Abstract
Most simian immunodeficiency virus (SIV), human immunodeficiency virus type 2 (HIV-2), and HIV-1 infection of host peripheral blood mononuclear cells (PBMCs) is CD4 dependent. In some cases, X4 HIV-1 chemotaxis is CD4 independent, and cross-species transmission might be facilitated by CD4-independent entry, which has been demonstrated for some SIV strains in CD4− non-T cells. As expected for CCR5-dependent virus, SIV required CD4 on rhesus and pigtail macaque PBMCs for infection and chemotaxis. However, SIV induced the chemotaxis of human PBMCs in a CD4-independent manner. Furthermore, in contrast to the results of studies using transfected human cell lines, SIV did not require CD4 binding to productively infect primary human PBMCs. CD4-independent lymphocyte and macrophage infection may facilitate cross-species transmission, while reacquisition of CD4 dependence may confer a selective advantage for the virus within new host species.
Recent evidence suggests that human immunodeficiency virus type 1 (HIV-1) arose from simian immunodeficiency virus strain cpz (SIV cpz) by transmission from chimpanzees to humans after prior cross-species transmission from monkeys to chimpanzees (9, 11). In contrast, all available evidence indicates that HIV-2 made the transition from monkeys to humans directly, without a chimpanzee intermediary (1, 2, 8, 10). These cross-species jumps raise questions about ongoing transmissions and the adaptations required of the virus during transition from one host species to another.
Our previous studies have shown that HIV-1 infection and envelope-induced chemotaxis can be dissociated with respect to CD4 dependence. Specifically, we (12) and others (16) have found that CXCR4 binding (X4) gp120 can induce chemotaxis without binding to CD4 or even requiring the presence of CD4 on the cell surface whereas infection by X4 HIV requires cell surface CD4. In contrast, both chemotaxis and infection by CCR5 binding (R5) HIV-1 envelope and virus, respectively, are CD4 dependent (12, 18). Regardless of the cell type infected, SIV preferentially exploits CCR5 rather than CXCR4 as a CD4-dependent coreceptor (3, 13–15), and many strains of SIV can utilize CCR5 on non-T cells in a CD4-independent manner (4, 5).
Given these diverse results and the role of coreceptor usage in restricting cross-species host range tropism, we set out to systematically compare SIV coreceptor requirements for viral infection and virus-induced chemotaxis in monkey and human cells. We chose two strains of SIV known to use CCR5 for entry into macaque cells: SIVmac239 is considered a prototypic virus infecting T-cell lines (such as CEMx174, in which we passage the virus), while SIV 17E-Fr is a chimeric recombinant of SIVmac239 containing the envelope of neurotropic SIV-17E (capable of infecting astrocytes, glial cells, and brain endothelial cells [6]). SIV 17E-Fr is dually tropic, and when passaged in CEMx174 cells it maintains its tropism for primary macaque macrophages (6). Despite envelope differences and distinct patterns of tropism in purified cell types, both SIVmac239 and SIV 17E-Fr infect monkey peripheral blood mononuclear cells (PBMCs) in vitro with high efficiency. In the present study, infection and chemotaxis were assessed in macaque and human cells.
MATERIALS AND METHODS
Cells and reagents.
Human and macaque (rhesus and pigtail) PBMCs were obtained by Ficoll-Hypaque centrifugation or by Percoll gradient centrifugation (Sigma Chemical Co., St. Louis, Mo.). The PBMCs were stimulated for 3 days with 5 μg of phytohemagglutinin (PHA), (GIBCO BRL, Gaithersburg, Md.) per ml in RPMI 1640 supplemented with 10% fetal calf serum (FCS) and 2 U of interleukin-2 (IL-2) (Boehringer Mannheim, Indianapolis, Ind.) per ml. CD4+ or CD8+ cells were enriched by negative depletion using, respectively, anti-CD8 or anti-CD4 magnetic beads (Dynal, Lake Success, N.Y.) at saturating concentrations. The purity of T-cell subsets was determined for each subset by flow cytometric analysis of cells immunostained with 20 μg of anti-CD4 and anti-CD8 monoclonal antibodies (MAbs) (Coulter Immunology, Hialeah, Fla.) per ml detected with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG) (Jackson Immunoresearch Laboratories, Bar Harbor, Maine). Anti-CXCR4 (12G5), anti-CCR5 (2D7), and HIV-blocking (Leu3a, SIM4) and HIV-nonblocking (SIM7) anti-CD4 MAbs were obtained from the National Institutes of Health AIDS Reagent Program. All were used at 10 μg/ml. Beta chemokines (RANTES, MIP1-β, and MIP-1α) and alpha chemokines (SDF1-α and SDF1-β) were from R&D Systems, Minneapolis, Minn., and were used at 200 ng/ml. Soluble CD4-Ig (Genentech Inc., South San Francisco, Calif.) was used at 10 μg/ml.
