ABSTRACT
The Burkholderia cepacia complex (Bcc) is a group of Gram-negative opportunistic bacteria often associated with fatal pulmonary infections in patients with impaired immunity, particularly those with cystic fibrosis (CF) and chronic granulomatous disease (CGD). Some Bcc strains are known to naturally produce pyomelanin, a brown melanin-like pigment known for scavenging free radicals; pigment production has been reported to enable Bcc strains to overcome the host cell oxidative burst. In this work, we investigated the role of pyomelanin in resistance to oxidative stress and virulence in strains J2315 and K56-2, two epidemic CF isolates belonging to the Burkholderia cenocepacia ET-12 lineage. We previously reported that a single amino acid change from glycine to arginine at residue 378 in homogentisate 1,2-dioxygenase (HmgA) affects the pigment production phenotype: pigmented J2315 has an arginine at position 378, while non-pigmented K56-2 has a glycine at this position. Herein, we performed allelic exchange to generate isogenic non-pigmented and pigmented strains of J2315 and K56-2, respectively, and tested these to determine whether pyomelanin contributes to the protection against oxidative stress in vitro as well as in a respiratory infection in CGD mice in vivo. Our results indicate that the altered pigment phenotype does not significantly impact these strains’ ability to resist oxidative stress with H2O2 and NO in vitro and did not change the virulence and infection outcome in CGD mice in vivo suggesting that other factors besides pyomelanin are contributing to the pathophysiology of these strains.
IMPORTANCE
The Burkholderia cepacia complex (Bcc) is a group of Gram-negative opportunistic bacteria that are often associated with fatal pulmonary infections in patients with impaired immunity, particularly those with cystic fibrosis and chronic granulomatous disease (CGD). Some Bcc strains are known to naturally produce pyomelanin, a brown melanin-like pigment known for scavenging free radicals and overcoming the host cell oxidative burst. We investigated the role of pyomelanin in Burkholderia cenocepacia strains J2315 (pigmented) and K56-2 (non-pigmented) and performed allelic exchange to generate isogenic non-pigmented and pigmented strains, respectively. Our results indicate that the altered pigment phenotype does not significantly impact these strains’ ability to resist H2O2 or NO in vitro and did not alter the outcome of a respiratory infection in CGD mice in vivo. These results suggest that pyomelanin may not always constitute a virulence factor and suggest that other features are contributing to the pathophysiology of these strains.
KEYWORDS: Burkholderia, pyomelanin, pigment, CGD
INTRODUCTION
Cystic fibrosis (CF) and chronic granulomatous disease (CGD) are hereditary diseases characterized by inflammatory complications and recurrent infections with opportunistic bacterial and fungal pathogens (1, 2). Members of the Burkholderia cepacia complex (Bcc), a group of at least 23 genetically and closely related bacterial species (3) that rarely cause disease in healthy individuals and are widely distributed in the natural environment, are among the select pathogens that infect both CF and CGD patients. Bcc is especially troublesome due to its inherent antibiotic resistance, capacity to survive in supposedly sterile hospital solutions, and ability to spread between patients leading to worse outcome (4–7). These infections can also result in fatal necrotizing pneumonia and septicemia known as “cepacia syndrome” (8).
Bcc is known to produce a multitude of virulence factors that enhance their pathogenicity in different stages of infection in host cells. Bcc has been shown to survive intracellularly in both professional and nonprofessional phagocytes. In particular, they have been detected inside airway and alveolar epithelial cells, neutrophils, and in pulmonary macrophages from resected lungs of CF patients undergoing transplant (9, 10). Interestingly, in macrophages, some Bcc isolates can survive and develop an intracellular niche and evade host immune killing using several mechanisms against oxidative stress, such as interfering with phagosome maturation and phago-lysosomal fusion (11), and delaying the phagosome acidification (12) leading to their resistance to oxidative damage, which acts as an important survival strategy for Bcc inside the host. The intracellular survival of Bcc within macrophages has been previously studied and documented in vitro in various models (11, 13, 14). The presence of the melanin-like pigment, known as pyomelanin, has been proposed to have scavenging properties and protect the bacteria during the respiratory burst (12).
Melanin constitutes a general class of complex high-molecular-weight polyphenolic heteropolymers that include eumelanin, pheomelanin, and pyomelanin. Melanin is capable of serving as an electron trap for reactive oxygen species due to its ability to exist in multiple oxidation states (15). Pyomelanin is a pigment that is commonly found in many systems of life, particularly bacteria and fungi. The pigment has been reported to efficiently scavenge reactive oxygen species and diminish the oxidizing stress of the host due to its high tolerance to hydrogen peroxide, thus promoting the survival of these pigmented organisms in their host (16).
We have noted a subset of Bcc isolates can produce pyomelanin under normal laboratory conditions (15). In previous in vitro studies of others, pyomelanin has been demonstrated to contribute to the increased resistance of B. cenocepacia strain C5424 to extracellular hydrogen peroxide, which is the main type of reactive oxygen species found in macrophages (13). In that same study, a non-pigmented isogenic mutant of C5424 was found to exhibit a higher sensitivity to reactive oxygen species killing. While potentially advantageous, as mentioned, the pyomelanin production phenotype is not found in every member of Bcc; in particular, it is present in B. cenocepacia J2315 and absent in B. cenocepacia K56-2, two closely related strains of the ET-12 lineage, which were both isolated from CF patients, calling into question the potential role of pyomelanin in infection.
