Abstract
Background:
Maternal cigarette smoking during pregnancy (MSDP) is associated with numerous adverse health outcomes in infants and children with potential lifelong consequences. Negative effects of MSDP on placental DNA methylation (DNAm), placental structure, and function are well established.
Objective:
Our aim was to develop biomarkers of MSDP using DNAm measured in placentas (), collected as part of the Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function double-blind, placebo-controlled randomized clinical trial conducted between 2012 and 2016. We also aimed to develop a digital polymerase chain reaction (PCR) assay for the top ranking cytosine–guanine dinucleotide (CpG) so that large numbers of samples can be screened for exposure at low cost.
Methods:
We compared the ability of four machine learning methods [logistic least absolute shrinkage and selection operator (LASSO) regression, logistic elastic net regression, random forest, and gradient boosting machine] to classify MSDP based on placental DNAm signatures. We developed separate models using the complete EPIC array dataset and on the subset of probes also found on the 450K array so that models exist for both platforms. For comparison, we developed a model using CpGs previously associated with MSDP in placenta. For each final model, we used model coefficients and normalized beta values to calculate placental smoking index (PSI) scores for each sample. Final models were validated in two external datasets: the Extremely Low Gestational Age Newborn observational study, ; and the Rhode Island Children’s Health Study, .
Results:
Logistic LASSO regression demonstrated the highest performance in cross-validation testing with the lowest number of input CpGs. Accuracy was greatest in external datasets when using models developed for the same platform. PSI scores in smokers only () were moderately correlated with maternal plasma cotinine levels. One CpG (cg27402634), with the largest coefficient in two models, was measured accurately by digital PCR compared with measurement by EPIC array ().
Discussion:
To our knowledge, we have developed the first placental DNAm-based biomarkers of MSDP with broad utility to studies of prenatal disease origins. https://doi.org/10.1289/EHP13838
Introduction
Maternal cigarette smoking during pregnancy (MSDP) is associated with increased risk of low birth weight, prematurity, and perinatal mortality.1–4 MSDP is also associated with greater risk of cardiovascular, metabolic, respiratory, and neurocognitive health outcomes in childhood and into adulthood.5–10 Despite smoking cessation efforts, of female smokers will continue to smoke during pregnancy, resulting in of all infants born in the United States exposed to cigarette smoking in utero.11,12
Quantification of the level and duration of exposure to MSDP can improve our understanding of the underlying mechanisms for the direct consequences of MSDP on fetal and childhood health and can also improve our ability to model other exposure–disease relationships by more accurately adjusting for MSDP as a covariate. However, MSDP is typically assessed based on self-report, which is subject to underreporting due to social stigma12,13 and does not accurately capture information on the intensity or frequency of smoking. Cotinine, the primary metabolite of nicotine, is the current gold standard chemical biomarker of daily smoking,14,15 but single or cross-sectional cotinine measurements may be unavailable. With a shorter half-life (8.8 h) during pregnancy,16,17 and the fluctuation in daily smoking rates among pregnant women,18 cotinine is an unreliable measure of cumulative prenatal smoke exposure.
Alternatively, robust DNA methylation (DNAm) signatures of prenatal exposure to maternal smoking may serve as molecular biomarkers, and it has been proposed that these DNAm-biomarkers can be used as a proxy for exposure when modeling exposure–outcome relationships.19–21 Several blood-based DNAm-biomarkers of prenatal MSDP exposure have been developed using samples collected at birth, in childhood, adolescence, and even in adulthood.22–26
We hypothesized that placental DNAm could be used to develop an accurate and more quantitative biomarker of MSDP based on several characteristics. First, the placenta regulates transfer of material between the mother and fetus, and some xenobiotics and chemicals, including nicotine, readily cross the placental barrier.27 Therefore, the placenta may provide a cumulative molecular record of exposure to maternal smoking throughout gestation. Second, the placenta contains a higher frequency of partially methylated domains relative to blood,28 and it has been suggested that cytosine–guanine dinucleotides (CpGs) sites with intermediate levels of methylation can be measured with greater precision than at extreme values (i.e., near or fully unmethylated and near or fully methylated).29,30 Third, placental DNAm is dysregulated with in utero exposure to environmental pollutants,31–33 and some of the greatest effect sizes have been reported in association with MSDP.33–36
To test this hypothesis, we measured placental DNAm genome-wide using the Illumina MethylationEPIC array platform in a subset of placentas (, from 24 never-smokers and 72 self-reported smokers) collected at delivery from the Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function (VCSIP) (NCT01723696) cohort. We applied four machine learning methods [logistic least absolute shrinkage and selection operator (LASSO) regression, logistic elastic net regression, random forest, and gradient boosting machine] to identify DNAm signatures that predict maternal smoking during pregnancy. We developed three final models using probes from the EPIC array, the 450K array, and a previous meta-analysis of MSDP. We also examined the association between the resulting placental smoking indices and cotinine levels, measured in plasma of pregnant smokers during different windows of gestation. Using digital polymerase chain reaction (PCR), we developed an inexpensive, high-throughput targeted assay for screening of prenatal smoking exposure in placental DNA. Last, we examined the biological relevance of CpGs in the placental smoking index (PSI) to help understand the role of DNAm in the pathway between smoke exposure and later health outcomes.
Methods
Study Populations
VCSIP.
The randomized controlled trial (RCT) cohort VCSIP recruited women with singleton pregnancies ( y old; wk gestation) with a history of current cigarette smoking and documented refusal/inability to quit37–39 from three centers in the United States (Oregon, Washington, and Indiana) between 2012 and 2016. The women were randomized to receive vitamin C () vs. placebo after a successful run-in trial for medication compliance that required 75% adherence and return for follow-up within 7–21 d. A total of 252 pregnant women who were smokers were randomized and 243 infants were available for study at delivery. The RCT was approved by each site’s institutional review board and monitored by a National Institutes of Health–appointed Data Safety Monitoring Board. A group of 33 pregnant women who were never-smokers were enrolled toward the end of the RCT as a reference group. We obtained written informed consent from all participants prior to enrollment.37 The training dataset consisted of a subset of 96 participants from this RCT with placental epigenome-wide DNA methylation data (24 never-smokers and 72 self-reported smokers). We excluded placentas from participants with gestational hypertension, preeclampsia, or preterm delivery ( wk), as well as placentas sampled h after delivery owing to concern over sample deterioration. We then prioritized placentas with RNA-sequencing (RNA-seq) data available () and 16 additional samples for inclusion on EPIC arrays (Figure S1).
Extremely Low Gestational Age Newborn.
The Extremely Low Gestational Age Newborn (ELGAN) observational study was established to learn more about medical and developmental deficits common in babies born very premature.40 Women giving birth before 28 wk gestation between 2002 and 2004 at 1 of the 14 participating ELGAN sites (in North Carolina, Michigan, Illinois, Connecticut, and Massachusetts) were invited to enroll. All protocols were approved by the institutional review board at each of the 14 participating sites and all participants provided written informed consent to participate.40 Within this cohort, sufficient placental DNA was available for methylation analysis from 426 participants.41 The present analysis included 399 participants (43 exposed to MSDP; 356 not exposed) with placental DNAm EPIC data and information on smoking status available through the Environmental influences on Child Health Outcomes (ECHO) consortium.42 Datasets were excluded if they were not available in the ECHO data portal or if they were missing information on smoking status.
Rhode Island Children’s Health Study.
The Rhode Island Children’s Health Study (RICHS) is a birth cohort of mother–child dyads in the Rhode Island and Southeastern Massachusetts area designed to study the role of the placenta in the effects of environmental exposures on children’s health, as previously described.43,44 Women and infants were enrolled between 2010 and 2013 from nonpathological pregnancies at term ( wk gestation), and the cohort was oversampled for infants classified as large or small for gestational age. All protocols were approved by the institutional review boards at the Women and Infants Hospital and Dartmouth College, and all participants provided written informed consent.43 In the present study, we included 237 participants with 450K DNAm data available on Gene Expression Omnibus (GEO) (GSE75248) and metadata available for maternal smoking during pregnancy (35 exposed; 202 not exposed). Datasets were excluded if they were missing information on maternal smoking status.
DNA Methylation Profiling
VCSIP.
Placentas were collected and processed by trained research staff using standardized protocols, as described previously.45 We analyzed 96 placentas and prioritized 80 samples used previously in transcriptome-wide analysis, as well as 16 additional placentas, to balance samples from RCT groups by sex and gestational age at delivery.45 In brief, of placental DNA was bisulfite converted and applied to Illumina Infinium Methylation EPIC BeadChips following the Infinium HD Methylation 15019521v01 protocol. Data normalization and quality control assessment were performed using the Chip Analysis Methylation Pipeline (ChAMP) package: non-CpG probes, probes with a beadcount (beadC) in at least 5% of samples, probes annotated to single nucleotide polymorphisms (SNPs),46 probes with a detection -value (detP) in one or more samples, cross-hybridizing probes,47 and probes on X/Y chromosomes were removed, and the remaining probes () were normalized via functional normalization.48
ELGAN.
The ELGAN Illumina EPIC dataset was accessed from the ECHO Cohort high-performance cluster.42 DNAm data was analyzed for quality and processed by the ECHO Data Analysis Center using a standard pipeline. Samples were removed in cases of discrepant sex, low overall intensity, duplicates, bisulfite intensity , probes with detP , or probes with beadC . Probes were removed in cases where of samples had detP , 1% of samples with beadC , cross-reactive probes, or probes with a SNP at the CpG site. The Noob method of normalization was applied to correct for probe and dye bias within an array/sample.49
RICHS.
The RICHS Illumina 450K dataset was obtained from the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) submission GSE75248. Sample collection and processing of samples and DNAm data has been previously described.50,51 Poor quality probes (detP ), sex-chromosome probes, and SNP-associated loci were removed, and the data was adjusted for type 1 and type 2 probe variation using functional normalization.48 The normalized beta matrix and phenotype matrix were downloaded from the GEO on 14 November 2019.
Smoking Variables
VCSIP.
In this RCT cohort, smoking status was confirmed based on repeated questionnaires, measures of urine cotinine, hair nicotine, and plasma nicotine metabolites collected at randomization ( 18.7 wk), at mid-gestation ( 26 wk), and at late-gestation ( 31.7 wk). At the randomization visit for our study, only 1 of the 72 self-reported smoking participants with placental DNAm had urine cotinine levels , a common cutoff to define active smoking.52
ELGAN.
