Abstract
Schistosomiasis is of medical and veterinary importance. Despite the critical situation of schistosomiasis in sub-Saharan Africa, few molecular epidemiological studies have been carried out to determine the role of animals in its transmission. In Mali, it has been over three decades since the last molecular study of animal schistosomes was carried out. It is now urgent to identify circulating strains of the parasite because of potential interactions with other schistosome species, which could complicate disease control. The aim of our work was to study the composition and genetic structure of schistosome populations collected from cattle. The prevalence of schistosome was 23.9%, with the prevalences of Schistosoma bovis (Sb) and S. curassoni (Sc) estimated at 12.6% and 9.8%, respectively. No hybrid strains or S. haematobium were found. The parasites displayed distinct geographical distribution with Sb dominant in Bamako (78.8% and 98% in Central Bamako Slaughterhouse and Sabalibougou Slaughterhouses, respectively) and Sc dominant in Kayes (95.3%). Of the 476 parasites with a complete genetic profile, 60.4% were pure Sc, and were mainly from Kayes. We identified two clusters at the site level (Fst of 0.057 and 0.042 for Sb and Sc, respectively). Cluster 1 was predominantly composed of pure Sb parasites and cluster 2 was mainly composed of pure Sc parasites, from Bamako and Kayes, respectively. Our study shows that cattle schistosomiasis remains endemic in Mali with S. bovis and S. curassoni. A robust genetic structure between the different schistosome populations was identified, which included two clusters based on the geographical distribution of the parasites.
Keywords: Schistosoma bovis, S. curassoni, Slaughterhouse, Cattle, Genetic structure, Mali
Abstract
La schistosomiase revêt une grande importance médicale et vétérinaire. Malgré la situation critique de la schistosomiase en Afrique subsaharienne, peu d’études épidémiologiques moléculaires ont été réalisées pour déterminer le rôle des animaux dans sa transmission. Au Mali, cela fait plus de trois décennies que la dernière étude moléculaire des schistosomes animaux a été réalisée. Il est désormais urgent d’identifier les souches circulantes du parasite en raison des interactions potentielles avec d’autres espèces de schistosomes, ce qui pourrait compliquer la lutte contre la maladie. Le but de notre travail était d’étudier la composition et la structure génétique des populations de schistosomes collectées chez des bovins. La prévalence des schistosomes était de 23,9 %, celles de Schistosoma bovis (Sb) et de S. curassoni (Sc) étant respectivement estimées à 12,6 % et 9,8 %. Aucune souche hybride ni S. haematobium n’ont été trouvés. Les parasites présentaient une répartition géographique distincte avec Sb dominant à Bamako (respectivement 78,8 % et 98 % aux Abattoirs Centraux de Bamako et aux Abattoirs de Sabalibougou) et Sc dominant à Kayes (95,3 %). Sur les 476 parasites ayant un profil génétique complet, 60,4 % étaient des Sc purs, et provenaient principalement de Kayes. Nous avons identifié deux clusters au niveau du site (Fst de 0,057 et 0,042 pour Sb et Sc, respectivement). Le groupe 1 était principalement composé de parasites Sb purs et le groupe 2 était principalement composé de parasites Sc purs, provenant respectivement de Bamako et de Kayes. Notre étude montre que la schistosomiase bovine reste endémique au Mali, avec S. bovis and S. curassoni. Une structure génétique robuste entre les différentes populations de schistosomes a été identifiée, comprenant deux groupes basés sur la répartition géographique des parasites.
Introduction
Schistosomiasis continues to be a major public health threat in tropical and subtropical areas. This parasitosis belongs to a group of Neglected Tropical Diseases which collectively affect 1 billion people worldwide, and of which schistosomiasis accounts for infections in 250 million people in 78 countries [5]. It was estimated that worldwide, approximately 165 million animals are also infected with the parasite, which causes hemorrhagic enteritis, anemia, cachexia and for most, death [10]. In Mali, compared to the numerous studies done on human schistosomiasis, livestock schistosomiasis has received little attention and the most recent studies were carried out more than 30 years ago, hence the need to update existing data [36]. Of the 23 species of Schistosoma known worldwide, only nine have been identified to be of significant veterinary importance, especially for ruminants in Asia and Africa; these include: Schistosoma mattheei; S. bovis (Sb); S. curassoni (Sc); S. spindale; S. indicum; S. nasale; S. incognitum; S. margrebowiei; and S. japonicum [4, 22, 35]. Of these, two species, Sb and Sc are native to West Africa, six are widely distributed in Central and Eastern Africa [22, 26, 28, 31, 36, 41, 42] and one, S. japonicum is native to Asia [40]. The prevalence of Sb and Sc has been reported in some countries [26, 28, 36, 41, 42]. A study by Rollinson [36] in 1990 reported a high prevalence of schistosomiasis in cattle across three West African countries, namely Gambia (S. bovis: 14.3–27%), Senegal (Sb plus Sc and hybrids: 0–41.3%) and Mali (Sb, Sc and hybrids) with 62.8% in Bamako and 85.1% in Mopti. The two most recent molecular epidemiology studies of animal schistosomes were carried out in Senegal and Cameroon [12, 21]. In Cameroon, a recent study showed a 19.5% prevalence of adult schistosome worms in cattle [12], and prevalences of 92% and 8% were reported for S. bovis and S. curassoni in cattle in Senegal, respectively [21]. The control or elimination of cattle schistosomiasis will contribute to a substantial increase in the weight of the animals, thus improving animal productivity, which in turn will increase the incomes of livestock farmers and the availability of quality meat for consumption.
The schistosome parasite has a complex life cycle. Individuals are dioecious rather than hermaphroditic, as is the case with most other trematodes like liver flukes or tapeworms. Gonochorism allows interspecific interactions between males and females within their definitive hosts. Today, parasite hybridization is an emerging public health problem, as the cohabitation of human populations and domestic or wildlife animals. Increased mobility of populations creates opportunity for new agents to combine, and concerning parasites, this includes new interactions between parasites of different lineages or species within individual hosts [20, 29, 39]. The genetic variation of schistosome species and populations over time and space is based on the hybridization phenomenon and can only be described through molecular epidemiological studies. While many studies on population genetics of schistosomes in humans have been conducted [35], only few studies exist for animal populations. Parasite hybridization is a biological phenomenon that is gaining renewed interest as new molecular tools have enabled hybridization to be more easily detected, and because the repercussions in the evolution of infections are still largely unknown [19]. Hybridization is known in some cases to increase the virulence of the parasite towards its vertebrate hosts and to allow the parasite to broaden its spectrum of intermediate and/or definitive hosts [19, 47]. The first cases of hybridization between schistosomes, notably between Sb and Sc in animals, were reported in Africa (Mali & Senegal) by Rollinson [36]. In Senegal, a recent study found a high occurrence of bidirectional hybridization between these animal schistosome species, and this is the second piece of conclusive evidence of natural hybridization between Sb and Sc [44]. Also in the Niger River valley region, transmission of human and bovine schistosomiasis has been identified, but it seems that the transmission of the different species was limited [32].
The only population genetic study carried out on livestock schistosome was done in Cameroon [12]. The present study aims to understand the genetic composition and the population genetic structure of schistosomes collected from cattle in Mali. Specifically, we proposed (i) to determine the prevalence of schistosomes in cattle in Kayes and Bamako slaughterhouses; (ii) to identify the schistosome species and the possible presence of S. haematobium group hybrid parasites using genetic markers (ITS, 18S and Cox1); and (iii) to describe the genetic structure of schistosomes in cattle based on microsatellite data.
