Abstract
Background
This study investigates the distribution and characteristics of linezolid and vancomycin susceptibilities among Enterococcus faecalis (E. faecalis) and Enterococcus faecium (E. faecium) and explores the underlying resistance mechanisms.
Methods
A total of 2842 Enterococcus clinical isolates from patients were retrospectively collected, and their clinical data were further analyzed. The minimum inhibitory concentrations (MICs) of vancomycin and linezolid were validated by broth dilution method. The resistance genes optrA, cfr, vanA, vanB and vanM were investigated using polymerase chain reaction (PCR). Housekeeping genes and resistance genes were obtianed through whole-genome sequencing (WGS).
Results
Of the 2842 Enterococcus isolates, 88.5% (2516) originated from urine, with E. faecium accounted for 60.1% of these. The vanA gene was identified in 27/28 vancomycin resistant Enterococcus (VRE) isolates, 4 of which carried both vanA and vanM genes. The remaining strain was vanM positive. The optrA gene was identified in all E. faecalis isolates among linezolid resistant Enterococcus (LRE). E. faecium showed a higher multiple antibiotic resistance index (MAR index) compared to E. faecalis. The multi-locus sequence typing (MLST) showed the sequence type of E. faecium mainly belongs to clonal complex (CC) 17, nearly E. faecalis isolates analyzed were differentiated into 7 characteristics of sequence types (STs), among which ST16 of CC16 were the major lineage.
Conclusion
Urine was the primary source of VRE and LRE isolates in this study. E. faecium showed higher levels of resistance compared to E. faecalis. OptrA gene was detected in 91.6% of LRE, which could explain linezolid resistance, and van genes were detected in all vancomycin resistant Enterococcus strains, while vanA was a key resistance mechanism in VRE identified in this study.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-024-01948-x.
Keywords: Vancomycin, Linezolid, Enterococcus faecalis, Enterococcus faecium, Antibiotic resistance
Introduction
Enterococcus faecalis (E. faecalis) and Enterococcus faecium (E. faecium) were major pathogens in healthcare-associated infections (HAIs), such as endocarditis, septicemia, urinary tract infections, and wound infections [1]. They possess a wealth of intrinsic resistance to cephalosporins, partial fluoroquinolones, aminoglycosides and so on. Meanwhile, Enterococcus isolates developed acquired resistance to β-lactam, aminoglycoside, tetracycline, erythromycin, chloramphenicol and rifampicin due to the widespread use of broad-spectrum antimicrobials [2]. In addition to intrinsic resistance and tolerance, enterococci have been extraordinarily successful at rapidly acquiring resistance to virtually any antimicrobial agent put into clinical use [3]. The plasticity of the Enterococcus genomes allowed them to rapidly respond and adapt to the environment by acquiring genetic determinants, which increased their ability to colonize and infect their host and cause diseases [4]. Meanwhile, the emergence of multidrug-resistant (MDR) Enterococcus had become a major public health threat (e.g. India, Japan) as it had limited the effective antimicrobial agents available to treat infections [5, 6], such as vancomycin resistant Enterococcus (VRE), linezolid resistant Enterococcus (LRE), and even the linezolid resistant vancomycin resistant Enterococcus (LRVRE), has increasingly challenged clinical treatments, as treatment options for VRE bacteremia were limited, the emergence of linezolid resistance as a result of selective pressure was of concern [7–10]. At present, van gene clusters were regarded as the most common mechanism of acquired vancomycin resistance [11], while cfr, optrA and mutation in 23s rRNA were recognized as prevalent mechanisms of linezolid resistance [12–14]. Recent studies have sought to establish a relationship between the phenotypic and genotypic drug resistance, or between the bacterial species and resistance genes in Enterococcus [15, 16], for instance, vanA type had been characterized by acquired resistance to high levels of both vancomycin and teicoplanin, of which vanA was mainly detected in vancomycin resistant Enterococcus faecium (VREfm), vanB type mediated resistance to vancomycin and had a broad MIC, but was sensitive to teicoplanin, vanM-positive VRE showed heterogeneous resistance to vancomycin and teicoplanin [17, 18]. Therefore, it is very necessary for continuous surveillance and understanding of antimicrobial resistance mechanisms in Enterococcus species to guide appropriate therapeutic strategies. This paper briefly elucidates the distribution of specimens, drug susceptibility phenotypes and molecular characteristics of E. faecium and E. faecalis isolates from patients between 2012 and 2021.
Materials and methods
Bacterial isolates
2842 non-duplicated clinical isolates of Enterococcus (including E. faecalis and E. faecium) from hospital of Zhejiang people’s armed police between 2012 and 2021 were analyzed. There were 75 LRE and 39 VRE. However, only 28 VRE and 12 LRE stains were successfully revived for further study, of which were confirmed using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems, Vitek MS (bioMerieux, France).
