Abstract
Quantitative analysis of the dynamic properties of thoraco-abdominal organs such as lungs during respiration could lead to more accurate surgical planning for disorders such as Thoracic Insufficiency Syndrome (TIS). This analysis can be done from semi-automatic delineations of the aforesaid organs in scans of the thoraco-abdominal body region. Dynamic magnetic resonance imaging (dMRI) is a practical and preferred imaging modality for this application, although automatic segmentation of the organs in these images is very challenging. In this paper, we describe an auto-segmentation system we built and evaluated based on dMRI acquisitions from 95 healthy subjects. For the three recognition approaches, the system achieves a best average location error (LE) of about 1 voxel for the lungs. The standard deviation (SD) of LE is about 1-2 voxels. For the delineation approach, the average Dice coefficient (DC) is about 0.95 for the lungs. The standard deviation of DC is about 0.01 to 0.02 for the lungs. The system seems to be able to cope with the challenges posed by low resolution, motion blur, inadequate contrast, and image intensity non-standardness quite well. We are in the process of testing its effectiveness on TIS patient dMRI data and on other thoraco-abdominal organs including liver, kidneys, and spleen.
Keywords: Lung, auto-segmentation, dynamic magnetic resonance imaging (dMRI), artificial intelligence, natural intelligence, deep neural networks, pediatric subjects, Thoracic Insufficiency Syndrome
Short Abstract
Quantitative analysis of dynamic properties of thoraco-abdominal organs during respiration could lead to more accurate surgical planning for certain disorders. This analysis can be done from semi-automatic delineations of lungs from dynamic dMRI acquisitions of the thoraco-abdominal region, although automatic segmentations of the organs in these images is challenging. In this paper, we describe an auto-segmentation system we built and evaluated based on dMRI acquisitions from 95 healthy subjects, which has yielded Dice Coefficient of about 0.95 in this very challenging problem of segmenting dynamic organs in a 4D MRI image.
1. INTRODUCTION
Thoracic Insufficiency Syndrome (TIS) [1] is an uncommon disorder in which there is inability of the thorax to support normal respiration or lung growth, leading to complications. In surgical planning for children with TIS, locations along the skeleton that should be connected with rods are identified so that abnormal growth and orientation of the osseous structures can be controlled with minimal complications. At present, this surgical planning is largely based on subjective qualitative intuition. Yet, quantitative comparisons of the dynamic properties of the thoraco-abdominal organs between children with TIS and healthy children, under free-breathing conditions, can give detailed insights for more accurate surgical planning. Such properties during respiration can be captured effectively with dynamic magnetic resonance imaging (dMRI), which does not involve radiation exposure, does not require special patient maneuvers or breathing control, and can be implemented readily on MRI scanners available in the community.
The aforementioned insights can be represented quantitatively from the semi-automatic delineations (segmentations) of the organs in dynamic MR images. To the best of our knowledge, methods dealing with multi-organ segmentation from dMRI acquisitions, especially of the thorax, do not exist. In general, MR images have lower spatial resolution compared to computed tomography (CT) images, and are prone to various artifacts and non-standardness, which are exacerbated in the case of dMRI, especially pediatric studies, all of which make multi-organ segmentation from dynamic MR images very challenging (see Figure 1). Therefore, the auto-segmentation system we present in this paper is novel.
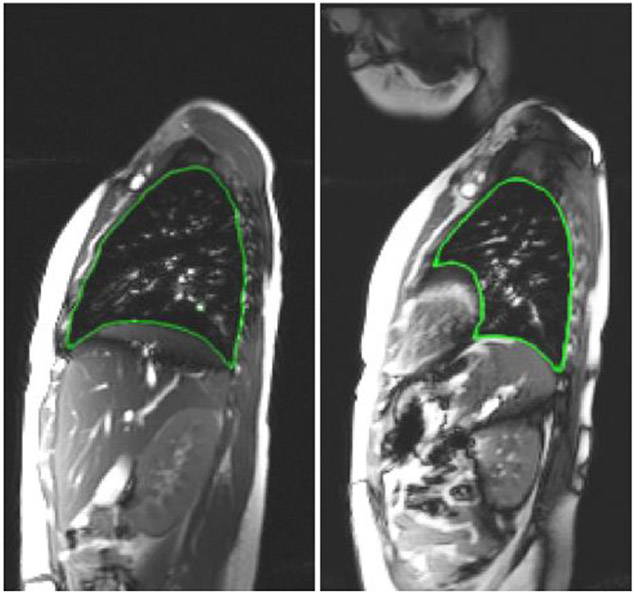
Figure 1.