Material and methods
Study sites
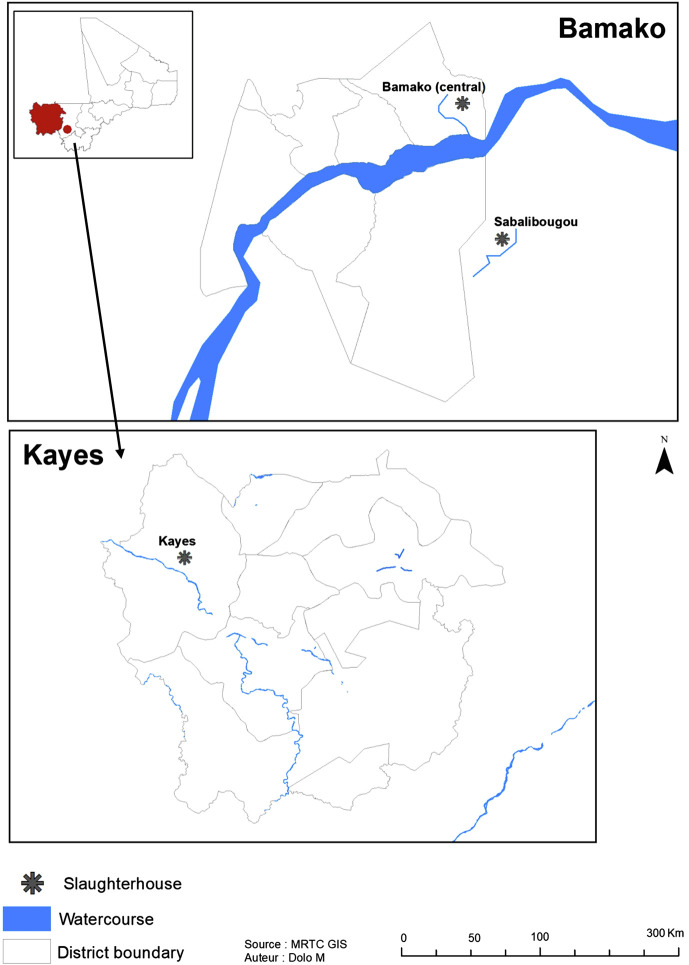
Mali ranks first in the West African Economic and Monetary Union (UEMOA) and second in the Economic Community of West African States (ECOWAS) zone in terms of bovine breeding, with an estimated 12,474,462 heads of cattle in its national herd [34]. A cross-sectional study was performed for two collections, in September (end of the rainy season) and December (cold dry season) 2021 in two areas: at the Central Bamako Slaughterhouse (CBS) and Sabalibougou Slaughterhouses (SS) in District of Bamako (Koulikoro region) and at the Kayes Slaughterhouse (KS) in Kayes region (Fig. 1). Unlike the two slaughterhouses located in the District of Bamako, where animals from all regions of the country converge, Kayes is an extensive livestock farming area. The two areas were specifically selected because they are known to be endemic for S. haematobium (Sh) [1], a human schistosome species. Sb and Sc are two schistosome species that infect animals [36]. In Mali, the Ministry of Livestock and Fisheries, through the National Veterinary Services Department, is the official body responsible for animal health. Anthrax, contagious bovine pneumonia, and plague of small ruminants are among the diseases managed by this service. No specific treatment is currently provided for schistosomiasis, even on small-scale dairy farms. Parasites were isolated and conditioned at the Department of Epidemiology and Parasitic Diseases (DEAP) of the Malaria Research and Training Center (MRTC) in Mali, and molecular manipulations were carried out at the Host-Pathogen-Environment Interactions (IHPE) laboratory in France.
Figure 1.
Map of Mali showing selected slaughterhouses.
Parasite collection
Collections of adult schistosomes were made between 0:00 am and 3:00 am from slaughtered cattle. Each collection session was followed by the identification of the animal from which the worms were collected, on a pre-established survey form. The site, the season, the number of worms recovered from each animal and the organ from which the worm was collected were recorded. In each slaughterhouse, intestines and ceca were randomly collected from slaughtered animals. The mesenteric veins surrounding the small intestines, and the veins which irrigate the ceca were carefully observed macroscopically by transparency for the presence of schistosome worms (whitish-colored worms through the vein). The worms were removed from the veins using forceps and placed in a 15 mL falcon tube containing physiological water, labelled with the bovine identifier and the removed organ. Once in the laboratory, the worms were isolated one by one using fine tweezers and stored individually in pure ethanol.
Extraction of genomic DNA (gDNA)
To extract genomic DNA (gDNA), the ethanol was first removed using a pipette, and then the tube containing the parasite was kept open and dried using the Speed Vac set at 14,000 rpm. Genomic DNA of 491 parasites was extracted using a DNeasy blood and tissue kit (QIAGEN, Hilden, Germany), following the manufacturer’s protocol, and the DNA was eluted in a total of 100 μL.
Rapid diagnostic polymerase chain reaction (RD-PCR) for Cox1 and tetraprimer amplification refractory mutations system – Polymerase chain reaction (T-ARMS-PCR) for ITS2 and 18S genotyping
Identification of Cox 1 profiles from 491 worms was done by Cox1 rapid diagnostic multiplex PCR (RD-PCR) with primers targeting two species groups: Sh and Sb or Sc [43]. In short, we used a universal reverse (Shmb.R) and two forward primers specific to three schistosome species: ShF (120 bp), SbF (260 bp – this band is the same for both Sc and Sb). Each PCR was carried out in a total volume of 10 μL containing 4 μL of ultra-pure water, 2 μL of buffer (Green GoTaq Flexi buffer, 5×; Promega; Madison, WI, USA), 0.6 μL of MgCl2 (Promega) at 25 mM, 0.2 μL of mixture of dNTPs (Promega) at 10 mM; 1 μL of 10× primer mix with 4 μL of each armor in 84 μL of ultra-pure water), 0.2 μL of GoTaq® Polymerase G2 Hot Start at 5 U (Promega), and 2 μL of DNA extract. After 45 cycles of amplification, the PCR products (Cox1) were visualized on a 2% agarose gel at 135 V for 40 min using the 100 bp DNA size marker (Promega) to estimate the size of the bands. We used the T-ARMS-PCR method developed in a recent study [32] to detect parasite nuclear genotypes (ITS/18S). Each partial PCR amplification of nuclear DNA was performed in a total volume of 25 μL, containing 3 μL of DNA template, 5 μL of GoTaq® Flexy buffer (Promega), 1, 5 μL of 25 mM MgCl2, 0.5 μL of a mixture of 8 prediluted primers (12 μM external primers and 15 μM for internal ones), 0.2 μL of dNTP solutions at 10 mM each, 0.5 μL of GoTaq® Taq polymerase G2 Hot Start (Promega) and 10.75 μL of milli-Q water. After 28 cycles of amplification, PCR products were examined on 1.8% agarose gels at 135 V for 40 min using the 100 bp DNA size marker (Promega) for size estimation. Four to six bands could be observed on the agarose gels, which allowed for the differentiation between Sc, Sb, Sh and all hybrid combinations [4].
Sanger sequencing
Cox1: Because the RD-PCR does not differentiate between Sc and Sb, we sequenced Cox 1 partial region (≈918 bp) for 188 parasites using a previously published protocol [23]. We considered the T-ARMS-PCR results as an indicator of sample selection for sequencing. We sequenced 107 and 81 parasites with either the Sb and Sc T-ARMS-PCR profile, respectively. The size fragment was checked on 5% agarose gel and PCR products were sent without purification to a subcontractor for sequencing (Genoscreen, Lille, France). Cox1 sequences were then compared to a reference sequence from GenBank (Accession number: AJ519521.1 and MT579424.1 for Sb and Sc, respectively).