Antibiotic susceptibility test
Antimicrobial susceptibility testing was conducted by BD PhoenixTM100 automatic microbial identification analyzer via BD Phoenix™ PMIC/ID-55. We used the broth dilution method to test the minimum inhibitory concentration (MIC) of 28 VRE to vancomycin and 12 LRE to linezolid according to the Clinical and Laboratory Standards Institute guidelines (CLSI M100 31th) [19], and considered vancomycin MIC: susceptible, ≤ 4 µg/mL; intermediate, 8–16 µg/mL; resistant, ≥ 32 µg/mL, However, linezolid MIC was considered as: susceptible, ≤ 2 µg/mL; intermediate, 4 µg/mL; resistant, ≥ 8 µg/mL. E. faecalis ATCC 29,212 was used as a quality control.
Detection of Vancomycin and linezolid antimicrobial resistance genes
28 VRE and 12 LRE isolates were all verified to be consistent with the results of standard biochemical reactions. We took common resistance genes vanA, vanB, and vanM gene for vancomycin resistance, and cfr and optrA for linezolid via polymerase chain reaction (PCR) [20]. The primers used in study were listed in Additional file 1.
Retrospective whole genome sequencing and analysis
We performed whole genome sequencing (WGS) on 28 VRE and 12 LRE isolates. Genomic DNA was extracted using a QIAamp DNA Micro Kit (QIAGEN, 56,304). The library was sequenced on a Novaseq 6000 platform (Illumina Inc., San Diego, CA, USA) and 150 bp paired-end reads were generated use default parameters. The detail of WGS was described in Additional file 2.
Statistical analysis
Using the WHONET 5.4 software, we conducted a statistical analysis total of 2842 Enterococcus strains in this study. Data analysis was performed using GraphPad Prism 8.0.2. Chi-square analysis was used to analyze the differences in the prevalence of the tested features in E. faecalis and E. faecium strains. The Mann Whitney test was used to analyze the amount of antibiotic resistance between linezolid resistant Enterococcus faecalis (LREfa) and VREfm (no statistical analysis was performed on vancomycin resistant Enterococcus faecalis (VREfa) and linezolid resistant Enterococcus faecium (LREfm) due to the presence of only one data point). P –value < 0.05 was considered statistically significant.
Results
Distribution of isolates
Over the past decade, 2842 Enterococcus isolates were detected. Among these, E. faecium (n = 1618, 56.9%) was more prevalent than E. faecalis (n = 1224, 43.1%). The majority of these Enterococci were isolated from urine specimens (2516/2842, 88.5%), followed by secretions (108/2842, 3.8%). Interestingly, E. faecium was found more frequently than E. faecalis in urine, pleural effusion, and ascites samples. However, in secretions, respiratory tract samples, pus, cerebrospinal (CSF), vaginal discharge, and blood, E. faecalis was detected in higher numbers than E. faecium (Table 1).
Table 1.
Samples distribution of Enterococcus isolates from 2012 to 2021
| Samples | E. faecalis | E. faecium | Total | |||
|---|---|---|---|---|---|---|
| Num. | Ratio (%) | Num. | Ratio (%) | Num. | Ratio (%) | |
| Urine | 1003 | 81.9 | 1513 | 93.5 | 2516 | 88.5 |
| Secretions | 82 | 6.7 | 26 | 1.6 | 108 | 3.8 |
| Sputum | 38 | 3.1 | 19 | 1.2 | 57 | 2.0 |
| Blood | 25 | 2.0 | 16 | 1.0 | 41 | 1.4 |
| Pus | 21 | 1.7 | 4 | 0.2 | 25 | 0.9 |
| Throat swab | 20 | 1.6 | 4 | 0.2 | 24 | 0.8 |
| Cerebrospinal fluid (CSF) | 8 | 0.7 | 5 | 0.3 | 13 | 0.5 |
| Catheter | 8 | 0.7 | 3 | 0.2 | 11 | 0.4 |
| Bile | 8 | 0.7 | 10 | 0.6 | 18 | 0.6 |
| Vaginal discharge | 7 | 0.6 | 1 | 0.1 | 8 | 0.3 |
| Pleural effusion and ascites | 4 | 0.3 | 17 | 1.1 | 21 | 0.7 |
| Total | 1224 | 100.0 | 1618 | 100.0 | 2842 | 100.0 |
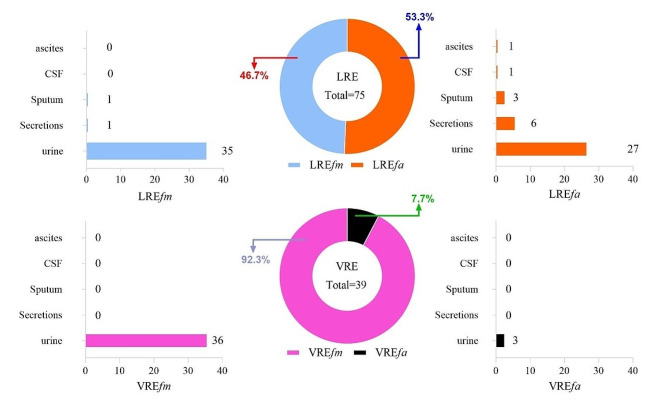
A total of 39 VRE and 75 LRE isolates were further analyzed (Fig. 1; Table 2). This screening including 36 VREfm and 3 VREfa isolates, all of which were found in urine specimens. Additionally, the study included 38 LREfa and 37 LREfm isolates, which were recovered from urine, sputum, secretions, and CSF.