Prenatal tobacco smoke exposure was defined at the time of enrollment based on maternal self-report of active smoking during pregnancy.53 Women were interviewed in their native language using a structured data questionnaire that included the question: “Did you smoke cigarettes during this pregnancy (, )?”53
RICHS.
We downloaded the metadata table for GSE75248 from the GEO and used the variable “smoking_status” as truth in our assessment of model performance, coded as “Yes,” “No,” or “unknown.” Subjects with DNAm and “smoking status” listed as “unknown” were not included in the present study.
Machine Learning Methods
Each machine learning method was performed via the Tidymodels library (Kuhn and Wickham) in R (version 4.0.3; R Development Core Team). The four methods evaluated in the present study include logistic LASSO regression54 from the glmnet engine,55 logistic elastic net regression56 from the glmnet engine, random forest57 from the ranger engine58 with importance set to impurity, and gradient boosting machine59 from the xgboost engine60 (Figure 1). CpGs beta values were centered and scaled for all modeling methods. For each method, we defined two model datasets: a) the entire set of EPIC probes, and b) the probes that overlap between EPIC and 450K platforms. For the LASSO regression, we defined an additional dataset: c) probes previously reported to be associated with MSDP in a meta-analysis by the Pregnancy And Childhood Epigenetics (PACE) consortium.35
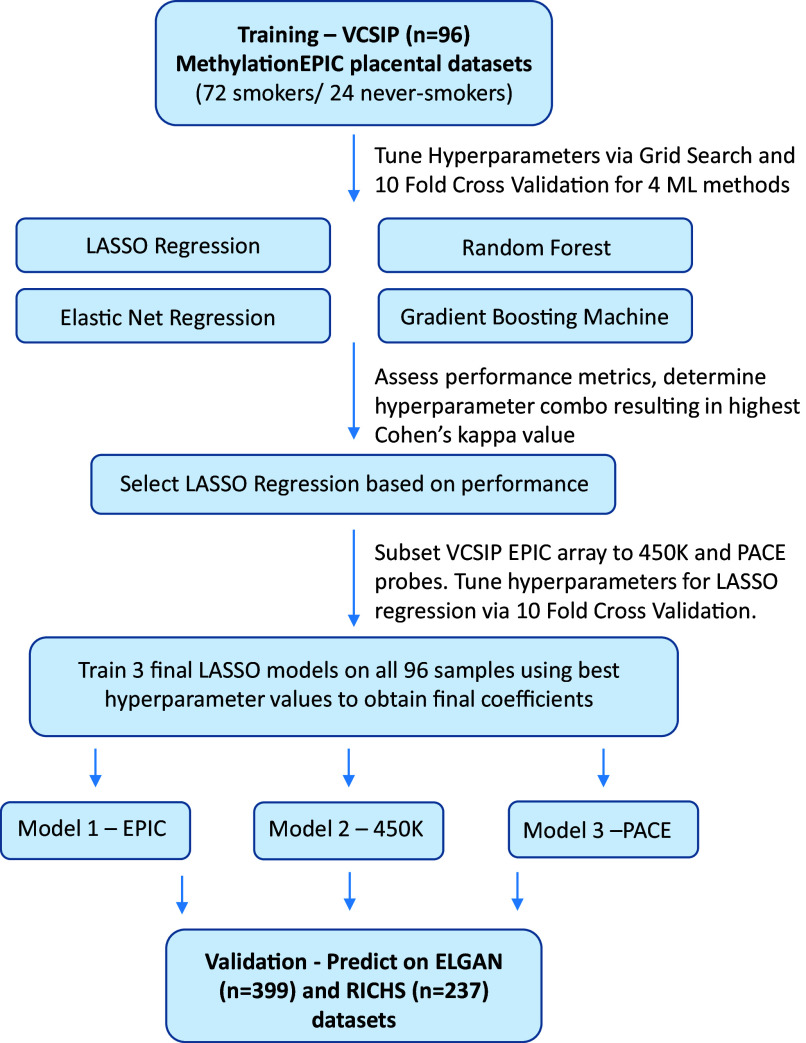
Figure 1.
Flowchart detailing the steps from training to final models. Initially, four machine learning methods for predicting smoking status in the VCSIP dataset underwent hyperparameter tuning via 10-fold cross-validation. Hyperparameters varied according to the machine learning method, but all methods included a hyperparameter “nCpG” that signifies the top CpGs ranked by -value following differential analysis with Limma. LASSO regression achieved among the highest kappa values while also providing a simpler interpretation than other methods. The VCSIP EPIC beta matrix was subset to probes overlapping the 450K array, as well as probes overlapping the PACE meta-analysis, and hyperparameter tuning was performed the same way on these datasets. Hyperparameter values resulting in the highest kappa value were used for a final round of training on all VCSIP samples for each dataset (EPIC, 450K overlaps, PACE overlaps) to obtain three final LASSO models. These final models were used to predict smoking status in two external placental DNA methylation datasets with known smoking exposure. Note: CpG, cytosine–guanine dinucleotide; ELGAN, Extremely Low Gestational Age Newborn; EPIC, Infinium MethylationEPIC array; LASSO, least absolute shrinkage and selection operator; ML, machine learning; PACE, Pregnancy and Childhood Epigenetics; RICHS, Rhode Island Children’s Health Study; VCSIP, Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function.
Variable Preselection
To subset the hundreds of thousands of CpGs for which we had methylation measurements prior to model training, we employed an unadjusted differentially methylated CpG analysis using the lmFit, contrasts.fit, and eBayes functions from the Limma library61 with the comparison set as “smoker vs. nonsmoker.” Following this statistical test, the top smoking associated CpGs with the lowest -values were selected, using “nCpGs” as a tunable hyperparameter (ranging from 10 to 1,000, in 10 intervals of 110). For LASSO and elastic net regression only, highly correlated top CpGs were removed according to a correlation cutoff of 0.75, and the remaining non-highly correlated top smoking-associated CpGs were used as features in the downstream model fitting process.
Hyperparameter Tuning
In machine learning, hyperparameters are user-defined values used to control the learning process and vary depending on the applied method. To find optimal hyperparameters values, we implemented a grid search along with 10-fold cross-validation stratified across smoking status for each unique hyperparameter value (or unique combination of values if there was more than one hyperparameter).
For logistic LASSO regression, the penalty parameter lambda was tuned via the grid_regular function from Tidymodels with a tune length set to 20 (tune length corresponds to the number of values to use). The nCpG parameter used 10 values evenly spaced between 10 and 1,000. For logistic elastic net regression, both the penalty parameter lambda and the mixture parameter alpha were tuned via the grid_regular function with tune length set to 10, and the nCpG parameter set to 5 values between 50 and 1,000. For random forest, two rounds of hyperparameter tuning were employed. First, the hyperparameters ntree and nCpG were tuned with ntree set to 10 values between 50 and 1,000, and nCpG set to 10 values between 10 and 1,000. Following this first round of tuning, the best performing values were selected for ntree and nCpG, and the mtry and min_n parameters were tuned. The mtry parameter was set to 20 values between 1 and 100, whereas the min_n parameter was set to 10 values between 1 and 15. The gradient boosting machine used three hyperparameters: a) nCpG, set to 5 values between 50 and 1,000; b) ntree, set to 20 values between 50 and 1,000; and c) depth, set to 10 values between 1 and 20.
Criteria for Model Selection
Following 10-fold cross-validation for each unique hyperparameter combination, we obtained performance metrics measuring model performance for each fold. These values were averaged across all 10 folds to obtain a mean performance metric for each hyperparameter combination (Excel Table S1). Cohen’s kappa is designed to better assess model performance on datasets with a class imbalance,62 such as in the VCSIP training dataset, which is majority smokers. Although Cohen’s kappa was the primary measure used to compare model performance, accuracy and area under the receiver operating characteristic (ROC) curve were also computed, and we prioritized models with a lower number of nCpGs (more parsimonious).
Final LASSO Models
Following hyperparameter tuning of LASSO models on the three datasets defined above, the hyperparameter values resulting in the highest kappa score were selected for the final models (Excel Table S1). These hyperparameter values were used for an additional round of training on the entire VCSIP dataset () to create three final LASSO models to be tested on external datasets as validation. Although the 450K dataset had two combinations resulting in the same highest kappa score, we selected the combination resulting in the fewest number of CpGs with nonzero coefficients.
Weighted Mean Calculation for Missing CpGs
When CpGs required by the model are missing from a dataset, users have the option to fill in missing beta values with the weighted mean of the training dataset (Excel Table S2). We employed a weighted approach owing to our training dataset being majority smokers, which may not be representative of or appropriate for all future datasets taken from the US population. The weighted mean was calculated by first determining the average beta value for smokers and nonsmokers separately. The average beta value for smokers was multiplied by 0.123 (based on previously reported prevalence of MSDP63), and the average in nonsmokers was multiplied by (1 – 0.123). Finally, these resulting numbers were summed per CpG to obtain the weighted mean beta value.
Final Model Exploration
Calculation of PSI score.
For each of the three final LASSO models depicted in Figure 1, a PSI score can be calculated for each sample using the model coefficients for the selected CpGs and beta values centered and scaled according to the VCSIP training data to produce predictions consistent with the trained model (https://github.com/ba-davis/PSI_final_models). We provide mean and standard deviation values for each CpG in each final model in the code repository and in Excel Table S2. The PSI score is equal to the normalized CpG beta value multiplied by the coefficient for the same CpG, and summed over all CpGs in the model (Excel Table S3).
Correlation of PSI with maternal plasma cotinine levels.
The smoking samples from the VCSIP dataset include measures of cotinine at three different time points: at randomization, at mid-gestation, and at late-gestation. We performed Pearson correlation between PSI score and cotinine concentration at each gestational window in the 72 smokers only (Excel Table S3). We did not include nonsmokers in our correlation analyses because the majority of our nonsmokers had cotinine levels at or below the limit of detection ().
Performance metrics.
ROC curves (Figure 2), area under the ROC curve (AUC), and partial AUC (pAUC) values were calculated from the predictions using the pROC library.64 AUC values were obtained from the roc function, whereas partial AUC values from the 90%–100% specificity interval were obtained from the auc function. Plots were generated with the plot.roc function. Percentage accuracy was calculated as the number correctly classified/total number of . Sensitivity was calculated as the number of predicted smokers/total number of . Specificity was calculated as the number of predicted nonsmokers/total number of .