18S: In order to double check several of the T-ARMS-PCR results, partial 18S regions (329 bp) were amplified and sequenced on 26 and 20, pure Sb and pure Sc profiles, as previously described [32]. The size fragment was checked on 5% agarose gel and PCR products were sent without purification to a subcontractor for sequencing (Genoscreen). The sequences obtained were compared with reference sequences (Accession number: AY157238.1 and AY157236.1 for Sb and Sc, respectively). At position 297, a C base identified Sb and a T base identified Sc [4].
Microsatellite genotyping
Microsatellite genotyping was performed on parasites collected from the three slaughterhouses. Only samples of parasites with a well-defined profile for each of the three genes (Cox1, ITS and 18S) were selected for microsatellite genotyping. The worms were individually genotyped using the PCR multiplex kit from QIAGEN and two panels of 8 loci: (Sh9, Sh3, C102, Sh1, Sh14, Sh6, C111, and Sh7) & (Sh13, Sh4, Sh11, Sh15, Sh2, Sh5, Sh10, and Sh12) [45]. The PCR mixture consisted of 5 μL of QIAGEN MM 2×, 1 μL of 10× microsatellite primer mix and 4 μL of DNA extract for a final volume of 10 μL. Thermal cycling was performed in a PerkinElmer 9600 Thermal Cycler (PerkinElmer, Waltham, MA, USA): pre-denaturation at 95 °C for 15 min followed by 40 cycles. Microsatellite PCR products were sent to Genoscreen for genotyping. After verification and correction using GeneMarker software, the data were then transferred to Excel to determine the position of the various loci.
Statistical analyses
Data were entered using Excel software. Calculations of prevalence of infection were performed using SPSS v 23.0 (IBM) software. Logistic regressions were used to infer the relationship between variables (slaughterhouse, season, and organ of sampling) and genetic profile. Lastly, p-values less than 0.05 were considered significant.
Genetic data analysis
Observed (Ho) and expected (He) heterozygosity, number of alleles (A), allelic richness (Ar), and inbreeding coefficient (Fis) at each microsatellite locus and for each population were calculated using FSTAT 2 software, 9.3.2 [16]. Population refers to the set of parasites collected from a given slaughterhouse (CBS, SS or KS). These parameters were compared according to the different populations or species using either Wilcoxon or Friedman pairwise rank tests followed by Nemenyi post-hoc tests.
First, population genetic structure was assessed using Fst indices [46]. Second, Genetix software [3] enabled us to visualize the potential genetic structure between individuals according to species using principal component analysis (PCA). Third, we evaluated the probability that the microsatellite database followed a defined number of clusters from K = 1 to K = 4 using Structure software [33]. We considered 10 runs for each trial, each of which consisted of a running period of 50,000 following by 250,000 MCMC iterations. The ΔK values were then calculated in R to determine the most likely number of clusters [13]. Finally, 10 additional runs were calculated for the best K using a running period of 50,000 and 500,000 iterations. The probability of each worm belonging to each cluster was averaged over the 10 runs and graphically represented using CLUMPP, version 1.1.2 [14] and DISTRUCT, version 1.1 [37].
Results
Schistosome prevalence in cattle
Animals slaughtered in slaughterhouses were mainly local Zebu breeds (Bos taurus indicus) and less commonly crossbreeds with N’dama (B. taurus taurus). Post-mortem examination of veins surrounding the small intestine and the cecum of cattle from Bamako and Kayes slaughterhouses revealed the presence of adult schistosome worms. Out of 398 cattle examined at the three slaughterhouses (two in Bamako and one in Kayes), 23.9% (95/398) were found to be infected by schistosomes (Table 1). Of these 95 cattle, we obtained the species of parasite involved for 89 individuals, with 50 animals infected by Sb parasites and 39 animals infected by Sc parasites. We can therefore estimate prevalences of 12.6% (50/398) and 9.8% (39/398) for Sb and Sc, respectively. Of note, 4/89 were co-infected by both species (1 from CBS and 3 from KS). The highest prevalence was recorded in KS (44.6%) compared to CBS (18.4%). More worms were recovered from the intestine (22.9%) compared to cecum veins (5%). No statistical difference in the prevalence was observed between seasons (p = 0.12). Sixteen bovines (6.5%) had parasites in both the small intestine and the cecum.
Table 1.
Number (%) of worms recovered in cattle in three transmission sites in Mali according to the season of collection and their localization (veins surrounding the small intestine or the cecum).
| Cattle examined | Cattle infected | p-value | Worms collected in veins surrounding small intestine | p-value | Worms collected in veins surrounding the caecum | p-value | ||
|---|---|---|---|---|---|---|---|---|
| Area | ||||||||
| Bamako | CBS | 140 | 31 (22.1) | p < 0.0001 | 28 (20.0) | p < 0.0001 | 3 (2.1) | p < 0.0001 |
| SS | 175 | 27 (15.4) | 27 (15.4) | 0 (0) | ||||
| Total | 315 | 58 (18.4) | 55 (17.5) | 3 (0.9) | ||||
| Kayes | KS | 83 | 37 (44.6) | 36 (43.4) | 17 (20.5) | |||
| Total | 83 | 37 (44.6) | 36 (43.4) | 17 (20.5) | ||||
| Season | ||||||||
| Rainy | 268 | 69 (25.7) | p = 0.12 | 65 (24.3) | p = 0.2 | 20 (7.5) | p < 0.0001 | |
| Dry | 130 | 26 (20.0) | 26 (20.0) | 0 (0) | ||||
| Total | 398 | 95 (23.9) | 91 (22.9) | 20 (5.0) |
CBS; Central Bamako Slaughterhouse; SS: Sabalibougou Slaughterhouses; KS: Kayes Slaughterhouse.
Schistosome species identification
Of the 616 adult worms collected, 491 were analyzed genetically. Correct genetic profiles for 491 and 476 worms were obtained using RD-PCR and T-ARMS-PCR, respectively. Results from these analyses indicated that the majority of the parasites originated from KS 58.4% (278/476) (Table 2). The T-ARMS-PCR revealed either Sc or Sb homozygous profiles and no hybrids were identified (Table 2). All 18S sequences (26 and 20 with Sb and Sc T-ARMS-PCR profile, respectively) confirmed the T-ARMS-PCR profiles. The RD-PCR revealed only the Sb/Sc profile and not the Sh profile. All 107 parasites with a homozygous Sb T-ARMS-PCR profile showed a typical Cox1 Sb sequence and all 81 parasites with a homozygous Sc T-ARMS-PCR profile showed a typical Cox1 Sc sequence. Because we randomly selected 40% of the parasites for sequencing, we assumed that the T-ARMS-PCR (SbxSb or ScxSc) profile reflected the Cox1 profile (Sb or Sc). Sb_SbxSb profiles were predominantly present in Bamako with 78.8% in CBS and 98% in SS, respectively. All haplotypes are available on GenBank with access numbers for Cox1: Sb (PP654220 to PP654290); Sc (PP654291 to PP654330) and 18S: Sb (PP654446, PP654447, PP654448); Sc (PP654443, PP654444, PP654445).
Table 2.
