Abstract
Herpes simplex virus type 1 (HSV-1) glycoprotein E (gE) functions as an immunoglobulin G (IgG) Fc binding protein and is involved in virus spread. Previously we studied a gE mutant virus that was impaired for IgG Fc binding but intact for spread and another that was normal for both activities. To further evaluate the role of gE in spread, two additional mutant viruses were constructed by introducing linker insertion mutations either outside the IgG Fc binding domain at gE position 210 or within the IgG Fc binding domain at position 380. Both mutant viruses were impaired for spread in epidermal cells in vitro; however, the 380 mutant virus was significantly more impaired and was as defective as gE null virus. gE mutant viruses were inoculated into the murine flank to measure epidermal disease at the inoculation site, travel of virus to dorsal root ganglia, and spread of virus from ganglia back to skin to produce zosteriform lesions. Disease at the inoculation and zosteriform sites was reduced for both mutant viruses, but more so for the 380 mutant virus. Moreover, the 380 mutant virus was highly impaired in its ability to reach the ganglia, as demonstrated by virus culture and real-time quantitative PCR. The results indicate that the domain surrounding amino acid 380 is important for both spread and IgG Fc binding and suggest that this domain is a potential target for antiviral therapy or vaccines.
Glycoprotein gE of herpes simplex virus type 1 (HSV-1) functions as a receptor for the Fc portion of immunoglobulin G (IgG) (FcγR) and plays a role in virus spread from cell to cell (5, 6, 10–12, 16). gE interacts with glycoprotein gI to form a noncovalent heterodimer complex (20, 21) that increases Fc binding affinity so that the gE-gI complex binds IgG monomers, whereas gE alone binds IgG aggregates but not monomers (14). Considerable information exists defining the gE domains involved in IgG Fc binding (3, 4, 13); however, less is known about gE domains involved in cell spread (34).
Recent in vivo studies have established that gE-mediated immune evasion contributes to virulence in the murine flank model (27). The experiments focused on NS-gE339, an FcγR-negative virus containing a 12-bp linker insertion that introduces four amino acids at gE position 339. In the absence of passively transferred human anti-HSV antibody, wild-type virus and NS-gE339 caused similar disease at the inoculation site. However, passive transfer of human anti-HSV IgG resulted in a much greater reduction in disease scores in animals infected with the gE mutant virus than in those infected with wild-type virus, establishing the potency of the HSV-1 FcγR in blocking antibody-mediated attack. The results support the concept that gE contributes to pathogenesis because of its participation in antibody bipolar bridging, which refers to the ability of gE to block activities mediated by the Fc domain of an IgG molecule bound by its Fab domain to HSV antigen (16, 27).
During primary infection, HSV-1 spreads within epithelial cells and into sensory neurons located in ganglia, where it either establishes latency or replicates and then travels from neurons back to skin or mucosa. gE contributes to the ability of virus to spread from inoculation site to ganglia and likely from ganglia back to skin, based on a number of reports (2, 10, 11, 27, 28). However, gE does not appear to be involved in spread from neuron to epithelial cells (25). In vitro, gE is required for cell-cell fusion (9), and gE localizes to tight junctions at points of cell-cell contact by interacting with junctional components (12). This localization is postulated to facilitate virus spread to adjacent cells. Mutant viruses lacking gE form small plaques in cell cultures, supporting a role for gE in spread (2, 10, 34). Studies of pseudorabies virus (PRV) in animal models show that gE mutant viruses spread through the central nervous system less vigorously than wild-type virus (1, 7, 15, 35, 37), supporting a role in virus spread for gE homologues in other herpesviruses.
Linker insertions are thought to cause local disruptions to protein structure and are helpful to define the function of a disrupted domain. We evaluated two viruses with linker insertion mutations at different sites in gE. One mutation adds four amino acids at gE position 210, which lies outside the domain for FcγR function, while the other mutation adds four amino acids and modifies one adjacent codon at position 380, which lies within the FcγR domain (3, 13). Compared with wild-type virus, both mutant viruses are impaired for spread in epithelial cells in vitro, exhibit decreased transit from skin to sensory ganglia in vivo, and cause less severe disease.
MATERIALS AND METHODS
Cells.
Vero and HaCaT cells (34) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, gentamicin, amphotericin B, l-glutamine, and HEPES buffer solution.
NS, NS-gEnull, NS-gE210, NS-gE380, and rescued NS-gE380 viruses.
Wild-type HSV-1 strain NS is a low-passage-number clinical isolate that was used for the generation of gE mutant viruses (8, 18). NS-gEnull virus was described previously (27) and was constructed by deleting 1.1 kb of gE corresponding to amino acids 124 to 508 and replacing this segment with a β-galactosidase reporter gene. NS-gE210 and NS-gE380 mutant viruses were prepared by genetic recombination using NS-gEnull DNA and gE plasmids containing in-frame XhoI linkers at gE amino acids 210 and 380 (3, 13). Recombinant viruses were identified by an immunoperoxidase assay using anti-gE monoclonal antibody (MAb) 1BA10 (17). Infected-cell monolayers were incubated with anti-gE MAb 1BA10 (1:100 dilution) followed by protein A-horseradish peroxidase (1:150 dilution). Plaques were scored for gE expression after incubation with a 4-chloro-1-naphthol substrate (Sigma Chemical Co., St. Louis, Mo.). Recombinant viruses were purified in Vero cells by two rounds of plaque picking followed by one round of purification at limiting dilution.
Rescued NS-gE380 (rNS-gE380) virus was prepared using NS-gE380 DNA and wild-type NS gE DNA obtained from pCMV3-gE (13). Recombinant viruses were screened by selecting large plaques in HaCaT cells and by measuring IgG Fc binding by flow cytometry (27). The rescued NS-gE380 virus was purified by plaque picking and limiting dilution.
Southern blot.