Virus infection.
SIV17E-Fr and SIVmac239 were passaged in CEMx174 cells and purified from cell culture supernatants by sucrose density gradient centrifugation. PBMCs were stimulated with 5 μg of PHA per ml, aliquoted into tubes at 107 cells/ml of RPMI 1640–10% FCS–IL-2, and pretreated with antibodies to CD4, CXCR4, or CCR5 or with alpha or beta chemokines for 30 min on ice. The cells were then infected with 103 50% tissue culture infective doses of SIV 17E-Fr or SIVmac239 for 2 h with shaking at 37°C. In one group, the virus was pretreated with soluble CD4-Ig for 30 min on ice before being used to infect cells. After infection, the cells were washed free of virus and plated at 2 × 106 per well in triplicate wells of a 24-well culture plate.
A 1-ml volume of supernatant was collected from a total volume of 2 ml and replenished with an equal volume of fresh medium on days 0, 3, and 7 postinfection, and 200 μl of that 1-ml volume was used for the p27 assay. The remaining supernatant was frozen at −80°C freezer. The cell cultures were replenished with 1 ml of fresh medium on day 7 and again harvested on days 10 and 14. Supernatants collected on days 0, 3, and 7 were assayed for p27 by enzyme immunoassay (EIA) (Coulter Immunology). In all wells, day 0 (2 h after the final wash) and day 3 supernatant samples assayed in parallel with day 7 samples contained p27 levels below the positive cutoff. Day 0 concentrations were invariably <5 pg/ml (the limit of assay detection), while day 3 samples were generally higher but always <20 pg/ml, the calculated cutoff value for reliable positivity using the manufacturer's guidelines. Day 10 and 14 values appeared to follow the same trends among groups as day 7 values, but the absolute values were higher and frequently off-scale for linear quantitation without further dilution.
Chemotaxis assay.
CD4+ and CD8+ PBMCs obtained after PHA–IL-2 activation were stained with MAbs to CD4, CD8, CXCR4, and CCR5, and their expression levels were determined by fluorescence-activated cell sorter analysis. Cells (20,000/well) were labeled with 5 μM calcein (Molecular Probes) and placed above 5 μm-pore-size filters overlying medium alone, chemokines, or virus in serial dilutions (0.1 to 100 ng/ml) in 96-well microchemotaxis chambers (Neuroprobe, Gaithesburg, Md.). After a 2-h incubation at 37°C, labeled cells in the lower chamber were read in fluorimeter at excitation and emission wavelengths of 480 and 530 nm, respectively. Migration index (MI) is expressed as (experimental medium cell fluorescence/medium cell fluorescence). The results are expressed as mean ± standard deviation for triplicate experiments. Because absolute numbers of migrating cells can vary substantially by donor and between experimental runs, results are normalized against migrating cells in control medium (RPMI 1640 plus 0.5% FCS) and expressed as the ratio of absolute numbers of cells, or MI.
RESULTS AND DISCUSSION
CD4-dependent infection and chemotaxis of macaque PBMCs by SIV.
Macrophage-tropic SIV 17E-Fr, T-cell-tropic SIVmac239, and recombinant SIVmac239 envelope were analyzed for their ability to induce chemotaxis of PHA–IL-2-activated PBMCs from pigtail macaques in a 96-well microchemotaxis chamber assay. Purified recombinant MIP1-β was used at 10 ng/ml as a positive control. Both strains of virus and the recombinant envelope protein induced dose-dependent chemotactic activity (data not shown) with maximal chemotaxis at 10 ng of p27/ml (Fig. 1A).