Fig. 1.
Characteristic of source distribution and strain composition between vancomycin resistant Enterococcus (VRE) and linezolid resistant Enterococcus (LRE). VRE and LRE were mainly detected in urine specimen, while the origin of LRE specimens was more extensive than VRE. VRE was dominated by E. faecium, whereas the detection of E. faecalis was comparable to E. faecium in LRE
Table 2.
Distribution of VRE and LRE
| Samples | LREfa | LREfm | VREfm | VREfa | ||||
|---|---|---|---|---|---|---|---|---|
| Num. | Ratio (%) | Num. | Ratio (%) | Num. | Ratio (%) | Num. | Ratio (%) | |
| Urine | 27 | 2.69 | 35 | 2.31 | 36 | 2.38 | 3 | 0.30 |
| Others | 11 | 4.98 | 2 | 1.90 | 0 | / | 0 | / |
| Total | 38 | 3.10 | 37 | 2.29 | 36 | 2.22 | 3 | 0.25 |
Note 1: Ratio of each strain in different samples was obtained by the following formula, urine sample as an example:
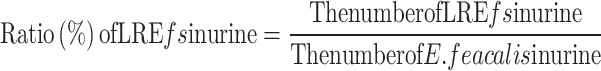 |
1 |
Note 2: Others represent secretions, sputum, blood, pus, throat swab, cerebrospinal fluid, catheter, bile, vaginal discharge, pleural effusion, and ascites, etc.
Abbreviations: Num., number; VRE, vancomycin resistant Enterococcus; LRE, linezolid-resistant Enterococcus ; VREfm, vancomycin resistant Enterococcus faecium; VREfa, vancomycin resistant Enterococcus faecalis; LREfa, linezolid resistant Enterococcus faecalis; LREfm, linezolid resistant Enterococcus faecium; Num., number; LREfs, linezolid resistant Enterococcus
Antimicrobial susceptibility testing
The resistance rates of E. faecalis and E. faecium were shown in Additional file 3. In both sample types, E. faecalis exhibited the highest resistance to rifampicin (812/1003, 81%; 186/221, 84.1%). Conversely, E. faecium showed high resistance frequencies to ciprofloxacin (1465/1513, 96.8%; 97/105, 92.1%).
All 28 VRE strains showed 100% resistance to vancomycin, with high resistance observed (MIC ≥ 256 µg/mL). Meanwhile, 12 LRE strains were verified to have a linezolid resistance phenotype, showing variable resistance to linezolid (8 µg/mL to 16 µg/mL) (Additional file 4, Additional file 5).
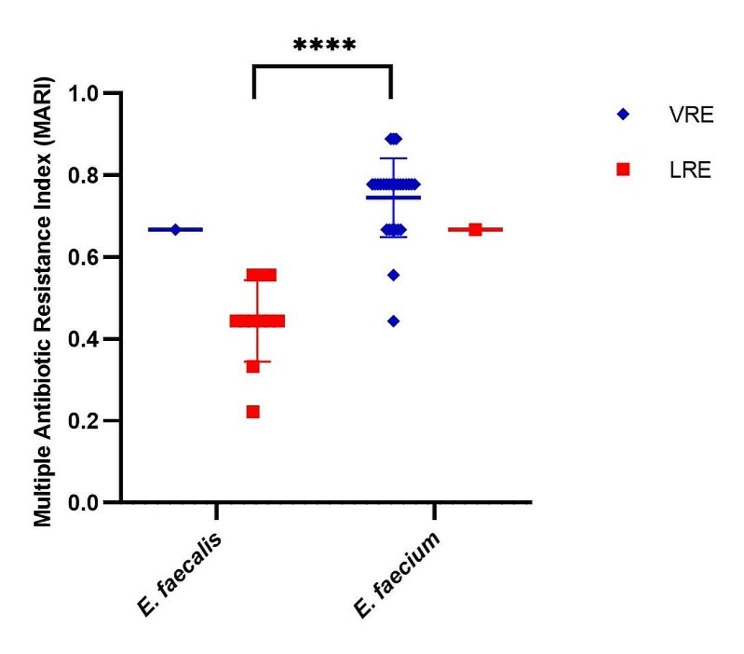
Multiple antibiotic resistance (MAR) index of VRE and LRE
The MAR (Multiple Antibiotic Resistance) index was used to analyse the resistance of 28 VRE and 12 LRE strains to 9 antibiotics (ampicillin, ciprofloxacin, rifampicin, penicillin, tetracycline, teicoplanin, vancomycin, nitrofurantoin, linezolid) using Eq. (1) in this study [21].