Pyomelanin is a natural polymer of homogentisic acid (HGA, 2,5-dihydroxyphenylacetic acid) synthesized through the L-tyrosine pathway, specifically due to the accumulation of HGA, which is synthesized via 4-hydroxyphenylpyruvate dioxygenase (4-HDDP). HGA is excreted and spontaneously auto-oxidizes to form benzoquinoneacetic acid, followed by self-polymerization to produce pyomelanin (15). Two essential enzymes in the synthesis of pyomelanin are HppD and HmgA. The hppD gene codes for a protein that is responsible for the conversion of 4-hydroxyphenylpyruvate to HGA. Homogentisate 1,2-dioxygenase, encoded by the hmgA gene, converts HGA to maleylacetoacetate. Our lab has previously reported that a single amino acid change from glycine (G; non-pigmented phenotype) to arginine (R; pigmented phenotype) at residue 378 of HmgA contributes to the pigment production phenotype. The G378R change renders HmgA non-functional, and the pathway stops at the intermediate molecule HGA, which ultimately results in pyomelanin production (15). While numerous studies had investigated the role of bacterial pyomelanin in vitro, particularly by deleting hppD or hmgA or complementing mutants (13, 16–20), crucially, there has been a gap in our understanding of the role of B. cenocepacia pyomelanin pigment in in vivo models that mimic human infection. We previously noted that J2315 has an R at position 378 of HmgA, while K56-2 has a G at this position. Our previous studies used trans complementation with a plasmid-borne wild-type copy of the hmgA gene from B. cenocepacia J2315 (pigmented) or K56-2 (non-pigmented); the resulting strains contained both the wild-type and a mutant copy of the hmgA gene. The altered pigmented phenotype of either strain did not confer any changes in the susceptibility to hydrogen peroxide (15). Here, we performed allelic exchange to generate isogenic non-pigmented and pigmented strains of J2315 and K56-2, respectively, and tested these to determine whether pyomelanin contributes to the protection of pigmented strains against oxidative stress in vitro as well as in a respiratory infection in CGD mice.
MATERIALS AND METHODS
Construction of hmgA mutants
B. cenocepacia J2315 and K56-2 were used as the parental strains to generate hmgA mutants. For cloning and conjugations, we used Escherichia coli DH5α and SM10, respectively. The hmgA gene was amplified from J2315 and K56-2 using the following primers: hmgA-F (5′ GGggtaccATGACGCTTGACCTGTCGAAAC 3′); hmgA-R (5′ ggaagcttTCATCGTTGCTCCGGATTGAAG 3′). The products were digested with KpnI and HindIII and ligated into similarly digested pEX18-Tc to generate pEX18-Tc::JhmgA and pEX18-Tc::KhmgA, respectively, and were electroporated into E. coli DH5α (21). Verified plasmids were then introduced into SM10 by electroporation. The donor SM10 strains containing the pEX18-Tc::JhmgA or pEX18-Tc::KhmgA plasmids were used for conjugations into K56-2 or J2315, respectively. For this, a mixture containing 100 µL of donor and 100 µL recipient was incubated on tryptic soy agar plates at room temperature for 1 hour and then moved to 37°C for 6 hours. Cells were resuspended and transconjugants were selected by plating on LB plates supplemented with 25 µg/mL of spectinomycin (to select against E. coli) and 300 µg/mL of tetracycline and incubated at 37°C for 3 days. Transconjugants were streaked on LB plates supplemented with 5% sucrose and incubated for 3 days at 37°C. Individual colonies were purified from the plates and tested for tetracycline sensitivity. Selected colonies were then analyzed for the correct hmgA integration by PCR and differential digest with AvaII.
Comparing growth and pigment production between strains
Three milliliters cultures of all four strains were grown shaking overnight at 37°C in Luria broth (LB) medium. The next day strains were back diluted to an (optical density) OD600 of 0.01 into 50 mL of LB media. Cultures were incubated at 37°C shaking, and measurements of the culture at OD600 and cell-free supernatants at OD480 [for growth and pigment production (15), respectively] were taken at various times, as indicated.
In vitro sensitivity and survival in the presence of reactive oxygen and nitrogen species
To test cultures for sensitivity to reactive oxygen and nitrogen species, bacteria were grown overnight at 37°C, shaking in LB medium. Late stationary phase cultures were back diluted to an OD600 of 0.2 and were incubated with various concentrations (1 mM to 200 mM) of the reactive oxygen species, hydrogen peroxide (H2O2) (Sigma-Aldrich), or the nitric oxide (NO) donor (16), sodium nitrosprusside (SNP) (Chem-ImpexInt’l Inc), in 96-well plates at 37°C for 18–20 hours. OD600 readings were taken every 20 minutes using a microplate reader (BioTek Instruments, Inc). The strains were incubated in duplicates, and the growth analysis was performed at each concentration of H2O2 and SNP.
To determine the survival in the presence of H2O2 and SNP, bacteria were treated with high concentrations of these species and plated for survival at fixed endpoints. For this, cultures were grown until late stationary phase and back diluted to an OD600 of 1.0. Strains were treated with 100 mM of either H2O2 or SNP. Aliquots were removed at 0-, 30-, 60-, and 120 minutes post-treatment, were serially diluted, and plated in duplicate on LB agar plates and incubated up to 48 hours at 37°C and counted for colony-forming units (CFU/mL). As a negative control, parallel untreated cultures of each strain were included, and they were aliquoted at the same indicated time points. The percent survival was calculated by dividing the CFU/mL at indicated fixed timepoints by parallel untreated cultures of each strain collected at the same timepoint.
CGD mouse lung infections
This animal study was conducted according to the guidelines of the Emory University Institutional Animal Care and Use Committee (IACUC), under approved protocol number PROTO201700441. The study was carried out according to the established guidelines and policies at Emory University School of Medicine, and recommendations in the Guide for Care and Use of Laboratory Animals, and local, state, and federal laws.