Multivariate analyses of adult worm species according to the site and the organ.
| Regions | Total | Sb_SbxSb | Sc_ScxSc | p | ||
|---|---|---|---|---|---|---|
| Bamako | CBS | Small Intestine | 95 | 75 (78.9) | 20 (21.1) | p < 0.0001 |
| Cecum | 4 | 3 (75) | 1 (25) | |||
| Total | 99 | 78 (78.8) | 21 (21.2) | |||
| SS | Small Intestine | 99 | 97 (98) | 2 (2) | p < 0.5 | |
| Cecum | 0 | 0 | 0 | |||
| Total | 99 | 97 (98.0) | 2 (2.0) | |||
| Kayes | KS | Small Intestine | 196 | 10 (5.1) | 186 (94.9) | p < 0.0001 |
| Cecum | 82 | 3 (3.7) | 79 (96.3) | |||
| Total | 278 | 13 (4.7) | 265 (95.3) | |||
| Total | 476 | 188 (39.5) | 288 (60.5) |
CBS; Central Bamako Slaughterhouse; SS: Sabalibougou Slaughterhouses; KS: Kayes Slaughterhouse.
Tropism of schistosome species
The vast majority of worms analyzed (81.9%) were localized in the veins surrounding the small intestine (Table 2). The small number of Sc_ScxSc parasites collected from Bamako were also principally localized in the veins surrounding the small intestine. In contrast, most parasites from KS exhibited an Sc_ScxSc profile (95.3%). The localization of these worms is more shared, with 82% (390/476) localized in the veins surrounding the small intestine.
Microsatellite analysis
Genetic diversity
Among the 476 parasites with complete mitochondrial and nuclear profiles, 166 were genotyped using 16 microsatellite markers. Parasite genetic diversity indices (He, Ho, A, Ar, and Fis) at each marker and for each slaughterhouse are presented in Table 3. The Friedman test showed a difference between populations for all calculated indices, He (χ2 = 8, df = 2, p = 0.011), Ar (χ2 = 8.375, df = 2, p = 0.015) and Fis (χ2 = 12.5, df = 3, p = 0.0017) (Table 3). The Nemenyi post-hoc tests revealed that the difference in Ar and He observed was significant between CBS and KS (p = 0.013 and p = 0.009, respectively). For Fis, a significant difference was observed between SS and KS (p = 0.002). Table 4 represents the overall gene diversity (He) and allelic richness (Ar) for Sb_SbxSb and Sc_ScxSc: mean He and Ar are both higher for Sb_SbxSb than for Sc_ScxSc, but the difference is only significant for He (W = 116, p = 0.21 for Ar, W = 116, p = 0.01 for He) (Table 4).
Table 3.
Genetic diversity indices of schistosome worms collected in cattle in three Malian slaughterhouses. Expected (He) and observed (Ho) heterozygosity, total number of alleles (A), allelic richness (Ar), and inbreeding coefficient (Fis) for each microsatellite locus and for each sampled site in Mali.
| Slaughterhouse | Sh9 | Sh3 | C102 | Sh1 | Sh14 | Sh6 | C111 | Sh7 | Sh13 | Sh4 | Sh11 | Sh15 | Sh2 | Sh5 | Sh10 | Sh12 | Moy | Std |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CBS (n = 55) | ||||||||||||||||||
| He exp | 0.84 | 0.89 | 0.89 | 0.90 | 0.65 | 0.76 | 0.71 | 0.91 | 0.92 | 0.89 | 0.62 | 0.40 | 0.95 | 0.72 | 0.83 | 0.64 | 0.78 | 0.12 |
| He Obs | 0.40 | 0.71 | 0.24 | 0.67 | 0.67 | 0.33 | 0.44 | 0.66 | 0.90 | 0.87 | 0.48 | 0.32 | 0.96 | 0.29 | 0.71 | 0.58 | 0.57 | 0.22 |
| A | 13 | 16 | 13 | 13 | 8 | 8 | 8 | 20 | 17 | 18 | 10 | 7 | 22 | 7 | 12 | 10 | 12.63 | 3.88 |
| Ar | 10.83 | 13.94 | 12.42 | 11.84 | 6.51 | 7.06 | 6.99 | 16.11 | 15.64 | 14.87 | 8.37 | 5.74 | 19.55 | 6.83 | 10.55 | 7.69 | 10.93 | 3.48 |
| Fis | 0.51 | 0.20 | 0.73 | 0.25 | −0.03 | 0.56 | 0.38 | 0.28 | 0.02 | 0.01 | 0.22 | 0.19 | −0.02 | 0.60 | 0.14 | 0.09 | 0.26 | 0.19 |
| SS (n = 42) | ||||||||||||||||||
| He exp | 0.75 | 0.87 | 0.88 | 0.90 | 0.67 | 0.76 | 0.67 | 0.88 | 0.93 | 0.89 | 0.52 | 0.23 | 0.93 | 0.68 | 0.76 | 0.58 | 0.74 | 0.14 |
| He Obs | 0.46 | 0.82 | 0.30 | 0.89 | 0.59 | 0.40 | 0.54 | 0.54 | 0.91 | 0.88 | 0.50 | 0.21 | 0.94 | 0.31 | 0.83 | 0.63 | 0.60 | 0.24 |
| A | 9 | 17 | 14 | 12 | 5 | 7 | 7 | 12 | 17 | 15 | 4 | 3 | 16 | 6 | 8 | 3 | 9.69 | 4.40 |
| Ar | 8.02 | 15.25 | 14.00 | 11.73 | 4.99 | 6.80 | 6.49 | 11.49 | 16.49 | 14.09 | 3.91 | 2.69 | 15.67 | 5.83 | 7.87 | 3.00 | 9.27 | 4.23 |
| Fis | 0.38 | 0.06 | 0.69 | 0.01 | 0.12 | 0.47 | 0.19 | 0.39 | 0.02 | 0.01 | 0.05 | 0.08 | −0.02 | 0.54 | −0.11 | −0.09 | 0.17 | 0.20 |
| KS (n = 69) | ||||||||||||||||||
| He exp | 0.56 | 0.83 | 0.69 | 0.16 | 0.36 | 0.27 | 0.22 | 0.21 | 0.94 | 0.91 | 0.81 | 0.19 | 0.82 | 0.28 | 0.68 | 0.83 | 0.55 | 0.27 |
| He Obs | 0.10 | 0.60 | 0.03 | 0.06 | 0.3 | 0.07 | 0.2 | 0.08 | 0.9 | 0.9 | 0.5 | 0.13 | 0.8 | 0.09 | 0.72 | 0.90 | 0.53 | 0.30 |
| A | 6 | 19 | 11 | 7 | 4 | 6 | 5 | 5 | 21 | 15 | 18 | 5 | 21 | 5 | 8 | 10 | 10.38 | 5.34 |
| Ar | 4.69 | 14.87 | 10.33 | 4.42 | 3.41 | 5.20 | 4.01 | 4.45 | 18.14 | 12.83 | 14.15 | 4.17 | 16.08 | 4.59 | 7.33 | 8.79 | 8.59 | 4.38 |
| Fis | 0.79 | 0.29 | 0.96 | 0.62 | 0.21 | 0.73 | 0.06 | 0.65 | 0.06 | −0.02 | 0.33 | 0.27 | 0.01 | 0.69 | −0.04 | −0.09 | 0.34 | 0.30 |
| Total (n = 166) | ||||||||||||||||||
| A | 15 | 25 | 19 | 14 | 8 | 10 | 10 | 22 | 24 | 20 | 18 | 7 | 26 | 8 | 13 | 11 | 15.63 | 5.58 |
| Ar | 8.66 | 15.29 | 14.02 | 11.00 | 5.26 | 7.05 | 5.96 | 13.08 | 17.39 | 13.70 | 10.79 | 4.36 | 19.61 | 7.02 | 10.50 | 7.95 | 10.73 | 3.63 |
Table 4.