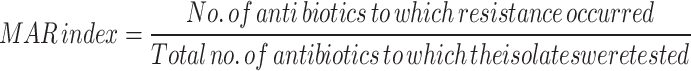 |
It could be found that the MAR index of LRE strains ranged between 0.2 and 0.7, while VREfm strains ranged between 0.4 and 0.9. VREfm strains exhibited higher resistance to antibiotics than LREfa strains (P<0.0001). All strains in this study were scored a MAR index of 0.3 or higher. Additionally, 19 out of 28 (67.9%) VRE strains were scored a MAR index of 0.8 or more. Therefore, we concluded that E. faecium showed extensive resistance to multiple evaluated antibacterial drugs and played larger MAR index values (Fig. 2, Additional file 6).
Fig. 2.
Multiple antibiotic resistance index (MAR index) of LRE and VRE. There was a significant difference between the median MAR index of LREfa strains and VREfm strains (P<0.0001)
The prevalence of drug resistance gene
van genes were detected in all VRE strains, and the prevalence of vanA among VRE isolates was 96.4% (27/28), with 26 VREfm and 1 VREfa, while the remaining 1 VREfm was found to carry a single resistance gene (vanM), and among 26 VREfm, 4 strains were found to carry 2 resistance genes (vanA and vanM). The prevalence of optrA among LRE isolates was 91.7% (11/12), all of which were detected in LREfa, however, no resistance gene designed in this study was detected in a phenotypically positive strain. Unfortunately, cfr was not identified in the study (Additional file 4, Additional file 5).
Characteristics of sequence types (STs)
In this study, multi-locus sequence typing (MLST) was performed for VRE and LRE based on the nucleotide sequences of seven housekeeping genes, respectively. Nine distinct STs were identified from 28 VRE isolates. The most prevalent type was ST78 (n = 17), which was prone to more variation in drug resistance gene profiles, followed by ST80 (n = 2), ST761 (n = 2), ST555 (n = 2). Additionally, there was only one strain each of ST262, ST789, ST6, ST17, and ST18. Meanwhile, seven STs were detected in 12 LRE strains, with ST1287 and ST16 dominating, each accounting for 25% (3/12). Additionally, ST256 (n = 1), ST409 (n = 1), ST480 (n = 1), ST911 (n = 1), and ST262 (n = 1) were also found in LRE strains (Additional file 4, Additional file 5).
Discussion
In this study, we provided data on the characterization of clinical Enterococcus isolates including species, specimen distribution, prevalence of resistance genes, resistance phenotypes, and MLST among VRE and LRE.
In accordance with previous findings that Enterococcus spp. were frequently linked to urinary tract infections (UTIs) [22], the majority of Enterococcus in this study were isolated from urine (88.5%), predominantly, E. faecium (1618/2842, 56.9%). E. faecium had an inherent tenacity to develop resistance to antibiotics and environmental stressors, providing an advantage to thrive in hospital environments [23–25], thus, E. faecium showed a higher contribution than E. faecalis except in Pleural effusion and ascites.
According to the definition of enterococcal antibiotic resistance by Magiorakos et al., this study included 5/11 classes of antibiotics and 7/17 classes of antibiotics: Fluoroquinolones (Ciprofloxacin), Penicillins (Penicillin G, Ampicillin), Tetracycline, Glycopeptides (Vancomycin, Teicoplanin), and Oxazolidinones (Linezolid). All isolates (100%) were classified as extensive drug resistant (XDR) bacteria (defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories) [26] (Additional file 6). Similar to our study, XDR strains were reported in 100% of E. faecium and E. faecalis in Egypt [27]. However, E. faecium was intrinsically more frequently reported as being more resistant to antibiotics, especially to vancomycin than E. faecalis [3]. And in our study, the detection of vancomycin resistance genes in 28 VRE strains also showed predominantly van-positive E. faecium (27 van-VREfm, 1 van-VREfa). Additionally, a MAR index of greater than 0.3 indicates that bacteria had already developed in an area in which antibiotics were often administered [21], in our study, the MAR index of 0.3 and more were recorded for the majority (97.5%) of LRE and VRE isolates. One isolate showed a 0.2 MAR index (LZD34), indicating certainly the link to heavy and uncontrolled use of antibiotics which might create a high antibiotic selective pressure in the hospital treatment. Therefore, the current isolates represent a high public health risk, which made many drugs unusable.