B. cenocepacia J2315 and K56-2 and isogenic mutants were grown from glycerol stocks on LB agar plates, and then a single colony was grown in LB broth and then incubated with shaking at 37°C for 4–5 hours until an OD600 of 0.08 was reached. Cultures were spun down and washed twice with phosphate-buffered saline (PBS). Inoculum was adjusted spectrophotometrically to obtain the desired challenge dose of 1 × 103 in a volume of 50 µL.
Mice deficient in the gp91phox subunit of the NADPH oxidase [B6.129S6-Cybbtm1Din/J (gp91phox−/−); Jackson Laboratory, Bar Harbor, ME] were bred and maintained at the Emory Division of Animal Resources (DAR). Eight- to 10-week-old animals gender matched were anesthetized by intraperitoneal injection of 0.2 mL of ketamine (10 mg/mL) and xylazine (0.5 mg/mL). Mice were infected by non-invasive intratracheal instillation of J2315 and K56-2 or isogenic mutant strains, as previously described (22). For colonization experiments, mice were euthanized at indicated time points, and whole lungs were collected aseptically and homogenized in 1 mL PBS. Lung homogenates were serially diluted and plated onto Pseudomonas isolation agar (BD Difco) for CFU determination. For survival experiments, mice were monitored up to 12 days post-infection, and animals that succumbed to infection or appeared to be under acute distress were humanely euthanized and were included in the reported results.
RESULTS
B. cenocepacia J2315 and K56-2 show distinct levels of virulence in a CGD mouse lung infection model
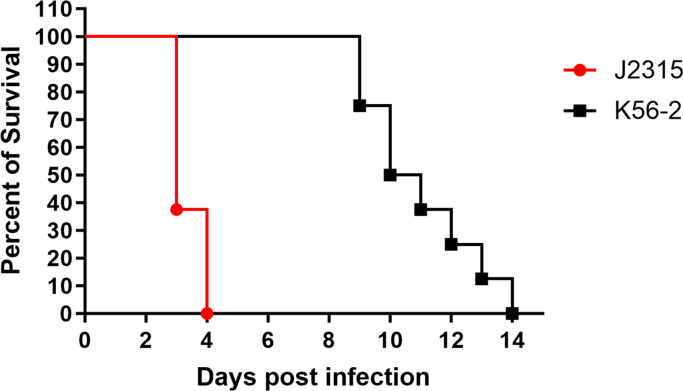
The ET-12 lineage B. cenocepacia strains, J2315 and K56-2, were tested for virulence in a respiratory infection model in CGD mice. We used gp91phox−/− mice and delivered 1 × 103 CFU of bacteria intratracheally. As seen in Fig. 1, all CGD mice infected with J2315 succumbed by day 4 post-infection. On the other hand, mice infected with K56-2 showed delayed time to death and animals started to succumb to infection starting at day 9 post-infection. These results indicate the J2315 is more virulent than K56-2 in this model of lung infection.
Fig 1.
B. cenocepacia J2315 and K56-2 show distinct levels of virulence in a CGD mouse lung infection model. gp91phox−/− (CGD) mice were infected intratracheally with ∼103 CFU/mouse of strains J2315 and K56-2. Mice were monitored for 14 days post-infection. Results are represented by Kaplan-Meier survival curves and were analyzed by log rank test. Results were compiled from two independent experiments, n = 8/strain.
A single amino acid change in HmgA is sufficient for pyomelanin production in B. cenocepacia
Our lab has previously reported that a single amino acid change from glycine to arginine at residue 378 of HmgA contributes to the pigmented phenotype. An arginine at residue 378 renders the HmgA protein nonfunctional and the pathway stops at the intermediate molecule homogentisic acid (HGA), which ultimately results in pyomelanin production. We found that J2315 had an arginine at position 378, while K56-2 had an glycine (15).
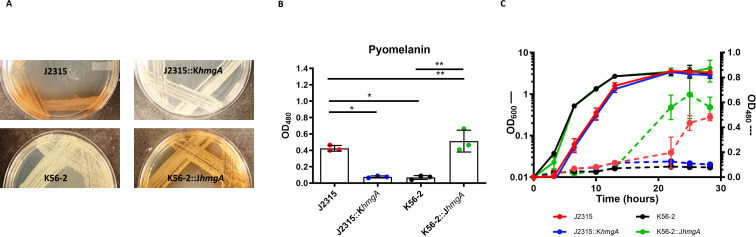
To determine whether this amino acid substitution in HmgA was sufficient for pyomelanin production in B. cenocepacia, we used allelic exchange to construct a J2315 strain containing the hmgA from K56-2 and a K56-2 strain containing the hmgA from J2315. This gave us two pairs of isogenic strains (J2315 and J2315::KhmgA as well as K56-2 and K56-2::JhmgA), which we used to determine the role of pyomelanin. After 48 hours of growth on solid LB media, J2315::KhmgA did not produce visible pigment, as compared to J2315, while K56-2::JhmgA did produce pigment as compared to K56-2 (Fig. 2A). We observed the same phenotypes of pyomelanin production as measured by the absorbance of the culture supernatant of all strains grown for 28 hours in liquid LB media (Fig. 2B).
Fig 2.
A single amino acid change in HmgA is sufficient for pyomelanin production in B. cenocepacia. (A) Colony phenotypes of wild-type J2315 (top) and K56-2 (bottom) and the designated isogenic mutants on LB agar media. Strains were incubated at 37°C for 48 hours. (B) Pyomelanin production in culture supernatant grown for 28 hours in LB. Cultures were normalized to an OD600 of 0.1, and supernatant was measured by OD480. (C) Growth and pigment production of all four strains incubated with shaking at 37°C in LB cultures. Growth was measured by OD600 on the left Y axis, pyomelanin production measured by OD480 on the right Y axis. Solid lines represent OD600 measurements and dashed lines represent OD480 measurements. For culture experiments, each point represents the mean of three independent cultures and error bars represent standard deviation. Data were analyzed by one-way ANOVA. *P < 0.05, **P < 0.01 by one-way ANOVA. Unlabeled comparisons were not statistically significant.