Allelic richness and genetic diversity for Sc_ScxSc and Sb_SbxSb genotypes.
| Allelic richness (Ar) | Gene diversity (He) | |||
|---|---|---|---|---|
| Sc_ScxSc | Sb_SbxSb | Sc_ScxSc | Sb_SbxSb | |
| Sh9 | 2.86 | 12.06 | 50% | 81% |
| Sh3 | 16.71 | 16.8 | 79% | 88% |
| C102 | 10 | 16.74 | 62% | 88% |
| Sh1 | 1 | 13.09 | – | 90% |
| Sh14 | 2.85 | 6.5 | 25% | 70% |
| Sh6 | 5.18 | 7.91 | 27% | 76% |
| C111 | 4.42 | 7.59 | 21% | 69% |
| Sh13 | 20.92 | 15.71 | 94% | 92% |
| Sh4 | 14.28 | 15.39 | 90% | 87% |
| Sh11 | 16.5 | 3.99 | 82% | 49% |
| Sh15 | 5.78 | 4 | 25% | 29% |
| Sh2 | 17.91 | 17.38 | 81% | 93% |
| Sh5 | 4.76 | 6.4 | 28% | 74% |
| Sh12 | 9.64 | 4.75 | 81% | 57% |
| Mean | 9.49 | 10.59 | 57% | 75% |
| SE | 1.76 | 1.39 | 8% | 5% |
Population genetic structure
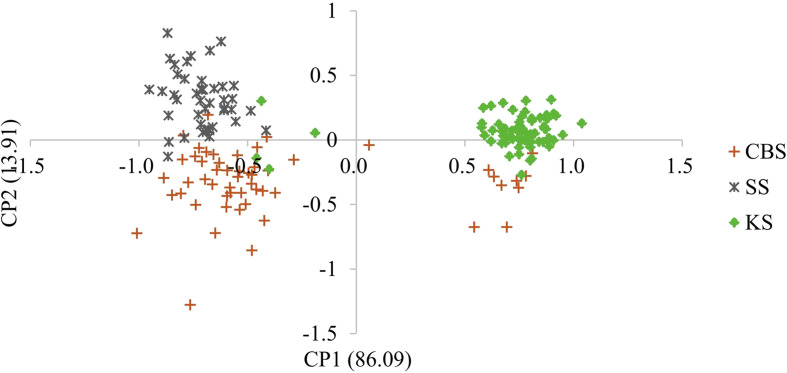
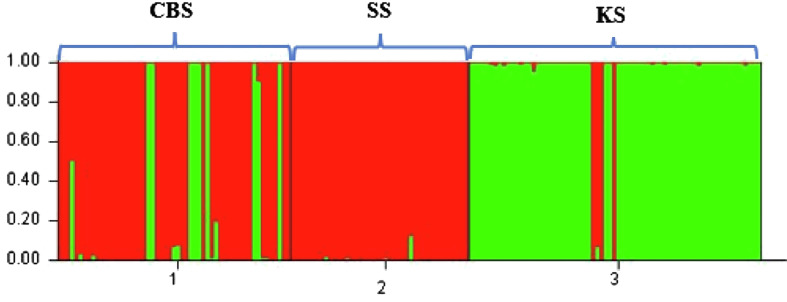
Whether we consider the PCA (Fig. 2) or Bayesian approach (Fig. 3), two groups were clearly identified. The first group is mainly composed of parasites from Bamako (CBS or CS) and the second group is mainly composed of parasites from Kayes (KS). According to the parasite genotyping, this structure is more a result of the differences between species than the sampling site. The few parasites from the Kayes group that clustered with the Bamako group are Sb_SbxSb, and conversely the few parasites from the Bamako group that clustered with the Kayes group are Sc_ScxSc parasites. The PCA first axes cluster the genotypes, and the second axes cluster the sites. The Bayesian analysis (Fig. 3) also shows that the vast majority of parasites are either assigned to cluster 1 and possess a Sb_SbxSb profile or to cluster 2 and possess a Sc_ScxSc profile. A single parasite from Bamako shows a 50/50 assignment probability between cluster 1 and 2. This parasite has an Sb_SbxSb profile in both PCR and sequencing and thus has not been identified as a hybrid.
Figure 2.
Principal component analysis (PCA) of schistosome worm parasite recovered from three slaughterhouses in Mali. Each point represents a parasite, and the color label corresponds to the population of origin (CSB, SS or KS). The first and second axes explain 86.09% and 13.91% of the genetic variation, respectively.
Figure 3.
Bar plot showing the population genetic structure of 491 schistosome worms collected in three slaughterhouses in Mali, using Structure software. Each column represents one worm. The colors show the proportion of contribution of each cluster to each genotype. The cluster structure is for K = 2. The geographic populations are CBS (1), SS (2) and KS (3).
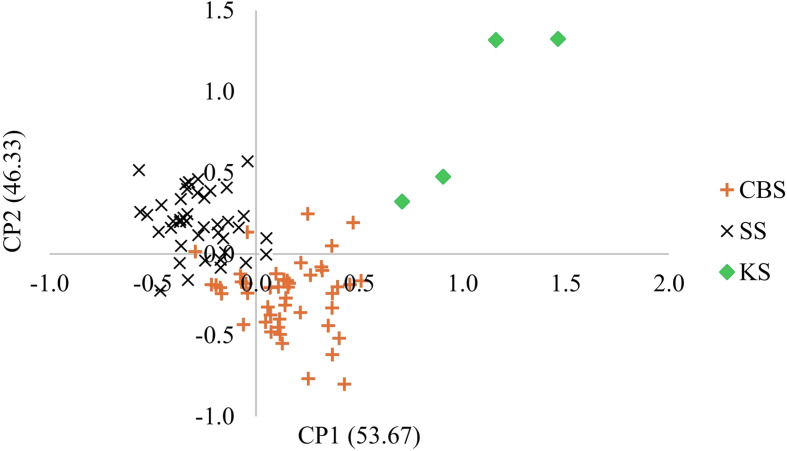
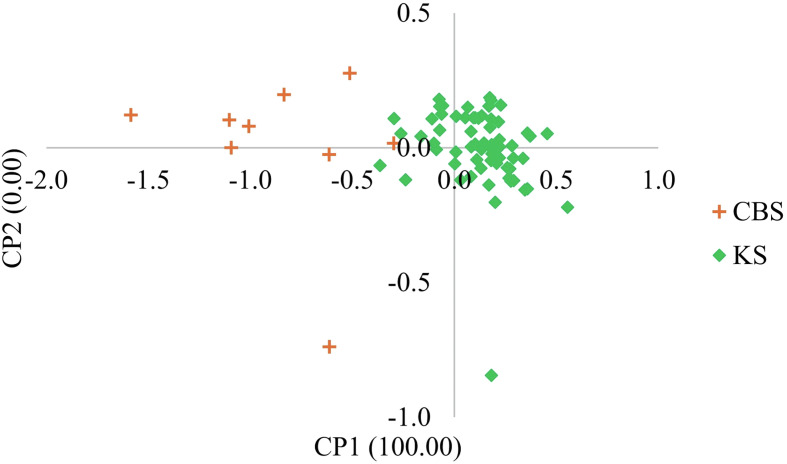
PCAs by species showed that regardless of the species, Sb (Fig. 4) or Sc (Fig. 5), the parasites are structured according to the collection site. Pairwise Fst value between Sb_SbxSb and Sc_ScxSc genotypes is 23%. Table 5 presents the Fst values between genotypes at different sites. Except for the Sb_SbxSb genotype between SS and CBS, the Fst values range between 4.2% and 5.7%.
Figure 4.