Some dMRI slices with the true boundary delineated for right lung (left figure) and left lung (right figure).
Dynamic MRI acquisitions are inherently four dimensional with the dimensions being time and space in the three spatial dimensions. In our dMRI acquisition, a sagittal plane MR image is first acquired continuously for a specified duration (typically over 10 respiratory cycles), then the next sagittal slice is captured for the next specified duration, and so on until the image of the entire thoraco-abdominal region is fully captured. However, to segment these organs, we first perform a 4D construction of the body region image representing the dynamic body region over one respiratory cycle via an optical flux strategy [2] and then segment the 3D organs in the 3D image corresponding to any specified respiratory phase, such as the end-inspiration (EI) and end-expiration (EE) time points.
In this paper, we present a system for auto-segmentation of the right lung and left lung in pediatric dMRI by utilizing our extensive experience and the tools we had previously developed for segmenting organs body-wide in computed tomography (CT) images [3-6]. The contributions in this paper are as follows.
We present a novel and unique system for the problem of multi-organ segmentation from dynamic MRI acquisitions of the thoraco-abdominal region. This challenging segmentation problem has not been addressed in the literature.
We have compared several combinations of recognition and delineation approaches toward building a practical and accurate system to be employed in the setting of thoracic insufficiency syndrome (TIS).
2. METHODS
In this section, we give a brief overview of the 4 modules which operate sequentially. They are referred to as AAR-R, DL-R, AAR-R-DL-R, and DL-D in subsections 2.1, 2.2, 2.3, and 2.4, respectively. We perform segmentation in two steps: a recognition step and a delineation step. In recognition, we try to get a rough idea of the location of the object of interest in the unseen image with the help of bounding boxes. In delineation, the approach marks the outline of the object of interest within the aforesaid bounding box. The first three modules (AAR-R, DL-R, and AAR-R-DL-R) are recognition steps while the last module (DL-D) is a delineation step.
2.1. AAR-R [3]:
The first step in AAR-R is the model building step. AAR-R takes as input from the user the order in which multiple objects of interest should be recognized in an unseen image. This hierarchy of objects is given as a tree. Objects at the parent nodes are recognized before the objects at the child nodes of the tree.
An object at a child node can be recognized with additional information that uses the recognition of its parent object. If we know that additional information such as relative anatomical position to the parent object, shape, pose, and textural properties of the objects are consistent in the images of the seen dataset, then the additional information can be encoded as fuzzy measures in a model which can be used later on during recognition of the object in an unseen image. The model building step involves encoding these fuzzy measures in a model.
The next step in AAR-R is the recognition step where we try to localize objects of interest in the unseen image. In the recognition step, after the root object has been recognized in the unseen image from its textural properties, the location of its child in the unseen image is guessed from the relevant fuzzy measures in the model. This is continued recursively until the objects at the leaves of the tree are recognized. In the aforesaid manner, the hierarchy and the model are used by the AAR approach to localize multiple objects in an unseen image. The output of AAR-R is a fuzzy mask for the object of interest in the unseen image. This fuzzy mask gives an estimate of the shape, pose, and location of the object in the unseen image.
2.2. DL-R [4]:
The DL-R approach, a recognition step, was primarily targeted at detecting thin sparse objects like the esophagus as well as non-sparse objects like the heart. In this paper, we discuss those design aspects of DL-R that pertain to segmentation of non-sparse objects. The design of the DL-R approach can be broken down into three modules: backbone network, neck network, and head network. We discuss these modules briefly in the next paragraphs.