To determine whether pyomelanin production would alter growth under these conditions, we performed growth curves in LB with each of the four strains. Over nearly 30 hours, there was no difference in the growth rate between the isogenic strains (K56-2 vs K56-2::JhmgA or J2315 vs J2315::KhmgA). However, we did note that the J2315 strains grew slower than the K56-2 strains under these conditions (Fig. 2C). When we monitored pigment production during the course of this experiments, no pigment was produced by K56-2 or J2315::KhmgA, while K56-2::JhmgA and J2315 both produced pigment as detected in the supernatant, with K56-2::JhmgA producing it between 12 and 24 hours and J2315 producing it after 24 hours (Fig. 2C).
These results demonstrate the functionality of HmgA, and by extension pigment production, does not alter growth in LB. These data also indicate that the production of pyomelanin in J2315 does not account for its decreased growth rate compared to K56-2. Finally, these results show that a single amino acid change (from glycine to arginine) in HmgA is responsible for the production of pigment in these strains.
Effect of pyomelanin production in protection against specific immune defense molecules in vitro
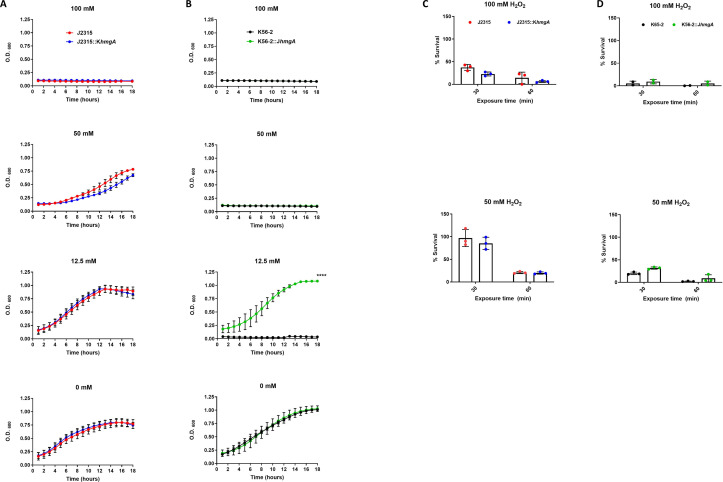
Previous work by Keith et al. (13) showed in B. cenocepacia C5424 that pyomelanin is associated with increased resistance to reactive oxygen species, like hydrogen peroxide (H2O2). However, whether this is a strain-specific phenomenon is unclear. To begin to determine whether pyomelanin production would alter resistance characteristics to specific immune response molecules in J2315 and K56-2, we performed similar studies on our strains. We monitored growth over 16 hours in various concentrations of H2O2 (0–100 mM). J2315 was more resistant to this treatment than K56-2. We found that J2315 showed growth inhibition at 100 mM, while K56-2 showed growth inhibition at 50 mM. However, the absence of pyomelanin did not seem to affect the resistance of J2315 mutant to H2O2, as the growth of J2315::KhmgA strain, under these conditions, was not altered compared to wild-type J2315 strain (Fig. 3A), which suggest that in this strain pyomelanin pigment is dispensable and other factors are playing a major role in the resistance of J2315 to H2O2. On the other hand, while the growth of the non-pigmented wild-type K56-2 strain was abolished at a concentration of 12.5 mM H2O2, K56-2::JhmgA showed increased growth at the same H2O2 concentration (Fig. 3B). These results suggest that under these experimental conditions, pyomelanin pigment is contributing to the resistance of K56-2::JhmgA to H2O2.
Fig 3.
Pyomelanin production and in vitro susceptibility to killing by hydrogen peroxide. Growth curves of wild-type and designated isogenic mutant of (A) J2315 and (B) K56-2 treated with various concentrations of hydrogen peroxide (H2O2). Stationary phase cultures were back diluted to an OD600 = 0.2 and were incubated with indicated concentrations of H2O2. Viable CFU counts of wild-type and isogenic mutants of (C) J2315 or (D) K56-2 were determined after treatment with indicated H2O2 concentrations for 30 or 60 minutes. For panels (C) and (D), the percent survival was calculated by dividing the CFU/mL at fixed timepoints (30 or 60 minutes) by parallel untreated cultures of each strain. For all experiments, the data are pooled from three independent experiments. Error bars represent standard deviation (SD). Data were analyzed by student’s t test. ****P < 0.0001. Unlabeled comparisons were not statistically significant.
To determine the impact of pyomelanin on the survival of the strains after short-term exposure to reactive oxygen, we treated the strains with high doses of H2O2 (50 and 100 mM) and plated for survival at fixed timepoints (30 and 60 minutes) post-exposure. There was a modest decrease in survival of the non-pigmented strains, but these differences were not statistically significant (Fig. 3C and D).
