Abstract
Adenovirus late mRNA export is facilitated by viral early proteins of 55 and 34 kDa. The 34-kDa protein contains a leucine-rich nuclear export signal (NES) similar to that of the human immunodeficiency virus Rev protein. It was proposed that the 34-kDa protein might facilitate the export of adenovirus late mRNA through a Rev-like NES-mediated export pathway. We have tested the role of NES-mediated RNA export during adenovirus infection, and we find that it is not essential for the expression of adenovirus late genes.
Two adenovirus (Ad) early proteins with molecular weights of 34,000 (34K) and 55,000 (55K) facilitate the export of late viral mRNA and inhibit the export of cellular mRNA during the late phase of infection (reviewed in reference 16). The 34K protein contains a Rev-like nuclear export signal (NES) and a nuclear retention signal (NRS) (see Fig. 1A); both signals are important mediators of 34K protein export in transfected cells (6). 34K forms a complex with 55K in infected cells (20). Recently 55K was shown to have RNA binding activity (12). Taken together, these observations led to the proposal that 34K mediates Ad RNA export by a Rev-like pathway (6, 16). In this model the 34K protein is present on viral mRNA export complexes, and the signal used for export is provided by the NES of 34K. One prediction of this model is that 34K constructs carrying lesions in their NESs will fail to rescue the late gene expression defect of a virus mutant lacking the 34K gene. Another prediction is that Ad late gene expression will be inhibited by the drug leptomycin B, which specifically interferes with the function of the recently identified receptor for the Rev-like NES, exportin-1 (8, 9, 15). Exportin-1 belongs to a family of import and export receptors that are responsible for translocating “cargo” through nuclear pores (17, 21). We have tested these predictions and find that Ad late gene expression is not dependent on a functional Rev-like NES from 34K. We also find no evidence for a substantial decrease in viral late gene expression when infected cells are cultured in the presence of leptomycin B. Our results indicate that a functional exportin-1-mediated export pathway is not critical for the expression of Ad late genes.
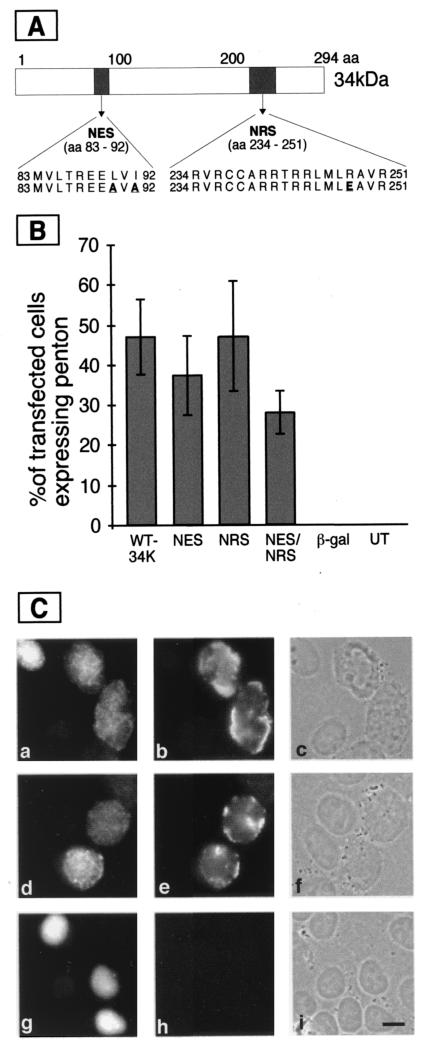
FIG. 1.