Principal component analysis (PCA) of Schistosoma bovis worms recovered from three slaughterhouses in Mali. Each point represents a parasite, and the color corresponds to the population of origin (CSB, SS or KS). The first and second axes explain 53.67% and 46.33% of the genetic variation, respectively.
Figure 5.
Principal component analysis (PCA) of Schistosoma curassoni worms recovered from three slaughterhouses in Mali. Each point represents a parasite, and the color corresponds to the population of origin (CSB, SS or KS). The first and second axes explain 100.00% and 0.00% of the genetic variation, respectively.
Table 5.
Pairwise estimates of Fst values for microsatellite DNA genotype (below the diagonal) of Sb and Sc parasite populations as a function of slaughterhouse.
| Sb_SbxSb | CBS | SS |
|---|---|---|
| SS | 0.0043 | – |
| KS | 0.044 | 0.057 |
| Sc_ScxSc | ||
| KS | 0.042 |
CBS; Central Bamako Slaughterhouse; SS: Sabalibougou Slaughterhouses; KS: Kayes Slaughterhouse.
Discussion
Our study focused on the epidemiology, molecular characterization, and population genetics of Schistosoma spp. that infect cattle in Mali. The study was conducted in abattoirs in two regions, the CBS and the SS in the District of Bamako and the KS. The results showed that the livestock schistosomiasis was endemic in the two regions with a global prevalence of 23.9%. The highest rate was observed in KS (44.6%) compared to CBS and SS (18.4%). The prevalence of Sc in our study was estimated at 9.8%. This prevalence is similar to that reported in Cameroon (10.03%) in domestic ruminants, using morphological egg criteria [30], but lower than that observed in Bamako (Mali) for cattle (30.8%) using an iso-enzymatic method [36]. Using a molecular approach, Sc was also reported in Barkedji and Linguère in Senegal, with an estimated prevalence of 73% in sheep, 84% in goats, and 8% in cattle [21]. The low prevalence of Sc in our study may be due to the final host examined (cattle). As previously observed in Senegal [21], Sc preferred goats and sheep as hosts. The prevalence of Sb in our study was estimated at 12.6%. The prevalence of Sb varies greatly from one study to another according to the host species concerned. In sheep, prevalence rates ranging from 4% to 20% have been observed in Senegal [21, 27]. In goats, a prevalence of 15% has been observed in Senegal [21]. Prevalence rates are usually higher in cattle, with 19.5% in Cameroon [12] and 92% in Senegal [21]. The presence of schistosomes is dependent on the presence of intermediate snail hosts. Unfortunately, too few studies exist on the distribution of snails in Africa, particularly those that transmit schistosomes that infect animals. In West African countries, the snail species that hosts Sb may be Bulinus truncatus, B. forskalii or B. globosus [25], whereas B. umbilicatus is described as the main host of Sc [11]. In Mali, no study has yet looked specifically at snails as intermediate hosts of Sc or Sb, even though various intermediate hosts of schistosomiasis have been documented (B. truncatus, B. globosus, B. umbilicatus, B. forskalii, and B. senegalensis) [9, 24]. The snail B. umbilicatus has only been identified in the Mopti region, but the authors of the study were unable to determine the species of schistosome it transmitted [24]. Despite the specific distribution of snails due to ecological factors, animals slaughtered in Kayes or Bamako can be moved over great distances (600 km separate the livestock supply areas of Mopti from Bamako and those of Kayes and Nara on the Mauritanian border), which can bring them into contact with different transmission sites. No significant variation in prevalence was found with respect to season. The seasonal movement of animals over large distances to find new pastures and water points (transhumance) in Mali promotes their exposure to a variety of infected water bodies.
During this study, we did not find any SbxSc hybrid species of bovine schistosome, nor any hybrid between ShxSb or ShxSc, as recently highlighted in Cameroon [12]. However, SbxSc hybrids have been reported in cattle in Senegal, Niger, and Mali [6, 36, 44] and sheep and goats, in Niger [7, 8]. The absence of hybrid parasites could be explained by the fact that the transmission sites we sampled were characterized by a high prevalence of either Sb or Sc or by a low rate of co-infected cattle. In the latter case, interspecific encounters are infrequent and hybridization unlikely. In addition, we collected adult parasites without identifying whether we had inter-specific pairs. Genotyping the eggs might have enabled us to identify first-generation hybrid eggs, as observed in Benin [38].
Regardless of the method used (Fst, PCA or Structure), there is strong structuring between Sb and Sc species, and weak structuring between slaughterhouses within the same species. Concerning the genetic difference between species, most published studies are based on sequence comparisons, and in particular, involve sequencing the cytochrome oxidase gene [18]. In the latter study, the authors measured a genetic difference of 6.1% between Sb and Sc, in Senegal. Based on microsatellite markers, we measured a genetic differentiation of 4.2% between these two species. Concerning the difference between populations within the same species, we measured an Fst of 0.43% between the CBS and SS abattoirs for Sb. These two abattoirs are about 18 km apart. This result is in agreement with the weak genetic structuring of Sb observed in Cameroon [12] and Côte d’Ivoire [15]. With only two sites, it is difficult to provide more general results on the genetic structure of this parasite species in Mali. As Mali is the leading livestock-producing country in the West African Economic and Monetary Union, it is natural for livestock to move across the country and even across borders in search of pasture, which encourages the spread of parasites. During this transhumance, it is often possible for herders to sell animals along the way [2]. Moreover, cattle trade between countries is universally present in West Africa. It is possible that some of the cattle sampled in this study came from countries bordering Mali. As Bamako is the country’s capital, it receives cattle from all areas, including Burkina-Faso, Niger, and even Senegal. All of these movements may be at the root of the weak structuring within the various species. The strong presence of Sc in the Kayes slaughterhouse is easily explained by its proximity to Mauritania and Senegal, where this species has already been reported [17, 44]. Being able to use the same panel of microsatellite markers on Sh, Sb, and Sc is an opportunity to carry out more studies on animal parasites and to infer their possible role in schistosome transmission to humans. For instance, our study shows that the heterozygosity rate is higher for Sb than for Sc. Variations in heterozygosity have already been observed in previous studies between Sb and Sh in Cameroon [12] and in Côte d’Ivoire [15].
Our work has a number of limitations. Because the worms were grouped by animal and by organ, we were unable to isolate the pairs directly in the abattoir and we were unable to obtain the genotypes/species of each pair. It is also incredibly difficult to collect parasites in abattoirs. The time allowed per animal is always very limited so as not to disrupt the slaughter line. In addition, we limited our study to cattle. It would have been interesting to also inspect sheep and goats.
Animal schistosomiasis is prevalent in cattle in the Kayes and Bamako abattoirs, with the highest prevalence found in Kayes. Our study focused on the genetic profile and diversity of schistosome adults collected from livestock in Mali. Two pure species were found (Sb and Sc). Parasites were well structured according to species, with Sb and Sc dominating in Bamako and Kayes, respectively. Further studies on the genome of animal schistosomes on a large scale and the potential impacts of hybrid strains on animals are required. Additional studies are also needed to expand our understanding in this field by increasing the sample size and by extending the surveys to other slaughterhouses and to farm livestock in Mali. In view of our findings, a strategic control program for cattle trematodes is warranted. An important component of such a control program is to water cattle in troughs rather than to let them water freely from rivers and ponds, in order to reduce fecal contamination.