Viral DNA was digested with restriction endonuclease NruI to excise gE protein coding sequences and XhoI to detect the presence of an XhoI linker in gE. DNA was electrophoresed in 1% agarose gels and transferred to Immobilon-S membranes (Millipore, Bedford, Mass.) that were probed with biotinylated gE (New England Biolabs, Beverly, Mass.) prepared from the 1.1-kb fragment deleted from the NS-gEnull virus.
Western blot.
Sucrose gradient-purified viruses were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 10% polyacrylamide gel under denaturing conditions and transferred to an Immobilon-P membrane (Millipore). gE was detected using anti-gE MAb 1BA10, goat anti-mouse IgG antibody conjugated to horseradish peroxidase, and enhanced chemiluminescence (Amersham Pharmacia Biotech Inc., Piscataway, N.J.).
Single-step growth curves.
Vero cells were infected at a multiplicity of infection (MOI) of 2 to 5 for 1 h at 37°C. The cells were washed three times with phosphate-buffered saline (PBS), fresh medium was added at time point 1 h, and at 1, 4, 8, 16, 20, and 24 h the flasks were frozen at −70°C. Flasks were later thawed, cells and medium were sonicated for 3 min on ice (Heatsystems Ultrasonics Inc., Plainview, N.Y.), and virus titers were measured by plaque assay on Vero cells.
Flow cytometry.
Vero cells were infected at an MOI of 2 for 12 to 16 h, and cells were dissociated using cell dissociation buffer (Life Technologies Inc., Rockville, Md.). gE expression at the cell surface was detected using anti-gE MAb 1BA10, while IgG Fc binding was measured using biotinylated nonimmune IgG. 1BA10 binding was detected using fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG, and IgG binding was measured using streptavidin–R-phycoerythrin (PE). Cells were fixed in 2% paraformaldehyde and observed with a FACScan flow cytometer (Becton-Dickinson, San Jose, Calif.).
Rosetting assays.
Sheep erythrocytes (ICN Pharmaceuticals, Inc., Costa Mesa, Calif.) were sensitized with subagglutinating concentrations of goat anti-sheep erythrocyte IgG (Life Technologies Inc.). Vero cells were infected at an MOI of 5 for 16 h and incubated with the IgG-coated erythrocytes for 1 h at 37°C. Cells were placed on a hemocytometer, and rosettes were counted at ×100 magnification. Infected cells that bound ≥4 erythrocytes were considered positive (8). Infected cells incubated with erythrocytes in the absence of antierythrocyte IgG served as controls.
In vitro cell spread assay.
HaCaT cells were grown to 80% confluence in six-well plates and inoculated with 25 to 50 PFU of wild-type or gE mutant virus per well for 1 h at 37°C (34). Cells were overlaid with low-melting-temperature agarose and DMEM. After 48 h, the agarose was removed, and the cells were stained for 5 min with Giemsa and rinsed with PBS. Plaque size was measured along perpendicular diameters using an eyepiece micrometer. Area was calculated as pi times radius squared, and the radius was calculated from the average of the perpendicular diameters.
Murine flank model.
The shaved right flanks of 5- to 6-week-old female BALB/c mice were denuded with depilatory cream. Twenty-four hours later, 5 × 105 PFU of virus in 10 μl of sterile PBS was applied to the denuded flank several millimeters from the spinal column (23, 27, 29). For some experiments, wild-type virus was incubated with 1% paraformaldehyde at 37°C for 2 h to inactivate the virus. The mice were scored for disease on days 3 to 8 postinfection as follows: 0 points for no disease, 0.5 point for swelling without vesicles, and 1 point for each vesicle or scab. The maximum daily score was 5 for inoculation site disease and 10 for zosteriform disease. The higher potential score for zosteriform disease was because of the larger skin area involved. In the case of confluent vesicles or scabs at the inoculation site, the score was assigned based on the size of the confluent lesion.
Harvesting of dorsal root ganglia was performed at 3, 5, and 7 days postinfection. The mice were euthanized, and the ganglia that innervate the inoculated skin were identified under 4× magnifying goggles. The ganglia were removed and homogenized in DMEM, and virus in the ganglia was titrated by plaque assay. For PCR experiments, ganglia were placed in RNAlater (Ambion Inc., Austin, Tex.), stored at 4°C for 24 h, and then frozen at −70°C.
Extraction of DNA and RNA and reverse transcription.
DNA and RNA were isolated from dorsal root ganglia using TRIzol reagent (Life Technologies-Gibco BRL, Grand Island, N.Y.). RNA was digested with RNase-free DNase I (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) for 45 min at 37°C followed by 5 min at 70°C to inactivate the enzyme. Complementary DNA was generated from 0.5 μg of total RNA using the Superscript preamplification kit (Life Technologies) and priming with oligo(dT) and random hexamers as described previously (31).
Real-time quantitative PCR analysis.
Reactions were performed in 50-μl volumes containing 2× TaqMan Universal PCR master mix (Perkin-Elmer, Norwalk, Conn.) and 5 μl of cDNA for detection of viral transcripts, 1 μl of cDNA for detection of cellular transcripts, or 100 ng of dorsal root ganglion DNA for detection of viral DNA. Reactions contained 200 nM TaqMan primers and 200 nM TaqMan probe. Primer pairs and probes are described in Table 1 and were designed using Primer Express software (Perkin-Elmer). Probes were labeled at the 5′ end with the fluorescent reporter dye Fam and at the 3′ end with the fluorescent quencher dye Tamra (Synthegen, Houston, Tex.) to allow direct detection of the PCR product. Real-time PCR amplification and detection were performed using an ABI 7700 sequence detector (PE Biosystems, Norwalk, Conn.). Relative copy number was calculated using a standard curve generated from purified HSV-1 (NS) viral DNA that was serially diluted in 10 ng of mouse genomic DNA (Clontech, Palo Alto, Calif.) per μl. Viral DNA was diluted to contain from 1 to 1 million copies in 2 μl and subjected to TaqMan PCR with each primer set to generate standard curves and evaluate relative primer sensitivity.
TABLE 1.
