Abstract
The epithelial lining of the small intestine mediates its absorptive and secretory function and thus is a critical component of human health. Regeneration and renewal of the epithelium is the result of proliferation of intestinal stem cells (ISCs). Many cell types and molecular factors are known to regulate the ability of ISCs to proliferate, including adjacent neighboring epithelial cells and the underlying, supportive stromal cells. The microbiome resides in the lumen of the small intestine and is in close contact with the epithelium. Due to its proximity to ISCs, it has been hypothesized that species within the microbiome have the capacity to regulate ISC proliferation and differentiation. This review highlights research that probes interactions between ISCs and the microbiome in the small intestine to detail the current understanding of microbial regulation of ISCs. Results from these studies provide important knowledge that can be exploited to identify therapeutic targets or develop novel preventative treatments to treat intestinal diseases.
Introduction:
The small intestinal epithelium is a single layer of diverse types of secretory and absorptive cells that mediate intestinal function. The epithelium is divided into two microdomains or compartments: the villus and the crypt. The crypt region is characterized by invaginating pocket-like structures that are surrounded by the underlying stroma, while the villus region protrudes into the intestinal lumen to increase surface area for nutrient absorption. The crypt is home to intestinal stem cells (ISCs), that undergo two types of cell division. Symmetric division generates new ISCs for self-renewal, and asymmetric division gives rise to daughter cells that migrate from the crypt region into the villus region, where they undergo differentiation to functional secretory and absorptive cells. ISCs continuously proliferate and differentiate, replacing the mature cells that line the villus region every three to five days. The microenvironment of the small intestinal crypt where ISCs reside is termed the “ISC niche.” Components of the ISC niche include the surrounding differentiated epithelial cells that remain in the crypt instead of migrating to the villus, and the underlying stromal cells, a heterogenous group of cells that reside in the connective tissue including mesenchymal, immune, neuronal, and vascular cells. The epithelial and stromal components of the ISC niche serve as a well-known, major source of signals that modulate ISC proliferation and differentiation (Santos et al., 2018, Greicius and Virshup, 2019). Recently, the human microbiome has emerged as another source of factors that regulate the function of ISCs. The advent of the Human Microbiome Project resulted in a wealth of information regarding the putative microbial composition in the small intestine (Human Microbiome Project, 2012), and a recent focus on the functionality of these organisms has resulted in the discovery that the microbiome and ISCs have an intimate relationship (Peck et al., 2017). This review will discuss the regulation of ISCs, the current understanding of the landscape of the small intestinal microbiome and its products, and the evidence that the microbiome is a source of signals for ISCs and thereby a major component of the ISC niche. It is important to comprehend how ISCs interact with the small intestinal microbiota and are influenced by bacterial products, as this knowledge enhances our understanding of the factors that assist in maintaining epithelial homeostasis and contribute to renewal and repair in the context of small intestinal damage.
Regulation of the Intestinal Stem Cell:
ISCs in the small intestine undergo continuous rounds of cell division approximately every 24 hours (Potten, 1998). As they divide in the base of the crypt, undifferentiated daughter cells migrate upwards through the transit-amplifying (TA) zone near the crypt-villus axis, where they continue to divide as they progress upwards towards the villus. As the cells move up the villus, they differentiate into their respective functional cell types, including enteroendocrine cells, which secrete hormones, enterocytes, which absorb nutrients, tuft cells, which perform chemosensory functions, and goblet cells, which secrete mucus. The exception to this upwards movement of differentiating cells are Paneth cells, which reside in the base of the crypts, where they secrete anti-microbial factors and shape the intestinal microbial composition (Figure 1) (Clevers, 2013, van der Flier and Clevers, 2009, Barker et al., 2012, Tian et al., 2011, Bevins and Salzman, 2011, Metcalfe et al., 2014, Clevers and Bevins, 2013). Apart from Paneth cells and a few enteroendocrine and tuft cells that remain in the crypt region, differentiated daughter cells migrate to the tip of the villus, where they are eventually shed into the small intestinal lumen by a process called anoikus. As ISCs are primarily responsible for this process of generating new daughter cells that differentiate into the mature intestinal cell types, regulation of ISCs is tightly controlled to ensure normal epithelial homeostasis and renewal, and conversely, is dysregulated in many small intestinal diseases.