Complementation of gene expression in E4 deletion mutant infection by different mutant constructs of 34K. (A) Map of the 34K protein. 34K is depicted as a rectangle with the positions of the amino acids of the protein shown above. NES and NRS regions are shaded, and the regions are expanded below to show the amino acid sequence. Mutant and wild-type expression constructs were a kind gift of Matthias Dobbelstein and are described in reference 6. The mutations present in the NES and NRS are in bold and underlined. The NES mutation has a leucine and an isoleucine at positions 90 and 92 changed to alanines. The NRS mutation has an arginine at position 248 changed to glutamic acid. (B) HeLa cells were transfected and infected with an E4 deletion mutant as described in the text. Fixed cells were stained with M45 to detect 34K, and they were also stained with a polyclonal serum to detect the viral penton base and fiber proteins. Transfected cells were identified by the presence of 34K and were then scored for the presence or absence of late proteins. The columns represent the percent of transfected cells that also contained late proteins. Abbreviations used for the different constructs: WT34K, wild-type 34K; NES, double point mutation L90A/I92A in the nuclear export signal; NRS, point mutation R248E in the nuclear retention signal; NES/NRS, L90A/I92-R248E double mutation in the nuclear export and retention signals; β-gal, construct expressing β-galactosidase; UT, infected cells that were not transfected. (C) Cells transfected with constructs expressing wild-type 34K (panels a through c), NES-mutated 34K (panels d through f), or green fluorescent protein (panels g through i). Following infection with H5dl 1011, cells were fixed and stained as described in Fig. 1B. Cells stained for the transfected protein are shown in panels a, d, and g; late viral proteins are shown in panels b, e, and h. The total number of cells in the microscope fields are visualized in panels c, f, and i. Bar, 10 μm.
Ad mutants lacking the E4 region are defective for the production of their late mRNA (2, 11, 23). Transiently expressed 34K can overcome the late gene expression defects exhibited by such mutants (14). We reasoned that a similar complementation assay could be used to assess the ability of 34K mutants to restore late gene expression in cells infected by an E4 mutant virus. To investigate the role of the 34K NES and NRS in viral late gene expression, HeLa cells were transfected with constructs expressing wild-type or mutant 34K proteins (Fig. 1A). For controls, we used either mock-transfected cells or cells transfected with constructs expressing either β-galactosidase or green fluorescent protein. At 24 h posttransfection, cells were infected at a multiplicity of 3 focus-forming units (FFU) of H5dl 1011 per cell (3). This mutant virus lacks the E4 sequences between the SmaI sites located at 33093 and 35355 and is missing the gene encoding the 34K polypeptide. At 24 h postinfection (hpi), cells were fixed and immunostained as described previously (4) with the monoclonal antibody M45 (18), to detect 34K, and with a polyclonal antiserum that detects the late proteins penton and fiber (a kind gift of Ulf Pettersson). We used secondary antibodies coupled to fluorescein isothiocyanate and Texas red (Southern Biotechnology Associates Inc., Birmingham, Ala.) to simultaneously visualize 34K and viral late proteins. We identified transfected cells that contained the 34K protein by microscopy. The fraction of these 34K-positive cells containing late proteins was then calculated (Fig. 1B). At least three different transfection/infection experiments were performed to obtain the data columns given for each plasmid. More than 100 transfected cells were scored in each experiment. About half of the cells transfected with the wild-type 34K expression construct and subsequently infected with H5dl 1011 had produced late proteins. β-Galactosidase expression constructs did not complement late gene expression of the mutant virus. No late gene expression was observed in cells that were infected by H5dl 1011 without transfection. These results indicate that a wild-type 34K expression construct significantly complements the late gene expression defect of cells infected by an E4 mutant. Next we tested the ability of 34K constructs carrying disruptions of the NES, NRS, or both to restore late gene expression in the same complementation assay. 34K containing two amino acid substitutions in the NES (L90A/I92A) is no longer able to direct the export of the 34K complex in heterokaryon assays (6). If the same signal is required to direct the export of viral mRNA, one might expect that a construct expressing 34K-L90A/I92A would fail to complement late gene expression. Surprisingly, the NES mutant construct produced a protein that complemented H5dl 1011 late gene expression as well as the wild-type 34K protein (Fig. 1B; compare panels b and e in Fig. 1C). Therefore, in our complementation assay, amino acid substitutions in the NES of 34K do not dramatically reduce the ability of 34K to rescue late protein production in E4 mutant-infected cells. Immunoblotting confirmed that similar levels of wild-type and NES mutant 34K proteins resulted in similar complementation of late protein levels (data not shown). Constructs producing a 34K NRS mutant (R248E) (6) also efficiently complemented late gene expression (Fig. 1B). A double mutant carrying substitutions in both the NES and NRS (L90A/I92A-R248E) (6) complemented late protein production to about 60% of the level of the wild-type 34K protein. This might reflect a distorted structure of the 34K protein with the NES-NRS double mutation that slightly inhibits its activity. We do not yet know the reason for the modest complementation defect seen with the double mutant. It should be noted however, that none of these mutations dramatically reduced late protein complementation. Our results suggest that other features of the 34K protein must provide its critical activity in promoting viral late gene expression.