Primers and probes used for PCR amplification of HSV-1 and genomic DNA or cDNA
| Primer | Location | Size (bp) | Sequence |
|---|---|---|---|
| gC (UL44) | 5′ | 19 | GATGCCGGTTTCGGAATTC |
| 3′ | 22 | CCCATGGAGTAACGCCATATCT | |
| Probe | 21 | ACCCGCATGGAGTTCCGCCTC | |
| US9 | 5′ | 17 | AGGCGGCCAACGACTTC |
| 3′ | 21 | TCGACGCCTTAATACCGACTG | |
| Probe | 19 | TCGTACGCATGGGCCGCCA | |
| ICP27 (UL54) | 5′ | 21 | CGCCAAGAAAATTTCATCGAG |
| 3′ | 19 | ACATCTTGCACCACGCCAG | |
| Probe | 20 | CTGGCCTCCGCCGACGAGAC | |
| GAPDHa (control gene) | 5′ | 23 | CAAGGTCATCCATGACAACTTTG |
| 3′ | 20 | GGCCATCCACAGTCTTCTGG | |
| Probe | 25 | ACCACAGTCCATGCCATCACTGCCA |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
RESULTS
Characterization of gE mutant viruses. (i) Southern blots to show proper construction of gE mutant viruses.
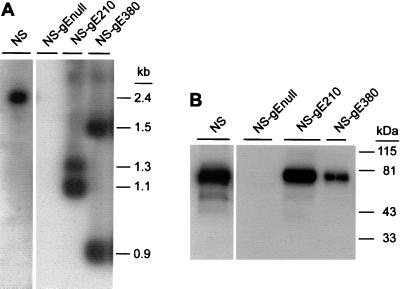
DNA from NS, NS-gEnull, NS-gE210, and NS-gE380 viruses was digested with NruI and XhoI to determine whether XhoI linkers had been incorporated into the mutant viruses (Fig. 1A). As expected, a 2.4-kb fragment representing gE DNA without an XhoI insert was detected in NS virus, and no gE DNA was detected in NS-gEnull virus. Two gE bands were present in NS-gE210 and NS-gE380 viruses: 1.1 and 1.3 kb for NS-gE210, and 0.9 and 1.5 kb for NS-gE380. The results indicate that the XhoI linkers were incorporated at the expected positions in the gE linker insertion mutant viruses.
FIG. 1.
(A) Southern blot of viral DNA from NS, NS-gEnull, NS-gE210, and NS-gE380 viruses digested with NruI and XhoI and probed to detect gE DNA. Two bands were detected for mutant viruses NS-gE210 and NS-gE380, indicating the presence of the XhoI linker. (B) Western blot of sucrose gradient-purified viruses to detect gE. Lane 1, NS; lane 2, NS-gEnull; lane 3, NS-gE210; lane 4, NS-gE380. Bands of approximately 80 kDa were detected in lanes loaded with NS, NS-gE210, and NS-gE380 viruses.
(ii) Western blots to show that mutant gE proteins are incorporated into virus.
Purified viruses were run on a 10% denaturing polyacrylamide gel (Fig. 1B). A broad band of approximately 80 kDa was detected in NS virus, and a similar band was present in NS-gE210 and NS-gE380 viruses. No gE protein was detected in NS-gEnull virus. The results indicate that recombinant proteins gE210 and gE380 are incorporated into virus.
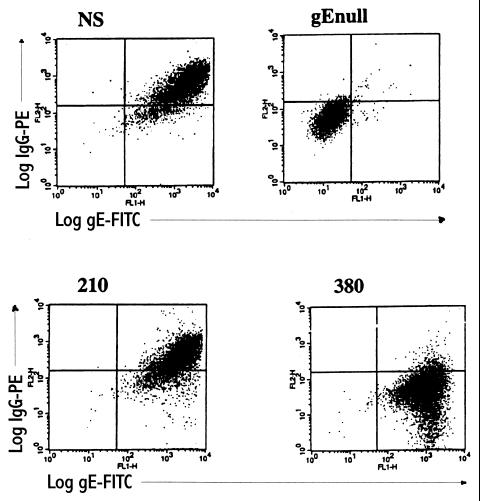
(iii) Flow cytometry to demonstrate gE expression and IgG Fc binding at the surface of infected cells.
NS, NS-gEnull, NS-gE210, and NS-gE380 viruses were evaluated for gE expression and Fc binding of nonimmune human monomeric IgG on infected Vero cells (Fig. 2). Cells infected with NS-gE210 and NS-gE380 viruses had levels of gE expression comparable to that with NS virus, while NS-gEnull-infected cells expressed no gE. Cells infected with NS-gE210 showed IgG binding comparable to that with NS, while NS-gE380- and NS-gEnull-infected cells demonstrated little IgG Fc binding. The results indicate that recombinant gE proteins are expressed at the cell surface and that the mutation at gE position 380 eliminates IgG Fc binding.
FIG. 2.
gE expression and monomeric IgG binding following infection of Vero cells with NS virus, NS-gEnull virus (labeled gEnull), NS-gE210 virus (labeled 210), or NS-gE380 virus (labeled 380). Each panel represents two-color immunofluorescence using anti-gE-FITC on the x axis and anti-HSV-1 human nonimmune IgG-PE on the y axis.
(iv) Rosetting assays to measure IgG Fc binding of gE mutant viruses.
Binding of monomeric nonimmune IgG to HSV-1-infected cells requires both gE and gI, while rosetting is mediated by gE alone (14). Rosetting assays were performed as further evidence that the mutations in NS-gE210 and NS-gE380 viruses were in gE. Rosettes formed around cells infected with NS (68%) and NS-gE210 viruses (78%), while few NS-gE380-infected cells formed rosettes (10%), and no cells infected with NS-gEnull virus formed rosettes. Taken together, the rosetting and IgG monomer binding results indicate that FcγR activity is intact for NS-gE210 virus, while it is disrupted for NS-gE380 virus because of the mutation in gE.
(v) Single-step growth curves.
Mutant viruses NS-gE210 and NS-gE380 were evaluated for replication kinetics. Both mutant viruses showed at least a 100-fold increase in titer by 20 h postinfection and were comparable to NS virus (results not shown).
