Abstract
Despite their successful implementation in the COVID-19 vaccines, lipid nanoparticles (LNPs) still face a central limitation in the delivery of mRNA payloads – endosomal trapping. Improving upon this inefficiency could afford improved drug delivery systems, paving the way toward safer and more effective mRNA-based medicines. Here, we present Polyphenolic Nanoparticle Platforms (PARCELs) as effective mRNA delivery systems. In brief, our investigation begins with a computationally guided structural analysis of 1825 discrete polyphenolic structural data points across 73 diverse small molecule polyphenols and 25 molecular parameters. We then generate structurally diverse PARCELs, evaluating their in vitro mechanism and activity, ultimately highlighting the superior endosomal escape properties of PARCELs relative to analogous LNPs. Finally, we examine the in vivo biodistribution, protein expression, and therapeutic efficacy of PARCELs in mice. In undertaking this approach, the goal of this study is to establish PARCELs as viable delivery platforms for safe and effective mRNA delivery.
Keywords: mRNA, lipid nanoparticles, polyphenol, endosomal escape, delivery platforms
Graphical Abstract
Maximizing therapeutic efficacy at the lowest possible dose is a fundamental objective of the drug delivery sciences.1–4 For example, this principle is true for mRNA-based therapies, which leverage LNP technologies as one way to reduce the required dose of mRNA-based drugs.5–11 Despite their promising role in preventing COVID-19 infections, LNPs suffer from a significant limitation – endosomal trapping.8,12–19 In essence, this process prevents mRNA from reaching the cytoplasm, thus hindering its translation into the desired therapeutic protein.20–23 Therefore, overcoming endosomal trapping is crucial for advancing non-viral vector-based mRNA delivery in biomedical applications.24–30
While many approaches exist to overcome endosomal trapping, one powerful strategy aims to develop novel material platforms whose structural features promote higher levels of endosomal escape.31–36 Development of these materials requires identifying molecular candidates from diverse and virtually infinite pools of chemical space that may improve endosomal escape. While many classes of these molecules exist, polyphenols (a class of naturally occurring small molecules found in nature) have emerged as a particularly promising group of molecules whose ability to interact with biological systems makes them attractive candidates in the drug delivery sciences.37–40 For example, polyphenols have been widely employed in addressing various diseases, including cardiovascular disease,41–43 Alzheimer’s and Parkinson’s disease,44–46 and cancer, amongst others, highlighting their potential utility as drug delivery agents.47–49 However, studies that leverage polyphenols to improve the efficacy of mRNA-based drugs, particularly from the standpoint of improving endosomal escape, currently remain underexplored.
Here, we present Polyphenolic Nanoparticle Platforms (PARCELs) as effective mRNA delivery systems (Figure 1). Our study begins with a computationally guided structural analysis of 1825 discrete polyphenolic structural data points to identify critical design parameters to incorporate into PARCEL. Informed by these data, we successfully formulate and characterize the mRNA delivery properties of PARCELs, ultimately evaluating their in vitro performance including intracellular (e.g., FLuc) and secreted (e.g., EPO) protein expression. To further the generalizability of these data, multiple types of mechanistic studies including cellular association, uptake mechanism, intracellular degradation and trafficking studies, and endosomal escape studies are then performed, ultimately highlighting that PARCEL has superior endosomal escape properties to analogous LNPs. Finally, we examine the in vivo biodistribution and protein expression of PARCELs in mice. In undertaking this approach, the goal of this study is to establish PARCEL as a viable platform for mRNA delivery, while more broadly highlighting the utility in synergizing techniques in structural analysis, formulation, and mechanism to afford better therapies.
Figure 1.
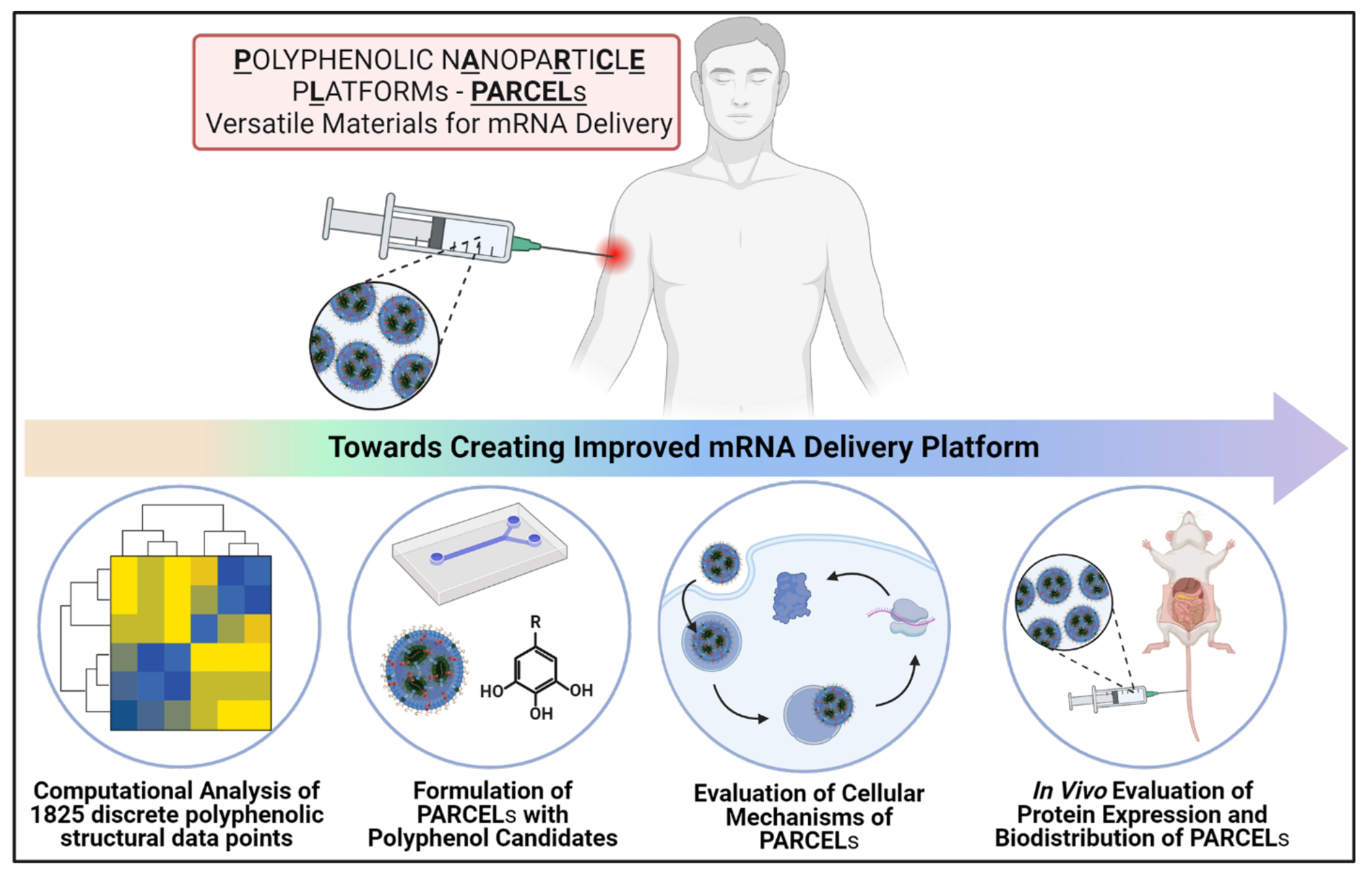
Schematic illustration of the overall concept of this manuscript – to develop and understand the functionality of PARCEL.