Figure 1: Landscape of the Intestinal Epithelium.
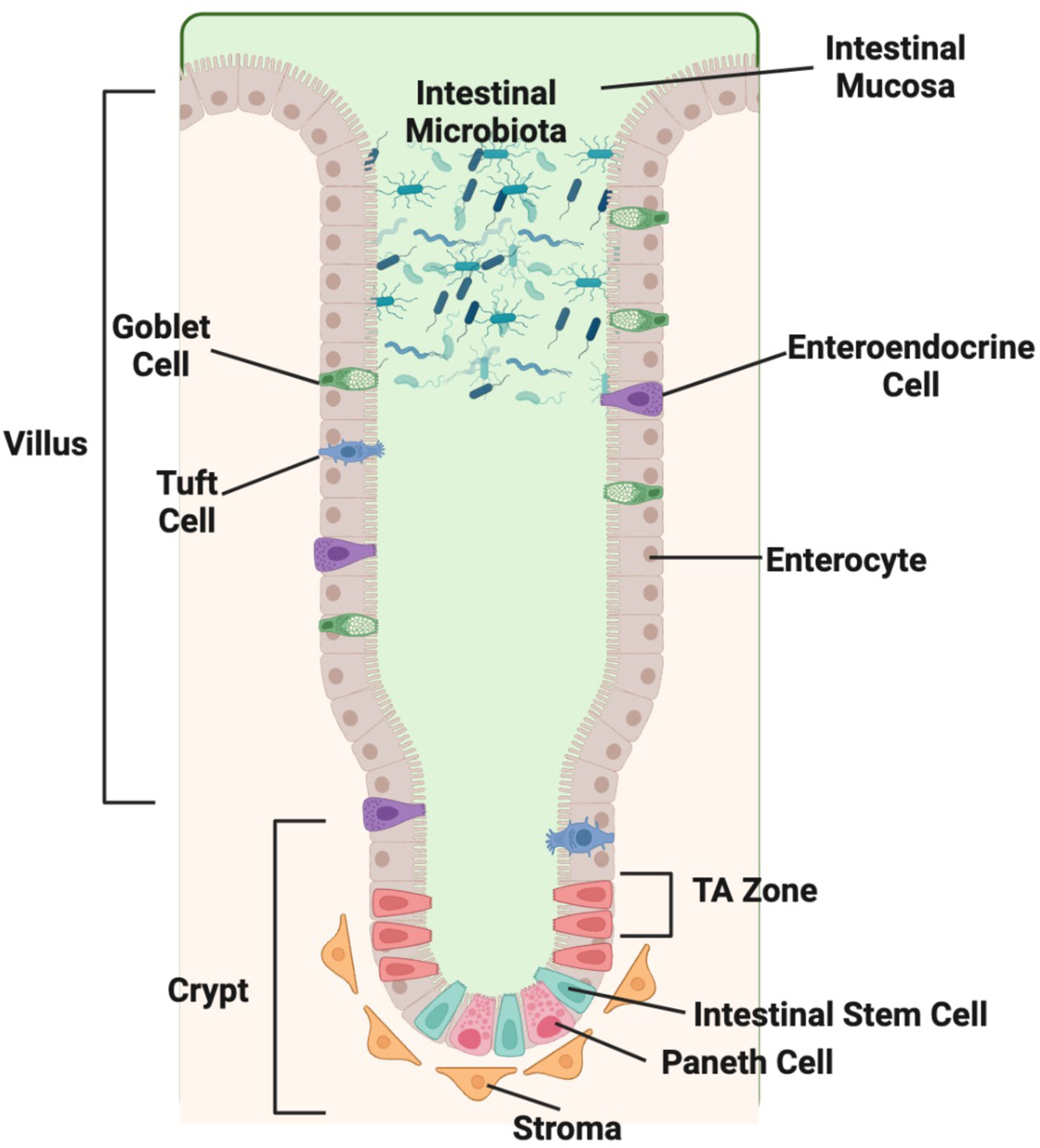
The intestinal epithelium is made up of diverse cell types organized into microdomains termed the crypt and villus. The microbiome lies adjacent to the intestinal epithelium in close contact with a mucous layer produced by the epithelium. The villus contains differentiated cells including enterocytes, goblet cells, enteroendocrine cells, and tuft cells. The crypt contains the undifferentiated intestinal stem cells (ISCs) and transient amplifying (TA) cells. The Paneth cells and few enteroendocrine and tuft cells reside alongside the ISC in the crypt. The stromal cells reside directly beneath the epithelium in the crypt region and provide essential regulatory factors to the ISC. Created in BioRender.