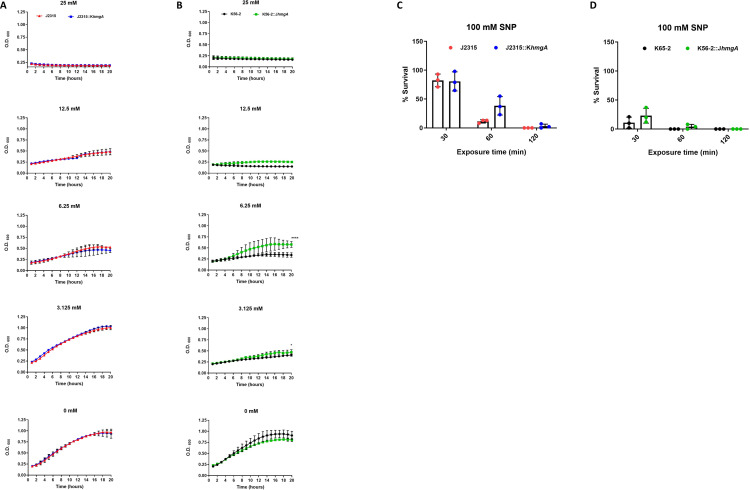
In order to test pyomelanin’s role in resistance to reactive nitrogen species, we exposed all four strains to sodium nitroprusside (SNP), a nitric oxide (NO) donor, and measured their growth over 20 hours. Similar to what we observed for the H2O2 treatment, we noted that J2315 was more resistant to SNP than K56-2, which was particularly evident at 12.5 mM SNP, comparing results in Fig. 4A and B. Additionally, we observed that nonpigmented J2315::KhmgA demonstrated similar growth at the various concentrations of SNP compared to wild-type J2315 (Fig. 4A). This suggests that both the pigmented and non-pigmented phenotypes of J2315 were equally resistant to the NO generated by SNP. Conversely, we noted that at a SNP concentration of 6.25 mM, pigmented K56-2::JhmgA demonstrated higher growth compared to nonpigmented wild-type K56-2 (Fig. 4B). These results suggest a potential protective role of pyomelanin pigment under these experimental conditions in K56-2.
Fig 4.
Effect of pyomelanin production in protection against reactive nitrogen species in vitro. Growth curves of wild-type and designated isogenic mutant of (A) J2315 and (B) K56-2 treated with various concentrations of sodium nitroprusside (SNP). Stationary phase cultures were back diluted to an OD600 = 0.2 and were incubated with indicated concentrations of SNP. Viable CFU counts of wild-type and isogenic mutants of (C) J2315 and (D) K56-2 were determined after treatment with indicated SNP concentrations for 30, 60, or 120 minutes. For panels (C) and (D), the percent survival was calculated by dividing the CFU/mL at fixed timepoints (30, 60, or 120 minutes) by parallel untreated cultures of each strain. For all experiments, the data are pooled from three independent experiments. Error bars represent standard deviation (SD). Data were analyzed by student’s t test. *P < 0.05, ****P < 0.0001. Unlabeled comparisons were not statistically significant.
When we treated cultures of bacteria with 100 mM SNP for 2 hours, K56-2::JhmgA showed a higher survival than K56-2 (Fig. 4D) although this difference was not statistically significant. Distinct from this were the results with the isogenic J2315 strains where J2315::KhmgA demonstrated a higher survival compared to J2315 (Fig. 4C). This result suggests that there may be an advantage of the pigment in vitro to protect against reactive nitrogen species in the K56-2 strain background; however, since J2315::KhmgA survives much better than K56-2, this suggests there are additional strain-specific defenses against reactive nitrogen species in J2315 (Fig. 4C and D).
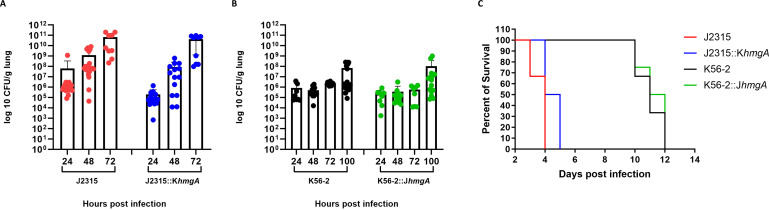
HmgA does not alter either the survival or the colonization in a CGD mouse lung infection
In order to determine whether pyomelanin plays a role during respiratory infection in CGD mice, we performed a non-invasive intratracheal infection of 103 CFU/mL of each strain into CGD mice lungs. After the infection, we harvested lung tissue at fixed time-points post-infection, and we observed no significant difference between the colonization level of J2315 and J2315::KhmgA (Fig. 5A), as well as between K56-2 and K56-2::JhmgA (Fig. 5B).
Fig 5.
Altered pigment production does not impact the virulence of J2315 or K56-2 in a CGD mouse lung infection. Mice were infected intratracheally with ∼103 CFU/mouse and the bacterial load in lung homogenates of mice infected with the wild-type or indicated isogenic mutants of (A) J2315 or (B) K6-2 was assessed. (C) A parallel group of mice were monitored for survival. For colonization experiments, mice were euthanized at indicated time points post-infection, and lungs were aseptically collected and homogenized in 1 mL PBS. All samples were plated for viable CFU on Pseudomonas isolation agar. Each dot represents the CFU from a single mouse. Error bars represent standard deviation (SD). Results were compiled from two independent experiments. For survival experiments, animals were monitored for 12 days post-infection. Results are represented by Kaplan-Meier survival curves and were analyzed by log rank test. Results were compiled from two independent experiments, n = 8/strain. Unlabeled comparisons were not statistically significant.
A parallel group of infected mice were monitored for survival. Our results indicate that the absence of the pigment in J2315::KhmgA did not seem to compromise the virulence of this strain. Mice infected with either J2315 and its non-pigmented mutant J2315::KhmgA succumbed to infection starting at day 4 post-infection (Fig. 5C). Similarly, mice infected with K56-2 or its pigmented mutant K56-2::JhmgA showed similar time to death as all mice succumbed to infection starting at day 9 post-infection (Fig. 5C). As the wild-type strains and their isogenic mutants showed no statistically significant difference in the killing of CGD mice, these results suggest a very minor, if any, contribution of HmgA function to virulence in this model of infection.