To further test the role of NES-mediated export in viral late gene expression we treated infected cells with the cytotoxin leptomycin B, which inhibits the function of exportin-1 (8, 9, 15). If 34K interacts with exportin-1 through its NES to mediate late-phase Ad mRNA export, we would expect Ad late gene expression to be sensitive to leptomycin B treatment. To address this question, HeLa cells grown on coverslips were infected with Ad2 or Ad5 (20 to 40 FFU/cell), and leptomycin B was added at 10 to 12 hpi to a final concentration of 10 nM. Prior to addition of the drug, some coverslips were removed from the culture and fixed for immunostaining to determine the number of late-phase cells present in the culture at the time leptomycin B was added. Infected cells cultured in the absence or presence of leptomycin B were fixed 6 h later and immunostained with antiserum raised against the viral late proteins penton and fiber. Cells were then scored for the presence of viral late proteins. Untreated cultures showed a 30 to 50% increase in cells expressing late proteins (Fig. 2A). Leptomycin B had no significant effect on this increase. To confirm that leptomycin B was functional we measured cell growth in the presence and absence of the drug over a 48-h period. The appearance of new cells was completely blocked when cultures were incubated with 10 nM leptomycin B (data not shown). This is consistent with other reports showing that leptomycin B inhibits cell cycle progression (25) and shows that the drug was effective at this concentration.
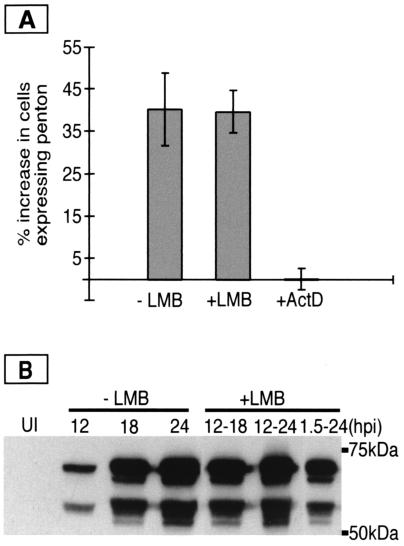
FIG. 2.
Late viral gene expression is not sensitive to leptomycin B treatment. (A) HeLa cells were grown, infected, fixed, and stained for viral late proteins as described in the text. Infected cells (Ad5, ∼20 FFU/cell) were either not treated (−LMB), treated for 6 h with 10 nM leptomycin B (+LMB), or treated with 5 μg of actinomycin D (+ActD) per ml between 12 and 18 hpi. More than 150 cells/coverslip were counted and scored for the presence of late viral proteins. The columns represent the mean values of four different experiments and show the percent increase in cells expressing late proteins. This was calculated by subtracting the percent late protein-positive cells at the time the drug was added to the culture from the percent late protein-positive cells in the culture following incubation with or without the drug. (B) HeLa cells were infected with Ad5 (∼20 FFU/cell) and were either untreated (−LMB) or treated with 10 nM leptomycin B between 12 and 18, 12 and 24, or 1.5 and 24 hpi (+LMB). Infected cells were harvested at 12, 18, and 24 hpi, and an equal amount of cell extract was electrophoresed on a sodium dodecyl sulfate–10% polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane and incubated with antiserum against viral late proteins for immunoblotting. The viral proteins penton (63.3 kDa) and fiber (62 kDa) were visualized by the ECL detection kit according to the manufacturer's instructions. LMB, leptomycin B; UI, uninfected.