Many markers have been used to identify ISCs, facilitating the study of ISC regulation. Currently in the small intestine, the most well-studied crypt ISC marker is a cell surface receptor named LGR5 (leucine-rich-repeat-containing G-protein-coupled receptor 5, also known as Gpr49). Other markers that have been associated with ISCs include OLFM4, KI67, PCNA, SOX9, ASCL2, and AXIN2 (Barker et al., 2012, Burclaff et al., 2022). Postulated quiescent or reserved stem cells – also called +4 ISCs – reside directly above the terminal Paneth cells in the beginning of the TA zone, and are marked by BMI1, HOPX, TERT, and LRIG1 (Burclaff et al., 2022, Duckworth, 2021, Yan et al., 2012). The existence of quiescent ISCs is controversial, and unlike the actively dividing LGR5+ ISCs, quiescent ISCs are not thought to be the primary source of epithelial renewal in homeostatic conditions, but are rather believed to play a major role in regenerative repair in the context of LGR5+ ISC damage, especially following radiation exposure (Andersson-Rolf et al., 2017, Metcalfe et al., 2014, Dheer and Young, 2021). Control of LGR5+ ISC proliferation and differentiation in the small intestine has been linked to conserved signaling molecules and pathways, including wingless/integrated (WNT), R-spondin, epidermal growth factor (EGF), Hedgehog, Notch, Gremlin, and bone morphogenetic proteins (BMPs) (Vanuytsel et al., 2013). Key factors in these pathways are provided to ISCs by various cells that comprise the ISC niche (Duckworth, 2021, Greicius and Virshup, 2019, Santos et al., 2018), namely Paneth cells, which reside in between the actively dividing LGR5+ ISCs (Figure 1) and provide WNT and Notch ligands to ISCs (Clevers and Bevins, 2013), and stromal cells, such as telocytes and myofibroblasts, which reside underneath the epithelium and serve as a source of WNTs, R-spondin proteins, and EGF (Greicius et al., 2018, Greicius and Virshup, 2019, Gregorieff et al., 2005, Kabiri et al., 2014). The canonical WNT pathway is the most well-established regulator of ISCs (Barker et al., 2008, Barker et al., 2007, Barker et al., 2012, Logan and Nusse, 2004, Katoh, 2007, Katoh and Katoh, 2007), and WNT proteins secreted from stromal or Paneth cells locally activate intracellular signaling cascades within neighboring ISCs in a paracrine fashion (Sato et al., 2011, Greicius and Virshup, 2019, Greicius et al., 2018, Kabiri et al., 2014, Katoh, 2007, Katoh and Katoh, 2007, Theodosiou and Tabin, 2003). WNT target genes play an indispensable role in maintaining stemness and promoting proliferation in the small intestine, and WNT signaling thereby serves as the master regulator of proliferation in the ISC niche (Logan and Nusse, 2004, Katoh and Katoh, 2007, Biechele and Moon, 2008). In addition to their role in small intestinal homeostasis, many of these proliferation-promoting factors, such as WNT, have been shown to play a vital role in ISC recovery after damage (Gehart and Clevers, 2019, Santos et al., 2018, Greicius and Virshup, 2019, Duckworth, 2021).