As mentioned above, linezolid was the only Food and Drug Administration (FDA)-approved antibiotic indicated for VRE infections [28–30]. Resistance mechanisms to linezolid include mutations in 23 S rRNA, alterations in ribosomal proteins (L3, L4, and L22), and acquisition of transferable resistance determinants such as cfr-like genes, optrA and poxtA, or cfr encoding 23 S rRNA methyltransferase [31, 32]. In this study, the optrA was the main resistant mechanisms to linezolid resistant Enterococcus (LRE). All E. faecalis, with MICs mainly at 8 µg/mL (7/11) and 16 µg/mL (4/11), compared to 0.5 to 2 µg/mL linezolid-sensitive strains, highlighting optrA’s significant role in resistance [33]. Notably, a linezolid resistant E. faecalis strain (MIC = 16 µg/mL) displayed no detectable resistance gene in our research, suggesting other resistance mechanisms.
MLST revealed nine STs among VRE strains and seven STs of LRE strains. As we know, ST78 has been reported to be the predominant ST in vanA- and vanM-type VREfm strains in China [34, 35]. And ST16 was the most frequent ST among LRE isolates [36]. The majority of VREfm underscore the prevalence reported previously [37], whereas the predominance of LREfa suggests that E. faecalis may develop resistance to linezolid more readily than E. faecium. Clonal complex 17 (CC17) was a polyclonal group consisting of multiple STs, the epidemiology of enterococcal infections has been attributed to the increased ability of a genogroup of E. faecium related to human pathogen designated CC17 to colonize the gastrointestinal tract of humans, cause severe diseases [38]. ST78, ST17, ST18 and ST80 belonged to CC17 in this study (21/28), and there was one vanM-type CC17, which explained the higher resistance level of E. faecium than E. faecalis in our study. Similar to our report, ST78 of CC17 E. faecium strains had been reported previously in hospitalized patients, while the majority ST type of CC17 in Rao’s study was ST80 [39, 40]. E. faecalis isolates analyzed were differentiated into nine STs, among which ST16 of CC16 were the major lineage, similar to Aung’s study in Northern Japan [6].
Continuous emergence of VRE, and their resistance to other antibiotics like daptomycin, linezolid, oritavancin, and the evolution of new resistance mechanisms pose a serious global health threat [41]. In this study, E. faecalis showed better susceptibility to ampicillin and nitrofurantoin, and vancomycin resistance in urine samples was primarily associated with E. faecium, making ampicillin and nitrofurantoin suitable for treating E. faecalis infection from urine in our hospital. In addition, linezolid still could be an option for treating VREfm, as there was a bias towards E. faecalis (both in our study and from other studies) [28, 42]. In this study, VRE strains were all high-level vancomycin-resistance (≥ 256 µg/mL), and the potential for vanM-positive E. faecium to develop vancomycin-resistance with prolonged treatment, enhanced surveillance for vanM is advised [11, 28]. The detection of optrA could indicate linezolid resistance, highlighting the need for PCR-based molecular assays in clinical laboratories to facilitate rapid diagnostics due to the variability and time-consuming nature of conventional culture-based methods [43]. Besides, in our hospital, the majority of patients are convalescing, requiring a long-term hospital care, it is important to strengthen the clinical management of patients with VRE or LRE, education of healthcare workers, implementation and observation of hand-washing practices; active surveillance cultures (cultures at hospital admission, weekly cultures, cultures of high-risk patients) and the consequent prompt isolation of VRE or LRE-positive patients; and early isolation of high-risk patients [44].
Conclusions
This study showed that E. faecium, particularly VRE strains were all obtained from urine, while the sources of LRE specimens were diverse. E. faecium showed higher levels of resistance owing to genetic characterization. vanA and vanM were currently detected in VRE, meanwhile optrA was found in LRE.
Vancomycin was the front-line agent for the treatment of ampicillin-resistant enterococcal infections or in patients with severe β-lactam allergies until the emergence and dissemination of VRE [43] and it was worth noting that some vanM-carrying enterococci exhibit phenotypic susceptibility to vancomycin, and could revert to a vancomycin-resistant phenotype. The absence of phenotypically vancomycin sensitive strains for vanM detection in our study might result in a decrease in the proportion of vanM carrying Enterococcus strains. From Rao’s research, there was a significant association between virulence genes and antibiotics due to the presence of mobile resistance and virulence determinants on conjugative transposon [5], but our study was lacking the detection and analysis of virulence. Above all, the vanM gene among vancomycin phenotypically sensitive strains and assess virulence.
Antibiotic resistance was a growing threat to human health, primarily driven by the overuse of antibiotics in clinical medicine. Clinically, drug resistance emerges after a series of antibiotic treatments, this trend was reflected not only in the increasing proportion of isolates resistant to multiple antibiotics, but also in the evolution of resistance to specific antibiotics.