A sagittal slice of the approximated 3D view is converted into a 3 channel image by mapping the pixel intensity to a number in each of three pre-defined intensity intervals. This three-channel slice is taken as input to the backbone network. The backbone network uses pre-trained model weights of classical neural networks like the ResNet [7] and the DenseNet [8]. Four feature maps (C2, C3, C4, and C5) are taken from the last four convolutional layers of the backbone network. These convolutional layers use strides of 4, 8, 16, and 32 pixels, respectively. The map C2 captures lower level textural information compared to C3, C4, and C5. The map C5 captures higher level contextual information from the 3 channel input image. These feature maps (C2, C3, C4, and C5) are taken as input to the neck network.
The design of the neck network is based on the Path Aggregation Network (PAN) [9] and the Dual Attention Network (DAN) [10]. The PAN uses bottom-up connections, top-down connections, and lateral connections to merge feature maps (C2, C3, C4, and C5) into maps referred to by Q4, Q5, and Q6. In addition to the PAN, DAN is used to create prediction maps which contain the information dependency across the spatial dimensions and the channel dimensions of the maps Q4, Q5, and Q6. The maps Q4, Q5, Q6 and the prediction maps are taken as input to the head network.
The head network recognizes the non-sparse organs with the maps Q4, Q5, and Q6 by associating them with anchor sizes 32 x 32, 64 x 64, and 128 x 128, respectively. This recognition step gives a rough idea of where the organs may be located. The recognition is further refined with the help of convolutional layers in the head network that use the prediction maps and the anchors. The convolutional layers take into account high-level semantic information from the prediction maps and predict the category and location of the non-sparse objects in an unseen image in the form of bounding boxes and their corresponding labels. The output of DL-R is a bounding box in those sagittal slices which are identified to contain the objects of interest. These bounding boxes come from the head network.
2.3. AAR-R-DL-R [4, 5]:
Each of the above two recognition modules, the natural intelligence approach like AAR-R and the artificial intelligence approach like DL-R, have their own distinct advantages and disadvantages. The DL-R approach can use textural information to accurately discern the location of the object of interest in the unseen image, but fails to localize the object if the textural information of the object in the unseen image is different from the textural information of the object over all images of the seen dataset. The AAR-R approach uses high-level anatomical knowledge of the thoraco-abdominal organs to localize the objects in the unseen image. This design feature of the AAR-R is able to localize the organ robustly even if textural information from the unseen image cannot provide reliable insights into the image or artifacts corrupt the image. Yet, the drawback of AAR-R is that the fuzzy mask for the objects of interest is usually coarse, meaning that the mask is larger in size compared to the actual size of the organ.
The objective of AAR-R-DL-R approach is to include the recognition output of AAR-R and the recognition output of DL-R for the same objects in the same unseen image to get a better localization of the objects of interest. The shape and pose of an object can be captured with the fuzzy mask by AAR-R and the location of an object can be captured with the position of the bounding box of DL-R. This is accomplished by shifting the fuzzy mask of AAR-R for a slice such that the centroid of the fuzzy mask coincides with the centroid of the bounding box that is obtained by DL-R for the same slice. This shifting of the fuzzy mask is actually done after some smoothing of the positions of the bounding boxes by DL-R, across all the slices of a subject. The shifted (refined) fuzzy mask is taken along with the input image (the sagittal slice) to another neural network (i-DL-R) whose design is similar to DL-R. The only difference between the design of DL-R and i-DL-R is that the former takes only the sagittal slice (in the form of a 3 channel image) as input while the latter takes the sagittal slice as well as the refined fuzzy mask as input (in the form of 3+1=4 channel image). The output of i-DL-R (and AAR-R-DL-R) is a set of refined bounding boxes which localize the objects of interest in every sagittal slice of the unseen dynamic MRI image.