The Microbiome:
The microbiome consists of more than 300 trillion bacteria, fungi, and viruses (Dave et al., 2012, Human Microbiome Project, 2012) that colonize the gastrointestinal tract. The small intestinal organisms are dynamic due to the changing conditions in the small intestine caused by intact of food, the intermittent secretion of enzymes and factors, and short transit time(Kastl et al., 2020). The composition of organisms varies by region with increasing amount and diversity from the proximal to distal portions (Kastl et al., 2020). Interactions between these organisms and ISCs could occur through two possible points of contact: direct communication between the organism and ISCs or communication with a niche cell that then indirectly signals ISCs (Peck et al., 2017, Savage and Blumershine, 1974, Nelson and Mata, 1970). Although tantalizing data suggests small intestinal microbes play a role in regulating ISCs, the underlying mechanisms that form the basis for microbiome-ISC interactions have not been elucidated. Filling this knowledge gap has been challenging, partly due to the lack of in vitro models in which microbe-epithelium interactions can be interrogated, the diversity of the gut microbiome and factors it produces, and the difficulty in pinpointing exactly what organisms are present in the small intestine. Most of the studies investigating small intestinal microbe-ISC interactions have utilized murine models. Although much has been learned from these studies, the composition of the murine microbiome is distinct from humans due to many factors, including host genetics, differences in the composition of outer intestinal mucosa layer that affects mucus-associated bacteria survival, lower intestinal pH values, and differences in diet (Arrieta et al., 2016, Hugenholtz and de Vos, 2018). Humans and mice share only 15% of intestinal bacteria lineages, and while murine models can provide valuable insights into ISC-microbiome interactions, the many differences between the human and mouse intestinal microbiomes highlight the need to study these interactions in a complimentary human model system (Arrieta et al., 2016). Human intestinal organoids provide a new model system in which human microbe-ISC interactions can be interrogated ex vivo, as they enable the culturing of human ISCs, model differentiation of the human epithelium, and facilitate the exploration of microbial-ISC interactions in a reductionist environment (Blutt et al., 2018, Foulke-Abel et al., 2014, Zachos et al., 2016). New advances in human organoid cultures will be addressed below.
The Landscape of the Intestinal Microbiome and Its Metabolites:
The composition of the small intestinal microbiome is highly variable based on the genetics of the individual and environmental factors such as age, diet, and antibiotic use (Hasan and Yang, 2019, Kurilshikov et al., 2021). The microbiome produces many factors that appear to modulate its functional role in the intestine, including short chain fatty acids (Boffa et al., 1992, Ichikawa and Sakata, 1997, Lee et al., 2017, Lee et al., 2018), tryptophan metabolites (Roager and Licht, 2018), polyamines, (Wang et al., 1991), secondary bile acids (Kozoni et al., 2000, Pai et al., 2004), vitamins (Lai et al., 2021), reactive oxygen species (Morris and Jasper, 2021), and hydrogen sulfide (Xing et al., 2020). Most of the research characterizing the overall intestinal microbiome has utilized 16S gene content analysis in stool, which is easily collected via non-invasive methods (Dave et al., 2012, Tang et al., 2020, Human Microbiome Project, 2012) but reveals very little about what might be present in the small intestine. Recent studies have revealed differences between the composition of the stool microbiome compared to the microbiome found at the intestinal epithelial surface (Vuik et al., 2019, Vasapolli et al., 2019). Differences in bacterial composition also exist between the luminal microbiota and the microbiota proximal to the epithelium, potentially because epithelial-associated organisms have unique properties allowing them to utilize nutrients and adhere to glycans within the mucous layer that coats the epithelial surface (Robbe et al., 2004, Juge, 2012). The composition of the microbiome also varies by region in the intestine and increases in diversity from the small intestine to the large intestine, with a smaller bacterial load and less-diverse microbiota associated within the upper duodenal regions of the small intestine and the largest numbers and greatest diversity of bacteria found in the terminal large intestine (Martinez-Guryn et al., 2019, Zhang et al., 2014). The diversity of the bacterial communities along the different regions of the intestine also contributes to the spectrum of products produced by microbes in each intestinal region, thereby affecting the local interactions of microbes and their metabolites with the epithelium (Wang et al., 2005, Stearns et al., 2011).
While researchers are making strides in gaining a deeper understanding of which bacteria inhabit the upper and lower regions of the human gastrointestinal tract, the composition of the human microbiota that colonizes the middle portions of the small intestine – the jejunum and upper ileum – has remained elusive (Dave et al., 2012, Tang et al., 2020, Kastl et al., 2020). Difficulties in assessing the microbiota in this region is mainly a result of technical challenges in sampling these regions through traditional endoscopy and colonoscopy methods, as the middle regions of the small intestine are not easily reached by scope equipment. Additionally, utilizing animal models to make these discoveries is limited by the substantial variations in microbial composition between animals and humans. Advancements in identifying which bacteria reside in the small intestine may be achieved by examining small intestinal tissues obtained through organ donation. By scraping the intestinal tissue, organ donor samples can be used to isolate microbes found in these regions from healthy individuals (Sartor, 2015).