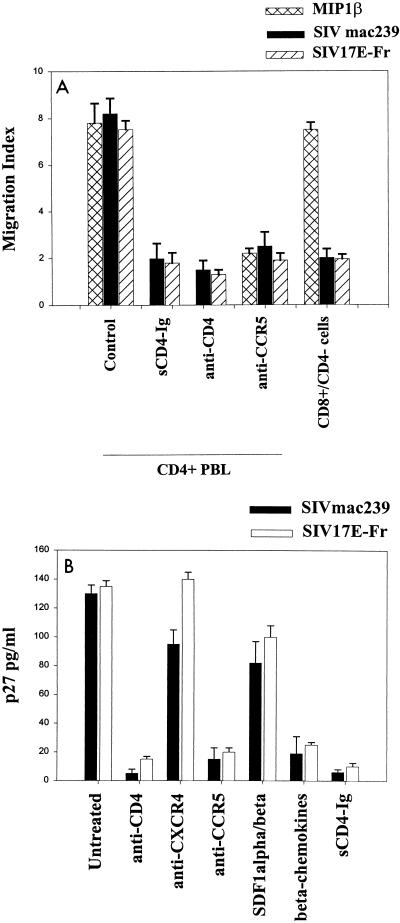
FIG. 1.
Exposure of PHA–IL-2-activated pigtail macaque PBMCs to SIVmac239 or SIV 17E-Fr causes chemotaxis (A) and infection (B). Chemotaxis to SIV or MIP1-β was inhibited by pretreatment of cells with 10 μg of anti-CD4 MAb (Leu3A) or anti-CCR5 MAb (2D7) per ml for 30 min on ice. Alternatively, purified virus was pretreated with sCD4-Ig (10 μg/ml) for 30 min on ice before use in the chemotaxis assay, as described in Materials and Methods. Macaque CD8+ cells isolated by negative depletion with anti-CD4 magnetic beads (>85% CD8+, <2% CD4+ cells) did not migrate in response to SIVmac239 or SIV 17E-Fr. CD4+ and CD8+ lymphocytes expressed comparable levels of CXCR4 and CCR5 and had similar migratory dose-response curves to SDF1-α and MIP1-β. Infection with 103 50% tissue culture infective doses of SIVmac239 or SIV 17E-Fr was inhibited by pretreatment of cells with 10 μg of MAbs to CD4 or CCR5 per ml or 200 ng of the beta chemokines (RANTES, MIP1-α and MIP1-β) per ml for 30 min on ice but not with anti-CXCR4 MAb (12G5) or 200 ng of the alpha chemokines (SDF1-α and SDF1-β) per ml. Treatment of purified virus with sCD4-Ig for 30 min on ice prior to infection of cells was also inhibitory. Supernatants were assayed for p27 on day 7. Results represent the mean ± standard error of the mean of three experiments. Similar trends were observed on days 10 and 14, although absolute values of p27 were higher.
Two approaches were used to determine the requirement for CD4 and CCR5 in this signaling process. In the first approach, various soluble reagents were added to cells and/or virus inocula at high concentrations to competitively inhibit envelope-receptor interactions. Chemotaxis assays were performed with activated PBMCs that were untreated (positive control), pretreated with anti-CD4, or pretreated with anti-CCR5 antibodies. CD4-specific (SIM4 or Leu3a) and CCR5-specific (2D7) antibodies that block virus binding and infection (blocking MAbs) had an inhibitory effect on the chemotactic property of SIV (Fig. 1A). In contrast, anti-CD4 (SIM7) and anti-CXCR4 (12G5) MAbs that do not interfere with virus binding or infection (nonblocking MAbs) did not inhibit the chemotactic response of PBMCs to SIV (data not shown). Consistent with an essential role for CD4 in SIV envelope-mediated signaling through CCR5 on pigtail macaque PBMCs, pretreatment of virus with 10 μg of soluble CD4-IgG fusion protein per ml was shown to have an inhibitory effect on SIV-induced chemotaxis (Fig. 1A).
The second approach to defining the role of CD4 in this combination of virus and macaque cells involved the removal of CD4+ cells from the PBMC and assessment of chemotaxis in the residual nonadherent population (85% CD8+, <2% CD4+). As seen from Fig. 1A, PBMCs depleted of CD4+ cells were unresponsive to both strains of SIV but migrated normally in response to MIP1-β. Recombinant SIVmac239 envelope paralleled SIV virions in inducing chemotaxis of pigtail macaque PBMCs in a CD4- and CCR5-dependent manner (data not shown).