2.4. DL-D [6]:
This module utilizes a network called ABCNet [6] which was originally designed to delineate the different types of body tissues: subcutaneous adipose tissue, visceral adipose tissue, skeletal muscle tissue, and skeletal tissue from low dose CT images of the body torso. The design of ABCNet is similar to an encoder-decoder architecture.
All units in the design of ABCNet are derivatives of BasicConv, which is comprised of four modules in succession: concatenation, batch normalization, activation, and convolution. Bottleneck is a special case of BasicConv with a convolutional kernel of 1x1x1. The ABCNet employs the technique of adopting connections between a layer and each of its successive layers as done in DenseBlock [8]. There are four DenseBlocks used in the encoder-decoder architecture of ABCNet. The DenseBlock in direct connection to the input image extracts low level information while the DenseBlock higher up in the encoder architecture extracts higher-level contextual information from the input image. Each DenseBlock of ABCNet is composed of several layers referred by Dense Layers, which are themselves composed of Bottleneck and a BasicConv with a kernel size of 3x3x3 in succession. The bottleneck, because of its lower convolutional kernel size, keeps the number of parameters low and simultaneously acts as a feature extractor through the normalization and activation functions of its BasicConv architecture.
The ABCNet uses a Dice coefficient-based loss function for training its model and uses patch-based training. The patches are randomly selected from the images in the seen dataset. The output of ABCNet is the outline of the objects of interest in the unseen image, as obtained from the decoder of ABCNet. Unlike existing encoder-decoder architectures (DeepMedic, Dense V-Net, V-Net, and 3D U-Net) which have typically 12 or 31 layers and 1 million or 80 million parameters, ABCNet has 118 layers with only 1.4 million parameters. The usage of ABCNet is thus attractive because of its deeper architecture with a fewer number of parameters.
3. RESULTS AND DISCUSSION
We have utilized dMRI acquisitions of 95 normal pediatric subjects from the Children’s Hospital of Philadelphia (CHOP) for evaluation of our thoraco-abdominal organ segmentation algorithms on these images. These acquisitions have been approved for our study by the Institutional Review Board of CHOP along with a Health Insurance Portability and Accountability Act Waiver. These sagittal dMRI acquisitions have a pixel resolution of 1.46 mm in the plane of the slice and 6.00 mm in between adjacent sagittal slices. The dataset from CHOP was gradually increasing in size with time based on availability of pediatric subjects for dMRI acquisitions. Therefore, in some experiments we utilized only those images which were available at the time of carrying out the experiment and not all the 95 subjects.
In the first experiment, we compare the three recognition approaches: AAR-R, DL-R, and AAR-R-DL-R based on the location error (LE). The LE expresses the average of the distance between the 2D centroid of the bounding box given by the recognition approach for the object in a slice and the 2D centroid of the tight fitting bounding box around the ground truth of the object in the same slice, over all slices in the 3D image. Images of 60 subjects have been used for modelbuilding of AAR-R and for training the DL-R and i-DL-R networks. Images of 21 subjects have been used for testing the AAR-R, DL-R, and AAR-R-DL-R approaches. At the time of carrying out this experiment, we had 81 (=60+21) subjects (which later gradually increased in number with collection of dMRI acquisitions of pediatric subjects in CHOP, to 95 subjects). The results are shown in Table 1.
Table 1:
The average location error (Avg. LE) in mm and standard deviation (SD) of LE for the recognition of left lung and right lung by AAR-R (A), DL-R (D), and AAR-R-DL-R (A-D) methods.
| Organ | Left Lung | Right Lung | ||||
|---|---|---|---|---|---|---|
| Method | A | D | A-D | A | D | A-D |
| Avg. LE (mm) | 28.04 | 6.11 | 6.98 | 16.55 | 5.92 | 6.16 |
| SD (mm) | 10.78 | 3.19 | 6.04 | 7.98 | 2.99 | 2.46 |
We observe that the performance of the DL-R approach is the best for the recognition of the left lung and right lung. Because of the promising performance of DL-R, we have adopted DL-R as the recognition approach in a separate experiment which evaluates the performance of ABCNet, a delineation approach. The ABCNet uses the bounding boxes from the DL-R as input during delineation of a thoraco-abdominal organ in an unseen image.