Abbreviations
- Sb
Schistosoma bovis
- Sc
Schistosoma curassoni
- ITS
Internal transcribed spacer
- Cox1
Cytochrome oxidase subunit 1 gene
- RD-PCR
Rapid diagnostic multiplex PCR
- T-ARMS-PCR
Tetra-amplification refractory mutation system PCR
- FTA
Flinders Technology Associates
- DNA
Deoxyribonucleic acid
- UEMOA
Union Économique et Monétaire Ouest Africaine
- ECOWAS
Economic Community of West African States
- PRODEVIM
Programme de Développement à l’Exportation de la Viande du Mali
- PCA
Principal Component Analysis
- CBS
Central Bamako Slaughterhouse
- SS
Sabalibougou Slaughterhouses
- KS
Kayes Slaughterhouse
- He
Expected heterozygosity
- Ho
Observed heterozygosity
- A
Allele
- Ar
Allelic richness
- Fis
Inbreeding coefficient
- B
Bulinus
Acknowledgments
We thank Dr Bonnie Webster for her constructive comments on the manuscript. We thank slaughterhouse managers and butchers for their frank collaboration. The authors would particularly like to thank: Amadou Dabo dit Boua for technical support in the field; Mathias Dolo and Kadra Koné, geographers; all slaughterhouse managers and butchers of all three sites; the staff of the MRTC (Malaria Research and Training Center); and the staff of the Faculty of Medicine and Dentistry and the Faculty of Pharmacy. We would like to thank the government of Mali for funding this research, and the IHPE laboratory, Univ. Montpellier, CNRS, Ifremer, Univ. Perpignan Via Domitia, Perpignan, France for molecular analyses.
Cite this article as: Diakité A, Agniwo P, Dabo A, Sidibé B, Savassi BAES, Akplogan A, Guindo H, Dembélé L, Ibikounlé M, Doumbo Niaré S, Tembely S & Boissier J. 2024. Population genetic structure of Schistosoma bovis and S. curassoni collected from cattle in Mali. Parasite 31, 36.
Edited by: Jean-Lou Justine
These authors contributed equally to this work.
Funding
This research was funded by the Competitive Fund for Research and Technological Innovation (FCRIT).
Conflicts of interest
The authors declare that they have no competing interests.
Data availability statement
All data generated or analyzed during this study are included in this published article.
Author contribution statement
P.A., A.D., B.J., N.S.D., D.L, S.T., I.M., participated in the design of the study, updated the research methodology and contributed to the writing of the final document. P.A., A.D., A.D., J.B., N.S.D., L.D., S.B., A.A., H.G., coordinated the field trial. P.A., B.A.E.S., provided laboratory handling and sequence correction. P.A., J.B., A.D., carried out the statistical analysis. B.A.E.S., L.D., M.I., A.D., S.T., J.B., reviewed the manuscript for submission.
Ethics approval
The global protocol covering both human and animal schistosomiasis study was reviewed and approved by the Institutional Ethics Committee of the Faculty of Medicine, Pharmacy and Dentistry of the University of Sciences, Techniques and Technologies of Bamako under the number 2018/71/CE/FMPOS. The officials of the veterinary services and the managers of the slaughterhouses were informed before any sampling. To avoid the disagreements caused by our presence inside the slaughterhouses and the loss of time caused by the examination of animals, the butchers were willing to cooperate with us over the course of this study.
References
- 1.Agniwo P, Sidibé B, Diakité AT, Niaré SD, Guindo H, Akplogan A, Ibikounlé M, Boissier J, Dabo A. 2023. Ultrasound aspects and risk factors associated with urogenital schistosomiasis among primary school children in Mali. Infectious Diseases of Poverty, 12(1), 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassett T. 2009. Mobile pastoralism on the brink of land privatization in Northern Côte d’Ivoire. Geoforum, 40(5), 756–766. [Google Scholar]
- 3.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme M. 2004. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Montpellier (France): Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II. Available from http://www.univmontp2.fr/~genetix/genetix/genetix.htm (accessed on 2022-07-20). [Google Scholar]
- 4.Blin M, Dametto S, Agniwo P, Webster BL, Angora E, Dabo A, Boissier J. 2023. A duplex tetra-primer ARMS-PCR assay to discriminate three species of the Schistosoma haematobium group: Schistosoma curassoni, S. bovis, S. haematobium and their hybrids. Parasites & Vectors, 16, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boissier J, Mouahid G, Moné H. 2019. Schistosoma spp, in water and sanitation for the 21st century: health and microbiological aspects of excreta and wastewater management (global water pathogen project), in Rose JB, Jiménez-Cisneros B, Editors. Part 3: Specific excreted pathogens: environmental and epidemiology aspects – Section 4: Helminths, Robertson L, Editor. E. Lansing, MI: Michigan State University, UNESCO. 10.14321/waterpathogens.45. [DOI] [Google Scholar]
- 6.Bremond P, Sellin B, Sellin E, Nameoua B, Labbo R, Theron A. 1993. Arguments for the modification of the genome (introgression) of the human parasite Schistosoma haematobium by genes from S. bovis, in Niger. Comptes Rendus de l’Académie des Sciences. Série III, Sciences de la Vie, 316(7), 667–670. [PubMed] [Google Scholar]
- 7.Brémond P. 1990. Application des techniques électrophorétiques à deux aspects de la biologie des populations de schistosomes africains : Caractérisation des parasites et de leurs hôtes intermédiaires; détection des schistosomes hybrides, in Les Schistosomes, in Conférence International sur la Situation Épidémiologique et Stratégies de Lutte contre les Schistosomiases en Afrique de l’Ouest. Bobo Dioulasso: IRD, OCCGE, p. 182–189. [Google Scholar]
- 8.Brémond P, Mouchet F, Chevallier P, Sellin E, Vera C, Sellin B. 1990. Flux génique entre Schistosoma bovis et S. curassoni au Niger. Bulletin de la Société Française de Parasitologie, 8, 708. [Google Scholar]
- 9.Coulibaly G, Madsen H. 1990. Seasonal density fluctuations of intermediate hosts of schistosomes in two streams in Bamako, Mali. Journal of African Zoology, 104(3), 201–212. [Google Scholar]
- 10.De Bont J, Vercruysse J. 1997. The epidemiology and control of cattle schistosomiasis. Parasitology Today, 13(7), 255–262. [DOI] [PubMed] [Google Scholar]
- 11.Diaw O, Vassiliadès G. 1987. Épidémiologie des schistosomoses du bétail au Sénégal. Revue d’Élevage et de Médecine Vétérinaire des Pays Tropicaux, 40(3), 265–274. [PubMed] [Google Scholar]
- 12.Djuikwo-Teukeng FF, Kouam Simo A, Allienne JF, Rey O, Njayou Ngapagna A, Tchuem-Tchuente LA, Boissier J. 2019. Population genetic structure of Schistosoma bovis in Cameroon. Parasites & Vectors, 12, 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14(8), 2611–2620. [DOI] [PubMed] [Google Scholar]
- 14.Francis RM. 2017. Pophelper: an R package and web app to analyse and visualize population structure. Molecular Ecology Resources, 17(1), 27–32. [DOI] [PubMed] [Google Scholar]
- 15.Giovanoli Evack J, Kouadio JN, Achi LY, Bonfoh B, N’Goran EK, Zinsstag J, Jürg Utzinger J, Balmer O. 2024. Genetic characterization of schistosome species from cattle in Côte d’Ivoire. Parasites & Vectors, 17, 122–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goudet J, Perrin N, Waser P. 2002. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Molecular Ecology, 11(6), 1103–1114. [DOI] [PubMed] [Google Scholar]