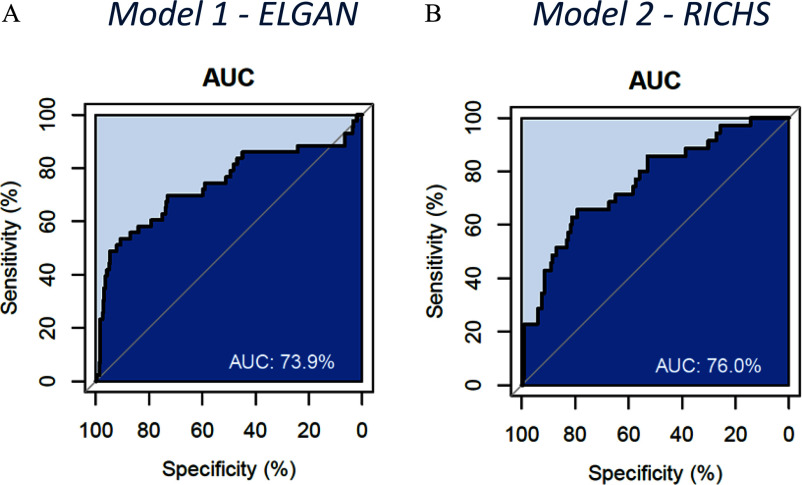
Figure 2.
ROC curves for (A) model 1, trained on EPIC probes using placental DNAm data from 72 smokers and 24 never-smokers in the VCSIP RCT and applied to the ELGAN dataset (), and (B) model 2, trained on 450K probes and applied to the RICHS dataset (). Note: AUC, area under the ROC curve; ELGAN, Extremely Low Gestational Age Newborn; EPIC, Infinium MethylationEPIC array; RCT, randomized controlled trial; RICHS, Rhode Island Children’s Health Study; ROC, receiver operating characteristic; VCSIP, Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function.
Biological importance/previous epigenome-wide association studies.
We used the online Epigenome-Wide Association Studies (EWAS) Open Platform (https://ngdc.cncb.ac.cn/ewas/) to examine the biological relevance and previous associations of probes selected in any one of our models.65 First, we input the list of probes selected in any of the three final LASSO models (Excel Table S2) into the EWAS Toolkit for enrichment of GO (gene ontology), KEGG (Kyoto Encyclopedia of Genes and Genomes) terms, and traits. Next, we looked up individual probes in the EWAS Atlas to compile a table of traits, studies, tissue sources, and publications previously associated with each probe.
Digital PCR
Primer design.
Digital PCR primers were designed using the DNA sequence 75 bp upstream and downstream from the target CpG (cg27402634) extracted from the University of Santa Cruz Genome Browser.66 Synthetic gblock DNA for the matching sequence was purchased from IGT and used for primer optimization. The DNA sequence was bisulfite converted in silico using MethPrimer followed by primer and probe design using Primer3Plus (Fprimer: AGTTTTTAGTAAACGTTTTTT; Rprimer: CTTCCCCTTTACCAATAA; Methylated probe: FAM- TGGATTATAGAcgTATTTTTGA; Nonmethylated probe: HEX- TGGATTATAGActTATTTTTGAT). Primers and probes were combined in a primer–probe mix as recommended [ forward primer; reverse primer; probe ( each if duplex)].
Assay validation.
Digital PCR was performed using standard reaction conditions recommended for the QIAcuity 8.5K 24-well plates and the QIAcuity 4X Probe PCR Master Mix (Qiagen): of Probe PCR Master Mix; of primer–probe mix; DNA; RNase-free water up to . Methylated and nonmethylated standards were used to generate a standard curve with samples at 20%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 80%, and 85% methylation, and run in duplicate on the QIAcuity Eight digital PCR instrument (Qiagen) with the following PCR conditions: 95° for 5 min, 40 cycles of (95° for 55 s, 52° for 40 s). Pearson correlation was used to compare concentration of standards to measured concentrations using digital PCR. Human placental DNA (; average of ) was bisulfite converted using the Methylamp DNA Modification Kit, as described in the manufacturer protocol (Epigentek). We performed digital PCR on the bisulfite product using the same reaction conditions as above, but in triplicate per sample. The coefficient of variation (CV) was calculated across replicates. Pearson correlation was used to compare mean results from digital PCR vs. EPIC array beta values for the same samples.
Results
Baseline Characteristics
Demographics of the participants included in the training and validation datasets are summarized in Table 1. Limited demographic data for the RICHS validation dataset participants were obtained from the metadata linked to the GEO submission (GSE75248). We did not perform statistical comparisons between datasets but, rather, describe the major known differences herein. First, given that the training dataset was an RCT cohort of pregnant smokers, the prevalence of maternal smoking based on self-report in participants with placental DNAm was 75%, whereas in the validation datasets the prevalence of smoking among participants with placental DNAm was 11% in the ELGAN and 15% in the RICHS cohorts. Second, given that the ELGAN cohort is a prospective cohort of extremely low gestational age newborns, the median gestational age of participants with DNAm was 25.68 wk compared with those in our training dataset, which excluded preterm deliveries from DNAm analysis [median wk]. The mean birthweight in ELGAN participants with DNAm data was compared with in the training dataset. Our training dataset also excluded participants with preeclampsia or gestational hypertension, whereas this was not an exclusion criterion for ELGAN. Several socioeconomic measures indicated lower socioeconomic status (e.g., fewer years of maternal education and lack of private health insurance) in the training data of majority smokers compared with ELGAN participants. The ELGAN dataset also included a higher proportion of participants that self-identified as Black or other race, as well as a higher proportion of self-described Hispanic or Latino ethnicity, relative to the training dataset, which predominantly included White/Caucasian participants.
Table 1.
Participant characteristics for the VCSIP study training and testing dataset and the ELGAN and RICHS validation datasets.
| Characteristic | VCSIP (training and testing) | ELGAN (EPIC validation) | RICHS (450K validation) |
|---|---|---|---|
| Maternal smoking—self-report (%) [ (%)] | |||
| Not exposed | 24 (25) | 356 (89) | 202 (85) |
| Exposed | 72 (75) | 43 (11) | 35 (15) |
| Maternal plasma cotinine (ng/mL)—smokers only () | |||
| Randomization | NA | NA | |
| Mid-gestation | NA | NA | |
| Late-gestation | NA | NA | |
| Maternal race [ (%)] | |||
| American Indian or Alaska Native | 1 (1) | NA | |
| Asian | 0 (0) | NA | |
| Black | 5 (5) | 111 (28) | NA |
| Multiple race | 7 (7) | 14 (4) | NA |
| Other race | 1 (1) | 19 (5) | NA |
| White | 82 (85) | 245 (61) | NA |
| Missing | 0 | NA | |
| Maternal ethnicity [ (%)] | |||
| Not Hispanic or Latino | 91 (96) | 364 (91) | NA |
| Hispanic or Latino | 4 (4) | 34 (9) | NA |
| Missing | 1 | 1 | NA |
| Maternal education [ (%)] | |||
| Master’s degree (MA, MS) and above (PhD, MD) | 8 (8) | 63 (16) | NA |
| Bachelor’s degree (BA, BS) | 14 (15) | 81 (21) | NA |
| Some college, no degree; associate’s degree (AA, AS); trade school | 16 (17) | 92 (24) | NA |
| High school degree, GED or equivalent | 21 (22) | 102 (26) | NA |
| Less than high school | 37 (39) | 51 (13) | NA |
| Missing | 0 | 10 | NA |
| Marital status [ (%)] | |||
| Married or living with a partner | 25 (26) | 310 (78) | NA |
| Single, never married; partnered (boyfriend or girlfriend), not living together | 29 (30) | 74 (19) | NA |
| Widowed; separated; divorced | 42 (44) | 15 (4) | NA |
| Private insurance [ (%)] | |||
| No | 62 (65) | 137 (34) | NA |
| Yes | 34 (35) | 262 (66) | NA |
| Gestational diabetes [ (%)] | |||
| No | 84 (88) | 362 (93) | NA |
| Yes | 12 (13) | 28 (7) | NA |
| Missing | 0 | 9 | NA |
| Gestational hypertension [ (%)]a | |||
| No | 96 (100) | 370 (93) | NA |
| Yes | 0 (0) | 29 (7) | NA |
| Missing | 0 | 0 | NA |
| Preeclampsia [ (%)]a | |||
| No | 96 (100) | 326 (82) | NA |
| Yes | 0 (0) | 73 (18) | NA |
| Missing | 0 | 0 | NA |
| Child sex [ (%)] | |||
| Female | 51 (53) | 189 (47) | NA |
| Male | 45 (47) | 209 (53) | NA |
| Unknown | 0 | 1 | NA |
| Gestational age [ (%)] | |||
| Mean | 39.21 | 25.68 | 39.36 |
| Median (min, max) | 39 (37, 42) | 26 (23, 27) | 39 (39, 40) |
| Preterm [ (%)]a | |||
| Preterm ( or CBI indicates preterm at birth) | 0 (0) | 399 (100) | 0 (0) |
| Term ( or CBI indicates not preterm at birth) | 96 (100) | 0 (0) | 237 (100) |
| Birth weight (g) [ (%)] | |||
| Mean | 3,328 | 832.61 | NA |
| Median (min, max) | 3,359 (2,376, 4,241) | 830 (420, 1,418) | NA |
| Missing | 1 | 0 | NA |
Note: For categorical variables we present the total per cohort above the columns and the and percentage for each category. When data is missing, we do not include the missing observations in calculation of percentages. For the ELGAN dataset (), we were restricted in reporting summary statistics and counts . For the RICHS dataset (), we were limited to metadata available in the GEO repository. AA, associate of arts; AS, associate of science; BA, bachelor of arts; BS, bachelor of science; CBI, child birth information; DNAm, DNA methylation; ELGAN, Extremely Low Gestational Age Newborn; EPIC, Infinium MethylationEPIC array; GA, gestational age; GED, General Educational Development; GEO, Gene Expression Omnibus; IQR, interquartile range; MA, master of arts; max, maximum; MD, doctor of medicine; min, minimum; MS, master of science; NA, not available; PhD, doctor of philosophy; RICHS, Rhode Island Children’s Health Study; VCSIP, Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function.
In VCSIP DNAm analysis (), we excluded placentas from patients with preeclampsia, preterm delivery, gestational hypertension, or collection h after delivery owing to concern of sample degradation.
Summary of Cross-Validation Metrics
EPIC beta matrix.