Cell-to-cell spread phenotypes of NS-gE210 and NS-gE380.
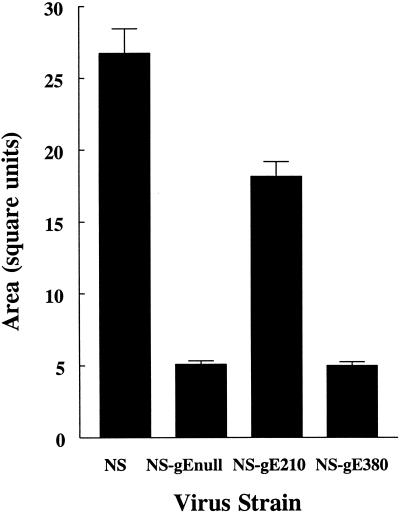
Plaque size formed by the gE mutant viruses was measured in HaCaT cells, an epidermal cell line derived from human keratinocytes (Fig. 3). NS virus plaques were significantly larger than those of each of the gE mutant viruses (P < 0.0001 comparing NS virus with NS-gEnull, NS-gE210, or NS-gE380 virus). Comparing the gE mutant viruses, NS-gE380 virus and NS-gEnull virus plaques were of comparable size (P = 0.77), and both were significantly smaller than NS-gE210 (P < 0.0001). These results indicate that the mutation at gE position 380 has a greater effect than the mutation at gE position 210 on virus spread in epidermal cells and that the mutation at gE position 380 inhibits epidermal cell spread comparable to the deletion in the gE null virus.
FIG. 3.
Mean area of plaques formed on HaCaT cells following infection by NS, NS-gEnull, NS-gE210, or NS-gE380 virus. Results are the mean ± standard error of the mean (SEM) for 50 plaques measured for each virus.
Disease in the murine flank model.
In the murine flank model, virus replicates at the inoculation site to produce vesicles that ulcerate and scab as they heal (29), which is similar to HSV-1 lesions of skin and mucocutaneous junctions in humans. In the flank model, virus spreads from the skin to the ganglia, where it replicates, infects additional neurons in the ganglia, and then travels along axons back to the skin to produce zosteriform disease in areas innervated by the neurons (29). Inoculation site disease first appears on day 3 and is localized to an area <1 cm in diameter. Zosteriform disease generally appears on day 5 and can be distinguished from inoculation site disease by its distribution on the flank over an area 3 to 5 cm in length and 1 to 2 cm in width (22). The number of lesions that develop at the inoculation and zosteriform sites directly correlates with the amount of virus injected (22).
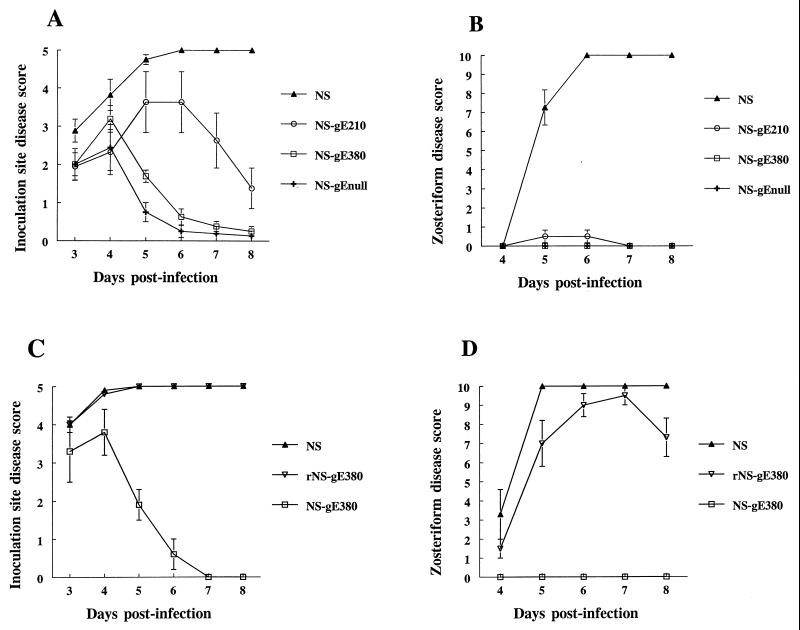
BALB/c mice were infected with 5 × 105 PFU of NS, NS-gEnull, NS-gE210, or NS-gE380 by scratch inoculation on denuded flanks. As reported previously (27), large differences in disease at the inoculation site were noted when comparing NS and NS-gEnull viruses (P < 0.0001 for days 5 to 8) (Fig. 4A). Disease scores for NS-gE380 virus were also significantly reduced compared with NS virus (P < 0.0001 for days 5 to 8), while differences between NS-gE210 and NS viruses were less striking but statistically significant on days 7 (P < 0.01) and 8 (P < 0.0001). NS-gE380 virus was almost as attenuated as NS-gEnull virus, since only on day 5 was a difference detected between these two mutant viruses (P < 0.01). Of note, disease scores among the viruses did not differ significantly on days 3 and 4, suggesting that each of the mutant viruses is capable of establishing infection at the site of inoculation and that the defect in virulence occurs subsequent to the initial infection. No lesions developed when mice were infected with paraformaldehyde-inactivated wild-type virus, which supports the conclusion that lesions appearing on days 3 and 4 after injection of NS-gE380 and NS-gEnull viruses represent active infection.
FIG. 4.
Disease scores at (A) the inoculation site and (B) the zosteriform site of mice infected with NS, NS-gE210, NS-gE380, or NS-gEnull virus. The results in panels A and B are means ± SEM for eight mice in each group. Disease scores at (C) the inoculation site and (D) the zosteriform site of mice infected with NS, rescued NS-gE380 (rNS-gE380), or NS-gE380 virus. The results in panels C and D are the means ± SEM for four mice in each group.