Given that polyphenols represent a large class of bioactive small molecules, we sought to begin our study by leveraging computationally guided approaches to select a representative class of diverse polyphenols for incorporation into PARCELs. Toward that end, we first generated 1825 discrete polyphenolic structural data points by analyzing 25 physiochemical properties of 73 unique polyphenols using Marvin Sketch (ChemoAxon) and visualized them as a heat map (Figure S1a, b, Table S1).50 Building on these analyses, we selected gallic acid (GA), catechin (CAT), epigallocatechin gallate (EGCG), and tannic acid (TA) as representative polyphenols for investigation in PARCEL due to their diverse structural features within the polyphenol family (Figure S1a, b). Principal component analysis (PCA) of our four selected polyphenols was then performed to project the 25-dimensional parameters into 2-dimensional space, highlighting the structural versatility across our selected polyphenols (Figure S1c).50
Each representative PARCEL was then formulated using microfluidic approaches by mixing an aqueous phase containing mRNA and an ethanol phase containing a clinically relevant ionizable lipid (either Moderna’s SM-102 or Pfizer/BioNTech’s ALC-0315),51–53 a phospholipid (DOPE),54 cholesterol,55,56 a PEG lipid (C14-PEG-2000),57 and a polyphenol (GA, CAT, EGCG or TA) (Figure 2a, b).14,35,40,58,59 The PARCELs were formulated at a ratio of 56/10/23/6/11 for SM-102/DOPE/cholesterol/C14-PEG-2000/polyphenol (Figure 2c, Figure S1d). The size, charge, and mRNA encapsulation efficiency for each PARCEL was reproducible, with sizes ranging from ~109 nm to ~154 nm (Figure 2d), PDI ranging from ~0.16 to ~0.28 (Figure 2e), zeta potentials ranging from ~−1.5 mV to ~1.0 mV (Figure 2f), and mRNA encapsulation efficiencies ranging from ~78.9% to ~92.4% (Figure 2g). It is noted that the size of PARCELs was similar to the LNP formulation and mRNA encapsulation efficiencies of PARCELs were greater than the LNP formulation. The pKa for each PARCEL was ranging from 6.8 to 7.4 (Figure S2).
Figure 2.
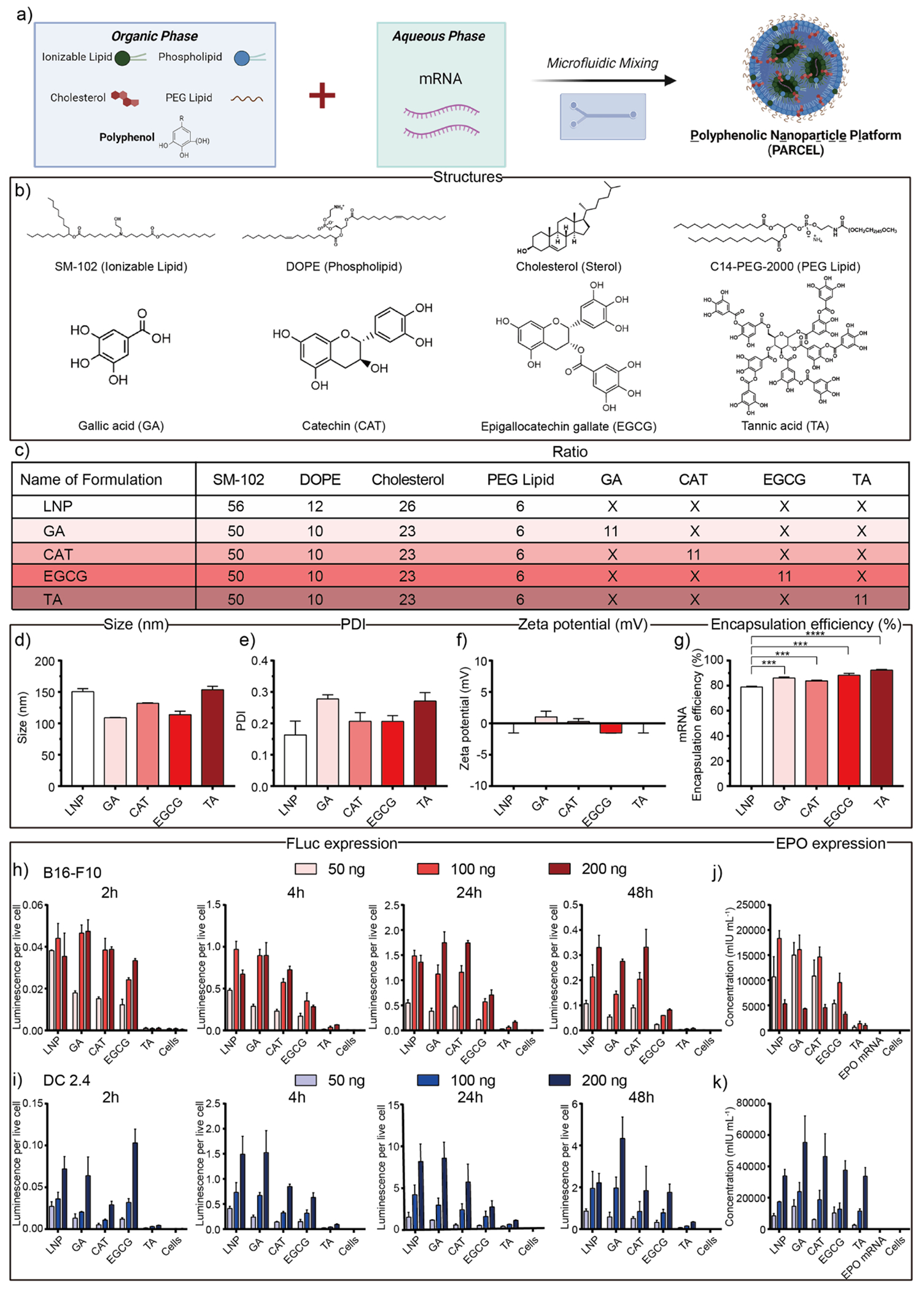
(a) Schematic illustration of mRNA-loaded PARCEL formulation via microfluidic chip. (b) Chemical structures of representative molecular excipients within mRNA-loaded PARCEL were used in this study. (c) Composition ratios for the formulation of mRNA-loaded PARCEL including GA, CAT, EGCG, and TA with the same weight ratio. (d) Size/Diameter, (e) PDI, (f) zeta potentials, and (g) mRNA encapsulation efficiency of PARCELs., (****p < 0.0001 and ***p < 0.001 with 95% of confidence level from unpaired t-test). In vitro FLuc expression of PARCELs treated on (h) B16-F10 and (i) DC 2.4 cells under 50, 100, and 200 ng mRNA dose per well across desired time (2, 4, 24, 48 h). In vitro EPO expression of PARCEL treated on (j) B16-F10 and (k) DC 2.4 cells under 50, 100, and 200 ng doses per well for 24 h. (All data presented as mean ± SD, n = 3).
Having evaluated the structure and the formulation of PARCELs, we then evaluated their in vitro efficacy by evaluating the protein expression in a dose-responsive (50, 100 and 200 ng) and a time-dependent (2, 4, 24 and 48 h) fashion using mRNA encoding for either firefly luciferase (FLuc, an intracellular protein) or human erythropoietin (EPO, a secreted protein) (Figure 2h–k, Table S2). Given the utility of mRNA therapies in cancer immunotherapy, we performed these studies on DC 2.4 cells (a dendritic cell line relevant as antigen presenting cells) and B16-F10 cells (a melanoma cell line). Upon collectively analyzing these data, several trends emerged. First, PARCELs were well-tolerated under each studied condition (Figure S3, S4). Second, FLuc and EPO expressions for each PARCEL were higher for DC 2.4 cells than for B16-F10 cells. Third, in the time-dependent scenario, FLuc expression for each PARCEL increased from 2 h to 24 h followed by a decrease in expression at 48 h (Figure 2h, i). Fourth, in a dose-responsive scenario, FLuc expression generally increased with FLuc mRNA dose from 50 ng to 200 ng (Figure 2h, i). Alternatively, treatment of B16-F10 cells and DC 2.4 cells with EPO mRNA PARCELs for 24 hours showed the highest EPO expression at 100 ng and 200 ng overall mRNA doses, respectively (Figure 2j, k). Finally, different PARCELs resulted in different levels of in vitro protein expression across both cell lines, highlighting the importance of polyphenol selection in PARCEL.