In summary, the effective combination of clinical strain’s sources, species identification, and drug resistance genotyping is of great significance for targeted clinical treatment. This approach aids healthcare providers and patients to make reasonable treatment decisions at the bedside. Besides, we will explore novel therapeutic strategies for combating multidrug-resistant Enterococcus infections in future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the clinical microbiology staff from all the hospitals that sent clinical Enterococcus isolates for this research.
Abbreviations
- CC
Clonal complex
- CSF
Cerebrospinal fluid
- CLSI
Clinical and Laboratory Standards Institute guidelines
- E.
Faecium Enterococcus faecium
- E.
Faecalis Enterococcus faecalis
- HAIs
Healthcare-associated infections
- LRE
Linezolid resistant Enterococcus
- LREfa
Linezolid resistant Enterococcus faecalis
- LREfm
Linezolid resistant Enterococcus faecium
- LREfs
Linezolid resistant Enterococcus
- LRVRE
Linezolid resistant vancomycin resistant Enterococcus
- MAR
Multiple antibiotic resistance
- MALDI-TOF
Matrix-assisted laser desorption/ionization time-of-flight
- MICs
Minimum inhibitory concentrations
- MLST
Multilocus sequence typing
- MS
Mass spectrometry
- Num.
Number
- PCR
Polymerase chain reaction
- STs
Characteristics of sequence types
- UTIs
Urinary tract infections
- VRE
Vancomycin resistant Enterococcus
- VREfm
Vancomycin resistant Enterococcus faecium
- VREfa
Vancomycin resistant Enterococcus faecalis
- WGS
Whole-genome sequencing
- XDR
Extensively drug-resistant
Author contributions
Qiang Shen: conceptualization, project administration, funding acquisition; Lihua Hu: data curation, resources; Xian Huang: original draft, software; Yiping Yin and Beilei Pan: validation; Xinyan Shi: review and editing the draft; Ping Pan: original draft, methodology; Long Sun: design of the work, review and editing the draft, data curation. All authors reviewed the manuscript.
Funding
This research was funded by Hangzhou agricultural and social development Scientific Research, grant number 20201203B151.
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Ethics approval and consent to participate
The authors declare that all the experimental research studies on strains, including the collection of strains, were carried out in accordance with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ping Pan and Long Sun contributed equally to this work.
Contributor Information
Lihua Hu, Email: hhx1819@sina.com.
Qiang Shen, Email: shenq215@163.com.
References
- 1.Fiore E, Van Tyne D, Gilmore MS. Pathogenicity Enterococci Microbiol Spectr. 2019;7(4). [DOI] [PMC free article] [PubMed]
- 2.Hollenbeck BL, Rice LB. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence. 2012;3(5):421–33. doi: 10.4161/viru.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristich CJ, Rice LB, Arias CA. Enterococcal infection—treatment and antibiotic resistance. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: from commensals to leading causes of drug resistant infection. Boston: Massachusetts Eye and Ear Infirmary; 2014. [PubMed] [Google Scholar]
- 4.Guzman Prieto AM, van Schaik W, Rogers MR, Coque TM, Baquero F, Corander J, et al. Global emergence and dissemination of Enterococci as nosocomial pathogens: attack of the clones? Front Microbiol. 2016;7:788. doi: 10.3389/fmicb.2016.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao C, Dhawan B, Vishnubhatla S, Kapil A, Das B, Sood S. Emergence of high-risk multidrug-resistant Enterococcus faecalis CC2 (ST181) and CC87 (ST28) causing healthcare-associated infections in India. Infect Genet Evolution: J Mol Epidemiol Evolutionary Genet Infect Dis. 2020;85:104519. doi: 10.1016/j.meegid.2020.104519. [DOI] [PubMed] [Google Scholar]
- 6.Aung MS, Urushibara N, Kawaguchiya M, Ohashi N, Hirose M, Kudo K et al. Antimicrobial Resistance, virulence factors, and genotypes of Enterococcus faecalis and Enterococcus faecium clinical isolates in Northern Japan: identification of optrA in ST480 E. faecalis. Antibiot (Basel Switzerland). 2023;12(1). [DOI] [PMC free article] [PubMed]
- 7.Smith TT, Tamma PD, Do TB, Dzintars KE, Zhao Y, Cosgrove SE, et al. Prolonged linezolid use is associated with the development of linezolid-resistant Enterococcus faecium. Diagn Microbiol Infect Dis. 2018;91(2):161–3. doi: 10.1016/j.diagmicrobio.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Solache M, Rice LB. The Enterococcus: a model of adaptability to its environment. Clin Microbiol Rev. 2019;32(2). [DOI] [PMC free article] [PubMed]
- 9.Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, et al. Infect Control Hosp Epidemiol. 2016;37(11):1288–301. Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. [DOI] [PMC free article] [PubMed]