The LASSO models exhibited high performance with mean kappa scores of . The mean performance scores for elastic net regression were similar to those of LASSO regression, but the top performing nCpG values were larger overall than those of the LASSO regression. The random forest and gradient boosting machine returned lower mean kappa scores, with top performing values of and 0.85, respectively (Excel Table S1).
450K beta matrix.
The same machine learning methods and hyperparameter tuning scheme were performed on the 450K beta matrix. Once again, the LASSO regression and the elastic net regression performed well with top mean kappa scores of , higher than the top mean kappa values from the hyperparameter tuning performed on the EPIC array beta matrix. Random forest resulted in slightly worse mean kappa scores than from the EPIC array beta matrix cross-validation, although the second round of tuning increased the top mean kappa score dramatically from 0.74 to 0.82. Gradient boosting machine reached a top mean kappa score of 0.84 (Excel Table S1).
PACE beta matrix.
Logistic LASSO regression hyperparameter tuning was performed on the sustained smoking CpGs obtained from the PACE meta-analysis,35 which overlapped the EPIC array beta matrix. The mean kappa scores were high following cross-validation, with a top mean kappa score of 0.95 (Excel Table S1).
Final Models
We selected the logistic LASSO regression for final model building owing to the high-performance metrics and the simplicity of model interpretation. Three final models were developed using the hyperparameter values resulting in the highest mean kappa score for each probe matrix (EPIC, 450K, PACE) using all 96 VCSIP samples to obtain model coefficients. This resulted in selection of 18 CpGs with nonzero coefficients for model 1 (EPIC), 21 CpGs for model 2 (450K), and 18 for model 3 (PACE–sustained). A total of 5 CpGs were shared for all three models and 4 CpGs in two models (Figure S2; Table 2). PSI scores had a similar overall distribution across datasets, with the exception of one extreme outlier in the ELGAN dataset with dramatically lower PSI in each of the three final LASSO models (Figure S3; Excel Table S3).
Table 2.
Five CpGs were selected in all three final LASSO models (EPIC, 450K, and PACE) to predict maternal smoking trained on placental DNAm data from 72 smokers and 24 never-smokers in the VCSIP randomized controlled trial.
| IlmnID | CHR | MAPINFO | Nearest gene | Feature—CpG island | Gene description |
|---|---|---|---|---|---|
| cg04233054 | 1 | 23112654 | EPHB2 | Body-shore | Ephrin type-B receptor 2 |
| cg27402634 | 3 | 156536860 | LEKR1 | IGR-shore | Leucine, glutamate, and lysine rich 1 |
| cg08103568 | 4 | 100737138 | DAPP1 | TSS1500-opensea | Dual adapter of phosphotyrosine and 3-phosphoinositides 1 |
| cg08621277 | 11 | 68271518 | SAPS3 | 5′UTR-opensea | Protein phosphatase 6 regulatory subunit 3 |
| cg07168214 | 17 | 7380112 | ZBTB4 | 5′UTR-shelf | Zinc finger and BTB domain containing 4 |
Note: The full list of selected CpGs is in Table S2. 450K, Illumina Methylation450K array; BTB, Broad-complex, Tramtrack, and Bric à brack; CHR, chromosome; CpG, cytosine–guanine dinucleotide; DNAm, DNA methylation; EPIC, Infinium MethylationEPIC array; IlmnID, unique identifier from the Illumina CG database; LASSO, least absolute shrinkage and selection operator; MAPINFO, Chromosomal coordinates of the CpG; PACE, Pregnancy And Childhood Epigenetics; VCSIP, Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function.
Correlation of PSI with maternal plasma cotinine levels at early-, mid-, and late-gestation.
Because the CpGs selected to calculate the PSI are implicated in biological processes affected by smoking, we hypothesized that the PSI scores would be correlated with smoking level even though the models were trained on binary exposure. The PSI scores for the VCSIP smoker samples () correlated with maternal cotinine levels, with lower PSI scores associated with lower cotinine levels. We did not include nonsmokers in our correlation analyses because the majority of our nonsmokers had cotinine levels at or below the limit of detection (LOD). The lowest correlation between each PSI score (one from each final LASSO model) and maternal cotinine was at mid-gestation (model–EPIC , model 2–450K , and model 3–PACE ), and the highest correlation for each PSI was at late-gestation (model 1 ; model 2 ; and model 3 ; Table 3; Excel Table S3). Cotinine correlation coefficients at randomization fell between those for mid- and late-gestation. If we replaced values below the LOD with 0.195, the correlation coefficients between PSI and cotinine increased to between .
Table 3.
Correlation of placental smoking index (PSI) with maternal plasma cotinine levels in VCSIP training data from smokers only ().
| Model | -Value | |
|---|---|---|
| Model 1–EPIC | ||
| Randomization | 0.241 | |
| Mid-gestation | 0.221 | |
| Late-gestation | 0.340 | |
| Model 2–450K | ||
| Randomization | 0.304 | |
| Mid-gestation | 0.234 | |
| Late-gestation | 0.397 | |
| Model 3–PACE | ||
| Randomization | 0.276 | |
| Mid-gestation | 0.231 | |
| Late-gestation | 0.432 | |
Note: PSI scores were calculated for each sample using the sum of model coefficients multiplied by normalized beta values. Maternal plasma cotinine was measured at randomization ( 18.7 wk), at mid-gestation ( 26 wk), and at late-gestation ( 31.7 wk) in the VCSIP training dataset for smokers. Maternal plasma cotinine level was either not measured or was below the limit of detection in nonsmokers and was therefore not included in the correlation analysis. -Value from Pearson correlation. 450K, Illumina Methylation450K array; EPIC, Infinium MethylationEPIC array; PACE, Pregnancy And Childhood Epigenetics; VCSIP, Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function.
External validation.
The three models were used to predict smoking status on two external datasets from ELGAN and RICHS. The performance metrics from these predictions are shown in Table 4. Model 1 (trained on all EPIC CpGs) achieved the highest accuracy and kappa values when predicting on the ELGAN dataset (Figure 2; Table 4), although the kappa values dropped significantly for all three models on the new datasets compared with the training dataset. Model 2 (trained on CpGs filtered for 450K probes) had the highest accuracy and kappa value for the RICHS dataset (Figure 2; Table 4), but also the lowest sensitivity with 12 of the 35 smokers being classified as nonsmokers. Model 3 (trained on CpGs previously associated with sustained smoking) ranked among the highest sensitivity and lowest specificity for both datasets, resulting in the greatest number of true positives and the also the most false positives. We additionally performed a sensitivity analysis of model performance in the ELGAN dataset after removing 76 patients with either preeclampsia or hypertension, to be more comparable to our original training dataset. The accuracy and specificity improved for all models, and the sensitivity was also improved in model 1, which is trained on data from the same EPIC platform (Table 4).
Table 4.
Model performance metrics (sensitivity, specificity, Cohen’s kappa, accuracy, AUC curve) for three final LASSO models (EPIC–18 CpGs, 450K–21 CpGs, and PACE–18 CpGs) to predict maternal smoking trained on placental DNAm data from 72 smokers and 24 never-smokers in the VCSIP randomized controlled trial based on application in the ELGAN EPIC dataset () and in the RICHS 450K dataset ().
| Dataset | CpGs selected () | CpGs available () | Accuracy | Sensitivity | Specificity | Kap | AUC | p_AUC |
|---|---|---|---|---|---|---|---|---|
| ELGAN validation dataset (; EPIC) | ||||||||
| Model 1–EPICa | 18 | 18 | 0.6015 | 0.7442 | 0.5843 | 0.1368 | 0.7390 | 0.6651 |
| Model 2–450K | 21 | 21 | 0.4987 | 0.7442 | 0.4691 | 0.0756 | 0.6661 | 0.6486 |
| Model 3–PACE | 18 | 18 | 0.3734 | 0.8605 | 0.3146 | 0.051 | 0.724 | 0.6239 |
| ELGAN validation dataset (; EPIC)b | ||||||||
| Model 1–EPICa | 18 | 18 | 0.6749 | 0.7429 | 0.6667 | 0.1958 | 0.7664 | 0.7477 |
| Model 2–450K | 21 | 21 | 0.5263 | 0.6857 | 0.5069 | 0.0729 | 0.6502 | 0.669 |
| Model 3–PACE | 18 | 18 | 0.4149 | 0.8571 | 0.3611 | 0.0672 | 0.7656 | 0.7113 |
| RICHS validation dataset (; 450K) | ||||||||
| Model 1–EPIC | 18 | 7 | 0.4093 | 0.9143 | 0.3217 | 0.0914 | 0.7683 | 0.6002 |
| Model 2–450Ka | 21 | 20 | 0.7173 | 0.6571 | 0.7277 | 0.2552 | 0.7598 | 0.6099 |
| Model 3–PACE | 18 | 16 | 0.3797 | 0.9143 | 0.2871 | 0.0756 | 0.6897 | 0.5855 |
| RICHS validation dataset (; 450K)—fill missing | ||||||||
| Model 1–EPIC | 18 | 18 | 0.8565 | 0.1714 | 0.9752 | 0.2047 | 0.7683 | 0.6002 |
| Model 2–450K | 21 | 21 | 0.7468 | 0.6571 | 0.7624 | 0.2944 | 0.7598 | 0.6099 |
| Model 3–PACE | 18 | 18 | 0.3966 | 0.8857 | 0.3119 | 0.0762 | 0.6897 | 0.5855 |
Note: When validation datasets were missing CpGs in a given model, we filled in missing beta values with the weighted mean of the VCSIP training dataset to avoid adding values of zero for missing CpGs. The weighted mean was calculated by first determining the average beta value for smokers and nonsmokers separately. The average beta value for smokers was multiplied by 0.123 (based on previously reported national prevalence of maternal smoking in pregnancy), and the average in nonsmokers was multiplied by (1 – 0.123). 450K, Illumina Methylation450K array; AUC, area under the ROC curve; CpG, cytosine–guanine dinucleotide; DNAm, DNA methylation; ELGAN, Extremely Low Gestational Age Newborn; EPIC, Infinium MethylationEPIC array; Kap, Cohen’s kappa; LASSO, least absolute shrinkage and selection operator; PACE, Pregnancy And Childhood Epigenetics; p_AUC, partial area under the ROC curve; RICHS, Rhode Island Children’s Health Study; VCSIP, Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function.
Results from application of platform-specific models to the same test platform.
Sensitivity analysis removing 76 patients with either preeclampsia or hypertension.