To explore the reasons for differences in protein expression, we next sought to explore several mechanistic studies to better understand mRNA delivery using each PARCEL. To begin, we quantified the cellular uptake of PARCELs in vitro using flow cytometry and confocal microscopy (Figure 3a–f, Figure S5, Table S3). To further complement these studies, we also sought to elucidate the specific endocytosis pathway and degradation properties of each PARCEL. In brief, these mechanism studies were performed by the inhibition of various endocytic pathways, specifically caveolin-mediated endocytosis, clathrin-dependent endocytosis, micropinocytosis, phagocytosis, and energy-dependent endocytosis (Figure 3g).60–62 In collectively analyzing these data, several trends emerged. First, TA PARCEL had lower cellular uptake compared to other PARCELs across multiple time points (Figure 3a, c). Second, the cellular uptake of PARCELs was time-dependent, and maximum uptake was observed at 24 h for B16-F10 cells and 4 h for DC 2.4 cells. Third, each PARCEL was degraded after 24 h in B16-F10 cells and 4 h in DC 2.4 cells (Figure 3a, c), as indicated through the decrease in geometric mean fluorescence intensity (GMFI) after 24 h for B16-F10 cells and 4 h for DC 2.4 cells using flow cytometry (Figure 3b, d) and confocal microscopy (Figure 3e, f). Finally, phagocytosis was shown to be the main mechanism for non-TA PARCEL uptake, while micropinocytosis and energy-dependent endocytosis were important mechanisms for TA PARCELs (Figure 3g, h). Taken in tandem, these results suggest that PARCELs are internalized in a time-dependent fashion, through a combination of endocytic pathways for respective PARCELs.
Figure 3.
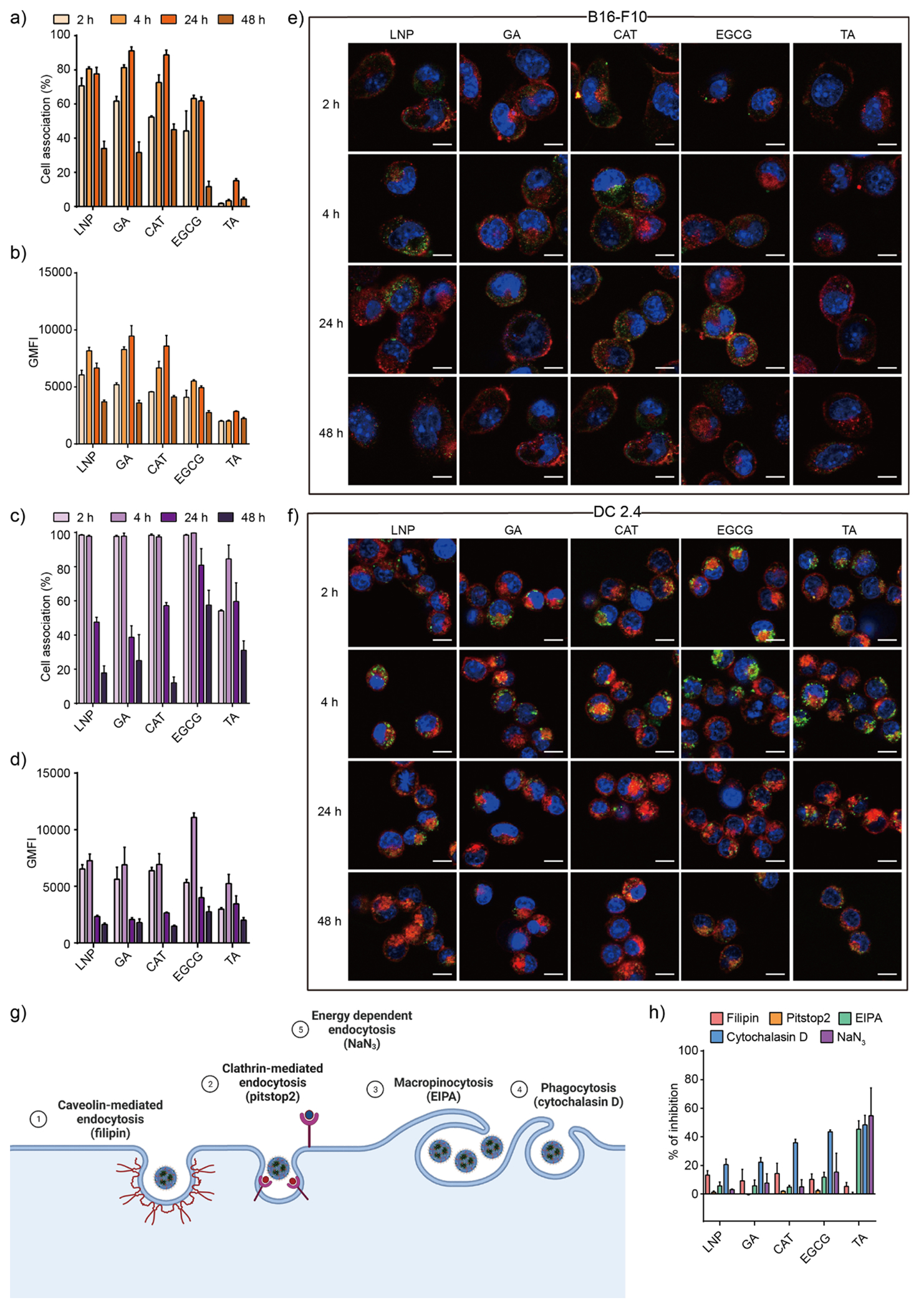
(a) Cellular uptake and (b) GMFI of B16-F10 cells treated with PARCEL at varying incubation times of 2, 4, 24, and 48 h. (c) Cellular uptake and (d) GMFI of DC 2.4 cells treated with PARCEL at varying incubation times of 2, 4, 24, and 48 h. Representative confocal microscopy images showing the intracellular trafficking of PARCEL in (e) B16-F10 and (f) DC 2.4 cells at varying incubation times of 2, 4, 24, and 48 h. Green: ATTO-488 labeled PARCEL; blue: nuclei; red: cell membrane. Scale bars are 10 μm. (g) Schematic illustration of the cell internalization mechanisms with corresponding related inhibitors used. (h) Study of the cell internalization mechanism of PARCEL by monitoring the cellular uptake efficiency in the presence of different endocytic inhibitors. Cells were treated with 500 ng mL−1 of PARCEL at 37 °C (All data presented as mean ± SD, n = 3).