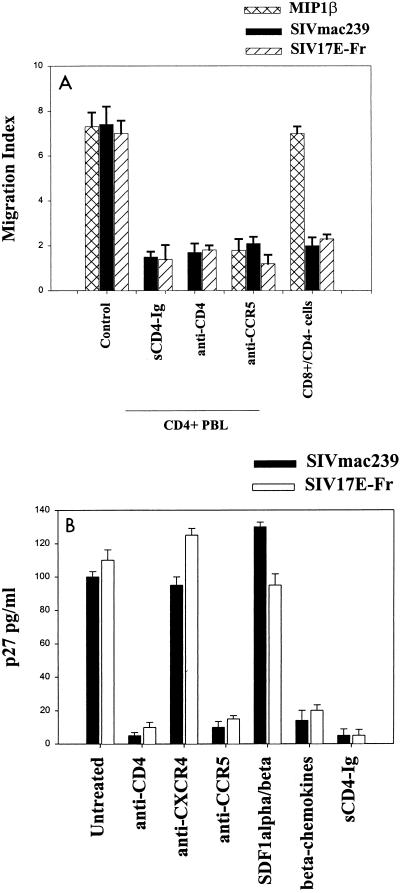
Similar requirements for CD4 and CCR5 were observed with experiments analyzing SIV infection of pigtail macaque PBMCs (Fig. 1B). In infection experiments, additional groups (cells pretreated with alpha and beta chemokines or CXCR4-specific MAb 12G5) were included to confirm expected coreceptor usage. Similar results were obtained when rhesus rather than pigtail macaque cells were used in the above chemotaxis and infection experiments (Fig. 2). Again, CD4-depleted rhesus macaque cells did not migrate in response to SIV virions (Fig. 2A) or envelope exposure (data not shown). Therefore, we found that SIVmac239 and SIV 17E-Fr attract and productively infect macaque PBMCs in a CD4-dependent, CCR5-dependent manner.
FIG. 2.
Exposure of PHA–IL-2-activated rhesus macaque PBMCs to SIVmac239 or SIV 17E-Fr causes chemotaxis (A) and infection (B). Reagents and assays of chemotaxis and infection were as described for Fig. 1.
CD4-independent infection and chemotaxis of human PBMCs by SIV.
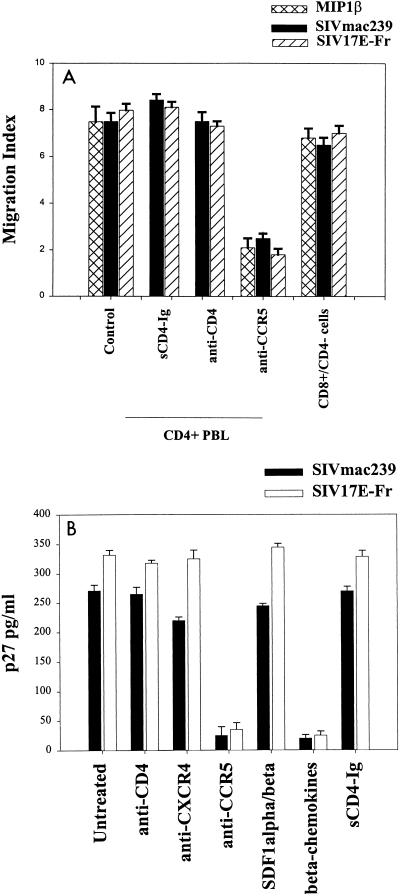
To determine if similar receptor requirements are retained by SIV in human cells, the chemotaxis and infection experiments described above were performed using PHA–IL-2-activated human PBMCs. Both SIVmac239 and SIV 17E-Fr induced dose-dependent chemotactic activity, illustrated at maximal chemotaxis with 10 ng of p27/ml in Fig. 3A. Neither SIV-induced chemotaxis (Fig. 3A) nor infection (Fig. 3B) of human PBMCs was inhibited by pretreatment of cells with any of the blocking or nonblocking anti-CD4 MAbs described above. However, the anti-CCR5 MAb was inhibitory for SIV-induced chemotaxis (Fig. 3A) and infection of human PBMCs (Fig. 3B). Additional groups that included cells pretreated with alpha or beta chemokines confirmed the requirement for CCR5 during SIV infection of human PBMCs (Fig. 3B).
FIG. 3.