- 17.Grétillat S. 1962. Une nouvelle zoonose, la “Bilharziose Ouest Africaine” à Schistosoma curassoni Brumpt, 1931 commune à l’homme et aux ruminants domestiques. Comptes Rendus Hebdomadaires des Séances de l’Académie des Sciences, 255(15), 1805. [PubMed] [Google Scholar]
- 18.Huyse T, Webster BL, Geldof S, Stothard JR, Diaw OT, Rollinson D. 2009. Bidirectional introgressive hybridization between a cattle and human schistosome species. PLoS Pathogens, 5(9), e1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King KC, Stelkens RB, Webster JP, Smith DF, Brockhurst MA. 2015. Hybridization in parasites: consequences for adaptive evolution, pathogenesis, and public health in a changing world. PLoS Pathogens, 11(9), e1005098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafferty KD. 2009. The ecology of climate change and infectious diseases. Ecology, 90(4), 888–900. [DOI] [PubMed] [Google Scholar]
- 21.Léger E, Borlase A, Fall C, Diouf N, Diop S, Yasenev L, Catalano S, Thiam CT, Ndiaye A, Emery A, Morrell A, Rabone M, Ndao M, Faye B, Rollinson D, Rudge WJ, Sène M, Webster JP. 2020. Prevalence and distribution of schistosomiasis in human, livestock, and snail populations in northern Senegal: a one health epidemiological study of a multi-host system. Lancet Planetary Health, 4(8), e330–e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Léger E, Webster JP. 2016. Hybridizations within the genus Schistosoma: implications for evolution, epidemiology and control. Parasitology, 144(1), 65–80. [DOI] [PubMed] [Google Scholar]
- 23.Lockyer AE, Olson PD, Ostergaard P, Rollinson D, Johnston DA, Attwood SW, Southgate VR, Horak P, Snyder SD, Le TH, Agatsuma T, McManus DP, Carmichael AC, Naem S, Littlewood DTJ. 2003. The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology, 126(3), 203–224. [DOI] [PubMed] [Google Scholar]
- 24.Madsen H, Coulibaly G, Furu P. 1987. Distribution of freshwater snails in the river Niger basin in Mali with special reference to the intermediate hosts of schistosomes. Hydrobiologia, 146, 77–88. [Google Scholar]
- 25.Moné H, Mouahid G, Morand S. 1999. The distribution of Schistosoma bovis Sonsino, 1876 in relation to intermediate host mollusc–parasite relationships. Advances in Parasitology, 44, 99–138. [DOI] [PubMed] [Google Scholar]
- 26.Mouchet F, Bremond P, Théron A. 1989. Preliminary observations on Schistosoma curassoni Brumpt, 1931 in Niger. Transactions of the Royal Society of Tropical Medicine and Hygiene, 83(6), 811. [DOI] [PubMed] [Google Scholar]
- 27.Ndao M, Belot J, Zinsstag J, Pfister K. 1995. Épidémiologie des helminthoses gastro-intestinales des petits ruminants dans la zone sylvo-pastorale au Sénégal. Veterinary Research, 26(2), 132–139. [PubMed] [Google Scholar]
- 28.Ndifon GT, Betterton C, Rollinson D. 1988. Schistosoma curassoni Brumpt, 1931 and S. bovis (Sonsino, 1876) in cattle in northern Nigeria. Journal of Helmintholology, 62(1), 33–34. [DOI] [PubMed] [Google Scholar]
- 29.Nichols GL, Andersson Y, Lindgren E, Devaux I, Semenza JC. 2014. European monitoring systems and data for assessing environmental and climate impacts on human infectious diseases. International Journal of Environmental Research and Public Health, 11(4), 3894–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ousseini S. 1990. Schistosomoses des ruminants domestiques au Cameroun septentrional. Thèse de Médecine vétérinaire de l’Université Cheikh Anta Diop, Dakar, Sénégal, p. 112. [Google Scholar]
- 31.Panzner U, Boissier J. 2021. Natural intra- and interclade human hybrid schistosomes in Africa with considerations on prevention through vaccination. Microorganisms, 9(7), 1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pennance T, Allan F, Emery A, Rabone M, Cable J, Garba AD, Hamidou AA, Webster JP, Rollinson D, Webster BL. 2020. Interactions between Schistosoma haematobium group species and their Bulinus spp. intermediate hosts along the Niger River Valley. Parasites & Vectors, 13, 268–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchard J, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.PRODEVIM. 2022. Mali meat export development program (PRODEVIM). Available from https://bamada.net/ La viande: Quatre usines de viande dans les starting blocs – Bamada.net (accessed 2023-06-12).
- 35.Rey O, Webster BL, Huyse T, Rollinson D, Van den Broeck F, Kincaid-Smith J, Onyekwere A, Boissier J. 2021. Population genetics of African Schistosoma species. Infection Genetics and Evolution, 89, 104727. [DOI] [PubMed] [Google Scholar]
- 36.Rollinson D, Southgate VR, Vercruysse J, Moore PJ. 1990. Observations on natural and experimental interactions between Schistosoma bovis and S. curassoni from West Africa. Acta Tropica, 47(2), 101–114. [DOI] [PubMed] [Google Scholar]
- 37.Rosenberg NA. 2004. Distruct: a program for the graphical display of population structure. Molecular Ecology Notes, 4(1), 137–138. [Google Scholar]
- 38.Savassi BAES, Mouahid G, Lasica C, Mahaman SK, Garcia A, Courtin D, Allienne JF, Ibikounlé M, Moné H. 2020. Cattle as natural host for Schistosoma haematobium (Bilharz, 1852) Weinland, 1858 x Schistosoma bovis Sonsino, 1876 interactions, with new cercarial emergence and genetic patterns. Parasitology Research, 119, 2189–2205. [DOI] [PubMed] [Google Scholar]
- 39.Shuman EK. 2010. Global climate change and infectious diseases. New England Journal of Medicine, 362(12), 1061–1063. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka H, Tsuji M. 1997. From discovery to eradication of Schistosomiasis in Japan: 1847–1996. International Journal for Parasitology, 27(12), 1465–1480. [DOI] [PubMed] [Google Scholar]
- 41.Vera C, Mouchet F, Bremond P, Sidiki A, Sellin E, Sellin B. 1992. Natural infection of Bulinus senegalensis by Schistosoma haematobium in a temporary pool focus in niger: characterization by cercarial emergence patterns. Transactions of the Royal Society of Tropical Medicine and Hygiene, 86(1), 62. [DOI] [PubMed] [Google Scholar]
- 42.Vercruysse J, Southgate VR, Rollinson D. 1984. Schistosoma curassoni brumpt, 1931 in sheep and goats in senegal. Journal of Natural History, 18(6), 969–976. [DOI] [PubMed] [Google Scholar]
- 43.Webster B, Rollinson D, Stothard J, Huyse T. 2010. Rapid diagnostic multiplex PCR (RD-PCR) to discriminate Schistosoma haematobium and S. bovis. Journal of Helminthology, 84(1), 107–114. [DOI] [PubMed] [Google Scholar]
- 44.Webster B, Diaw O, Seye M, Webster J, Rollinson D. 2013. Introgressive hybridization of Schistosoma haematobium group species in Senegal: species barrier break down between ruminant and human schistosomes. PLoS Neglected Tropical Diseases, 7(4), e2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webster BL, Rabone M, Pennance T, Webster JP. 2015. Development of novel multiplex microsatellite polymerase chain reactions to enable high-throughput population genetic studies of Schistosoma haematobium. Parasites & Vectors, 8, 432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weir B, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution, 38, 1358–1370. [DOI] [PubMed] [Google Scholar]
- 47.Wright CA, Southgate VR. 1976. Hybridization of schistosomes and some of its implications, in Genetic aspects of host-parasite interactions. Taylor A, Muller R, Editors. Oxford: Blackwell Scientific, p. 55–86. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.