Following cellular internalization/uptake studies, we sought to understand how well each PARCEL could escape endosomal trapping. In brief, endosomal escape studies were performed by incubating nuclei and endo/lysosome labeled DC 2.4 cells with ATTO-488 labeled FLuc mRNA PARCELs and performing confocal microscopy to analyze the colocalization of PARCELs [Figure 4a–c; In these confocal images, cell nuclei are blue, mRNA-loaded PARCEL are green, and endo/lysosomes are red; yellow (i.e. colocalization of the green and red signals) suggests that the mRNA PARCEL remain trapped in endosomes]. As a benchmark, confocal imaging was also performed on cells incubated with analogous LNPs. As a quantifiable endosomal escape metric for each PARCEL, the Pearson Coefficient Correlation (PCC) was also determined (where a PCC value of 0 indicates complete endosomal escape and a PCC value of 1 indicates no endosomal escape).63,64 To provide further insight into the endosomal escape properties of each PARCEL, we also investigated the buffering capacity of each PARCEL given that buffering capacity may relate to endosomal escape (Figure 4d).64 To further add depth to our understanding of endosomal escape, “label-free” approaches for each PARCEL were also investigated (Figure 4e–g). In brief, these “label-free” studies were performed using enzyme inhibition/brightfield imaging studies with bafilomycin A1 (a molecule that inhibits proton sponge aided endosomal escape by inhibiting V-ATPases)65,66 and calcein (a membrane-impermeable dye that remains entrapped within intact endosomes but becomes distributed throughout cells if endo/lysosomes are ruptured), in which the calcein was directly added to the cells followed by adding the PARCEL or the bafilomycin A1 (Figure 4e–g).
Figure 4.
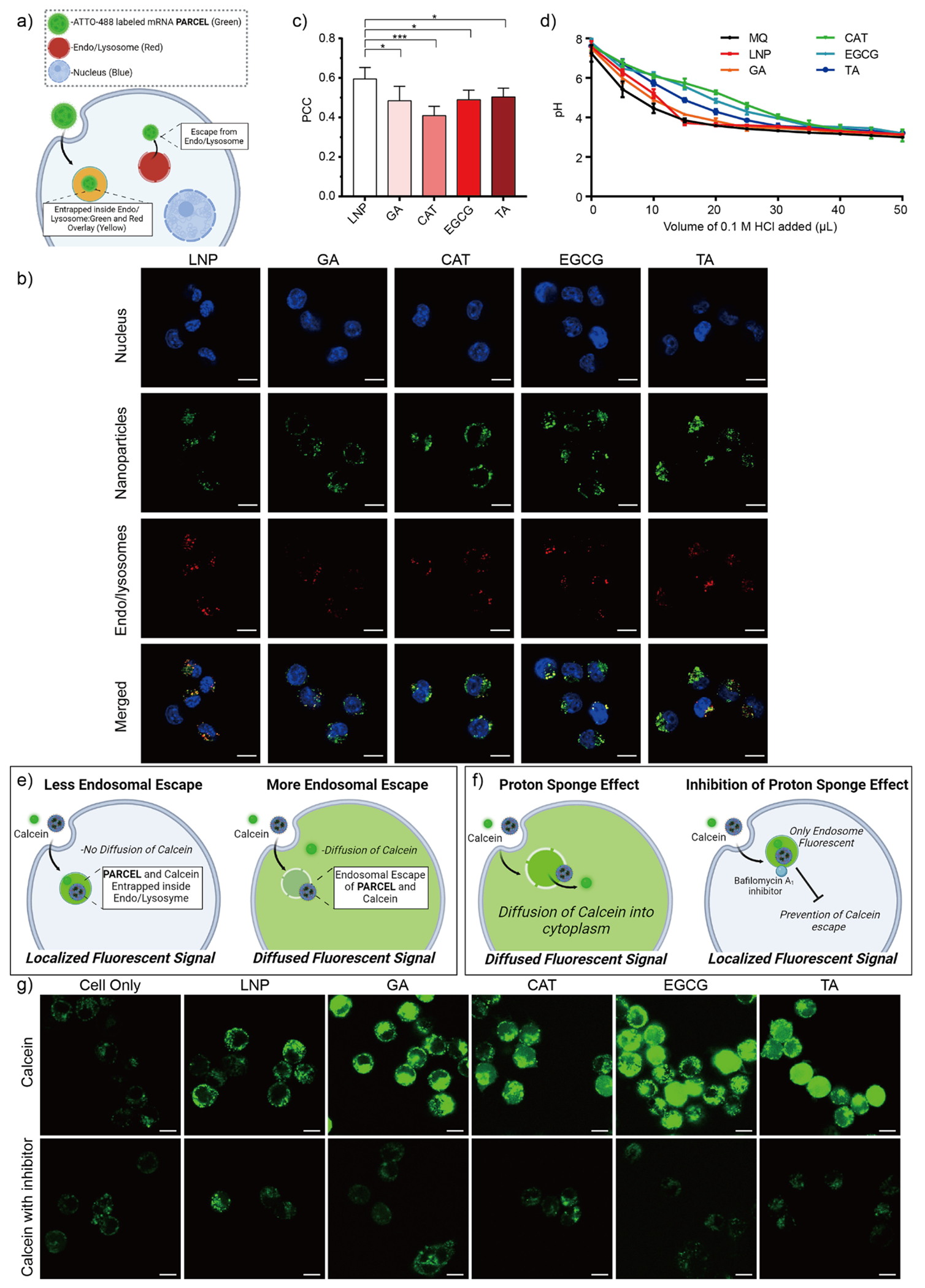
(a) Schematic illustration of our endosomal escape studies using a lysotracker confocal imaging assay. (b) Representative confocal images of DC 2.4 cells treated with ATTO-488 labeled PARCEL (green). Endo/lysosomes (red) were stained with LysoTracker Deep Red. Nuclei (blue) were stained with Hoechst 33342. Scale bars are 10 μm. (c) Pearson Correlation Coefficient (PCC) analysis of ATTO-488 labeled PARCEL (Data presented as the mean ± SD, ***p < 0.001 and *p < 0.05 with 95% of confidence level from unpaired t-test, Figure S6). (d) Titration curves of PARCEL in suspensions as a function of HCl. Schematic illustration of our endosomal escape studies using (e) a calcein assay and (f) a proton sponge effect assay using bafilomycin A1 for the termination of the inflow of H+ and Cl−. (g) Representative confocal images of DC 2.4 cells incubated with calcein and PARCEL in the absence (top row) and presence (bottom row) of inhibitor bafilomycin A1 for 4 h at 37 °C. PARCELs were not fluorescently labeled to avoid interference with the calcein signal. Scale bars are 10 μm. (All data presented as mean ± SD, n = 3).
Upon analyzing these mechanistic data, several findings were observed. First, PARCELs had better endosomal escape than analogous LNPs as observed by lesser colocalization (i.e., less yellow color) in confocal microscopy images (Figure 4b) and lower PCC values (Figure 4c). Second, the pH value of CAT, EGCG, and TA PARCEL samples gradually decreased with the addition of HCl, as compared to the GA PARCEL, LNP, and MilliQ water (as a control), suggesting that different PARCEL can differentially buffer protons which may be important for endosomal escape (Figure 4d). Third, diffuse fluorescence of calcein dye was observed when cells were incubated with PARCEL compared to LNP (upper row, Figure 4g) further suggesting the superior endosomal escape properties of PARCELs as compared to analogous mRNA LNPs. Fourth, punctuated fluorescence was observed in cells treated with bafilomycin A1 and PARCELs (bottom row, Figure 4g), suggesting that the ‘proton sponge effect’ could potentially be one of the mechanisms for triggering endosomal escape of PARCEL. Taken in tandem, these results highlight PARCELs as versatile mRNA carriers with tunable endosomal escape properties.