To be certain that our assay would detect a decrease in the availability of RNA from the nuclear compartment, we performed experiments in which infected cells were cultured in the presence of 5 μg of actinomycin D per ml from 12 to 18 hpi. Actinomycin D rapidly inhibits further synthesis of RNA when it is present in the culture medium. When no new viral RNA is synthesized, viral mRNA export is also necessarily blocked. We saw less than a 3% increase in the percent of late protein containing cells in cultures treated with actinomycin D (Fig. 2A). Thus, treatment with actinomycin D, which is expected to prevent Ad late RNA synthesis and thereby export, severely reduced the number of new late protein-expressing cells, while treatment with leptomycin B had no inhibitory effect. These results suggested that Ad late gene expression, including RNA export to the cytoplasm, occurred normally in leptomycin B-treated cells. To confirm that leptomycin B does not inhibit Ad late gene expression we measured the levels of viral late proteins in treated and untreated cultures by immunoblotting. HeLa cells were infected with Ad5 and treated with leptomycin B from 12 to 18, 12 to 24, or 1.5 to 24 hpi. Cells were harvested at the times indicated and an equal amount of cell extract was used for immunoblotting (Fig. 2B). Late proteins were detected by using rabbit antiserum raised against penton and fiber and enhanced chemiluminescence detection (ECL Western blotting analysis system; Amersham Pharmacia Biotech). Fluorescing signals were detected by exposing autoradiography film, and the signals were analyzed by image analysis with the NIH Image program (version 1.61, National Institutes of Health, Bethesda, Md.; http://rsb.info.nih.gov/nih-image). In the absence of leptomycin B we observed a sixfold increase in the levels of late proteins between 12 and 18 hpi and a ninefold increase between 12 and 24 hpi. Culturing cells with leptomycin B had no inhibitory effect on late protein production when treatment was from 12 to 18 hpi (a sevenfold increase in late proteins) or from 12 to 24 hpi (a ninefold increase in late proteins). Extended incubations with leptomycin B from 1.5 to 24 hpi had a modest (twofold) effect on the accumulation of viral late proteins. This could indicate a secondary effect of interfering with other exportin-1-mediated functions, such as cellular snRNA export (8). We conclude that leptomycin B does not have a significant effect on Ad late gene expression when the drug is added after the onset of the late phase. Taking this together with results obtained in the complementation assay, we find no requirement either for the 34K NES or for a functional NES-mediated export pathway during Ad late gene expression.
Since the 34K NES is not required for complementing late gene expression, we sought to determine if 34K was indeed shuttling between the nucleus and the cytoplasm in infected cells. Previous work (6) has established that 34K shuttles in transfected cells, but it is not known if the protein shuttles during viral infection. To address this question, HeLa cells infected with Ad5 were fused with uninfected HeLa cells by treatment with a 50% solution of polyethylene glycol (PEG) (5). Similar cell fusion studies have been used to investigate the shuttling of other proteins (1, 19). Cytoplasmic fusion in the presence of PEG created multinucleated cells (data not shown). We reasoned that if 34K was able to shuttle from an infected to an uninfected nucleus within multinucleated cells created by PEG fusion, then the total number of 34K-stained nuclei per microscope field should increase in coverslips prepared from PEG-treated cultures. To perform these experiments we infected HeLa cells with Ad5 for 1.5 h (20 FFU/cell). Infected cells were washed extensively to remove the inoculum, and culture plates were treated with trypsin to release cells from the monolayers. Infected and uninfected cells were mixed at a ratio of about 1:20 and plated onto 3.5-cm dishes containing coverslips. After 14 h in culture, cells were treated with medium containing cycloheximide (CHM; 100 μg/ml) for 1 h to inhibit protein synthesis prior to fusion. This step is critical because we wished to detect movement of 34K from infected to uninfected nuclei rather than the movement of newly synthesized 34K from the shared cytoplasm to uninfected nuclei. Following fusion induced by a 1-min treatment with 50% PEG, the cells were cultured for an additional 2.5 h in the presence of CHM and then fixed and processed for immunostaining with antibodies against 34K and viral late proteins. We performed controls in which the same procedure was followed but without inducing cell fusion. We determined the number of 34K-positive nuclei per field from coverslips that were and were not treated with PEG.
