growth differentiation factor 15 in lung disease and senescence: potential role across the lifespan. Front Med. 2020;7:594137. doi: 10.3389/fmed.2020.594137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wan Y, Fu J. GDF15 as a key disease target and biomarker: linking chronic lung diseases and ageing. Mol Cell Biochem. 2023;479:453–466. doi: 10.1007/s11010-023-04743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Jiang W, Wang L, Lingappan K. Sex-specific differences in the modulation of growth differentiation factor 15 (GDF15) by hyperoxia in vivo and in vitro: role of Hif-1α. Toxicol Appl Pharm. 2017;332:8–14. doi: 10.1016/j.taap.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang G, Wen B, Deng Z, Zhang Y, Kolesnichenko OA, Ustiyan V, et al. Endothelial progenitor cells stimulate neonatal lung angiogenesis through FOXF1-mediated activation of BMP9/ACVRL1 signaling. Nat Commun. 2022;13:2080. doi: 10.1038/s41467-022-29746-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collaco JM, McGrath-Morrow SA, Griffiths M, Chavez-Valdez R, Parkinson C, Zhu J, et al. Perinatal inflammatory biomarkers and respiratory disease in preterm infants. J Pediatrics. 2022;246:34–39.e3. doi: 10.1016/j.jpeds.2022.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oak P, Hilgendorff A. The BPD trio? Interaction of dysregulated PDGF, VEGF, and TGF signaling in neonatal chronic lung disease. Mol Cell Pediatrics. 2017;4:11. doi: 10.1186/s40348-017-0076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Course CW, Lewis PA, Kotecha SJ, Cousins M, Hart K, Watkins WJ, et al. Characterizing the urinary proteome of prematurity-associated lung disease in school-aged children. Respir Res. 2023;24:191. doi: 10.1186/s12931-023-02494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bose CL, Dammann CEL, Laughon MM. Bronchopulmonary dysplasia and inflammatory biomarkers in the premature neonate. Arch Dis Child Fetal Neonatal Ed. 2008;93:F455. doi: 10.1136/adc.2007.121327. [DOI] [PubMed] [Google Scholar]
- 73.Zasada M, Suski M, Bokiniec R, Szwarc-Duma M, Borszewska-Kornacka MK, Madej J, et al. Comparative two time-point proteome analysis of the plasma from preterm infants with and without bronchopulmonary dysplasia. Ital J Pediatr. 2019;45:112. doi: 10.1186/s13052-019-0676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lassus P, Heikkilä P, Andersson LC, von Boguslawski K, Andersson S. Lower concentration of pulmonary hepatocyte growth factor is associated with more severe lung disease in preterm infants. J Pediatr. 2003;143:199–202. doi: 10.1067/S0022-3476(03)00297-X. [DOI] [PubMed] [Google Scholar]
- 75.Wagner BD, Babinec AE, Carpenter C, Gonzalez S, O’Brien G, Rollock K, et al. Proteomic profiles associated with early echocardiogram evidence of pulmonary vascular disease in preterm infants. Am J Respir Crit Care Med. 2018;197:394–397. doi: 10.1164/rccm.201703-0654LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guasti L, Silvennoinen S, Bulstrode NW, Ferretti P, Sankilampi U, Dunkel L. Elevated FGF21 leads to attenuated postnatal linear growth in preterm infants through GH resistance in chondrocytes. J Clin Endocrinol Metab. 2014;99:E2198–E2206. doi: 10.1210/jc.2014-1566. [DOI] [PubMed] [Google Scholar]
- 77.Spencer R, Maksym K, Hecher K, Maršál K, Figueras F, Ambler G, et al. Ultrasound and biochemical predictors of pregnancy outcome at diagnosis of early-onset fetal growth restriction. Medrxiv. 2023;2023:298. doi: 10.1172/JCI169199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qiu Q, Bell M, Lu X, Yan X, Rodger M, Walker M, et al. Significance of IGFBP-4 in the development of fetal growth restriction. J Clin Endocrinol Metab. 2012;97:E1429–E1439. doi: 10.1210/jc.2011-2511. [DOI] [PubMed] [Google Scholar]