The gE mutant viruses each produced markedly reduced zosteriform disease (Fig. 4B), while NS virus caused extensive zosteriform disease (P < 0.0001 for each mutant virus compared with NS virus on days 5 to 8). Mice infected with NS-gE210 virus had very limited zosteriform disease, with only two of eight animals developing skin lesions, while animals infected with NS-gE380 or NS-gEnull virus had no zosteriform lesions. Mortality was also less for gE mutant viruses than for NS, since death occurred in five of eight animals infected with NS virus, but in none infected with NS-gEnull, NS-gE210, or NS-gE380 virus (P < 0.01). The results indicate that both NS-gE210 and NS-gE380 linker insertion mutant viruses cause less disease than wild-type virus in the murine flank model; however, NS-gE380 is the more attenuated of the two and is almost as impaired as NS-gEnull virus.
Because of the markedly reduced disease caused by NS-gE380 virus, a rescued virus was constructed to further confirm the importance of the gE380 region in pathogenesis. The phenotype of the rescued virus was comparable to that of wild-type virus when evaluated by Southern blot, Western blot, assays for FcγR activity, and spread in HaCaT cells (results not shown). In the murine flank model, the rescued virus produced disease scores that were significantly higher than those with NS-gE380 virus on days 5 to 8 postinfection (P < 0.001) (Fig. 4C and D). The rescued virus completely restored inoculation site disease to wild-type levels (Fig. 4C) and almost completely restored disease at the zosteriform site (Fig. 4D). The method used to construct the rescued virus likely explains the minor differences in disease caused by rescued and wild-type viruses. The rescued virus was constructed using DNA derived from a single virus particle, while DNA cloning was not used to prepare NS virus. Therefore, the wild-type virus represents a more heterogeneous population of virions. The key conclusion from the results shown in Fig. 4 is that the mutation at gE position 380 accounts for markedly reduced disease at the inoculation and zosteriform sites.
Virus titers in dorsal root ganglia.
Dorsal root ganglia of mice infected with NS, NS-gEnull, NS-gE210, or NS-gE380 virus were harvested at 3 and 5 days postinfection to evaluate infectious virus. NS and NS-gE210 viruses were recovered from ganglia at day 3 postinfection, while NS-gEnull and NS-gE380 viruses were not detected (Table 2). Ganglia harvested at day 5 postinfection showed a >500-fold increase in titer of NS virus. In contrast, ganglia from mice infected with NS-gE380 or NS-gEnull virus remained negative, and ganglia harvested from mice infected with NS-gE210 virus were negative at the later time point as well, suggesting that only small numbers of NS-gE210 virus reached the ganglia. Additional ganglia were harvested at 7 days postinfection from three mice infected with NS-gE380 to determine if virus was delayed in reaching the ganglia. Virus cultures remained negative. The results suggest that both NS-gE210 and NS-gE380 viruses are impaired in their ability to travel from skin to ganglia but that NS-gE380 virus is the more impaired of the two.
TABLE 2.
Virus titers in dorsal root ganglia of mice infected with NS, NS-gEnull, NS-gE210, or NS-gE380a
| Virus | Mean titer (PFU/ml) ± SEM
|
|
|---|---|---|
| Day 3 | Day 5 | |
| NS | 79 ± 20 | 30,125 ± 11,208 |
| NS-gEnull | ≤3 | ≤3 |
| NS-gE210 | 28 ± 5 | ≤3 |
| NS-gE380 | ≤3 | ≤3 |
Results are the means ± SEM for dorsal root ganglia from three mice at each time point except for NS at day 5, which represents six mice. The sensitivity of the assay to quantify virus titers is ≤3 PFU/ml of ganglion tissue.
Quantifying DNA copy number in dorsal root ganglia.
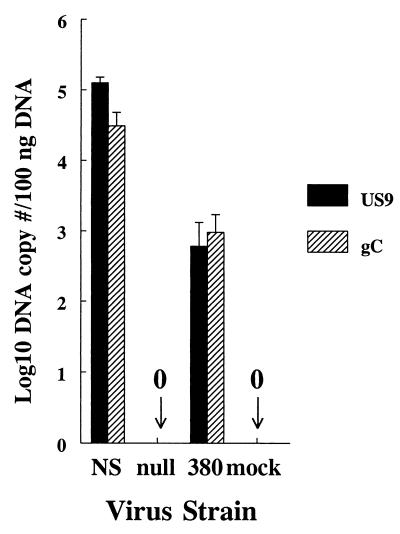
As further evidence of reduced infection of ganglia by NS-gE380 virus, experiments were performed using more sensitive techniques than virus culture. At 5 days postinfection, ganglia were harvested from mice infected with NS, NS-gE380, or NS-gEnull virus and mock-infected controls. Quantitative PCR assays were performed to detect HSV-1 DNA from two distinct regions of the genome, US9 and gC (Fig. 5). No DNA was detected in ganglia removed from mock-infected animals or from those infected with NS-gEnull virus. Viral DNA was detected in ganglia from all three NS-gE380-infected animals; however, the copy number was significantly lower than that of NS for both genes tested, 2 to 3 log10 (234-fold) lower at the US9 gene (P = 0.003), and 1 to 2 log10 (32-fold) lower at the gC gene (P = 0.008). The results suggest that NS-gEnull virus fails to reach the ganglia, while NS-gE380 virus is capable of spreading to the ganglia, but the amount of viral DNA reaching the ganglia is greatly reduced compared with NS virus.
FIG. 5.
Quantifying HSV-1 DNA in dorsal root ganglia of mice infected with NS, NS-gEnull (labeled null), or NS-gE380 (labeled 380) virus and mock-infected controls. Quantitative real-time PCR was performed on DNA extracted from the ganglia using primers for US9 or gC DNA. Results represent copy numbers per 100 ng of DNA and are the mean ± SEM for ganglia from three mice in each group.
Quantifying RNA copy number in dorsal root ganglia.