The RICHS 450K dataset was missing CpGs required for each of the three models. As a strategy to fill in the missing beta values for the required CpGs, we supplied the weighted mean beta value from the training data (Excel Table S2), with beta values from smokers and nonsmokers weighted according to the national average of women smokers of 12.3% (see the “Methods” section). Model 1 was built on EPIC data and therefore had the most missing CpGs in the RICHS 450K dataset (11 CpGs), and it also showed the largest improvement after filling in missing CpGs. The accuracy, kappa, and specificity increased, whereas the sensitivity greatly decreased due to the majority of the smokers being incorrectly classified as nonsmokers. Models 2 and 3 had only 1 or 2 CpGs missing, and each showed modest changes in performance metrics after filling in missing CpGs (Table 4).
Biological relevance of selected CpGs.
All CpGs selected in our three models were examined using the EWAS Toolkit for previous associations with biological traits, GO terms, and KEGG pathways. Of the five CpGs selected in all three models (Table 2), cg27402634 and cg08103568 were previously associated with personal smoking in blood from adults (Excel Table S4),67,68 cg07168214 has been associated with preeclampsia and preterm birth,69–71 cg04233054 with preterm birth and ancestry72,73 in placental DNA specifically, and cg08621277 with asthma status in airway epithelial cell DNA.74 A total of seven CpGs in any model have been previously associated with personal smoking, smoking cessation, or electronic cigarette use (Table S4). Hypergeometric testing identified enrichment of several traits with relevant biology among model CpGs, including preeclampsia, aging, preterm birth, asthma, maternal lead exposure, smoking cessation, and Down syndrome. The missMethyl package identified greatest enrichment in the GO term “negative regulation of bone development” and in the KEGG pathway “VEGF signaling pathway” (Figure S4; Excel Table S5).
Digital PCR
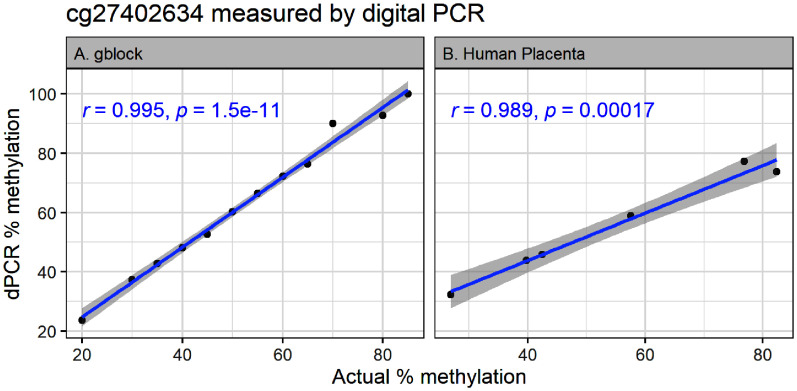
We developed a digital PCR assay for the top ranked CpG (cg27402634) associated with MSDP in both our EPIC and 450K models using the QIAcuity platform and fluorescently labeled probes designed to separately measure methylated and nonmethylated copies of DNA. In standard curve analysis of gblock DNA ranging from 20% to 85% methylation, the CV between replicates was between 1% and 16%, with higher CVs at the top of the standard curve. The correlation between actual and measured concentrations was 0.995 (Figure 3A; Excel Table S6). We next measured percent methylation in a subset of our human placental DNA samples by digital PCR and compared that with beta values measured by EPIC array. Again, the CV between replicates was low (between 4% and 10%) and the measured concentrations were highly correlated with EPIC measurements in the same samples (; Figure 3B; Excel Table S7).
Figure 3.
The top CpG ranked by absolute value of coefficient for all placental smoking index models was cg27402634. Measurement of cg27402634 methylation by digital PCR shows linear fit over (A) a wide range of synthetic DNA standards () and (B) high correlation with measurements in obtained from EPIC arrays () for the same samples from VCSIP. -Values are from Pearson correlation. Numeric data can be found in Excel Tables S6 and S7. Note: CpG, cytosine–guanine dinucleotide; dPCR, digital polymerase chain reaction; EPIC, Infinium MethylationEPIC array; VCSIP, Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function.
Discussion
In the present study, we developed three sets of placental DNAm-biomarkers for exposure to MSDP. We tested our models in two external datasets: a) a preterm cohort (i.e., ELGAN) with a median age of delivery of 26 wk, and b) a birth cohort (i.e., RICHS) in the Rhode Island and Southeastern Massachusetts area composed predominantly of nonsmokers based on self-report. Our model trained on all EPIC probes had 60% accuracy, 74% sensitivity, and 58% specificity when applied to the ELGAN EPIC dataset, and our model trained on 450K probes had 71% accuracy, 66% sensitivity, and 72% specificity when applied to the RICHS 450K dataset.
Although our models were trained on smoking status for classification of exposure, correlation of our DNAm-based PSI with maternal cotinine levels measured at different windows of gestation suggests that these placental DNAm-biomarkers may be useful as a continuous variable to account for smoke exposure, as previously suggested.21 Of note, when we explored the CpGs included in our models for biological relevance using the EWAS Atlas,65 we observed enrichment for CpGs previously associated with maternal lead exposure, which is a known chemical of concern in cigarette smoke, given that lead concentrations in maternal plasma are negatively correlated with infant birth weight, length, and head circumference.75 We also demonstrated enrichment of CpGs associated with preeclampsia, preterm birth, asthma, and Down syndrome, suggesting that our PSI may not only be a biomarker of exposure but also a predictor of adverse outcomes later in childhood because MSDP increases risk for these outcomes. Interestingly, the median PSI scores were higher in the placentas from preterm ELGAN nonsmokers than for the VCSIP and RICHS nonsmokers that were delivered at term, and therefore CpGs in the PSI may also be associated with preterm delivery or intrauterine growth restriction.
Previous research in the field of DNAm-biomarkers of prenatal smoke exposure have been primarily developed and tested using DNAm measured in blood specimens and a priori selection of CpGs identified in previous studies.22–24,76 In 2016, Ladd-Acosta et al. developed a maternal smoking classification model22 using 26 CpG loci previously associated with prenatal smoking in infant cord blood,77 and this model was able to classify prenatal smoke exposure in childhood blood with 81% accuracy within the same training dataset.22 That model included several covariates, such as maternal age, ancestry, maternal education, and cell type proportions, which may improve accuracy but decrease generalizability to external datasets missing this information.
In 2017, Reese et al. developed a blood-based DNAm score24 for sustained prenatal smoke exposure trained on a total of 1,057 cord blood datasets from the Norwegian Mother and Child Cohort Study (MoBA). The Reese model used raw beta values without covariates to increase reproducibility and generalizability, and the final model consisted of 28 CpGs with 91% accuracy, 58% sensitivity, and 97% specificity in a smaller batch of samples from the same cohort (). Because these performance metrics are from application to a subpopulation of the same larger cohort, they are likely to be higher than in an external dataset.24 To test this hypothesis, we applied the Reese cord blood model to 96 cord blood datasets from our training cohort (i.e., VCSIP), and calculated performance metrics of 69% accuracy, 58% sensitivity, and 100% specificity.
The PACE consortium performed a meta-analysis for the association between maternal smoking in pregnancy and newborn blood DNA methylation using Illumina 450K BeadChip data from 13 cohorts () and identified 568 Bonferroni-significant CpGs. The PACE consortium additionally examined 5 cohorts of older children, and identified 19 CpGs associated with prenatal smoke exposure at Bonferroni significance.78 Richmond et al. later tested whether a DNA methylation score derived by combining methylation values at these 568 or 19 CpGs, separately, could predict prenatal smoke exposure in samples collected from adults in the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort ().76 Prediction performance was assessed by AUC and resulted in values of 0.69 for the 568 CpG score and 0.72 for the 19 CpGs score.76 Of note, both newborn and child samples from the ALSPAC cohort were included in the original PACE meta-analyses and therefore may overlap with adult participants used for testing. In comparison, the AUC values calculated in the present study for prenatal smoke exposure models trained on placental DNAm from the VCSIP cohort () and validated in two independent datasets (ELGAN, ; RICHS, ) ranged from 0.66 to 0.77, and model performance was greatest when validation data included the same platform/probe selection as the training method (model 1, EPIC AUC in ; model 2, 450K AUC in ).
Most recently, Rauschert et al. tested several machine learning approaches to develop a DNAm score for prenatal smoke exposure trained on blood datasets collected from adolescents and adults.23 The best performing Rauschert model consisted of 204 CpGs, developed with elastic net regression, and exhibited 73%–83% accuracy when applied to external datasets.23 In the 96 cord blood datasets available from the VCSIP cohort, the prenatal smoke score generated using the Rauschert model is highly correlated with scores from the Reese model, with comparable performance (64% accuracy, 53% sensitivity, and 96% specificity). In the present study, we used a similar analysis approach as Rauschert et al. to identify the best performing machine learning method for this dataset. Therefore, our model accuracies in external datasets of the same platform (model 1, EPIC accuracy in ; model 2, 450K accuracy in ) are comparable or higher than previous models when applied to external data. When we used our own cohort data to generate ROC curves for each model, the internal accuracy for classification was 100%. If we trained our models on the probes identified by PACE meta-analysis in placenta, they then had much lower performance.