- 79.Voller SB, Chock S, Ernst LM, Su E, Liu X, Farrow KN, et al. Cord blood biomarkers of vascular endothelial growth (VEGF and sFlt-1) and postnatal growth: a preterm birth cohort study. Early Hum Dev. 2014;90:195–200. doi: 10.1016/j.earlhumdev.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim MJ, Romero R, Kim CJ, Tarca AL, Chhauy S, LaJeunesse C, et al. Villitis of unknown etiology is associated with a distinct pattern of chemokine up-regulation in the feto-maternal and placental compartments: implications for conjoint maternal allograft rejection and maternal anti-fetal graft-versus-host disease. J Immunol. 2009;182:3919–3927. doi: 10.4049/jimmunol.0803834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Romero R, Chaemsaithong P, Chaiyasit N, Docheva N, Dong Z, Kim CJ, et al. CXCL10 and IL-6: markers of two different forms of intra-amniotic inflammation in preterm labor. Am J Reprod Immunol. 2017;78:e12685. doi: 10.1111/aji.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sellmer A, Henriksen TB, Palmfeldt J, Bech BH, Astono J, Bennike TB, et al. The patent ductus arteriosus in extremely preterm neonates is more than a hemodynamic challenge: new molecular insights. Biomol. 2022;12:1179. doi: 10.3390/biom12091179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olsson KW, Larsson A, Jonzon A, Sindelar R. Exploration of potential biochemical markers for persistence of patent ductus arteriosus in preterm infants at 22–27 weeks’ gestation. Pediatr Res. 2019;86:333–338. doi: 10.1038/s41390-018-0182-x. [DOI] [PubMed] [Google Scholar]
- 84.Waleh N, Seidner S, McCurnin D, Giavedoni L, Hodara V, Goelz S, et al. Anatomic closure of the premature patent ductus arteriosus: the role of CD14+/CD163+ mononuclear cells and VEGF in neointimal mound formation. Pediatr Res. 2011;70:332–338. doi: 10.1203/PDR.0b013e3182294471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu C, Su X, Chen Y, Xu Y, Wang Z, Mo X. Proteomics analysis of plasma protein changes in patent ductus arteriosus patients. Ital J Pediatr. 2020;46:64. doi: 10.1186/s13052-020-00831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng Y, Zhu X, Linghu D, Xu Y, Liang J. Serum levels of cytokines in infants treated with conbercept for retinopathy of prematurity. Sci Rep-uk. 2020;10:12695. doi: 10.1038/s41598-020-69684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sato T, Kusaka S, Shimojo H, Fujikado T. Simultaneous analyses of vitreous levels of 27 cytokines in eyes with retinopathy of prematurity. Ophthalmology. 2009;116:2165–2169. doi: 10.1016/j.ophtha.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 88.Rathi S, Jalali S, Patnaik S, Shahulhameed S, Musada GR, Balakrishnan D, et al. Abnormal complement activation and inflammation in the pathogenesis of retinopathy of prematurity. Front Immunol. 2017;8:1868. doi: 10.3389/fimmu.2017.01868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.The Royal College of Ophthalmologists. Treating Retinopathy of Prematurity in the UK. The Royal College of Ophthalmologists; 2022
- 90.Rivera JC, Holm M, Austeng D, Morken TS, Zhou Ellen T, Beaudry-Richard A, et al. Retinopathy of prematurity: inflammation, choroidal degeneration, and novel promising therapeutic strategies. J Neuroinflamm. 2017;14:165. doi: 10.1186/s12974-017-0943-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klerk DH, Plösch T, Verkaik-Schakel RN, Hulscher JBF, Kooi EMW, Bos AF. DNA methylation of TLR4, VEGFA, and DEFA5 is associated with necrotizing enterocolitis in preterm infants. Front Pediat. 2021;9:630817. doi: 10.3389/fped.2021.630817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olaloye OO, Liu P, Toothaker JM, McCourt BT, McCourt CC, Xiao J, et al. CD16+CD163+ monocytes traffic to sites of inflammation during necrotizing enterocolitis in premature infants. J Exp Med. 2021;218:e20200344. doi: 10.1084/jem.20200344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pammi M, Suresh G. Enteral lactoferrin supplementation for prevention of sepsis and necrotizing enterocolitis in preterm infants. Cochrane Db Syst Rev. 2020;3:CD007137. doi: 10.1002/14651858.CD007137.pub6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Satrom KM, Ennis K, Sweis BM, Matveeva TM, Chen J, Hanson L, et al. Neonatal hyperglycemia induces CXCL10/CXCR3 signaling and microglial activation and impairs long-term synaptogenesis in the hippocampus and alters behavior in rats. J Neuroinflamm. 