Building on the previous data, we finally sought to establish the in vivo delivery properties of each PARCEL. Briefly, Black 6 mice were treated with each PARCEL delivering mRNA encoding for FLuc (Figure 5a–c, Table S4) or EPO (Figure 5d, Table S5) via intravenous (IV) administration. Tolerability studies including histological evaluation (Figure 5e), liver and kidney function blood tests (Figure 5f) within complete blood paneling (Figure S7, S8), and weight loss studies (Figure S9, S10) were also evaluated for each PARCEL. Upon analyzing these data, several trends were observed. First, EGCG and TA PARCELs displayed higher FLuc expression than LNP as suggested by the increased FLuc signal in comparative IVIS imaging on the resected organs of treated mice (Figure 5a, c). Second, the increases in FLuc expression occurred without altering the innate biodistribution of each studied PARCEL (Figure 5b). Third, EGCG and TA PARCELs also increased the amount of EPO expression secreted into the blood of treated mice (Figure 5d). Fourth, each PARCEL was well tolerated as analyzed by histology (Figure 5e), weight retention (Figure S9, S10), and complete blood paneling data including normal alkaline phosphatase (ALP), alanine transaminase (ALT), aspartate transferase (AST), blood urea nitrogen (BUN) and creatinine (CREAT) levels which are markers of liver and kidney function (Figure 5f). Taken in tandem, these results suggest that EGCG and TA PARCEL had better in vivo protein expression than analogous LNP formulations, highlighting their potential for therapeutic mRNA delivery.
Figure 5.
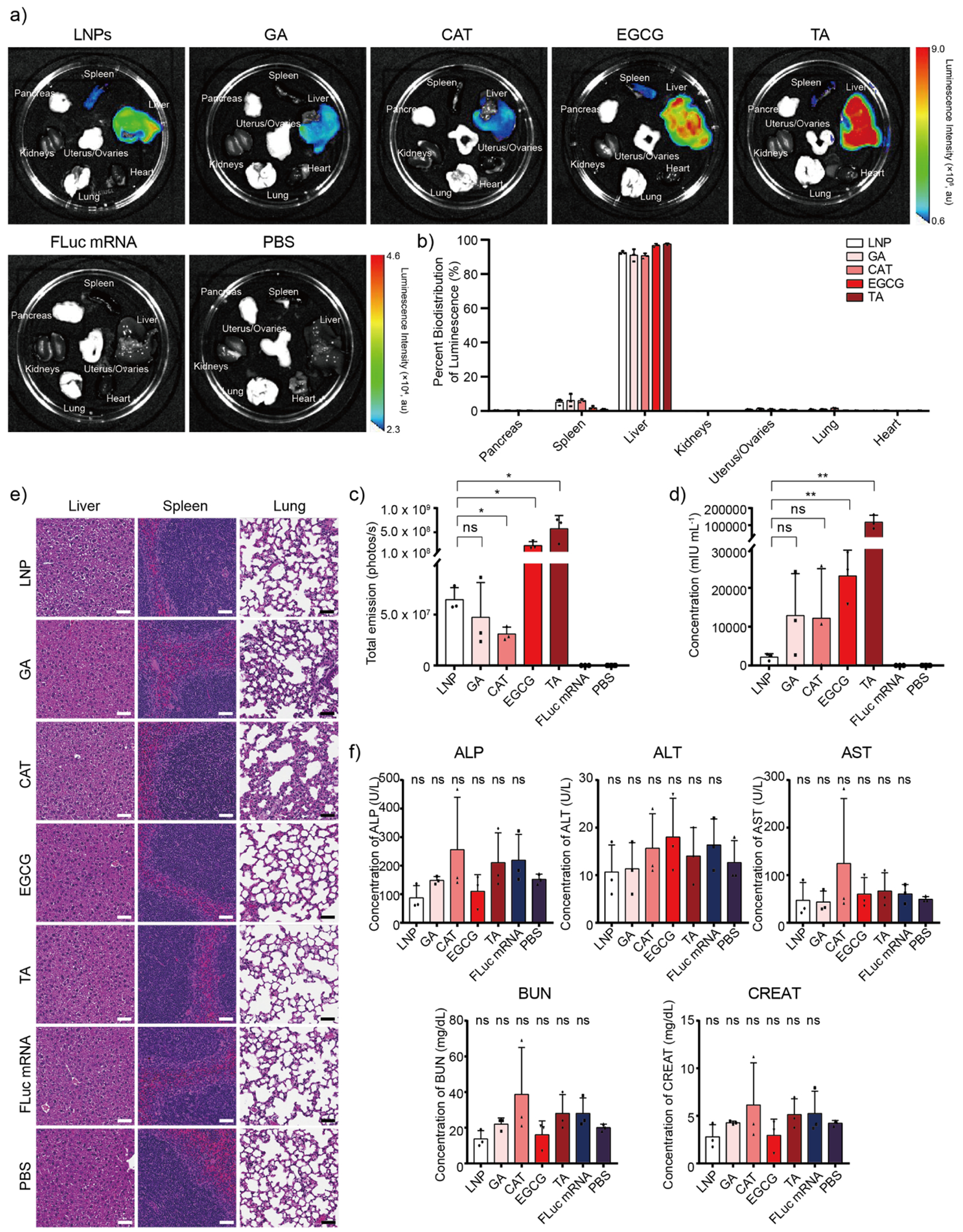
(a) Representative luminescence biodistribution of PARCEL encapsulated with FLuc mRNA ex vivo (n = 3) for each group via intravenous injection. Mice injected with naked FLuc mRNA and PBS were used as controls. (b) Associated percent of bioluminescence and (c) total luminescence of FLuc mRNA encapsulated PARCEL across various organs including the pancreas, spleen, liver, kidneys, uterus/ovaries, lung, and heart. (d) Human EPO concentration after the injection of EPO mRNA encapsulated PARCEL for 24 h. Mice injected with naked EPO mRNA and PBS were used as controls. The concentration of human erythropoietin was characterized by Human EPO ELISA kits following the manufacturer’s protocol. (e) Representative histology images of the liver, spleen, and lung of mice after treatment with FLuc mRNA encapsulated PARCEL via IV injection routes (n = 3). Scale bars are 50 μm. (f) ALP, ALT, AST, BUN, and CREAT blood testing results after the IV injection of FLuc mRNA encapsulated PARCEL (ns > 0.05 with 95% confidence level from unpaired t-test with PBS group, and all data presented as mean ± SD, n = 3).
In this report, we provide computationally guided, formulation-driven, and mechanism-driven studies to realize the development of PARCELs as a safe and effective mRNA delivery platform. In brief, we demonstrate their effectiveness as an mRNA delivery system by evaluating their physiochemical properties including the size, PDI, charge, encapsulation efficiency, as well as the mechanisms behind their cellular performance such as endocytosis and endosomal escape. Furthermore, our research also showed that TA PARCEL exhibited the best in vivo efficacy on both intracellular and secreted protein expression. Future work will be directed toward assessing the utility of PARCEL in the field of cancer immunotherapies and furthering the therapeutic utility of PARCEL for mRNA delivery. Taken collectively, the goal of this study was to establish PARCEL as a viable platform for mRNA delivery with superior endosomal escape properties to analogous LNPs while more broadly highlighting the utility of synergizing techniques in structural analysis, formulation, and mechanism to afford better therapies for the study and prevention of disease using mRNA.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by an NIH National Institute of Biomedical Imaging and Bioengineering award (1R21EB034942-01). This work was also supported by the NC Translational and Clinical Sciences (NC TraCS) Institute which is supported by the NIH National Center for Advancing Translational Sciences (NCATS) award 1K12TR004416-01. We also thank Cassie Pham, Mia Evangelista, and Rani Sellers in the Pathology Services Core for expert technical assistance with Histopathology and Digital Pathology. The PSC is supported in part by an NCI Center Core Support Grant (5P30CA016080-42). All animal studies were approved by the UNC Institutional Animal Care and Use Committee, were consistent with local, state, and federal regulations as applicable, and were supported within the UNC Lineberger ASC at the University of North Carolina at Chapel Hill which is supported in part by an NCI Center Core Support Grant (CA16086) to the UNC Lineberger Comprehensive Cancer Center. Microscopy was performed at the UNC Neuroscience Microscopy Core (RRID: SCR 019060), supported, in part, by funding from the NIH-NINDS Neuroscience Center Support Grant P30 NS045892 and the NIH-NICHD Intellectual and Developmental Disabilities Research Center Support Grant P50 HD103573. Research reported in this publication was supported in part by the North Carolina Biotech Center Institutional Support Grant 2017-IDG-1025 and by the National Institutes of Health 1UM2AI30836-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Figure 1, Figure 2a, Figure 3g, Figure 4a, 4e, 4f, and TOC figure created with BioRender.com.