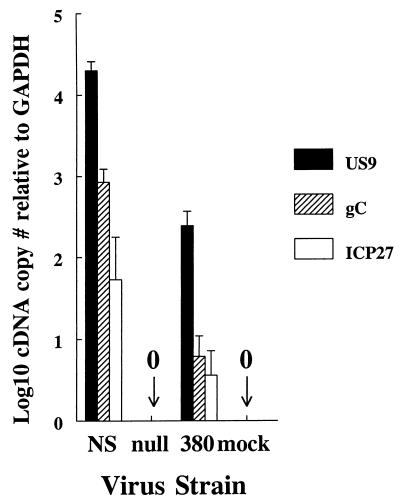
The same ganglia as above were evaluated for expression of HSV-1 RNA to determine whether transcription occurs in the ganglia. HSV-1 transcripts to an immediate-early gene, ICP27, an early gene, US9, and a late gene, gC, were detected in ganglia from mice infected with NS virus but not in ganglia from mice infected with NS-gEnull virus or mock-infected controls (Fig. 6). Ganglia from mice infected with NS-gE380 virus had detectable RNA; however, levels were lower than for NS virus. Differences between the two viruses were between 1 and 2 log10 (81-fold) for US9 transcripts (P = 0.0008), 2 and 3 log10 (138-fold) for gC transcripts (P = 0.002), and 1 and 2 log10 (16-fold) for ICP27 transcripts (P = 0.12, not significant). The fact that differences in ICP27 transcripts did not reach statistical significance likely reflects the small number of ICP27 transcripts detected and the relatively small number of mice analyzed. The important conclusion from these results is that NS-gE380 virus reaches the ganglia at reduced levels compared with NS virus and that NS-gE380 virus is transcriptionally active.
FIG. 6.
Real-time PCR quantification of cDNA generated from HSV-1 RNA in dorsal root ganglia of mice infected with NS, NS-gEnull (labeled null), or NS-gE380 (labeled 380) virus and mock-infected controls. RNA was quantified for an immediate-early gene, ICP27, an early gene, US9, and a late gene, gC. The number of viral transcripts was calculated in comparison with glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a control transcript expressed constitutively in cells. Results are the means ± SEM for ganglia from three mice in each group.
DISCUSSION
We previously reported that a four-amino-acid linker insertion mutation at gE amino acid 339 eliminates IgG Fc binding while having little effect on virus spread in epidermal cells in vitro (27, 34). A similar four-amino-acid linker inserted at gE position 406 had no effect on IgG Fc binding or virus spread (27, 34). We now report that a mutation at gE position 210 maintains IgG Fc binding activity but impairs virus spread in vitro, while a mutation at gE position 380 results in a virus that is defective in both IgG Fc binding and virus spread. These results indicate that different domains of gE are involved in IgG Fc binding and spread and that the domain surrounding gE amino acid 380 is important for both gE functions.
gE mutant viruses NS-gE210 and NS-gE380 both caused reduced disease compared with NS. One possible explanation for the impaired function of NS-gE210 and NS-gE380 viruses is that the gE proteins are grossly malformed. This explanation seems unlikely for several reasons. First, the mutations in gE210 and gE380 lie outside the cysteine-rich domain of the molecule (24), decreasing the likelihood that the tertiary structure of the protein is markedly disrupted. Second, gE is expressed at the surface of NS-gE210- and NS-gE380-infected cells and is incorporated into the virion, which suggests that the protein structure is not severely malformed. Third, the mutant gE 210 and 380 proteins are recognized by a panel of MAbs, including one that is sensitive to changes in gE conformation (3). Fourth, mutations at gE positions 210, 339, 380, and 406 affect gE functions differently (summarized in Table 3), suggesting that linker insertion mutations disrupt limited regions of the gE molecule rather than globally altering gE structure.
TABLE 3.
Comparison of gE mutant virus phenotypes with NS, the wild-type (WT) parental straina
| gE mutant | IgG Fc binding | Epidermal spread in vitro | Inoculation site disease | Spread to gangliab | Zosteriform disease |
|---|---|---|---|---|---|
| NS-gEnull | None | Much reduced | Much reduced | None | None |
| NS-gE210 | WT levels | Reduced | Reduced | Reduced | Much reduced |
| NS-gE339 | Much reduced | WT levels | WT levels | ND | Slightly reduced |
| NS-gE380 | Much reduced | Much reduced | Much reduced | Much reduced | None |
| NS-gE406 | WT levels | WT levels | WT levels | ND | WT levels |
Spread to ganglia was measured by virus titers and/or DNA and RNA quantitative PCR. ND, not done; however, the virus must reach the ganglion since the virus spreads along axons innervated by the ganglion to cause zosteriform disease. Much reduced, no infectious virus was detected in the ganglia by viral titration; however, viral DNA and RNA were detected by quantitative real-time PCR.
In previous studies with the murine flank model, we used passive transfer of human anti-HSV IgG to demonstrate the important role of the HSV-1 FcγR in pathogenesis. Passive transfer of human IgG was used to evaluate FcγR activity because human IgG binds to the HSV-1 FcγR, while murine IgG does not (19, 27). The observation that the FcγRs of NS-gE380 and NS-gE339 viruses are impaired to a similar extent in vitro suggests that NS-gE380 virus is likely to be very susceptible to anti-HSV IgG in vivo. However, disease caused by NS-gE380 virus was already so reduced, probably because of impaired viral spread, that antibody passive transfer experiments were not pursued to determine whether the FcγR− phenotype of NS-gE380 virus further affects virulence.
NS-gEnull DNA and RNA were not detected in dorsal root ganglia, which suggests that gE is essential for virus to travel from skin to ganglia in the murine flank model. NS-gE210 infectious virus was detected in ganglia at reduced levels, while NS-gE380 infectious virus was not detected at all, indicating that the gE mutations affected the ability of the viruses to reach the ganglia. NS-gE210 caused some zosteriform disease; therefore, this mutant virus is capable of spreading from ganglia to skin. In contrast, NS-gE380 virus did not produce zosteriform disease. This result can be explained in part by reduced titers of NS-gE380 virus reaching the ganglia; however, defects in NS-gE380 virus spread from ganglia to skin may be another explanation. Future studies using models of latency and reactivation in rabbits or guinea pigs may help define whether NS-gE380 virus is defective in spread from ganglia to skin.