DISCUSSION
B. cenocepacia J2315 was isolated from a CF patient in Edinburgh in 1989 who was the index case for a patient-to-patient outbreak that spread to Toronto (4). B. cenocepacia K56-2 was isolated from a CF patient in Toronto (23). These isolates belong to the same clonal complex, the ET-12 lineage (24). J2315 was the first B. cenocepacia strain sequenced (25), while K56-2 has been used more extensively for molecular genetic studies because of its high transformation rate (26). Interestingly, previous studies have shown that these two strains are distinct in their virulence. Specifically, various in vitro models such as zebrafish (27), Caenorhabditis elegans (28), Galleria mellonella (29), and alfalfa (30) showed that K56-2 was more virulent than J2315. However, whether J2315 and K56-2 would show differential virulence in a model that more closely mimics the clinical condition in humans was not clear. Since B. cenocepacia is a prominent pathogen in CGD, causing serious and deadly pneumonia, we used a lung infection in a CGD mouse (gp91phox−/−) and delivered bacteria via the intratracheal route (22). This is the same mouse reported previously by Sousa et al. (31); however, our method of administration of the bacteria used a non-surgical intratracheal delivery, while Sousa et al. (31) used a mid-line cervical incision to deliver bacteria to the trachea. They found that CGD mice given 103 CFU of J2315 showed 100% mortality within 3 days (31). This was similar to what we observed in our initial infection (Fig. 1A). While Sousa et al. (31) did not assess the virulence of K56-2 in this model, they did test H111; mice infected with H111 also showed 100% mortality, but this took 6 days to be observed. Since we had previously noted H111 was non-pigmented (and has an R at position 378 of HmgA) (15), we hypothesized that expression of pigment might be associated with increased virulence. To the best of our knowledge, the virulence of J2315 and K56-2 has not been directly assessed and compared any murine model of respiratory infection. Here, we took advantage of the two ET-12 isolates, K56-2 and J2315, and confirmed that J2315 was more virulent than K56-2 (Fig. 1A).
In order to test whether the difference in virulence between J2315 and K56-2 in the CGD mouse was attributable to the function of HmgA, we generated isogenic strains where the hmgA gene from one strain was exchanged with the other; as anticipated J2315::KhmgA was non-pigmented, while K56-2::JhmgA was pigmented. Similar to our previous observation (15), we saw that J2315 and K56-2::JhmgA produced more pyomelanin when cultures were supplemented with 0.02% tyrosine, suggesting that pyomelanin production occurs through the tyrosine assimilation pathway, similar to other bacterial species. Furthermore, neither J2315::KhmgA nor K56-2 produced pigment even when cultures were supplemented with tyrosine.
Surprisingly, we observed no difference in the virulence of either of the strains vs their isogenic mutants in the CDG mouse model. This is distinctly different from what was observed in studying an hmgA deletion mutant in Pseudomonas aeruginosa PAO1 in CD1 mice: the PAO1 ∆hmgA strain made pyomelanin, and it was found that this pigmented strain was more virulent following intratracheal infection (32). Similarly, an hppD deletion mutant of the pigmented Bordetella parapertussis resulted in a non-pigmented mutant that was less able to colonize the lungs of C57BL/6J mice after intranasal infection (19).
In testing the susceptibility to reactive oxygen and nitrogen species, we observed a subtle difference in the survival of J2315 vs J2315::KhmgA in the presence of either H2O2 or SNP. This is different from what was observed by Keith et al. with an hppD mutant of the pigmented strain C5424 (13). They observed that the non-pigmented C5424 mutant was more sensitive to H2O2 compared to the wild-type pigmented parent. More in keeping with the results of Keith et al., we did observe that the pigmented version of K56-2, K56-2::JhmgA, showed increased resistance to H2O2 (at 12.5 mM) and some resistance to SNP (at levels below 12.5 mM). Altogether these results suggest that the importance of pigmentation for virulence and resistance to reactive oxygen and nitrogen species may be dependent on the bacterial species and/or strain background.
In order to determine what genes might be responsible for the different virulence of J2315 and K56-2, we compared the genomes of these strains using Rapid Annotation using Subsystem Technology. We found that J2315 encodes over 700 unique predicted open reading frames. Of particular interest is an error prone lesion bypass DNA polymerase V (UmuC) that is annotated in J2315 but does not appear to be present in the K56-2 genome. In E. coli, UmuC is a known SOS-induced repair polymerase that can respond to, and repair, damaged DNA (33). This type of repair polymerase could potentially contribute to the increased protection that J2315 exhibits to NO compared to K56-2. The impact of UmuC on the pathogenesis of J2315 vs K56-2 would require additional experiments and is the topic of future investigations.
Previous studies have examined the transcriptional impact of reactive oxygen treatment to J2315, and they reported an increased expression of genes related to oxidative stress response via the transcriptional regulator OxyR within 30 minutes of H2O2 treatment (34). The J2315 genome specifically encodes the katB gene, encoding catalase, which can breakdown hydrogen peroxide; this gene is absent from K56-2 (25, 35). The presence of factors that protect against oxidative stress may be contributing to the higher resistance of J2315 compared to K56-2 that we have observed in our experiments.
One limitation of the CGD mouse model is that, while this mouse is deficient in producing reactive oxygen species through NADPH, it is still capable of producing reactive nitrogen species, such as NO. While it might be suggested that mice defective in the ability to produce NO (iNOS−/−) would uncover the potential role of the pyomelanin pigment in the absence of reactive nitrogen species and presence of reactive oxygen species (36), previous studies with these iNOS−/− mice showed that they were as resistant as wild-type mice to Bcc infection (36, 37).