2018;15:82. doi: 10.1186/s12974-018-1121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Groselj-Grenc M, Ihan A, Derganc M. Neutrophil and monocyte CD64 and CD163 expression in critically Ill neonates and children with sepsis: comparison of fluorescence intensities and calculated indexes. Mediat Inflamm. 2008;2008:202646. doi: 10.1155/2008/202646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kingsmore SF, Kennedy N, Halliday HL, Velkinburgh JCV, Zhong S, Gabriel V, et al. Identification of diagnostic biomarkers for infection in premature neonates. Mol Cell Proteomics. 2008;7:1863–1875. doi: 10.1074/mcp.M800175-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zilow EP, Hauck W, Linderkamp O, Zilow G. Alternative pathway activation of the complement system in preterm infants with early onset infection. Pediatr Res. 1997;41:334–339. doi: 10.1203/00006450-199703000-00005. [DOI] [PubMed] [Google Scholar]
- 98.Mohammed A, Okwor I, Shan L, Onyilagha C, Uzonna JE, Gounni AS. Semaphorin 3E regulates the response of macrophages to lipopolysaccharide-induced systemic inflammation. J Immunol. 2020;204:128–136. doi: 10.4049/jimmunol.1801514. [DOI] [PubMed] [Google Scholar]
- 99.Leviton A, Allred EN, Fichorova RN, O’Shea TM, Fordham LA, Kuban KKC, et al. Circulating biomarkers in extremely preterm infants associated with ultrasound indicators of brain damage. Eur J Paediatr Neuro. 2018;22:440–450. doi: 10.1016/j.ejpn.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schultz SJ, Aly H, Hasanen BM, Khashaba MT, Lear SC, Bendon RW, et al. Complement component 9 activation, consumption, and neuronal deposition in the post-hypoxic–ischemic central nervous system of human newborn infants. Neurosci Lett. 2005;378:1–6. doi: 10.1016/j.neulet.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 101.Morales DM, Townsend RR, Malone JP, Ewersmann CA, Macy EM, Inder TE, et al. Alterations in protein regulators of neurodevelopment in the cerebrospinal fluid of infants with posthemorrhagic hydrocephalus of prematurity. Mol Cell Proteom. 2012;11:M1110.11973. doi: 10.1074/mcp.M111.011973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ochoa TJ, Sizonenko SV. Lactoferrin and prematurity: a promising milk protein? Biochem Cell Biol. 2017;95:22–30. doi: 10.1139/bcb-2016-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Che X, Hornig M, Bresnahan M, Stoltenberg C, Magnus P, Surén P, et al. Maternal mid-gestational and child cord blood immune signatures are strongly associated with offspring risk of ASD. Mol Psychiatr. 2022;27:1527–1541. doi: 10.1038/s41380-021-01415-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leviton A, Joseph RM, Fichorova RN, Allred EN, Taylor HG, O’Shea TM, et al. Executive dysfunction early postnatal biomarkers among children born extremely preterm. J Neuroimmune Pharm. 2019;14:188–199. doi: 10.1007/s11481-018-9804-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Allred EN, Dammann O, Fichorova RN, Hooper SR, Hunter SJ, Joseph RM, et al. Systemic inflammation during the first postnatal month and the risk of attention deficit hyperactivity disorder characteristics among 10 year-old children born extremely preterm. J Neuroimmune Pharm. 2017;12:531–543. doi: 10.1007/s11481-017-9742-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aly H, Khashaba M, Nada A, Hasanen B, McCarter R, Schultz S, et al. The role of complement in neurodevelopmental impairment following neonatal hypoxic-ischemic encephalopathy. Am J Perinatol. 2009;26:659–665. doi: 10.1055/s-0029-1220793. [DOI] [PubMed] [Google Scholar]
- 107.Limbrick DD, Morales DM, Shannon CN, Wellons JC, Kulkarni AV, Alvey JS, et al. Cerebrospinal fluid NCAM-1 concentration is associated with neurodevelopmental outcome in post-hemorrhagic hydrocephalus of prematurity. PLoS ONE. 2021;16:e0247749. doi: 10.1371/journal.pone.0247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Voegele AF, Jerković L, Wellenzohn B, Eller P, Kronenberg F, Liedl KR, et al. Characterization of the vitazmin E-binding properties of human plasma afamin. Biochemistry. 2002;41:14532–14538. doi: 10.1021/bi026513v. [DOI] [PubMed] [Google Scholar]