Anti-CD4 MAb and sCD4-Ig do not inhibit chemotaxis (A) or infection (B) of activated human CD4+ PBMCs with SIVmac239 or SIV 17E-Fr, whereas anti-CCR5 MAb inhibits chemotaxis (A) and infection (B). Infection with both viruses was also inhibited by beta chemokines, but not by SDF1-α, SDF1-β, or anti-CXCR4 MAb. Reagents and assays were as described for Fig. 1. Human CD8+ cells isolated by negative depletion with anti-CD4 magnetic beads (>85% CD8+, <2% CD4+ cells) migrated in response to SIVmac239 or SIV 17E-Fr as well as to MIP1-β.
To further establish surface CD4 as nonessential on human cells for SIV envelope-mediated signaling through CCR5, human PBMCs were immunomagnetically depleted of CD4+ cells and the remaining cells (<2% CD4+, 85% CD8+) were used in chemotaxis assays with recombinant envelopes and purified virions. Flow cytometry revealed comparable expression by activated CD4+ and CD8+ cells of CXCR4 or CCR5 and similar dose-response curves of CD4+ and CD8+ cells migrating toward SDF1-α or MIP1-β (data not shown). The <2% CD4+ cells remaining in the CD4-depleted population cannot account for the observed high migration indices, since MIs of >6 represent migration of more than half the input cells (Fig. 3A). For both strains of SIV and recombinant SIVmac239 envelope, chemotaxis of CD8+ cells was comparable to that of whole PBMCs. Thus, CD4 was not necessary for the SIV strains to induce chemotaxis or productively infect human PBMCs (Fig. 3). This contrasts with the CD4 requirement for CCR5-mediated infection or chemotaxis of pigtail and rhesus macaque PBMCs (Fig. 1 and 2).
Our results have implications with respect to the evolution of HIV-2 and HIV-1 from SIV and SIVcpz, respectively. Specifically, CD4-independent infection of PBMCs might have facilitated initial cross-species transmission, while subsequent reacquisition of CD4 dependence for infection of lymphocytes may have ultimately conferred a selective entry or growth advantage. This would account for the observed CD4 dependence of SIV in macaque PBMCs and of HIV-2 in human PBMCs. Facilitation of cross-species transmission by CD4 independence could occur at two levels. First, it would obviate the need for virus to bind equally well to the new CD4 molecule of the new host, which might have important differences at the binding site. This would permit virus to enter CD4-positive cells without necessarily binding well to their surface CD4. Second, it raises the possibility of entry into CD4-negative cells as initial or temporary hosts for early replication.
Evidence to date from other laboratories indicates that, at a molecular level, SIV envelope binding to human CD4 domains occurs with good affinity (7, 17). Therefore, CD4 independence presumably would not be required for nonhuman-primate-to-human transmission of the SIV strains studied here. However, we do not know the binding capacity of the originally transmitted ancestral SIV for the originally infected human host CD4. If the affinity was low, CD4 independence may have been important. Alternatively, CD4-independent infection of CD4+ human PBMCs may simply be an epiphenomenon related to the ability of transmitted SIV to infect CD4− cells. This latter aspect may have been even more important in facilitating interspecies jumps and may be reflected in the relative ease with which many strains of SIV and HIV-2 can infect brain capillary endothelial cells and other CD4− cells (4).
At first blush, our results are not concordant with those of Edinger et al. (5), who found that rhesus macaque CCR5 was generally more supportive than human CCR5 of CD4-independent infection with various SIV strains, due to a single amino acid difference in the N terminus. In particular, these authors found that SIVmac239 envelope-bearing virus was highly dependent on the presence of CD4, irrespective of whether human or rhesus macaque CCR5 was coexpressed, while SIV 17E-Fr entered at a moderately reduced level in the absence of surface CD4, using rhesus CCR5 more effectively than it used human CCR5.
The explanation for these different results probably lies in the choice of target cells. We used primary, untransformed PBMCs from humans and macaques throughout, while Edinger et al. (5) used 293T cell lines transiently transfected with various highly expressing chemokine receptor plasmids. The entry requirements for tumor cell lines overexpressing transfected receptors may differ from those for viral entry into primary activated lymphocytes. In addition, the previous viral challenge used envelope chimeras from various SIV strains on a luciferase-expressing NL-LucR-E-backbone whereas ours used cultured infectious virions with p27 production as the infection readout.