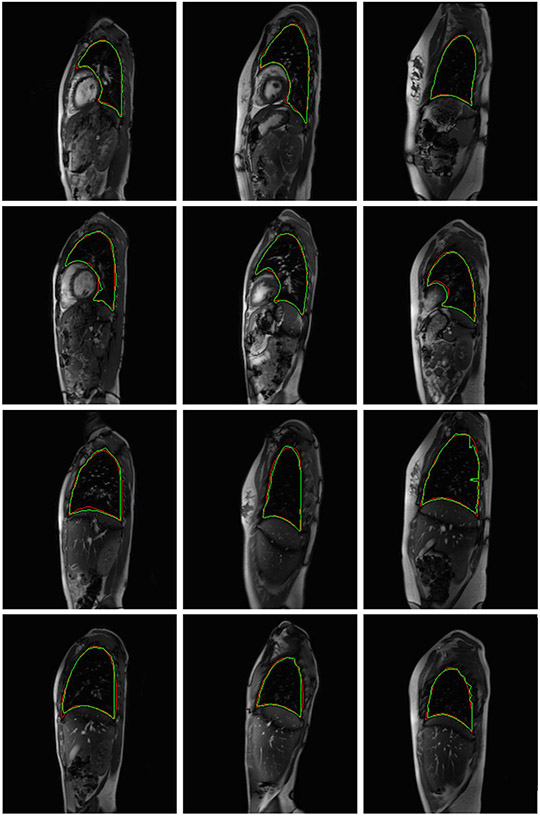
Seventy-two subjects were used in the training set, 10 subjects were used in the validation set, and 13 subjects were used for testing the ABCNet. In this experiment, we utilized the dMRI acquisitions of all the 95 (=72+10+13) subjects. The performance of ABCNet (DL-D) in terms of average (and standard deviation of) Dice coefficients is shown for the left lung and the right lung in Table 2. The results in the last column of Table 2 show excellent DC for the healthy subjects. In Figure 2, in the top (bottom) two rows, we have shown six sagittal slices where the delineations of the left (right) lung by the proposed auto-segmentation algorithm in green outline and the expert human tracer in red outline have been superimposed. No two sagittal slices in Figure 2 for the left lung (or right lung) belong to the same dMRI acquisition. We intend to adopt the DL-R approach for the recognition step and the ABCNet for the delineation step of the thoraco-abdominal organ segmentation from sagittal dMRI acquisition of patients with TIS.
Table 2:
The average Dice coefficients (Avg. DC) and the corresponding standard deviations (SD) for delineation of left lung and right lung by ABCNet.
| Organ | Left lung | Right lung |
|---|---|---|
| Avg. DC | 0.948 | 0.958 |
| SD | 0.021 | 0.008 |
Figure 2:
Twelve slices which depict the delineations of the left lung (top two rows) and right lung (bottom two rows) by the proposed auto-segmentation algorithm and the expert human tracer in green and red outlines respectively.
4. CONCLUSIONS
In this paper, we have developed an automated segmentation set up for the right lung and the left lung in dynamic MRI sagittal acquisitions of normal pediatric subjects. We have performed the segmentation in two steps: recognition and delineation. We have compared two artificial intelligence (DL-R and AAR-R-DL-R) modules and one natural intelligence module (AAR-R) for recognition. We have evaluated one artificial intelligence module (ABCNet) for delineation. We observe that (1) the simple artificial intelligence recognition approach (DL-R) performed the best compared to the simple natural intelligence recognition approach (AAR-R). The hybrid intelligence recognition approach (AAR-R-DL-R) for recognition showed competitive performance with respect to DL-R with no statistically significant difference (p=0.3645). (2) The delineation results for the lungs by ABCNet from sagittal dynamic MRI (dMRI) acquisitions of the thoraco-abdominal region of healthy subjects are excellent given the extreme difficulties of segmenting these objects in the dMRI images. We are further investigating this system for the segmentation of the thoraco-abdominal organs in sagittal dMRI acquisitions of patients with thoracic insufficiency syndrome (TIS).