- 109.Paetau S, Rolova T, Ning L, Gahmberg CG. Neuronal ICAM-5 inhibits microglia adhesion and phagocytosis and promotes an anti-inflammatory response in LPS stimulated microglia. Front Mol Neurosci. 2017;10:431. doi: 10.3389/fnmol.2017.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Skeel A, Yoshimura T, Showalter SD, Tanaka S, Appella E, Leonard EJ. Macrophage stimulating protein: purification, partial amino acid sequence, and cellular activity. J Exp Med. 1991;173:1227–1234. doi: 10.1084/jem.173.5.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kronenberg F, Kollerits B, Kiechl S, Lamina C, Kedenko L, Meisinger C, et al. Plasma concentrations of afamin are associated with the prevalence and development of metabolic syndrome. Circ Cardiovasc Genet. 2018;7:822–829. doi: 10.1161/CIRCGENETICS.113.000654. [DOI] [PubMed] [Google Scholar]
- 112.Montez JK, Bromberger JT, Harlow SD, Kravitz HM, Matthews KA. Life-course socioeconomic status and metabolic syndrome among midlife women. J Gerontol: Ser B. 2016;71:1097–1107. doi: 10.1093/geronb/gbw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Castagné R, Delpierre C, Kelly-Irving M, Campanella G, Guida F, Krogh V, et al. A life course approach to explore the biological embedding of socioeconomic position and social mobility through circulating inflammatory markers. Sci Rep. 2016;6:25170. doi: 10.1038/srep25170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leviton A, Allred EN, Dammann O, Joseph RM, Fichorova RN, O’Shea TM, et al. Socioeconomic status and early blood concentrations of inflammation-related and neurotrophic proteins among extremely preterm newborns. PLoS ONE. 2019;14:e0214154. doi: 10.1371/journal.pone.0214154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chybowska AD, Gadd DA, Cheng Y, Bernabeu E, Campbell A, Walker RM, et al. Epigenetic contributions to clinical risk prediction of cardiovascular disease. Circ Genom Precis Med. 2024;17:004265. doi: 10.1161/CIRCGEN.123.004265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cheng Y, Gadd DA, Gieger C, Monterrubio-Gómez K, Zhang Y, Berta I, et al. Development and validation of DNA methylation scores in two European cohorts augment 10-year risk prediction of type 2 diabetes. Nat Aging. 2023;3:450–458. doi: 10.1038/s43587-023-00391-4. [DOI] [PubMed] [Google Scholar]
- 117.Smith HM, Moodie JE, Monterrubio-Gómez K, Gadd DA, Hillary RF, Chybowska AD, et al. Epigenetic scores of blood-based proteins as biomarkers of general cognitive function and brain health. Clin Epigenet. 2024;16:46. doi: 10.1186/s13148-024-01661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cross B, Turner R, Pirmohamed M. Polygenic risk scores: an overview from bench to bedside for personalised medicine. Front Genet. 2022;13:1000667. doi: 10.3389/fgene.2022.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12:44. doi: 10.1186/s13073-020-00742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yousefi PD, Suderman M, Langdon R, Whitehurst O, Smith GD, Relton CL. DNA methylation-based predictors of health: applications and statistical considerations. Nat Rev Genet. 2022;23:369–383. doi: 10.1038/s41576-022-00465-w. [DOI] [PubMed] [Google Scholar]
- 121.Suarez A, Lahti J, Lahti-Pulkkinen M, Girchenko P, Czamara D, Arloth J, et al. A polyepigenetic glucocorticoid exposure score at birth and childhood mental and behavioral disorders. Neurobiol Stress. 2020;13:100275. doi: 10.1016/j.ynstr.2020.100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Blostein FA, Fisher J, Dou J, Schneper L, Ware EB, Notterman DA, et al. Polymethylation scores for prenatal maternal smoke exposure persist until age 15 and are detected in saliva in the Fragile Families and Child Wellbeing cohort. Epigenetics. 2022;17:2223–2240. doi: 10.1080/15592294.2022.2112815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Mckinnon K, Conole ELS, Vaher K, Binkowska J, Sullivan G, Hillary R, 2023. The relationship between socioeconomic status, preterm birth and systemic inflammation using DNA methylation proxies. OSF. [DOI]
Supplementary Materials
Data Availability Statement
DNA methylation data are available to researchers subject to the terms of the Data Access Policy: https://www.ed.ac.uk/centre-reproductive-health/tebc/about-tebc/for-researchers/data-access-collaboration.