Footnotes
Conflict of Interest
The authors declare no other competing financial interests.
Supporting Information
Materials, experimental procedures (synthesis and characterization of PARCEL protocol, quantification of in vitro FLuc expression protocol, EPO protein concentration protocol, cell viability protocol, cellular association protocol, intracellular trafficking protocol, endocytosis mechanism protocol, endosomal escape protocol, endosomal escape with bafilomycin A1 protocol, buffering capacity protocol, in vivo injection protocol, in vivo blood collection protocol), computational analysis, 1H NMR, cell viability data, size, charge, encapsulation data, gating strategies for flow cytometer, confocal microscopy images for PCC analysis, complete blood test data, weight gain data, meaning of physicochemical properties of polyphenols.
REFERENCES
- (1).Vargason AM; Anselmo AC; Mitragotri S The Evolution of Commercial Drug Delivery Technologies. Nat. Biomed. Eng 2021, 5 (9), 951–967. DOI: 10.1038/s41551-021-00698-w. [DOI] [PubMed] [Google Scholar]
- (2).Paunovska K; Loughrey D; Dahlman JE Drug Delivery Systems for RNA Therapeutics. Nat. Rev. Genet 2022, 23 (5), 265–280. DOI: 10.1038/s41576-021-00439-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Tibbitt MW; Dahlman JE; Langer R Emerging Frontiers in Drug Delivery. J. Am. Chem. Soc 2016, 138 (3), 704–717. DOI: 10.1021/jacs.5b09974. [DOI] [PubMed] [Google Scholar]
- (4).Zhang Q; Ma D; Wu F; Standage-Beier K; Chen X; Wu K; Green AA; Wang X Predictable Control of RNA Lifetime Using Engineered Degradation-Tuning RNAs. Nat. Chem. Biol 2021, 17 (7), 828–836. DOI: 10.1038/s41589-021-00816-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).VanKeulen-Miller R; Fenton OS Messenger RNA Therapy for Female Reproductive Health. Mol. Pharm 2024. DOI: 10.1021/acs.molpharmaceut.3c00803. [DOI] [PubMed] [Google Scholar]
- (6).Hou X; Zaks T; Langer R; Dong Y Lipid Nanoparticles for MRNA Delivery. Nat. Rev. Mater 2021, 6 (12), 1078–1094. DOI: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Eygeris Y; Gupta M; Kim J; Sahay G Chemistry of Lipid Nanoparticles for RNA Delivery. Acc. Chem. Res 2022, 55 (1), 2–12. DOI: 10.1021/acs.accounts.1c00544. [DOI] [PubMed] [Google Scholar]
- (8).Chaudhary N; Weissman D; Whitehead KA MRNA Vaccines for Infectious Diseases: Principles, Delivery and Clinical Translation. Nat. Rev. DrugDiscov 2021, 20 (11), 817–838. DOI: 10.1038/s41573-021-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lokugamage MP; Vanover D; Beyersdorf J; Hatit MZC; Rotolo L; Echeverri ES; Peck HE; Ni H; Yoon J-K; Kim Y; Santangelo PJ; Dahlman JE Optimization of Lipid Nanoparticles for the Delivery of Nebulized Therapeutic MRNA to the Lungs. Nat. Biomed. Eng 2021, 5 (9), 1059–1068. DOI: 10.1038/s41551-021-00786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).McKay PF; Hu K; Blakney AK; Samnuan K; Brown JC; Penn R; Zhou J; Bouton CR; Rogers P; Polra K; Lin PJC; Barbosa C; Tam YK; Barclay WS; Shattock RJ Self-Amplifying RNA SARS-CoV-2 Lipid Nanoparticle Vaccine Candidate Induces High Neutralizing Antibody Titers in Mice. Nat. Commun 2020, 11 (1), 3523. DOI: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Labouta HI; Langer R; Cullis PR; Merkel OM; Prausnitz MR; Gomaa Y; Nogueira SS; Kumeria T Role of Drug Delivery Technologies in the Success of COVID-19 Vaccines: A Perspective. Drug Deliv. Transl. Res 2022, 12 (11), 2581–2588. DOI: 10.1007/s13346-022-01146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Herrera M; Kim J; Eygeris Y; Jozic A; Sahay G Illuminating Endosomal Escape of Polymorphic Lipid Nanoparticles That Boost MRNA Delivery. Biomater. Sci 2021, 9 (12), 4289–4300. DOI: 10.1039/d0bm01947j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kulkarni JA; Witzigmann D; Thomson SB; Chen S; Leavitt BR; Cullis PR; van der Meel R The Current Landscape of Nucleic Acid Therapeutics. Nat. Nanotechnol 2021, 16 (6), 630–643. DOI: 10.1038/s41565-021-00898-0. [DOI] [PubMed] [Google Scholar]
- (14).Ma Y; Fenton OS Tannic Acid Lipid Nanoparticles Can Deliver Messenger RNA Payloads and Improve Their Endosomal Escape. Adv. Therap 2023, 2200305. DOI: 10.1002/adtp.202200305. [DOI] [Google Scholar]
- (15).Andresen JL; Fenton OS Nucleic Acid Delivery and Nanoparticle Design for COVID Vaccines. MRS Bull. 2021, 46 (9), 832–839. DOI: 10.1557/s43577-021-00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Mitchell MJ; Billingsley MM; Haley RM; Wechsler ME; Peppas NA; Langer R Engineering Precision Nanoparticles for Drug Delivery. Nat. Rev. Drug Discov 2021, 20 (2), 101–124. DOI: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hajj KA; Ball RL; Deluty SB; Singh SR; Strelkova D; Knapp CM; Whitehead KA Branched-Tail Lipid Nanoparticles Potently Deliver MRNA In Vivo Due to Enhanced Ionization at Endosomal PH. Small 2019, 15 (6), e1805097. DOI: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- (18).Akinc A; Maier MA; Manoharan M; Fitzgerald K; Jayaraman M; Barros S; Ansell S; Du X; Hope MJ; Madden TD; Mui BL; Semple SC; Tam YK; Ciufolini M; Witzigmann D; Kulkarni JA; van der Meel R; Cullis PR The Onpattro Story and the Clinical Translation of Nanomedicines Containing Nucleic Acid-Based Drugs. Nat. Nanotechnol 2019, 14 (12), 1084–1087. DOI: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- (19).Wang X; Liu S; Sun Y; Yu X; Lee SM; Cheng Q; Wei T; Gong J; Robinson J; Zhang D; Lian X; Basak P; Siegwart DJ Preparation of Selective Organ-Targeting (SORT) Lipid Nanoparticles (LNPs) Using Multiple Technical Methods for Tissue-Specific MRNA Delivery. Nat. Protoc 2023, 18 (1), 265–291. DOI: 10.1038/s41596-022-00755-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sun B; Wu W; Narasipura EA; Ma Y; Yu C; Fenton OS; Song H Engineering Nanoparticle Toolkits for MRNA Delivery. Adv. Drug Deliv. Rev 2023, 200, 115042. DOI: 10.1016/j.addr.2023.115042. [DOI] [PubMed] [Google Scholar]
- (21).Reichmuth AM; Oberli MA; Jeklenec A; Langer R; Blankschtein D MRNA Vaccine Delivery Using Lipid Nanoparticles. Ther. Deliv 2016, 7 (5), 319–334. DOI: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ma Y; Li S; Lin X; Chen Y Bioinspired Spatiotemporal Management toward RNA Therapies. ACS Nano 2023, 17 (24), 24539–24563. DOI: 10.1021/acsnano.3c08219. [DOI] [PubMed] [Google Scholar]