Although NS-gE380 infectious virus was not recovered from ganglia, viral DNA and RNA were detected, which likely reflects differences in sensitivity of viral culture and PCR assays. From the PCR and viral titer results, we estimate that the sensitivity of the quantitative DNA PCR assay is 2 to 3 log10 greater than that of virus culture and the sensitivity of the RNA PCR assay is from 0.5 to 2.5 log10 greater than that of virus culture, and the range of sensitivity depends upon which gene is being evaluated. The high specificity of DNA and RNA quantitative PCR was verified by the lack of PCR signal in mock-infected animals.
Experiments with PRV in the rat eye model defined a gE mutant virus that was normal for spread yet impaired for virulence, suggesting that spread and virulence are distinct functions mediated by gE (32, 33). Mutant viruses NS-gE210 and NS-gE380 differ from the PRV mutant virus in that a single mutation at either gE amino acid 210 or 380 affected both spread and virulence. However, it is possible that mutations in other gE domains could affect virulence without modifying spread. In fact, the mutation at gE amino acid 339 may be an example of such a mutation, since the virus lacks virulence because the mutation affects FcγR activity rather than virus spread (27).
The mechanism(s) by which HSV-1 gE mutations modify spread in HaCaT cells or reduce infection of ganglia was not defined. Possibilities include that gE mutations modify the normal routes of virus travel within epidermal or neuronal cells, that gE is involved in release of virus into synaptic spaces, and that gE is required for efficient transport of virus from one cell to another (10, 11, 12, 34). The murine eye model of infection allows analysis of HSV-1 spread along neural pathways without first infecting epidermal cells; therefore, this model can be used to better define the role of gE in neuronal transport independent of epidermal infection (30). Electron microscopy studies of HSV-1 infection of human fetal neurons in vitro demonstrated a role for microtubules in nucleocapsid, tegument, and glycoprotein axonal transport; however, a role for gE was not evaluated (26). The issue of which domains on gE are involved in spread has been examined in several studies using in vitro models (34, 36). Our current study extends the in vitro results by evaluating the role of gE in virus spread in vivo and defines the importance of extracellular domains around gE amino acids 210 and 380.
We previously performed dose-response studies in BALB/c mice using inocula that varied by 100-fold, ranging from 5 × 103 PFU to 5 × 105 PFU of NS virus (27). At 5 × 103 PFU, the cumulative disease score at the inoculation site from days 3 to 8 postinfection was 15.8 ± 5.8 (27), while the zosteriform disease score was 18.4 ± 8.1 (H. Friedman et al., unpublished data). For comparison purposes, we recalculated the results shown in Fig. 4 as cumulative disease scores for days 3 to 8. NS-gE210 virus at 5 × 105 PFU produced a disease score of 15.5 ± 3.4 at the inoculation site and 1 ± 0.7 at the zosteriform site. Therefore, the mutation at gE position 210 modified disease severity by more than 100-fold. The mutation at gE position 380 had an even greater effect on virulence, since an inoculum of 5 × 105 PFU produced less inoculation site disease than NS-gE210 virus (cumulative disease score of 8.1 ± 0.6) and no zosteriform disease. These findings raise the possibility that gE domains at 210 and 380 may be valuable targets for vaccines or antiviral chemotherapy. Small synthetic molecules may be targeted to these domains to disrupt gE function, or the 210 or 380 domain could be used as an immunogen to produce antibodies that block gE activities. These interventions may diminish the ability of the virus to spread within the host, and by targeting the 380 domain, the additional advantage of blocking HSV-1 evasion of antibody attack may be realized.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI 33063 from the National Institute of Allergy and Infectious Diseases.
We thank Ron Collman for help with the graphics for Fig. 1.
REFERENCES
- 1.Babic N, Klupp B, Brack A, Mettenleiter T C, Ugolini G, Flamand A. Deletion of glycoprotein E reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology. 1996;219:279–284. doi: 10.1006/viro.1996.0247. [DOI] [PubMed] [Google Scholar]
- 2.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI, or the putative gJ. J Gen Virol. 1994;75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 3.Basu S, Dubin G, Basu M, Nguyen V, Friedman H M. Characterization of regions of herpes simplex virus type 1 glycoprotein E involved in binding the Fc domain of monomeric IgG and in forming a complex with glycoprotein I. J Immunol. 1995;154:260–267. [PubMed] [Google Scholar]
- 4.Basu S, Dubin G, Nagashunmugam T, Basu M, Goldstein L T, Wang L, Weeks B, Friedman H M. Mapping regions of herpes simplex virus type 1 glycoprotein I required for formation of the viral Fc receptor for monomeric IgG. J Immunol. 1997;158:209–215. [PubMed] [Google Scholar]
- 5.Baucke R B, Spear P G. Membrane proteins specified by herpes simplex viruses. V. Identification of an Fc-binding glycoprotein. J Virol. 1979;32:779–789. doi: 10.1128/jvi.32.3.779-789.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell S, Cranage M, Borysiewicz L, Minson T. Induction of immunoglobulin G Fc receptors by recombinant vaccinia viruses expressing glycoprotein E and I of herpes simplex virus type 1. J Virol. 1990;64:2181–2186. doi: 10.1128/jvi.64.5.2181-2186.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Card J P, Levitt P, Enquist L W. Different patterns of neuronal infection after intracerebral injection of two strains of pseudorabies virus. J Virol. 1998;72:4434–4441. doi: 10.1128/jvi.72.5.4434-4441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cines D B, Lyss A P, Bina M, Corkey R, Kefalides N A, Friedman H M. Fc and C3 receptors induced by herpes simplex virus on cultured human endothelial cells. J Clin Investig. 1982;69:123–128. doi: 10.1172/JCI110422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis-Poynter N, Bell S, Minson T, Browne H. Analysis of the contribution of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J Virol. 1994;68:7586–7590. doi: 10.1128/jvi.68.11.7586-7590.