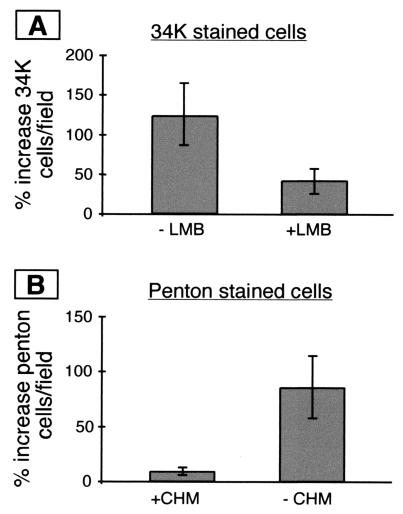
When cell fusions were created by PEG treatment we saw a 120% increase in the number of 34K-positive nuclei per field (Fig. 3A, left column). There were more than twice as many 34K-positive nuclei when infected and uninfected cells were fused than when they were not fused. These data provide evidence that 34K can be exported from the nuclei of infected cells. Interestingly, when cells were treated with the drug leptomycin B (10 nM for 1 h prior to and after PEG treatment) to inhibit NES-mediated export, we observed a threefold reduction in 34K protein shuttling (Fig. 3A, right column). Leptomycin B did not affect the number of fused cells in the culture (data not shown). Since 34K shuttling was sensitive to leptomycin B, it is likely that 34K uses the NES-mediated export pathway in infected cells, at least in part. To test the accuracy of the cell fusion system for measuring protein export, we stained coverslips for viral late proteins that are not expected to show nuclear export activity. We reasoned that in the absence of late protein export, there should be little or no increase in the number of late protein-positive nuclei in fused cells relative to the number in unfused cells. Indeed, coverslips treated with CHM and PEG showed less than a 10% increase in the number of late protein-positive nuclei relative to the number obtained from coverslips that were not treated with PEG (Fig. 3B, left column). This increase was markedly less than the 120% increase seen when cells were stained for 34K (Fig. 3A, left column). As a further control, fusions were performed in the absence of CHM. In this experiment, we expected that newly synthesized late proteins would be imported into the nuclei of uninfected cells when cell fusions were created by PEG treatment. Indeed, in the absence of CHM we saw an 85% increase in the number of late protein-positive nuclei in fused cells relative to unfused cells (Fig. 3B, right column). These results indicate that cell fusion did occur but that only newly synthesized late proteins could move into uninfected nuclei within the fusion. We saw no evidence of late protein export in the presence of CHM. This suggests that the 120% increase in 34K-positive nuclei observed in PEG-treated cells cultured in the presence of CHM is due to nuclear export. To rule out the possibility that 34K shuttling in the cell fusion assay was due to passive diffusion through the nuclear membrane rather than energy dependent transport, fused and unfused infected cells were placed at 4°C to inhibit active transport mechanisms. We did not observe any cytoplasmic staining in unfused cells or any increase in the number of 34K-stained nuclei in cell fusions when they were incubated at 4°C for 4 h (data not shown). We conclude that the observed shuttling of 34K is due to an active transport mechanism rather than passive diffusion.