In conclusion, the presence of pyomelanin is generally recognized for its role in resistance to reactive compounds produced as a host response to infection (38). Thus, the detection of pyomelanin-producing pathogens from clinical and environmental samples may be concerning (39). However, our data suggest that pyomelanin alone may not always constitute a virulence factor.
ACKNOWLEDGMENTS
We thank John J. Varga, Ph.D. for his assistance with the bioinformatic analysis of J2315 vs K56-2.
This work was supported, in part, by a grant from the National Institutes of Health, R21-AI103653 to J.B.G. S.C.F. was supported, in part, by the NIH IRACDA Fellowships in Research and Science Teaching (FIRST) program at Emory University.
Contributor Information
Joanna B. Goldberg, Email: joanna.goldberg@emory.edu.
Silvia T. Cardona, University of Manitoba, Winnipeg, Manitoba, Canada
REFERENCES
- 1. Cystic Fibrosis Foundation . 2021. Patient registry annual data report 2021
- 2. Nauseef WM, Clark RA. 2019. Intersecting stories of the phagocyte NADPH oxidase and chronic granulomatous disease. Methods Mol Biol 1982:3–16. doi: 10.1007/978-1-4939-9424-3_1 [DOI] [PubMed] [Google Scholar]
- 3. Zlosnik JEA, Henry DA, Hird TJ, Hickman R, Campbell M, Cabrera A, Laino Chiavegatti G, Chilvers MA, Sadarangani M. 2020. Epidemiology of Burkholderia infections in people with cystic fibrosis in Canada between 2000 and 2017. Ann Am Thorac Soc 17:1549–1557. doi: 10.1513/AnnalsATS.201906-443OC [DOI] [PubMed] [Google Scholar]
- 4. Govan JRW, Doherty CJ, Nelson JW, Brown PH, Greening AP, Maddison J, Dodd M, Webb AK. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15–19. doi: 10.1016/0140-6736(93)91881-L [DOI] [PubMed] [Google Scholar]
- 5. Speert DP, Bond M, Woodman RC, Curnutte JT. 1994. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J Infect Dis 170:1524–1531. doi: 10.1093/infdis/170.6.1524 [DOI] [PubMed] [Google Scholar]
- 6. Rhodes KA, Schweizer HP. 2016. Antibiotic resistance in Burkholderia species. Drug Resist Updat 28:82–90. doi: 10.1016/j.drup.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tavares M, Kozak M, Balola A, Sá-Correia I. 2020. Burkholderia cepacia complex bacteria: a feared contamination risk in water-based pharmaceutical products. Clin Microbiol Rev 33:e00139-19. doi: 10.1128/CMR.00139-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daccò V, Alicandro G, Consales A, Rosazza C, Sciarrabba CS, Cariani L, Colombo C. 2023. Cepacia syndrome in cystic fibrosis: a systematic review of the literature and possible new perspectives in treatment. Pediatr Pulmonol 58:1337–1343. doi: 10.1002/ppul.26359 [DOI] [PubMed] [Google Scholar]
- 9. Mesureur J, Feliciano JR, Wagner N, Gomes MC, Zhang L, Blanco-Gonzalez M, van der Vaart M, O’Callaghan D, Meijer AH, Vergunst AC. 2017. Macrophages, but not neutrophils, are critical for proliferation of Burkholderia cenocepacia and ensuing host-damaging inflammation. PLoS Pathog 13:e1006437. doi: 10.1371/journal.ppat.1006437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schwab U, Abdullah LH, Perlmutt OS, Albert D, Davis CW, Arnold RR, Yankaskas JR, Gilligan P, Neubauer H, Randell SH, Boucher RC. 2014. Localization of Burkholderia cepacia complex bacteria in cystic fibrosis lungs and interactions with Pseudomonas aeruginosa in hypoxic mucus. Infect Immun 82:4729–4745. doi: 10.1128/IAI.01876-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lamothe J, Thyssen S, Valvano MA. 2004. Burkholderia cepacia complex isolates survive intracellularly without replication within acidic vacuoles of Acanthamoeba polyphaga. Cell Microbiol 6:1127–1138. doi: 10.1111/j.1462-5822.2004.00424.x [DOI] [PubMed] [Google Scholar]
- 12. Lamothe J, Huynh KK, Grinstein S, Valvano MA. 2007. Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacteria-containing vacuoles. Cell Microbiol 9:40–53. doi: 10.1111/j.1462-5822.2006.00766.x [DOI] [PubMed] [Google Scholar]
- 13. Keith KE, Killip L, He P, Moran GR, Valvano MA. 2007. Burkholderia cenocepacia C5424 produces a pigment with antioxidant properties using a homogentisate intermediate. J Bacteriol 189:9057–9065. doi: 10.1128/JB.00436-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin DW, Mohr CD. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect Immun 68:24–29. doi: 10.1128/IAI.68.1.24-29.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gonyar LA, Fankhauser SC, Goldberg JB. 2015. Single amino acid substitution in homogentisate 1,2-dioxygenase is responsible for pigmentation in a subset of Burkholderia cepacia complex isolates. Environ Microbiol Rep 7:180–187. doi: 10.1111/1758-2229.12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodríguez-Rojas A, Mena A, Martín S, Borrell N, Oliver A, Blázquez J. 2009. Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology (Reading) 155:1050–1057. doi: 10.1099/mic.0.024745-0 [DOI] [PubMed] [Google Scholar]