Previous blood-based scores consist of a minimum of 19 CpGs and up to 568 CpGs, which could be cost prohibitive for measuring by targeted analysis in large studies. Given the large effect sizes of MSDP on placental DNAm, we postulated that a placental-based DNAm biomarker may contain fewer CpGs that could be measured inexpensively using digital PCR. As proof of principle, a digital PCR assay using just the single top ranked CpG associated with MSDP in all three of our models showed accuracy and linearity over a wide range of methylation levels and agreed with measurements performed on the Illumina EPIC array. Digital PCR assays have similarly been developed to detect, for example, current smoking and alcohol use.79,80 and have broad utility in clinical, forensic, and research applications.81 The present study provides evidence that digital PCR assays also have the potential to predict exposure to maternal smoking in utero in additional placental biospecimens in the absence of funds for genome-wide measurements. Further work is needed to determine whether this single-CpG assay can accurately predict exposure alone, similar to an assay available for screening in blood.80
Strengths and Limitations
To the best of our knowledge, the present study is the first known attempt to take advantage of substantial and robust placental DNA methylation signatures in response to cigarette smoke exposure to develop a novel objective DNAm biomarker, with great utility in identifying exposure-related health outcomes. Reliance on self-report of smoking often leads to information bias and pregnant persons are more likely to underreport their smoking. The gold standard nicotine metabolite, cotinine, is often not measured in large cohort studies, and reflects only very recent exposure.15 In contrast, a methylation biomarker in the placenta may reflect the cumulative exposure to that tissue and the fetus during pregnancy and allow for more reliable classification of smoking exposure when assessing impacts of this exposure on health outcomes. In addition, methylation biomarkers can be used to adjust for confounding introduced by prenatal smoke exposure when studying other exposure–outcome relationships.82 Therefore, given the decreasing cost of array- and sequencing-based technologies to measure epigenome-wide DNA methylation, our placental DNAm-based biomarkers can be used in existing placental DNAm datasets as either a proxy for exposure or as a covariate to adjust for the effects of MSDP in the absence of reliable exposure data. Our study also demonstrates that it is possible to measure methylation levels at biomarker CpGs using digital PCR at low cost. Because the effect size of prenatal smoke exposure on placental DNAm are substantial in comparison with effect sizes in blood and because of the unique methylation landscape of placental DNAm, a placental biomarker may have more quantitative precision than blood-based biomarkers.83
Additional strengths of our study include the extensive smoking history available for the VCSIP cohort (i.e., self-report, cigarettes per day, cotinine, hair nicotine) across the entire gestation period, as well as testing of our models in two cohorts with very different patient demographics from our training cohort. Our model performance in these diverse populations was comparable to previous models developed in blood, which supports the potential generalizability of our placental smoking indices to diverse populations. In addition, we developed separate models trained on EPIC and 450K CpGs so that models exist for both platforms. A third model we developed that was restricted to CpGs previously associated with maternal smoking in a placental meta-analysis of 450K data35 had lower accuracy in both of our test datasets but had higher sensitivity to predict true exposed samples as exposed.
Limitations of our study include a relatively small sample size compared with previous studies used to develop prenatal smoke biomarkers in blood. However, having a larger percentage of smokers in our cohort compared with previous studies allowed for correlation of our PSI with maternal cotinine levels over a broad range of exposure levels at three periods during gestation. We acknowledge that our correlation coefficients were relatively low (between 0.22 and 0.43) because we did not have cotinine measurements available for most of the nonsmokers in our study and those we had were below the LOD. Therefore, these correlations are conservative given that they include the narrow range of PSI scores and cotinine values only for active smokers. If we impute missing cotinine levels for nonsmokers with the LOD, these correlations increase to between 0.5 and 0.6. In addition, cotinine levels only reflect recent smoking, which is expected to fluctuate over the course of pregnancy. We also cannot separate cotinine resulting from cigarette smoking from other nicotine delivery products, such as electronic cigarettes.
In addition, the specificity of our models when applied to datasets other than the dataset used for training were low, suggesting that our models are best applied to datasets on the same platform and with limited missing CpGs. Ideally, we would advise using platform-specific models to limit the number of missing CpGs, given that imputing the weighted mean for missing CpGs may perform poorly if the proportion of missingness is high or if the external dataset is not representative of the general population (i.e., smoking prevalence is very high or low). Although this imputation approach may not be optimal, the alternate approach of deleting missing CpGs would be more biased given that it is implicitly imputing missing beta values as 0. An additional caveat in our validation datasets is that we relied on self-report of any smoking in pregnancy as our “truth” in assessing model performance. However, we know that some pregnant smokers will successfully quit during pregnancy or reduce to low use and that some pregnant women will not report smoking on medical records. Last, we acknowledge that the participant characteristics in the validation cohorts available for this study are distinct from our population of mostly active smokers and that predictions for new participants may be influenced by this dissimilarity. We would expect prediction performance in datasets with different baseline characteristics from the training set to be an underestimate of true model performance. We were especially impressed by the performance of our EPIC PSI model in the ELGAN validation dataset, which consisted of participants who delivered extremely preterm, whereas our training data were all from term placentas. Performance metrics improved with exclusion of ELGAN participants with either preeclampsia or gestational hypertension. Therefore, future studies to refine a placental DNAm biomarker should include a larger, more general population that includes a wider range of smoking behaviors, pregnancy complications, and sociodemographic characteristics.
Conclusions
Results from the present study have broad utility to studies of prenatal disease origins. First, a placental DNAm smoking index could be used as a proxy for in utero smoke exposure to assess its association with health outcomes or to adjust for MSDP when modeling other exposure–disease relationships. Second, we demonstrate that targeted digital PCR assays (based on the PSI loci) may be used if placenta was collected, but funds are not available for epigenome-wide DNAm analysis. Finally, by examining the loci that comprise the PSI, we may improve our mechanistic understanding of the effects of maternal smoking during pregnancy on later health outcomes.
Supplementary Material
Acknowledgments
We thank the families who participated in the Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function (VCSIP), Extremely Low Gestational Age Newborn (ELGAN), and Rhode Island Children’s Health Study (RICHS) studies, along with the research and clinical staff for cohort coordination, sample, and data collection. We also thank our Environmental influences on Child Health Outcomes (ECHO) Colleagues; the medical, nursing, and program staff; and the children and families participating in the ECHO cohorts. We also acknowledge the contribution of the following ECHO Program collaborators: ECHO Components—Coordinating Center: Duke Clinical Research Institute, Durham, North Carolina: Smith PB, Newby LK; Data Analysis Center: Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland: Jacobson LP; Research Triangle Institute, Durham, North Carolina: Catellier DJ; Person-Reported Outcomes Core: Northwestern University, Evanston, Illinois: Gershon R, Cella D. ECHO Awardees and Cohorts—Baystate Children’s Hospital, Springfield, MA: Vaidya R; Beaumont Children’s Hospital, Royal Oak, MI: Obeid R; Boston Children’s Hospital, Boston, MA: Rollins C; East Carolina University, Brody School of Medicine, Greenville, NC: Bear K; Corewell Health, Helen DeVos Children’s Hospital, Grand Rapids, MI: Pastyrnak S; Michigan State University College of Human Medicine, East Lansing, MI: Lenski M; Tufts University School of Medicine, Boston, MA: Singh R; University of Chicago Comer Medicine, Chicago, IL: Msall M; University of Massachusetts Chan Medical School, Worcester, MA: Frazier J; Atrium Health Wake Forest Baptist, Winston Salem, NC: Gogcu S; Yale School of Medicine, New Haven, CT: Montgomery A; Boston Medical Center, Boston, MA: Kuban K, Douglass L, Jara H; Boston University, Boston, MA: Joseph R.
This work was supported by the National Institutes of Health (NIH) through an Opportunities and Infrastructure Fund (OIF) subaward from the ECHO Program (U2COD023375 to Shorey).
Ethics approval and consent to participate: Study protocols were approved by the individual institutional review boards at each site (detailed in the “Methods” section). Written informed consent or parent’s/guardian’s permission was obtained along with child assent as appropriate, for ECHO-wide Cohort Data Collection Protocol participation and for participation in specific cohorts.
Funding acknowledgments and disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Research reported in this publication was supported by the ECHO Program, Office of the Director, NIH, under awards U2COD023375 (Coordinating Center/Shorey-Kendrick’s Opportunities and Infrastructure Fund), U24OD023382 (Data Analysis Center), U24OD023319 with co-funding from the Office of Behavioral and Social Science Research (Patient Reported Outcomes Core), UH3OD023287 (Breton), UH3OD023288 (McEvoy), UH3OD023342 (Lyall), UH3OD023348 (O’Shea), the National Heart, Lung, and Blood Institute (NHLBI; R01 HL105447 and R01 HL 105460) with co-funding from the Office of Dietary Supplements and by P51 OD011092565. Additional support from the Oregon Clinical Translational Research Institute funded by the National Center for Advancing Translational Sciences (UL1TR000128).
Authors’ contributions: conceptualization—L.E.S. and E.R.S.; data acquisition—L.E.S., C.V.B., E.G., M.O., E.R.S., R.J.S., R.S.T., and C.T.M.; data interpretation: L.E.S.K., B.D., L.G., C.V.B., R.F., E.G., R.J.S., M.O., and E.R.S.; formal analysis—L.E.S., B.D., and L.G.; methodology—L.E.S., B.D., and L.G.; statistical oversight—L.G. and B.P.; data coordination—A.V. and C.D.M.; supervision—B.P., C.T.M., R.S.T., and E.R.S.; and writing—L.E.S. wrote all drafts of the manuscript and made revisions based on feedback from all coauthors and the ECHO Publication Committee. All authors have critically reviewed and revised the manuscript, agreed to the published version of the manuscript, and agree to be accountable for all aspects of the work.
Clinical trial registration: ClinicalTrials.gov, NCT01723696. Registered 6 November 2012. https://clinicaltrials.gov/ct2/show/record/NCT01723696.
Data sharing: The raw and processed DNAm data used for model development are publicly accessible through the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) via accession series GSE169598. The source functions and reference data needed to apply these models to external datasets are available at https://github.com/ba-davis/PSI_final_models.
Conclusions and opinions are those of the individual authors and do not necessarily reflect the policies or views of EHP Publishing or the National Institute of Environmental Health Sciences.