Acknowledgements:
This research was supported by a grant from the National Institutes of Health R01HL150147.
References:
- [1].Tong Y, Udupa JK, Mcdonough JM, Wileyto EP, Capraro A, Wu C, Ho S, Galagedera N, Talwar D, Mayer OH, Torigian DA, Campbell RM. "Quantitative dynamic thoracic MRI: application to thoracic insufficiency syndrome." Radiology 292: 206–213, 2019, 10.1148/radiol.2019181731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hao Y, Udupa JK, Tong Y, Wu C, Li H, McDonough JM, Anari JB, Torigian DA, Cahill PJ. OFx: A method of 4D image construction from free-breathing non-gated MRI slice acquisitions of the thorax via optical flux. Medical Image Analysis, 72: 102088, 2021, 10.1016/j.media.2021.102088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Udupa JK, Odhner D, Zhao L, Tong Y, Matsumoto MMS, Ciesielski KC, Falcao AX, Vaideeswaran P, Ciesielski V, Saboury B, Mohammadianrasanani S, Sin S, Arens R, Torigian DA. "Body-wide hierarchical fuzzy modeling, recognition, and delineation of anatomy in medical images." Medical Image Analysis 18 (5):752–771, 2014, 10.1016/j.media.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jin C, Udupa JK, Zhao L, Tong Y, Odhner D, Pednekar G, Nag S, Lewis S, Poole N, Mannikeri S, Govindasamy S, Singh A, Camaratta J, Owens S, Torigian DA. "Object recognition in medical images via anatomy-guided deep learning." Medical Image Analysis 81, 2022, 10.1016/j.media.2022.102527. [DOI] [PubMed] [Google Scholar]
- [5].Udupa JK, Liu T, Jin C, Zhao L, Odhner D, Tong Y, Agrawal V, Pednekar GV, Nag S, Kotia T, Goodman M, Wileyto EP, Mihailidis D, Lukens JN, Berman AT, Stambaugh J, Lim T, Chowdary R, Jalluri D, Jabbour SK, Kim S, Reyhan M, Robinson C, Thorstad W, Choi JI, Press R, Simone CB, Camaratta J, Owens S, Torigian DA. “Combining natural and artificial intelligence for robust automatic anatomy segmentation: Application in neck and thorax auto-contouring for radiation therapy planning”. Medical Physics, accepted, 10.1002/mp.15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu T, Pan J, Torigian DA, Xu P, Miao Q, Tong Y, Udupa JK. "ABCNet: A new efficient 3D dense-structure network for segmentation and analysis of body tissue composition on body-torso-wide CT images." Medical Physics 47 (7): 2986–2999, 2020, 10.1002/mp.14141. [DOI] [PubMed] [Google Scholar]
- [7].He K, Zhang X, Ren S, Sun J. "Deep Residual Learning for Image Recognition." IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 770–778, 2016, 10.48550/arXiv.1512.03385. [DOI] [Google Scholar]
- [8].Huang G, Liu Z, Maaten LVD, Weinberger KQ. "Densely Connected Convolutional Networks." IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2261–2269, 2017, 10.48550/arXiv.1608.06993. [DOI] [Google Scholar]
- [9].Liu S, Qi L, Qin H, Shi J, Jia J. "Path Aggregation Network for Instance Segmentation." IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 8759–8768, 2018, 10.1109/CVPR.2018.00913 [DOI] [Google Scholar]
- [10].Fu J, Liu J, Tian H, Li Y, Bao Y, Fang Z, Lu H. "Dual attention network for scene segmentation." IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 3146–3154, 2019. 10.48550/arXiv.1809.02983. [DOI] [Google Scholar]