- 10.Egan SA, Shore AC, O’Connell B, Brennan GI, Coleman DC. Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: high prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J Antimicrob Chemother. 2020;75(7):1704–11. doi: 10.1093/jac/dkaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Qu T, Wang D, Chen Y, Fu Y, Yang Q, et al. Characterization of vanM carrying clinical Enterococcus isolates and diversity of the suppressed vanM gene cluster. Infect Genet Evolution: J Mol Epidemiol Evolutionary Genet Infect Dis. 2019;68:145–52. doi: 10.1016/j.meegid.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 12.Bender JK, Cattoir V, Hegstad K, Sadowy E, Coque TM, Westh H, et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: towards a common nomenclature. Drug Resist Updates. 2018;40:25–39. doi: 10.1016/j.drup.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Gagetti P, Faccone D, Ceriana P, Lucero C, Menocal A, Argentina GL, et al. Emergence of optra-mediated linezolid resistance in clinical isolates of Enterococcus faecalis from Argentina. J Global Antimicrob Resist. 2023;35:335–41. doi: 10.1016/j.jgar.2023.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Sordo M, Grilo T, Freire S, Rodrigues B, Bouvier M, Poirel L, et al. Rapid culture-based LNZ test for detection of linezolid susceptibility/resistance in staphylococci and enterococci. Diagn Microbiol Infect Dis. 2023;107(3):116058. doi: 10.1016/j.diagmicrobio.2023.116058. [DOI] [PubMed] [Google Scholar]
- 15.Yang JX, Liu CW, Wu FW, Zhu L, Liang GW. Molecular characterization and biofilm formation ability of Enterococcus faecium and Enterococcus faecalis bloodstream isolates from a Chinese tertiary hospital in Beijing. Int Microbiology: Official J Span Soc Microbiol. 2023. [DOI] [PMC free article] [PubMed]
- 16.Turner AM, Lee JYH, Gorrie CL, Howden BP, Carter GP. Genomic insights into last-line Antimicrobial Resistance in Multidrug-Resistant Staphylococcus and Vancomycin-resistant Enterococcus. Front Microbiol. 2021;12:637656. doi: 10.3389/fmicb.2021.637656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munita JM, Bayer AS, Arias CA. Evolving resistance among Gram-positive pathogens. Clin Infect Diseases: Official Publication Infect Dis Soc Am. 2015;61(Suppl 2):S48–57. doi: 10.1093/cid/civ523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorrie C, Higgs C, Carter G, Stinear TP, Howden B. Genomics of Vancomycin-resistant Enterococcus faecium. Microb Genomics. 2019;5(7). [DOI] [PMC free article] [PubMed]
- 19.Humphries R, Bobenchik AM, Hindler JA, Schuetz AN. Overview of changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J Clin Microbiol. 2021;59(12):e0021321. doi: 10.1128/JCM.00213-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma X, Zhang F, Bai B, Lin Z, Xu G, Chen Z, et al. Linezolid Resistance in Enterococcus faecalis Associated with urinary tract infections of patients in a Tertiary hospitals in China: resistance mechanisms, virulence, and risk factors. Front Public Health. 2021;9:570650. doi: 10.3389/fpubh.2021.570650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shakir ZM, Alhatami AO, Ismail Khudhair Y, Muhsen Abdulwahab H. Antibiotic Resistance Profile and multiple antibiotic resistance index of Campylobacter species isolated from Poultry. Arch Razi Inst. 2021;76(6):1677–86. doi: 10.22092/ari.2021.356400.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang L, Huang C, Yan Y, Sun L, Li H. Urinary tract infection etiological profiles and antibiotic resistance patterns varied among different age categories: a retrospective study from a Tertiary General Hospital during a 12-Year period. Front Microbiol. 2021;12:813145. doi: 10.3389/fmicb.2021.813145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X, Willems RJL, Friedrich AW, Rossen JWA, Bathoorn E. Enterococcus faecium: from microbiological insights to practical recommendations for infection control and diagnostics. Antimicrob Resist Infect Control. 2020;9(1):130. doi: 10.1186/s13756-020-00770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park B, Min YH. In vitro synergistic effect of retapamulin with erythromycin and quinupristin against Enterococcus faecalis. J Atibiotics. 2020;73(9):630–5. doi: 10.1038/s41429-020-0312-7. [DOI] [PubMed] [Google Scholar]
- 25.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–27. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 26.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infection: Official Publication Eur Soc Clin Microbiol Infect Dis. 2012;18(3):268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 27.Osman K, Zolnikov TR, Badr J, Naim H, Hanafy M, Saad A, et al. Vancomycin and florfenicol resistant Enterococcus faecalis and Enterococcus faecium isolated from human urine in an Egyptian urban-rural community. Acta Trop. 2020;201:105209. doi: 10.1016/j.actatropica.2019.105209. [DOI] [PubMed] [Google Scholar]