References
- 1.Anderson TM, Lavista Ferres JM, Ren SY, Moon RY, Goldstein RD, Ramirez JM, et al. . 2019. Maternal smoking before and during pregnancy and the risk of sudden unexpected infant death. Pediatrics 143(4):e20183325, PMID: 30858347, 10.1542/peds.2018-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.del Rocio Berlanga M, Salazar G, Garcia C, Hernandez J. 2002. Maternal smoking effects on infant growth. Food Nutr Bull 23(Suppl 3):142–145, PMID: 12362783, 10.1177/15648265020233S128. [DOI] [PubMed] [Google Scholar]
- 3.Kalinka J, Hanke W, Szymczak W. 1996. Risk factors of intrauterine growth retardation: a study of an urban population in Poland. Cent Eur J Public Health 4(3):192–196, PMID: 8884056. [PubMed] [Google Scholar]
- 4.Horta BL, Victora CG, Menezes AM, Halpern R, Barros FC. 1997. Low birthweight, preterm births and intrauterine growth retardation in relation to maternal smoking. Paediatr Perinat Epidemiol 11(2):140–151, PMID: 9131707, 10.1046/j.1365-3016.1997.d01-17.x. [DOI] [PubMed] [Google Scholar]
- 5.Albuquerque CA, Smith KR, Johnson C, Chao R, Harding R. 2004. Influence of maternal tobacco smoking during pregnancy on uterine, umbilical and fetal cerebral artery blood flows. Early Hum Dev 80(1):31–42, PMID: 15363837, 10.1016/j.earlhumdev.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Gishti O, Jaddoe VWV, Felix JF, Reiss I, Steegers E, Hofman A, et al. . 2015. Impact of maternal smoking during pregnancy on microvasculature in childhood. The Generation R Study. Early Hum Dev 91(10):607–611, PMID: 26298032, 10.1016/j.earlhumdev.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Slotkin TA, Seidler FJ, Spindel ER. 2011. Prenatal nicotine exposure in rhesus monkeys compromises development of brainstem and cardiac monoamine pathways involved in perinatal adaptation and sudden infant death syndrome: amelioration by vitamin C. Neurotoxicol Teratol 33(3):431–434, PMID: 21320590, 10.1016/j.ntt.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki K, Nakai K, Hosokawa T, Oka T, Okamura K, Sakai T, et al. . 2006. Association of maternal smoking during pregnancy and infant neurobehavioral status. Psychol Rep 99(1):97–106, PMID: 17037455, 10.2466/pr0.99.1.97-106. [DOI] [PubMed] [Google Scholar]
- 9.Rizzo G, Capponi A, Pietrolucci ME, Arduini D. 2009. Effects of maternal cigarette smoking on placental volume and vascularization measured by 3-dimensional power Doppler ultrasonography at 11+0 to 13+6 weeks of gestation. Am J Obstet Gynecol 200(4):415.e1–e5, PMID: 19070830, 10.1016/j.ajog.2008.10.041. [DOI] [PubMed] [Google Scholar]
- 10.McEvoy CT, Spindel ER. 2017. Pulmonary effects of maternal smoking on the fetus and child: effects on lung development, respiratory morbidities, and life long lung health. Paediatr Respir Rev 21:27–33, PMID: 27639458, 10.1016/j.prrv.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin JA, Osterman MJK, Driscoll AK. 2023. Declines in cigarette smoking during pregnancy in the United States, 2016–2021. NCHS Data Brief 458:1–8, PMID: 36723453. [PubMed] [Google Scholar]
- 12.Tong VT, Dietz PM, Farr SL, D’Angelo DV, England LJ. 2013. Estimates of smoking before and during pregnancy, and smoking cessation during pregnancy: comparing two population-based data sources. Public Health Rep 128(3):179–188, PMID: 23633733, 10.1177/003335491312800308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howland RE, Mulready-Ward C, Madsen AM, Sackoff J, Nyland-Funke M, Bombard JM, et al. . 2015. Reliability of reported maternal smoking: comparing the birth certificate to maternal worksheets and prenatal and hospital medical records, New York City and Vermont, 2009. Matern Child Health J 19(9):1916–1924, PMID: 25676044, 10.1007/s10995-015-1722-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benowitz NL, Dains KM, Dempsey D, Herrera B, Yu L, Jacob P III.. 2009. Urine nicotine metabolite concentrations in relation to plasma cotinine during low-level nicotine exposure. Nicotine Tob Res 11(8):954–960, PMID: 19525206, 10.1093/ntr/ntp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benowitz NL, St Helen G, Nardone N, Cox LS, Jacob P III.. 2020. Urine metabolites for estimating daily intake of nicotine from cigarette smoking. Nicotine Tob Res 22(2):288–292, PMID: 30852610, 10.1093/ntr/ntz034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dempsey D, Jacob P III, Benowitz NL. 2002. Accelerated metabolism of nicotine and cotinine in pregnant smokers. J Pharmacol Exp Ther 301(2):594–598, PMID: 11961061, 10.1124/jpet.301.2.594. [DOI] [PubMed] [Google Scholar]
- 17.Taghavi T, Arger CA, Heil SH, Higgins ST, Tyndale RF. 2018. Cigarette consumption and biomarkers of nicotine exposure during pregnancy and postpartum. Addiction 113(11):2087–2096, PMID: 29920836, 10.1111/add.14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondracki AJ. 2019. Prevalence and patterns of cigarette smoking before and during early and late pregnancy according to maternal characteristics: the first national data based on the 2003 birth certificate revision, United States, 2016. Reprod Health 16(1):142, PMID: 31519184, 10.1186/s12978-019-0807-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajaprakash M, Dean LT, Palmore M, Johnson SB, Kaufman J, Fallin DM, et al. . 2023. DNA methylation signatures as biomarkers of socioeconomic position. Environ Epigenet 9(1):dvac027, PMID: 36694711, 10.1093/eep/dvac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladd-Acosta C, Fallin MD. 2019. DNA methylation signatures as biomarkers of prior environmental exposures. Curr Epidemiol Rep 6(1):1–13, PMID: 31032172, 10.1007/s40471-019-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladd-Acosta C. 2015. Epigenetic signatures as biomarkers of exposure. Curr Environ Health Rep 2(2):117–125, PMID: 26231361, 10.1007/s40572-015-0051-2. [DOI] [PubMed] [Google Scholar]
- 22.Ladd-Acosta C, Shu C, Lee BK, Gidaya N, Singer A, Schieve LA, et al. . 2016. Presence of an epigenetic signature of prenatal cigarette smoke exposure in childhood. Environ Res 144(pt A):139–148, PMID: 26610292, 10.1016/j.envres.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rauschert S, Melton PE, Heiskala A, Karhunen V, Burdge G, Craig JM, et al. . 2020. Machine learning-based DNA methylation score for fetal exposure to maternal smoking: development and validation in samples collected from adolescents and adults. Environ Health Perspect 128(9):097003, PMID: 32930613, 10.1289/EHP6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reese SE, Zhao S, Wu MC, Joubert BR, Parr CL, Håberg SE, et al. . 2017. DNA methylation score as a biomarker in newborns for sustained maternal smoking during pregnancy. Environ Health Perspect 125(4):760–766, PMID: 27323799, 10.1289/EHP333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, et al. . 2015. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Hum Mol Genet 24(8):2201–2217, PMID: 25552657, 10.1093/hmg/ddu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shenker NS, Ueland PM, Polidoro S, van Veldhoven K, Ricceri F, Brown R, et al. . 2013. DNA methylation as a long-term biomarker of exposure to tobacco smoke. Epidemiology 24(5):712–716, PMID: 23867811, 10.1097/EDE.0b013e31829d5cb3. [DOI] [PubMed] [Google Scholar]
- 27.Wickström R. 2007. Effects of nicotine during pregnancy: human and experimental evidence. Curr Neuropharmacol 5(3):213–222, PMID: 19305804, 10.2174/157015907781695955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder DI, Schmidt RJ, Crary-Dooley FK, Walker CK, Ozonoff S, Tancredi DJ, et al. . 2016. Placental methylome analysis from a prospective autism study. Mol Autism 7:51, PMID: 28018572, 10.1186/s13229-016-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez-Jimenez N, Allard C, Bouchard L, Perron P, Bustamante M, Bilbao JR, et al. . 2019. Comparison of Illumina 450K and EPIC arrays in placental DNA methylation. Epigenetics 14(12):1177–1182, PMID: 31250700, 10.1080/15592294.2019.1634975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansell G, Gorrie-Stone TJ, Bao Y, Kumari M, Schalkwyk LS, Mill J, et al. . 2019. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genomics 20(1):366, PMID: 31088362, 10.1186/s12864-019-5761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loke YJ, Muggli E, Nguyen L, Ryan J, Saffery R, Elliott EJ, et al. . 2018. Time- and sex-dependent associations between prenatal alcohol exposure and placental global DNA methylation. Epigenomics 10(7):981–991, PMID: 29956547, 10.2217/epi-2017-0147. [DOI] [PubMed] [Google Scholar]
- 32.Cai J, Zhao Y, Liu P, Xia B, Zhu Q, Wang X, et al. . 2017. Exposure to particulate air pollution during early pregnancy is associated with placental DNA methylation. Sci Total Environ 607–608:1103–1108, PMID: 28724248, 10.1016/j.scitotenv.2017.07.029. [DOI] [PubMed] [Google Scholar]
- 33.Rousseaux S, Seyve E, Chuffart F, Bourova-Flin E, Benmerad M, Charles MA, et al. . 2020. Immediate and durable effects of maternal tobacco consumption alter placental DNA methylation in enhancer and imprinted gene-containing regions. BMC Med 18(1):306, PMID: 33023569, 10.1186/s12916-020-01736-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortillo M, Marsit CJ. 2023. Select early-life environmental exposures and DNA methylation in the placenta. Curr Environ Health Rep 10(1):22–34, PMID: 36469294, 10.1007/s40572-022-00385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Everson TM, Vives-Usano M, Seyve E, Cardenas A, Lacasaña M, Craig JM, et al. . 2021. Placental DNA methylation signatures of maternal smoking during pregnancy and potential impacts on fetal growth. Nat Commun 12(1):5095, PMID: 34429407, 10.1038/s41467-021-24558-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardenas A, Lutz SM, Everson TM, Perron P, Bouchard L, Hivert MF. 2019. Mediation by placental DNA methylation of the association of prenatal maternal smoking and birth weight. Am J Epidemiol 188(11):1878–1886, PMID: 31497855, 10.1093/aje/kwz184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McEvoy CT, Milner KF, Scherman AJ, Schilling DG, Tiller CJ, Vuylsteke B, et al. . 2017. Vitamin C to decrease the effects of smoking in pregnancy on infant lung function (VCSIP): rationale, design, and methods of a randomized, controlled trial of vitamin C supplementation in pregnancy for the primary prevention of effects of in utero tobacco smoke exposure on infant lung function and respiratory health. Contemp Clin Trials 58:66–77, PMID: 28495620, 10.1016/j.cct.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McEvoy CT, Shorey-Kendrick LE, Milner K, Schilling D, Tiller C, Vuylsteke B, et al. . 2020. Vitamin C to pregnant smokers persistently improves infant airway function to 12 months of age: a randomised trial. Eur Respir J 56:1902208, PMID: 32616589, 10.1183/13993003.02208-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McEvoy CT, Shorey-Kendrick LE, Milner K, Schilling D, Tiller C, Vuylsteke B, et al. . 