Fomsgaard et al. (7) reported that several strains of SIV used CD4 binding regions from African green monkeys, pigtail macaques, or humans indiscriminately when transfected into macaque-derived transformed CMMT cells, in spite of predicted structural differences. Nevertheless, when untransformed PBMCs are used, interspecies differences in CD4 and CCR5 may be sufficient to account for our results. In addition to initial binding constraints, CCR5- and/or CD4-mediated signal transduction may play important roles in viral entry into primary (as opposed to immortalized) cells, in which case small interspecies differences in coreceptor sequences or downstream transduction proteins could also contribute to restricted tropism.
In any case, preliminary unpublished data suggest that CD4 independence is a transient phenomenon during long-term in vitro passage, suggesting a major selective advantage for targeting of CD4+ host cells, even in the absence of an immune response. We are currently examining the relative rates of infection in CD4+ and CD4− PBMCs during the earliest rounds in vitro infection to determine the degree of replicative advantage conferred upon SIV by integration into CD4+ lymphocytes and macrophages.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants R21-A144725 and RO1-AI31806 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Chen Z, Telfer P, Gettie A, Reed P, Zhang L, Ho D D, Marx P A. Genetic characterization of a new west African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Z, Luckay A, Sodora D L, Telfer P, Reed P, Gettie A, Kanu J M, Sadek R F, Yee J, Ho D D, Zhang L, Marx P A. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of SIV-infected sooty mangabeys. J Virol. 1997;71:3953–3960. doi: 10.1128/jvi.71.5.3953-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edinger A L, Mankowski J L, Doranz B J, Marguiles B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edinger A L, Blanpain C, Kunstman K J, Wolinsky S M, Parmentier M, Doms R W. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flaherty M T, Hauer D A, Mankowski J L, Zink M C, Clements J E. Molecular and biological characterization of a neurovirulent molecular clone of simian immunodeficiency virus. J Virol. 1997;71:5790–5798. doi: 10.1128/jvi.71.8.5790-5798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fomsgaard A, Johnson P R, Nielsen C, Novembre F J, Hansen J, Goldstein S, Hirsch V M. Receptor function of CD4 structures from African green monkey and pig-tail macaque for simian immunodeficiency virus, SIVsm, SIVagm, and human immunodeficiency virus type-1. Viral Immunol. 1995;8:121–133. doi: 10.1089/vim.1995.8.121. [DOI] [PubMed] [Google Scholar]
- 8.Gao F, Yue L, White A T, Pappas P G, Barchue J, Hanson A P, Greene B M, Sharp P M, Shaw G M, Hahn B H. Human infection by genetically diverse SIVsm-related HIV-2 in West Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 9.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peters M, Shaw G M, Sharp P M, Hahn B H. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch V M, Olmstead R A, Murphy-Corb M, Purcell R H, Johnson R R. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 11.Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- 12.Iyengar S, Schwartz D H, Hildreth J E K. T cell-tropic HIV gp120 mediates CD4 and CD8 cell chemotaxis through CXCR4 independent of CD4: implications for HIV pathogenesis. J Immunol. 1999;162:6263–6267. [PubMed] [Google Scholar]
- 13.Kirchhoff F, Pohlmann S, Hamacher M, Means R E, Kraus T, Uberla K, Marzio P D. Simian immunodeficiency virus variants with differential T-cell and macrophage tropism use CCR5 and an unidentified cofactor expressed in CEMX174 cells for efficient entry. J Virol. 1997;71:6509–6516. doi: 10.1128/jvi.71.9.6509-6516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcon L, Choe H, Martin K A, Farzan N, Ponath P D, Wu W, Newman N, Gerard C, Gerard N P, Sodroski J. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marx P A, Chen Z. The function of simian chemokine receptors in the replication of SIV. Semin Immunol. 1998;10:215–223. doi: 10.1006/smim.1998.0135. [DOI] [PubMed] [Google Scholar]
- 16.Misse D, Cerruti M, Noraz N, Jourdan P, Favero J, Devauchelle G, Yssel H, Taylor N, Veas F. A CD4-independent interaction of human immunodeficiency virus-1 gp120 with CXCR4 induces their cointernalization, cell signaling, and T-cell chemotaxis. Blood. 1999;93:2454–2462. [PubMed] [Google Scholar]
- 17.Sattentau Q U, Clapham P R, Weiss R A, Beverley P C, Montagneir L, Alhalabi M F, Gluckman J C, Klatzmann D. The human and simian immunodeficiency virus HIV-1, HIV-2, and SIV interact with similar epitopes on their cellular receptor, the CD4 molecule. AIDS. 1988;2:101–105. doi: 10.1097/00002030-198804000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor. Nature. 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]