FIG. 3.
34K shuttles during Ad infection and leptomycin B inhibits its shuttling. Cell fusions with PEG between infected and uninfected HeLa cells were made as described in the text. Following fusion and fixation, the number of 34K- or penton-positive nuclei was scored. The percent increase in 34K or penton-positive nuclei was calculated by dividing the average number of stained cells per field on coverslips that were treated with PEG by the average number of stained cells per field on coverslips that were not treated with PEG. This ratio was expressed as a percentage. LMB, leptomycin B.
Our results indicate that 34K uses the exportin-1-mediated export pathway during protein shuttling in infected cells (Fig. 3). These data are in agreement with observations made by Dobbelstein et al. (6) for NES-mediated 34K shuttling in transfected cells. However, we have no evidence that exportin-1-mediated export is critical for late viral mRNA export. Leptomycin B did not inhibit viral late gene expression (Fig. 2), and an intact 34K NES was not essential to complement late gene expression of a defective Ad E4 mutant (Fig. 1). We have not tested the role of exportin-1-mediated RNA export in specific aspects of early gene expression or DNA replication. Weigel and Dobbelstein (22) found that transfected cells producing 34K with a mutated NES have a reduced ability to rescue both DNA replication and late gene expression of a defective E4 mutant compared with transfected cells producing wild-type 34K. Mutants lacking E4 have variable effects on DNA replication (2, 23). At low multiplicities of infection, E4 mutants have delayed onset of viral replication and can show substantial DNA replication defects (11, 23), while the requirement of E4 products for normal DNA replication is often not observed at higher multiplicities of infection (2, 13, 24). This could explain why Weigel and Dobbelstein observed a role for the 34K NES in viral replication and late gene expression while we did not. If our transfected cells were infected with higher levels of E4 mutant virus, this may have compensated for the lack of an intact 34K in promoting the efficient onset of replication. Nevertheless, we have found that following the onset of DNA replication in wild-type infected cells, inhibiting the exportin-1-mediated export pathway with leptomycin B had no effect on continued late protein production (Fig. 2) despite the observation that it reduced 34K shuttling threefold (Fig. 3). These data show that once the late phase is achieved, exportin-1-mediated RNA export is not critical for continued late RNA export and late gene expression.
Although leptomycin B treatment reduced 34K shuttling in infected cells threefold, the 30% residual shuttling activity is still significant. It is possible that 34K can access another nuclear export pathway via its interactions with other proteins or as part of a viral RNP complex that could use signals from other RNA binding proteins to direct RNP export. We do not yet know how Ad exports its processed mRNAs to the cytoplasmic compartment. Infection inhibits the export of most newly synthesized cellular RNA (7). This could indicate a virus-induced block in a critically important export pathway used by many cellular mRNAs, while virus late mRNAs use a separate pathway. Alternatively, Ad mRNP complexes may have exclusive access to the cellular mRNA export pathway in late-phase infected cells. In this model, viral regulatory factors would subvert the cellular export machinery for the export of viral mRNAs. There is evidence that the viral 55K protein can interact with a cellular hnRNP (10), providing some support for the idea that interactions could “recruit” a critical export factor to viral mRNP complexes. It remains to be determined if this hnRNP protein is an essential factor in the selective export of viral late mRNA during the late phase of infection.
Acknowledgments
C. Rabino and A. Aspegren contributed equally to this work.
We are very grateful to P. Hearing for providing the M45 antibody, Ulf Pettersson for antiserum against viral late proteins, M. Dobbelstein for the 34K mutant expression constructs, and B. Wolff for leptomycin B. We thank D. Cox, J. Stevenson, and U. Pettersson for critically reading the manuscript.
A.A. was supported by stipends from the American-Scandinavian Foundation and the Fulbright Commission. This research was supported by grants from the Swedish Medical Research Council, the Swedish Cancer Fund, and the National Cancer Institute (grant CA8211101) and by grants from Uppsala and Miami Universities.