- (23).Fenton OS; Olafson KN; Pillai PS; Mitchell MJ; Langer R Advances in Biomaterials for Drug Delivery. Adv. Mater 2018, e1705328. DOI: 10.1002/adma.201705328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Neill B; Romero AR; Fenton OS Advances in Nonviral MRNA Delivery Materials and Their Application as Vaccines for Melanoma Therapy. ACS Appl. Bio Mater 2023. DOI: 10.1021/acsabm.3c00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Oberli MA; Reichmuth AM; Dorkin JR; Mitchell MJ; Fenton OS; Jaklenec A; Anderson DG; Langer R; Blankschtein D Lipid Nanoparticle Assisted MRNA Delivery for Potent Cancer Immunotherapy. Nano Lett. 2017, 17 (3), 1326–1335. DOI: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Guevara ML; Persano F; Persano S Advances in Lipid Nanoparticles for MRNA-Based Cancer Immunotherapy. Front. Chem 2020, 8, 589959. DOI: 10.3389/fchem.2020.589959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pack DW; Hoffman AS; Pun S; Stayton PS Design and Development of Polymers for Gene Delivery. Nat. Rev. Drug Discov 2005, 4 (7), 581–593. DOI: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- (28).Merkel OM; Kissel T Nonviral Pulmonary Delivery of SiRNA. Acc. Chem. Res 2012, 45 (7), 961–970. DOI: 10.1021/ar200110p. [DOI] [PubMed] [Google Scholar]
- (29).Frederiksen LSF; Zhang Y; Foged C; Thakur A The Long Road Toward COVID-19 Herd Immunity: Vaccine Platform Technologies and Mass Immunization Strategies. Front. Immunol 2020, 11, 1817. DOI: 10.3389/fimmu.2020.01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Narasipura EA; VanKeulen-Miller R; Ma Y; Fenton OS Ongoing Clinical Trials of Nonviral SiRNA Therapeutics. Bioconjug. Chem 2023, 34 (7), 1177–1197. DOI: 10.1021/acs.bioconjchem.3c00205. [DOI] [PubMed] [Google Scholar]
- (31).Pei D; Buyanova M Overcoming Endosomal Entrapment in Drug Delivery. Bioconjug. Chem 2019, 30 (2), 273–283. DOI: 10.1021/acs.bioconjchem.8b00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Yanez Arteta M; Kjellman T; Bartesaghi S; Wallin S; Wu X; Kvist AJ; Dabkowska A; Székely N; Radulescu A; Bergenholtz J; Lindfors L Successful Reprogramming of Cellular Protein Production through MRNA Delivered by Functionalized Lipid Nanoparticles. Proc Natl Acad Sci USA 2018, 115 (15), E3351–E3360. DOI: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Rui Y; Wilson DR; Tzeng SY; Yamagata HM; Sudhakar D; Conge M; Berlinicke CA; Zack DJ; Tuesca A; Green JJ High-Throughput and High-Content Bioassay Enables Tuning of Polyester Nanoparticles for Cellular Uptake, Endosomal Escape, and Systemic in Vivo Delivery of MRNA. Sci. Adv 2022, 8 (1), eabk2855. DOI: 10.1126/sciadv.abk2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Coolen A-L; Lacroix C; Mercier-Gouy P; Delaune E; Monge C; Exposito J-Y; Verrier B Poly(Lactic Acid) Nanoparticles and Cell-Penetrating Peptide Potentiate MRNA-Based Vaccine Expression in Dendritic Cells Triggering Their Activation. Biomaterials 2019, 195, 23–37. DOI: 10.1016/j.biomaterials.2018.12.019. [DOI] [PubMed] [Google Scholar]
- (35).Han Y; Zhou J; Hu Y; Lin Z; Ma Y; Richardson JJ; Caruso F Polyphenol-Based Nanoparticles for Intracellular Protein Delivery via Competing Supramolecular Interactions. ACS Nano 2020, 14 (10), 12972–12981. DOI: 10.1021/acsnano.0c04197. [DOI] [PubMed] [Google Scholar]
- (36).Winkeljann B; Keul DC; Merkel OM Engineering Poly- and Micelleplexes for Nucleic Acid Delivery - A Reflection on Their Endosomal Escape. J. Control. Release 2023, 353, 518–534. DOI: 10.1016/j.jconrel.2022.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Singla RK; Dubey AK; Garg A; Sharma RK; Fiorino M; Ameen SM; Haddad MA; Al-Hiary M Natural Polyphenols: Chemical Classification, Definition of Classes, Subcategories, and Structures. J. AOAC Int 2019, 102 (5), 1397–1400. DOI: 10.5740/jaoacint.19-0133. [DOI] [PubMed] [Google Scholar]
- (38).Kumar N; Goel N Phenolic Acids: Natural Versatile Molecules with Promising Therapeutic Applications. Biotechnol Rep (Amst) 2019, 24, e00370. DOI: 10.1016/j.btre.2019.e00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Feng Y; Li P; Wei J Engineering Functional Mesoporous Materials from Plant Polyphenol Based Coordination Polymers. Coord. Chem. Rev 2022, 468, 214649. DOI: 10.1016/j.ccr.2022.214649. [DOI] [Google Scholar]
- (40).Qu Y; Ju Y; Cortez-Jugo C; Lin Z; Li S; Zhou J; Ma Y; Glab A; Kent SJ; Cavalieri F; Caruso F Template-Mediated Assembly of DNA into Microcapsules for Immunological Modulation. Small 2020, 16 (37), e2002750. DOI: 10.1002/smll.202002750. [DOI] [PubMed] [Google Scholar]
- (41).Quiñones M; Miguel M; Aleixandre A Beneficial Effects of Polyphenols on Cardiovascular Disease. Pharmacol. Res 2013, 68 (1), 125–131. DOI: 10.1016/j.phrs.2012.10.018. [DOI] [PubMed] [Google Scholar]
- (42).Kishimoto Y; Tani M; Kondo K Pleiotropic Preventive Effects of Dietary Polyphenols in Cardiovascular Diseases. Eur. J. Clin. Nutr 2013, 67 (5), 532–535. DOI: 10.1038/ejcn.2013.29. [DOI] [PubMed] [Google Scholar]
- (43).Sanches-Silva A; Testai L; Nabavi SF; Battino M; Pandima Devi K; Tejada S; Sureda A; Xu S; Yousefi B; Majidinia M; Russo GL; Efferth T; Nabavi SM; Farzaei MH Therapeutic Potential of Polyphenols in Cardiovascular Diseases: Regulation of MTOR Signaling Pathway. Pharmacol. Res 2020, 152, 104626. DOI: 10.1016/j.phrs.2019.104626. [DOI] [PubMed] [Google Scholar]
- (44).Colizzi C The Protective Effects of Polyphenols on Alzheimer’s Disease: A Systematic Review. Alzheimers Dement (N Y) 2019, 5, 184–196. DOI: 10.1016/j.trci.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Singh A; Tripathi P; Yadawa AK; Singh S Promising Polyphenols in Parkinson’s Disease Therapeutics. Neurochem. Res 2020, 45 (8), 1731–1745. DOI: 10.1007/s11064-020-03058-3. [DOI] [PubMed] [Google Scholar]