- 28.Pfaller MA, Cormican M, Flamm RK, Mendes RE, Jones RN. Temporal and Geographic Variation in Antimicrobial susceptibility and resistance patterns of Enterococci: results from the SENTRY Antimicrobial Surveillance Program, 1997–2016. Open forum infectious diseases. 2019;6(Suppl 1):S54–62. [DOI] [PMC free article] [PubMed]
- 29.Mercuro NJ, Davis SL, Zervos MJ, Herc ES. Combatting resistant enterococcal infections: a pharmacotherapy review. Expert Opin Pharmacother. 2018;19(9):979–92. doi: 10.1080/14656566.2018.1479397. [DOI] [PubMed] [Google Scholar]
- 30.Miller WR, Murray BE, Rice LB, Arias CA. Resistance in vancomycin-resistant Enterococci. Infect Dis Clin N Am. 2020;34(4):751–71. doi: 10.1016/j.idc.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bender JK, Cattoir V, Hegstad K, Sadowy E, Coque TM, Westh H et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: towards a common nomenclature. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy. 2018;40:25–39. [DOI] [PubMed]
- 32.Sharkey LK, Edwards TA, O’Neill AJ. ABC-F Proteins Mediate Antibiotic Resistance through Ribosomal Protection. mBio. 2016;7(2):e01975. doi: 10.1128/mBio.01975-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dejoies L, Boukthir S, Péan de Ponfilly G, Le Guen R, Zouari A, Potrel S, et al. Performance of commercial methods for linezolid susceptibility testing of Enterococcus faecium and Enterococcus faecalis. J Antimicrob Chemother. 2020;75(9):2587–93. doi: 10.1093/jac/dkaa180. [DOI] [PubMed] [Google Scholar]
- 34.Sun HL, Liu C, Zhang JJ, Zhou YM, Xu YC. Molecular characterization of Vancomycin-resistant enterococci isolated from a hospital in Beijing, China. J Microbiol Immunol Infect. 2019;52(3):433–42. doi: 10.1016/j.jmii.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Sun J, Guo Y, Lin D, Guo Q, Hu F, et al. High prevalence of vanM in Vancomycin-resistant Enterococcus faecium isolates from Shanghai, China. Antimicrobial agents and chemotherapy. Antimicrob Agents Chemother. 2015;59(12):7795–8. doi: 10.1128/AAC.01732-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hua R, Xia Y. Molecular Epidemiology and mechanisms of 43 low-level linezolid-resistant Enterococcus faecalis strains in Chongqing. China Annals Lab Med. 2019;39(1):36–42. doi: 10.3343/alm.2019.39.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou W, Zhou H, Sun Y, Gao S, Zhang Y, Cao X, et al. Characterization of clinical enterococci isolates, focusing on the Vancomycin-resistant enterococci in a tertiary hospital in China: based on the data from 2013 to 2018. BMC Infect Dis. 2020;20(1):356. doi: 10.1186/s12879-020-05078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Permana B, Harris PNA, Runnegar N, Lindsay M, Henderson BC, Playford EG, et al. Using Genomics to investigate an outbreak of vancomycin-resistant Enterococcus faecium ST78 at a large Tertiary Hospital in Queensland. Microbiol Spectr. 2023;11(3):e0420422. doi: 10.1128/spectrum.04204-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Yuan Y, Tang M, Liu L, Yang K, Liu J. Phenotypic and genetic characteristics of Vancomycin-resistant Enterococcus faecium. Microb Pathog. 2019;128:131–5. doi: 10.1016/j.micpath.2018.12.046. [DOI] [PubMed] [Google Scholar]
- 40.Rao C, Dhawan B, Vishnubhatla S, Kapil A, Das B, Sood S. Clinical and molecular epidemiology of Vancomycin-resistant Enterococcus faecium bacteremia from an Indian tertiary hospital. Eur J Clin Microbiol Infect Dis. 2021;40(2):303–14. doi: 10.1007/s10096-020-04030-3. [DOI] [PubMed] [Google Scholar]
- 41.Xavier BB, Coppens J, De Koster S, Rajakani SG, Van Goethem S, Mzougui S et al. Novel Vancomycin resistance gene cluster in Enterococcus faecium ST1486, Belgium, June 2021. Euro Surveill. 2021;26(36). [DOI] [PMC free article] [PubMed]
- 42.Sabzi N, Moniri R, Sehat M, Fathizadeh H, Nazari-Alam A. Antimicrobial effect of silver and gold nanoparticles in combination with linezolid on Enterococcus biofilm. Iran J Microbiol. 2022;14(6):863–73. doi: 10.18502/ijm.v14i6.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan A, Miller WR. Antimicrobial susceptibility testing for Enterococci. J Clin Microbiol. 2022;60(9):e0084321. doi: 10.1128/jcm.00843-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tacconelli E, Cataldo MA. Vancomycin-resistant enterococci (VRE): transmission and control. Int J Antimicrob Agents. 2008;31:99–106. doi: 10.1016/j.ijantimicag.2007.08.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.