REFERENCES
- 1.Bear J, Tan W, Zolotukhin A S, Tabernero C, Hudson E A, Felber B K. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol Cell Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bridge E, Ketner G. Redundant control of adenovirus late gene expression by early region 4. J Virol. 1989;63:631–638. doi: 10.1128/jvi.63.2.631-638.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridge E, Medghalchi S, Ubol S, Leesong M, Ketner G. Adenovirus early region 4 and viral DNA synthesis. Virology. 1993;193:794–801. doi: 10.1006/viro.1993.1188. [DOI] [PubMed] [Google Scholar]
- 4.Bridge E, Carmo-Fonseca M, Lamond A, Pettersson U. Nuclear organization of splicing small nuclear ribonucleoproteins in adenovirus-infected cells. J Virol. 1993;67:5792–5802. doi: 10.1128/jvi.67.10.5792-5802.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson R L, Gerald P S. Improved techniques for the induction of mammalian cell hybridization by polyethylene glycol. Somatic Cell Genet. 1976;2:165–176. doi: 10.1007/BF01542629. [DOI] [PubMed] [Google Scholar]
- 6.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flint S J. Regulation of adenovirus mRNA formation. Adv Virus Res. 1986;31:169–228. doi: 10.1016/s0065-3527(08)60264-x. [DOI] [PubMed] [Google Scholar]
- 8.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 9.Fukuda M, Asano S, Nakamura T, Adachi M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 10.Gabler S, Schütt H, Groitl P, Wolf H, Shenk T, Dobner T. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J Virol. 1998;72:7960–7971. doi: 10.1128/jvi.72.10.7960-7971.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horridge J J, Leppard K N. RNA-binding activity of the E1B55-kilodalton protein from human adenovirus type 5. J Virol. 1998;72:9374–9379. doi: 10.1128/jvi.72.11.9374-9379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang M-M, Hearing P. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol. 1989;63:2605–2615. doi: 10.1128/jvi.63.6.2605-2615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ketner G, Bridge E, Virtanen A, Hemström C, Pettersson U. Complementation of adenovirus E4 mutants by transient expression of E4 cDNA and deletion plasmids. Nucleic Acids Res. 1989;17:3037–3048. doi: 10.1093/nar/17.8.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 16.Leppard K N. Regulated RNA processing and RNA transport during adenovirus infection. Semin Virol. 1998;8:301–307. [Google Scholar]
- 17.Mattaj I W. Snail mail to the nucleus. Nature. 1999;399:208–210. doi: 10.1038/20322. [DOI] [PubMed] [Google Scholar]
- 18.Obert S, O'Connor R J, Schmid S, Hearing P. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol Cell Biol. 1994;14:1333–1346. doi: 10.1128/mcb.14.2.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 20.Sarnow P, Hearing P, Anderson C W, Halbert D N, Shenk T, Levine A J. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 22.Weigel S, Dobbelstein M. The nuclear export signal within the E4orf6 protein of adenovirus type 5 supports virus replication and cytoplasmic accumulation of viral mRNA. J Virol. 2000;74:764–772. doi: 10.1128/jvi.74.2.764-772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberg D H, Ketner G. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J Virol. 1986;57:833–838. doi: 10.1128/jvi.57.3.833-838.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoder S S, Berget S M. Role of adenovirus type 2 early region 4 in the early-to-late switch during productive infection. J Virol. 1986;60:779–781. doi: 10.1128/jvi.60.2.779-781.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida M, Nishikawa M, Nishi K, Abe K, Horinouchi S, Beppu T. Effects of leptomycin B on the cell cycle of fibroblasts and fission yeast cells. Exp Cell Res. 1990;187:150–156. doi: 10.1016/0014-4827(90)90129-x. [DOI] [PubMed] [Google Scholar]