2019. Oral vitamin C (500 mg/d) to pregnant smokers improves infant airway function at 3 months (VCSIP). a randomized trial. Am J Respir Crit Care Med 199(9):1139–1147, PMID: 30522343, 10.1164/rccm.201805-1011OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Shea TM, Allred EN, Dammann O, Hirtz D, Kuban KCK, Paneth N, et al. . 2009. The ELGAN study of the brain and related disorders in extremely low gestational age newborns. Early Hum Dev 85(11):719–725, PMID: 19765918, 10.1016/j.earlhumdev.2009.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark J, Martin E, Bulka CM, Smeester L, Santos HP, O’Shea TM, et al. . 2019. Associations between placental CpG methylation of metastable epialleles and childhood body mass index across ages one, two and ten in the Extremely Low Gestational Age Newborns (ELGAN) cohort. Epigenetics 14(11):1102–1111, PMID: 31216936, 10.1080/15592294.2019.1633865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knapp EA, Kress AM, Parker CB, Page GP, McArthur K, Gachigi KK, et al. . 2023. The Environmental Influences on Child Health Outcomes (ECHO)-wide cohort. Am J Epidemiol 192(8):1249–1263, PMID: 36963379, 10.1093/aje/kwad071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marsit CJ, Maccani MA, Padbury JF, Lester BM. 2012. Placental 11-beta hydroxysteroid dehydrogenase methylation is associated with newborn growth and a measure of neurobehavioral outcome. PLoS One 7(3):e33794, PMID: 22432047, 10.1371/journal.pone.0033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Everson TM, Armstrong DA, Jackson BP, Green BB, Karagas MR, Marsit CJ. 2016. Maternal cadmium, placental PCDHAC1, and fetal development. Reprod Toxicol 65:263–271, PMID: 27544570, 10.1016/j.reprotox.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shorey-Kendrick LE, McEvoy CT, O’Sullivan SM, Milner K, Vuylsteke B, Tepper RS, et al. . 2021. Impact of vitamin C supplementation on placental DNA methylation changes related to maternal smoking: association with gene expression and respiratory outcomes. Clin Epigenetics 13(1):177, PMID: 34538263, 10.1186/s13148-021-01161-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou W, Laird PW, Shen H. 2017. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res 45(4):e22, PMID: 27924034, 10.1093/nar/gkw967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nordlund J, Båcklin CL, Wahlberg P, Busche S, Berglund EC, Eloranta ML, et al. . 2013. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol 14(9):r105, PMID: 24063430, 10.1186/gb-2013-14-9-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, et al. . 2014. Functional normalization of 450K methylation array data improves replication in large cancer studies. Genome Biol 15(12):503, PMID: 25599564, 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Triche TJ Jr, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. 2013. Low-level processing of Illumina Infinium DNA methylation BeadArrays. Nucleic Acids Res 41(7):e90, PMID: 23476028, 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maccani JZJ, Koestler DC, Lester B, Houseman EA, Armstrong DA, Kelsey KT, et al. . 2015. Placental DNA methylation related to both infant toenail mercury and adverse neurobehavioral outcomes. Environ Health Perspect 123(7):723–729, PMID: 25748564, 10.1289/ehp.1408561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paquette AG, Houseman EA, Green BB, Lesseur C, Armstrong DA, Lester B, et al. . 2016. Regions of variable DNA methylation in human placenta associated with newborn neurobehavior. Epigenetics 11(8):603–613, PMID: 27366929, 10.1080/15592294.2016.1195534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silva AI, Camelo A, Madureira J, Reis AT, Machado AP, Teixeira JP, et al. . 2022. Urinary cotinine assessment of maternal smoking and environmental tobacco smoke exposure status and its associations with perinatal outcomes: a cross-sectional birth study. Environ Res 203:111827, PMID: 34363802, 10.1016/j.envres.2021.111827. [DOI] [PubMed] [Google Scholar]
- 53.Venkatesh KK, Leviton A, Fichorova RN, Joseph RM, Douglass LM, Frazier JA, et al. . 2021. Prenatal tobacco smoke exposure and neurological impairment at 10 years of age among children born extremely preterm: a prospective cohort. BJOG 128(10):1586–1597, PMID: 33682301, 10.1111/1471-0528.16690. [DOI] [PubMed] [Google Scholar]
- 54.Tibshirani R. 1996. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B Methodol 58(1):267–288, 10.1111/j.2517-6161.1996.tb02080.x. [DOI] [Google Scholar]
- 55.Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33(1):1–22, PMID: 20808728, 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou H, Hastie T. 2005. Regularization and variable selection via the elastic net. J R Stat Soc Ser B Methodol 67(2):301–320, 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
- 57.Breiman L. 2001. Random forests. Mach Learn 45:5–32, 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 58.Wright MN, Ziegler A. 2017. ranger: a fast implementation of random forests for high dimensional data in C++ and R. J Stat Softw 77(1):1–17, 10.18637/jss.v077.i01. [DOI] [Google Scholar]
- 59.Friedman JH. 2001. Greedy function approximation: a gradient boosting machine. Ann Stat 29(5):1189–1232, 10.1214/aos/1013203451. [DOI] [Google Scholar]
- 60.Chen T, Guestrin C. 2016. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Krishnapuram B, ed. 13–17 August 2016. New York, NY: Association for Computing Machinery, 785–794. 10.1145/2939672.2939785. [DOI] [Google Scholar]
- 61.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. . 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47, PMID: 25605792, 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cohen J. 1960. A coefficient of agreement for nominal scales. Educ Psychol Meas 20(1):37–46, 10.1177/001316446002000104. [DOI] [Google Scholar]
- 63.Tong VT, Jones JR, Dietz PM, D’Angelo D, Bombard JM, Centers for Disease Control and Prevention. 2009. Trends in smoking before, during, and after pregnancy—Pregnancy Risk Assessment Monitoring System (PRAMS), United States, 31 sites, 2000–2005. MMWR Surveill Summ 58(4):1–29, PMID: 19478726. [PubMed] [Google Scholar]
- 64.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. . 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77, PMID: 21414208, 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li M, Zou D, Li Z, Gao R, Sang J, Zhang Y, et al. . 2019. EWAS Atlas: a curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res 47(D1):D983–D988, PMID: 30364969, 10.1093/nar/gky1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Navarro Gonzalez J, Zweig AS, Speir ML, Schmelter D, Rosenbloom KR, Raney BJ, et al. . 2021. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res 49(D1):D1046–D1057, PMID: 33221922, 10.1093/nar/gkaa1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sikdar S, Joehanes R, Joubert BR, Xu CJ, Vives-Usano M, Rezwan FI, et al. . 2019. Comparison of smoking-related DNA methylation between newborns from prenatal exposure and adults from personal smoking. Epigenomics 11(13):1487–1500, PMID: 31536415, 10.2217/epi-2019-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, et al. . 2016. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet 9(5):436–447, PMID: 27651444, 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tilley SK, Martin EM, Smeester L, Joseph RM, Kuban KCK, Heeren TC, et al. . 2018. Placental CpG methylation of infants born extremely preterm predicts cognitive impairment later in life. PLoS One 13(3):e0193271, PMID: 29513726, 10.1371/journal.pone.0193271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson SL, Leavey K, Cox BJ, Robinson WP. 2018. Mining DNA methylation alterations towards a classification of placental pathologies. Hum Mol Genet 27(1):135–146, PMID: 29092053, 10.1093/hmg/ddx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herzog EM, Eggink AJ, Willemsen SP, Slieker RC, Wijnands KPJ, Felix JF, et al. . 2017. Early- and late-onset preeclampsia and the tissue-specific epigenome of the placenta and newborn. Placenta 58:122–132, PMID: 28962690, 10.1016/j.placenta.2017.08.070. [DOI] [PubMed] [Google Scholar]
- 72.Yuan V, Price EM, Del Gobbo G, Mostafavi S, Cox B, Binder AM, et al. . 2019. Accurate ethnicity prediction from placental DNA methylation data. Epigenetics Chromatin 12(1):51, PMID: 31399127, 10.1186/s13072-019-0296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Goede OM, Lavoie PM, Robinson WP. 2017. Cord blood hematopoietic cells from preterm infants display altered DNA methylation patterns. Clin Epigenetics 9:39, PMID: 28428831, 10.1186/s13148-017-0339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nicodemus-Johnson J, Myers RA, Sakabe NJ, Sobreira DR, Hogarth DK, Naureckas ET, et al. . 2016. DNA methylation in lung cells is associated with asthma endotypes and genetic risk. JCI Insight 1(20):e90151, PMID: 27942592, 10.1172/jci.insight.90151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chelchowska M, Ambroszkiewicz J, Jablonka-Salach K, Gajewska J, Maciejewski TM, Bulska E, et al. . 2013. Tobacco smoke exposure during pregnancy increases maternal blood lead levels affecting neonate birth weight. Biol Trace Elem Res 155(2):169–175, PMID: 23934137, 10.1007/s12011-013-9775-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richmond RC, Suderman M, Langdon R, Relton CL, Davey Smith G. 2018. DNA methylation as a marker for prenatal smoke exposure in adults. Int J Epidemiol 47(4):1120–1130, PMID: 29860346, 10.1093/ije/dyy091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Joubert BR, Håberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. . 2012. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 120(10):1425–1431, PMID: 22851337, 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. . 2016. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet 98(4):680–696, PMID: 27040690, 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Philibert R, Miller S, Noel A, Dawes K, Papworth E, Black DW, et al. . 2019. A four marker digital PCR toolkit for detecting heavy alcohol consumption and the effectiveness of its treatment. J Insur Med 48(1):90–102, PMID: 31609642, 10.17849/insm-48-1-1-1.1. [DOI] [PubMed] [Google Scholar]
- 80.Philibert R, Dogan M, Noel A, Miller S, Krukow B, Papworth E, et al. . 2018. Dose response and prediction characteristics of a methylation sensitive digital PCR assay for cigarette consumption in adults. Front Genet 9:137, PMID: 29740475, 10.3389/fgene.2018.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu M, Heinzerling TJ, Grady WM. 2018. DNA methylation analysis using droplet digital PCR. Methods Mol Biol 1768:363–383, PMID: 29717454, 10.1007/978-1-4939-7778-9_21. [DOI] [PubMed] [Google Scholar]
- 82.Yousefi PD, Suderman M, Langdon R, Whitehurst O, Davey Smith G, Relton CL. 2022. DNA methylation-based predictors of health: applications and statistical considerations. Nat Rev Genet 23(6):369–383, PMID: 35304597, 10.1038/s41576-022-00465-w. [DOI] [PubMed] [Google Scholar]
- 83.Schroeder DI, Blair JD, Lott P, Yu HOK, Hong D, Crary F, et al. . 2013. The human placenta methylome. Proc Natl Acad Sci USA 110(15):6037–6042, PMID: 23530188, 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.