- (46).Kujawska M; Jodynis-Liebert J Polyphenols in Parkinson’s Disease: A Systematic Review of in Vivo Studies. Nutrients 2018, 10 (5). DOI: 10.3390/nu10050642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Asensi M; Ortega A; Mena S; Feddi F; Estrela JM Natural Polyphenols in Cancer Therapy. Crit. Rev. Clin. Lab. Sci 2011, 48 (5–6), 197–216. DOI: 10.3109/10408363.2011.631268. [DOI] [PubMed] [Google Scholar]
- (48).Zhou Y; Zheng J; Li Y; Xu D-P; Li S; Chen Y-M; Li H-B Natural Polyphenols for Prevention and Treatment of Cancer. Nutrients 2016, 8 (8). DOI: 10.3390/nu8080515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zhang Z; Sang W; Xie L; Li W; Li B; Li J; Tian H; Yuan Z; Zhao Q; Dai Y Polyphenol-Based Nanomedicine Evokes Immune Activation for Combination Cancer Treatment. Angew. Chem. Int. Ed 2021, 60 (4), 1967–1975. DOI: 10.1002/anie.202013406. [DOI] [PubMed] [Google Scholar]
- (50).Mei K-C; Stiepel RT; Bonacquisti E; Jasiewicz NE; Chaudhari AP; Tiwade PB; Bachelder EM; Ainslie KM; Fenton OS; Nguyen J Single-Tailed Heterocyclic Carboxamide Lipids for Macrophage Immune-Modulation. Biomater. Sci 2023, 11 (8), 2693–2698. DOI: 10.1039/d2bm01804g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Schoenmaker L; Witzigmann D; Kulkarni JA; Verbeke R; Kersten G; Jiskoot W; Crommelin DJA MRNA-Lipid Nanoparticle COVID-19 Vaccines: Structure and Stability. Int. J. Pharm 2021, 601, 120586. DOI: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Tenchov R; Bird R; Curtze AE; Zhou Q Lipid Nanoparticles—From Liposomes to MRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15 (11), 16982–17015. DOI: 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- (53).Pirkis J; John A; Shin S; DelPozo-Banos M; Arya V; Analuisa-Aguilar P; Appleby L; Arensman E; Bantjes J; Baran A; Bertolote JM; Borges G; Brečić P; Caine E; Castelpietra G; Chang S-S; Colchester D; Crompton D; Curkovic M; Deisenhammer EA; Du C; Dwyer J; Erlangsen A; Faust JS; Fortune S; Garrett A; George D; Gerstner R; Gilissen R; Gould M; Hawton K; Kanter J; Kapur N; Khan M; Kirtley OJ; Knipe D; Kolves K; Leske S; Marahatta K; Mittendorfer-Rutz E; Neznanov N; Niederkrotenthaler T; Nielsen E; Nordentoft M; Oberlerchner H; O’Connor RC; Pearson M; Phillips MR; Platt S; Plener PL; Psota G; Qin P; Radeloff D; Rados C; Reif A; Reif-Leonhard C; Rozanov V; Schlang C; Schneider B; Semenova N; Sinyor M; Townsend E; Ueda M; Vijayakumar L; Webb RT; Weerasinghe M; Zalsman G; Gunnell D; Spittal MJ Suicide Trends in the Early Months of the COVID-19 Pandemic: An Interrupted Time-Series Analysis of Preliminary Data from 21 Countries. Lancet Psychiatry 2021, 8 (7), 579–588. DOI: 10.1016/S2215-0366(21)00091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Zuhorn IS; Bakowsky U; Polushkin E; Visser WH; Stuart MCA; Engberts JBFN; Hoekstra D Nonbilayer Phase of Lipoplex-Membrane Mixture Determines Endosomal Escape of Genetic Cargo and Transfection Efficiency. Mol. Ther 2005, 11 (5), 801–810. DOI: 10.1016/j.ymthe.2004.12.018. [DOI] [PubMed] [Google Scholar]
- (55).Lu JJ; Langer R; Chen J A Novel Mechanism Is Involved in Cationic Lipid-Mediated Functional SiRNA Delivery. Mol. Pharm 2009, 6 (3), 763–771. DOI: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Allen TM; Cullis PR Liposomal Drug Delivery Systems: From Concept to Clinical Applications. Adv. Drug Deliv. Rev 2013, 65 (1), 36–48. DOI: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- (57).Mui BL; Tam YK; Jayaraman M; Ansell SM; Du X; Tam YYC; Lin PJ; Chen S; Narayanannair JK; Rajeev KG; Manoharan M; Akinc A; Maier MA; Cullis P; Madden TD; Hope MJ Influence of Polyethylene Glycol Lipid Desorption Rates on Pharmacokinetics and Pharmacodynamics of SiRNA Lipid Nanoparticles. Mol. Ther. Nucleic Acids 2013, 2, e139. DOI: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Ma Y; Fenton OS An Efficacy and Mechanism Driven Study on the Impact of Hypoxia on Lipid Nanoparticle Mediated MRNA Delivery. J. Am. Chem. Soc 2023, 145 (20), 11375–11386. DOI: 10.1021/jacs.3c02584. [DOI] [PubMed] [Google Scholar]
- (59).Ma Y; Fenton OS A Unified Strategy to Improve Lipid Nanoparticle Mediated MRNA Delivery Using Adenosine Triphosphate. J. Am. Chem. Soc 2023, 145 (36), 19800–19811. DOI: 10.1021/jacs.3c05574. [DOI] [PubMed] [Google Scholar]
- (60).Qu Y; De Rose R; Kim C-J; Zhou J; Lin Z; Ju Y; Bhangu SK; Cortez-Jugo C; Cavalieri F; Caruso F Supramolecular Polyphenol-DNA Microparticles for In Vivo Adjuvant and Antigen Co-Delivery and Immune Stimulation. Angew. Chem. Int. Ed 2023, 62 (12), e202214935. DOI: 10.1002/anie.202214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).McLendon PM; Fichter KM; Reineke TM Poly(Glycoamidoamine) Vehicles Promote PDNA Uptake through Multiple Routes and Efficient Gene Expression via Caveolae-Mediated Endocytosis. Mol. Pharm 2010, 7 (3), 738–750. DOI: 10.1021/mp900282e. [DOI] [PubMed] [Google Scholar]
- (62).Sahay G; Querbes W; Alabi C; Eltoukhy A; Sarkar S; Zurenko C; Karagiannis E; Love K; Chen D; Zoncu R; Buganim Y; Schroeder A; Langer R; Anderson DG Efficiency of SiRNA Delivery by Lipid Nanoparticles Is Limited by Endocytic Recycling. Nat. Biotechnol 2013, 31 (7), 653–658. DOI: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).French AP; Mills S; Swarup R; Bennett MJ; Pridmore TP Colocalization of Fluorescent Markers in Confocal Microscope Images of Plant Cells. Nat. Protoc 2008, 3 (4), 619–628. DOI: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- (64).Chen J; Li J; Zhou J; Lin Z; Cavalieri F; Czuba-Wojnilowicz E; Hu Y; Glab A; Ju Y; Richardson JJ; Caruso F Metal-Phenolic Coatings as a Platform to Trigger Endosomal Escape of Nanoparticles. ACS Nano 2019, 13 (10), 11653–11664. DOI: 10.1021/acsnano.9b05521. [DOI] [PubMed] [Google Scholar]
- (65).Hu Y; Litwin T; Nagaraja AR; Kwong B; Katz J; Watson N; Irvine DJ Cytosolic Delivery of Membrane-Impermeable Molecules in Dendritic Cells Using PH-Responsive Core-Shell Nanoparticles. Nano Lett. 2007, 7 (10), 3056–3064. DOI: 10.1021/nl071542i. [DOI] [PubMed] [Google Scholar]
- (66).Dröse S; Altendorf K Bafilomycins and Concanamycins as Inhibitors of V-ATPases and P-ATPases. J. Exp. Biol 1997, 200 (Pt 1), 1–8. DOI: 10.1242/jeb.200.1.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