- 17. Wang H, Qiao Y, Chai B, Qiu C, Chen X. 2015. Identification and molecular characterization of the homogentisate pathway responsible for pyomelanin production, the major melanin constituents in Aeromonas media WS. PLoS One 10:e0120923. doi: 10.1371/journal.pone.0120923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noorian P, Hu J, Chen Z, Kjelleberg S, Wilkins MR, Sun S, McDougald D. 2017. Pyomelanin produced by Vibrio cholerae confers resistance to predation by Acanthamoeba castellanii. FEMS Microbiol Ecol 93:fix147. doi: 10.1093/femsec/fix147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hiramatsu Y, Nishida T, Nugraha DK, Sugihara F, Horiguchi Y. 2021. Melanin produced by Bordetella parapertussis confers a survival advantage to the bacterium during host infection. mSphere 6:e0081921. doi: 10.1128/mSphere.00819-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Q, Yang X, Lin L, Wu S, Wang P, Wei W, Wei D. 2023. Pyomelanin production via heterologous expression of 4-hydroxyphenylpyruvate dioxygenase (HPPD) and construction of HPPD inhibitor screening model. J Biosci Bioeng 135:93–101. doi: 10.1016/j.jbiosc.2022.10.005 [DOI] [PubMed] [Google Scholar]
- 21. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/s0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- 22. Revelli DA, Boylan JA, Gherardini FC. 2012. A non-invasive intratracheal inoculation method for the study of pulmonary melioidosis. Front Cell Infect Microbiol 2:164. doi: 10.3389/fcimb.2012.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Darling P, Chan M, Cox AD, Sokol PA. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect Immun 66:874–877. doi: 10.1128/IAI.66.2.874-877.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baldwin A, Mahenthiralingam E, Thickett KM, Honeybourne D, Maiden MCJ, Govan JR, Speert DP, Lipuma JJ, Vandamme P, Dowson CG. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J Clin Microbiol 43:4665–4673. doi: 10.1128/JCM.43.9.4665-4673.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holden MTG, Seth-Smith HMB, Crossman LC, Sebaihia M, Bentley SD, Cerdeño-Tárraga AM, Thomson NR, Bason N, Quail MA, Sharp S, et al. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 191:261–277. doi: 10.1128/JB.01230-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mahenthiralingam E, Coenye T, Chung JW, Speert DP, Govan JRW, Taylor P, Vandamme P. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol 38:910–913. doi: 10.1128/JCM.38.2.910-913.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vergunst AC, Meijer AH, Renshaw SA, O’Callaghan D. 2010. Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infect Immun 78:1495–1508. doi: 10.1128/IAI.00743-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cardona ST, Wopperer J, Eberl L, Valvano MA. 2005. Diverse pathogenicity of Burkholderia cepacia complex strains in the Caenorhabditis elegans host model. FEMS Microbiol Lett 250:97–104. doi: 10.1016/j.femsle.2005.06.050 [DOI] [PubMed] [Google Scholar]
- 29. Seed KD, Dennis JJ. 2008. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect Immun 76:1267–1275. doi: 10.1128/IAI.01249-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bernier SP, Silo-Suh L, Woods DE, Ohman DE, Sokol PA. 2003. Comparative analysis of plant and animal models for characterization of Burkholderia cepacia virulence. Infect Immun 71:5306–5313. doi: 10.1128/IAI.71.9.5306-5313.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sousa SA, Ulrich M, Bragonzi A, Burke M, Worlitzsch D, Leitão JH, Meisner C, Eberl L, Sá-Correia I, Döring G. 2007. Virulence of Burkholderia cepacia complex strains in gp91phox-/- mice. Cell Microbiol 9:2817–2825. doi: 10.1111/j.1462-5822.2007.00998.x [DOI] [PubMed] [Google Scholar]
- 32. Menon ND, Penziner S, Montaño ET, Zurich R, Pride DT, Nair BG, Kumar GB, Nizet V. 2022. Increased innate immune susceptibility in hyperpigmented bacteriophage-resistant mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother 66:e0023922. doi: 10.1128/aac.00239-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tippin B, Pham P, Goodman MF. 2004. Error-prone replication for better or worse. Trends Microbiol 12:288–295. doi: 10.1016/j.tim.2004.04.004 [DOI] [PubMed] [Google Scholar]
- 34. Peeters E, Sass A, Mahenthiralingam E, Nelis H, Coenye T. 2010. Transcriptional response of Burkholderia cenocepacia J2315 sessile cells to treatments with high doses of hydrogen peroxide and sodium hypochlorite. BMC Genomics 11:90. doi: 10.1186/1471-2164-11-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x [DOI] [PubMed] [Google Scholar]
- 36. Segal BH, Ding L, Holland SM. 2003. Phagocyte NADPH oxidase, but not inducible nitric oxide synthase, is essential for early control of Burkholderia cepacia and Chromobacterium violaceum infection in mice. Infect Immun 71:205–210. doi: 10.1128/IAI.71.1.205-210.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mardiney M, Jackson SH, Spratt SK, Li F, Holland SM, Malech HL. 1997. Enhanced host defense after gene transfer in the murine p47phox-deficient model of chronic granulomatous disease. Blood 89:2268–2275. doi: 10.1182/blood.V89.7.2268 [DOI] [PubMed] [Google Scholar]
- 38. Pavan ME, López NI, Pettinari MJ. 2020. Melanin biosynthesis in bacteria, regulation and production perspectives. Appl Microbiol Biotechnol 104:1357–1370. doi: 10.1007/s00253-019-10245-y [DOI] [PubMed] [Google Scholar]
- 39. Nosanchuk JD, Casadevall A. 2003. The contribution of melanin to microbial pathogenesis. Cell Microbiol 5:203–223. doi: 10.1046/j.1462-5814.2003.00268.x [DOI] [PubMed] [Google Scholar]