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingwell K S, Brunetti C R, Hendricks R L, Tang Q, Tang M, Rainbow A J, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingwell K S, Doering L C, Johnson D C. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol. 1995;69:7087–7098. doi: 10.1128/jvi.69.11.7087-7098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingwell K S, Johnson D C. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J Virol. 1998;72:8933–8942. doi: 10.1128/jvi.72.11.8933-8942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubin G, Basu S, Mallory D L, Basu M, Tal-Singer R, Friedman H M. Characterization of domains of herpes simplex virus type 1 glycoprotein E involved in Fc binding activity for immunoglobulin G aggregates. J Virol. 1994;68:2478–2485. doi: 10.1128/jvi.68.4.2478-2485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubin G, Frank I, Friedman H M. Herpes simplex virus type 1 encodes two Fc receptors which have different binding characteristics for monomeric immunoglobulin G (IgG) and IgG complexes. J Virol. 1990;64:2725–2731. doi: 10.1128/jvi.64.6.2725-2731.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enquist L W, Dubin J, Whealy M E, Card J P. Complementation analysis of pseudorabies virus gE and gI mutants in retinal ganglion cell neurotropism. J Virol. 1994;68:5275–5279. doi: 10.1128/jvi.68.8.5275-5279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank I, Friedman H M. A novel function of the herpes simplex virus type 1 Fc receptor: participation in bipolar bridging of antiviral immunoglobulin G. J Virol. 1989;63:4479–4488. doi: 10.1128/jvi.63.11.4479-4488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman H M, Cohen G H, Eisenberg R J, Seidel C A, Cines D B. Glycoprotein C of herpes simplex virus 1 acts as a receptor for the C3b complement component on infected cells. Nature. 1984;309:633–635. doi: 10.1038/309633a0. [DOI] [PubMed] [Google Scholar]
- 18.Friedman H M, Wang L, Fishman N O, Lambris J D, Eisenberg R J, Cohen G H, Lubinski J. Immune evasion properties of herpes simplex virus type 1 glycoprotein gC. J Virol. 1996;70:4253–4260. doi: 10.1128/jvi.70.7.4253-4260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson P J, Myhre E B, Blomberg J. Specificity of Fc receptors induced by herpes simplex virus type 1: comparison of immunoglobulin G from different animal species. J Virol. 1985;56:489–494. doi: 10.1128/jvi.56.2.489-494.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson D C, Feenstra V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol. 1987;61:2208–2216. doi: 10.1128/jvi.61.7.2208-2216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson D C, Frame M C, Ligas M W, Cross A M, Stow N G. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol. 1988;62:1347–1354. doi: 10.1128/jvi.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubinski J, Wang L, Mastellos D, Sahu A, Lambris J D, Friedman H M. In vivo role of complement-interacting domains of herpes simplex virus type 1 glycoprotein gC. J Exp Med. 1999;190:1637–1646. doi: 10.1084/jem.190.11.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lubinski J M, Wang L, Soulika A M, Burger R, Wetsel R A, Colten H, Cohen G H, Eisenberg R J, Lambris J D, Friedman H M. Herpes simplex virus type 1 glycoprotein gC mediates immune evasion in vivo. J Virol. 1998;72:8257–8263. doi: 10.1128/jvi.72.10.8257-8263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGeoch D J, Dolan A, Donald S, Rixon F J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1984;181:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 25.Mikloska Z, Sanna P P, Cunningham A L. Neutralizing antibodies inhibit axonal spread of herpes simplex virus type 1 to epidermal cells in vitro. J Virol. 1999;73:5934–5944. doi: 10.1128/jvi.73.7.5934-5944.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miranda-Saksena M, Armati P, Boadle R A, Holland D J, Cunningham A L. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J Virol. 2000;74:1827–1839. doi: 10.1128/jvi.74.4.1827-1839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagashunmugam T, Lubinski J, Wang L, Goldstein L T, Weeks B S, Sundaresan P, Kang E H, Dubin G, Friedman H M. In vivo immune evasion mediated by the herpes simplex virus type 1 immunoglobulin G Fc receptor. J Virol. 1998;72:5351–5359. doi: 10.1128/jvi.72.7.5351-5359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajcani J, Hergert U, Kaermer H C. Spread of herpes simplex virus strains SC16, ANG, ANGpath, and its gC and gE mutants in DBA/2 mice. Acta Virol. 1990;34:305–320. [PubMed] [Google Scholar]
- 29.Simmons A, Nash A A. Zosteriform spread of herpes simplex virus as a model of recrudescence and its use to investigate the role of immune cells in prevention of recurrent disease. J Virol. 1984;52:816–821. doi: 10.1128/jvi.52.3.816-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun N, Cassell M D, Perlman S. Anterograde, transneuronal transport of herpes simplex virus type 1 strain H129 in the murine visual system. J Virol. 1996;70:5405–5413. doi: 10.1128/jvi.70.8.5405-5413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tal-Singer R, Lasner T M, Podrzucki W, Skokotas A, Leary J J, Berger S L, Fraser N W. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J Virol. 1997;71:5268–5276. doi: 10.1128/jvi.71.7.5268-5276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tirabassi R S, Enquist L W. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J Virol. 1999;73:2717–2728. doi: 10.1128/jvi.73.4.2717-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6464. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weeks B S, Sundaresan P, Nagashunmugam T, Kang E, Friedman H M. The herpes simplex virus-1 glycoprotein E (gE) mediates IgG binding and cell-to-cell spread through distinct gE domains. Biochem Biophys Res Commun. 1997;235:31–35. doi: 10.1006/bbrc.1997.6720. [DOI] [PubMed] [Google Scholar]
- 35.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H-J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wisner T, Brunetti C, Dingwell K, Johnson D C. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J Virol. 2000;74:2278–2287. doi: 10.1128/jvi.74.5.2278-2287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zsak L, Zuckermann F, Sugg N, Ben-Porat T. Glycoprotein gI of pseudorabies virus promotes cell fusion and virus spread via direct cell-to-cell transmission. J Virol. 1992;66:2316–2325. doi: 10.1128/jvi.66.4.2316-2325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]