Role of the Microbiome on Intestinal Stem Cell Regulation:
The idea that small intestinal microbiome plays a role in modulating ISC activity results from its proximity to the epithelium and ability to secrete factors that can modulate ISC biology. Indirect evidence for microbial regulation of ISCs originated from observations by Gordon and Bruckner-Kardoss in 1961 that germ-free mice showed reduced intestinal surface area compared to that of conventionally raised mice (Gordon and Bruckner-Kardoss, 1961). Many other studies have observed the phenomena of reduced villus height, crypt depth, and mitotic indices in the small intestine of germ-free or antibiotic-treated mice when compared to mice with a unaltered microbiome (Lesher et al., 1964, Greig et al., 2018, Khoury et al., 1969), suggesting that the microbiome can control epithelial regeneration and renewal. Further support for a link between the microbiome and ISCs comes from reports that colonization with microbes restores normal intestinal epithelial histology in germ-free mice, rats, and Drosophila (Gordon and Bruckner-Kardoss, 1961, Buchon et al., 2009a, Banasaz et al., 2002). Evidence for direct interactions between microbes and ISCs has also emerged from studies by Lee et al. where microbially produced lactate was shown to enhance proliferation in the murine small intestine via the stimulation of the LGR5 receptor on ISCs (Lee et al., 2018). It remains to be seen whether this activation of LGR5 ISCs occurs because microbially derived lactate interacts directly with ISCs or indirectly via the stimulation of other cell types of the ISC niche (Table 1). Although an in-depth mechanism of how the microbiome regulates ISCs remains to be fully elucidated, evidence from the literature suggests that a complex network of microbes and their metabolites are clearly involved in regulating ISC activity through various niche pathways, in the context of both homeostasis and damage.
Table 1.
Summary of the known effects of microbial species and strains on ISC regulation.
| Microbe / Strain | Model | Effects on ISCs | Mechanisms | Reference | |
|---|---|---|---|---|---|
| Akkermansia muciniphila | |||||
| ATCC BAA-835 and AK32 | Normal mouse, radiation and methotrexate mouse damage model | Stimulates proliferation of Lgr5+ ISCs, promotes differentiation to Paneth cells and goblet cells, promotes gut repair following damage | Activates Wnt/β-catenin pathway by promoting Wnt3 secretion | Kim et al. (2021), Duan et al. (2023) | |
| Bacillus subtilis | |||||
| JNFE0126 | DSS-induced mouse colitis model | Stimulates proliferation, increases Lgr5+ ISC numbers | Protects ISCs from inflammatory injury; rebalances intestinal microbiota | Zhang et al. (2020) | |
| Bacteroides fragilis | |||||
| ZY-312 | DSS-induced mouse colitis model | Promotes colonic proliferation | Promotes colonic mucosal regeneration in colitis via IL-22-induced STAT3 phosphorylation | Zhang et al. (2023) | |
| 086-5443-2-2 | Human colonic epithelial HT29/C1 cells | Promotes proliferation and c-myc expression | Activates β-catenin/TCF signaling pathway via | Wu et al. (2003) | |
| Erwinia carotovora carotovora 15 | |||||
| Ecc15 | Ecc15-infected Drosophila model | Stimulates ISC proliferation and differentiation to enteroendocrine cells | JAK-STAT pathway induction of ISC proliferation via Upd3 release from enterocytes | Buchon et al. (2009b), Liu et al. (2022) | |
| Lactobacillus acidophilus | |||||
| ATCC4356 | Salmonella typhimurium-infected mouse intestinal organoids | Inhibits proliferation, inhibits excessive differentiation of Paneth cells and goblet cells | Inhibits overactivation of Wnt/β-catenin pathway | Lu et al. (2020) | |
| Lactobacillus casei | |||||
| ATCC334 | Radiation-induced rat injury model | Stimulates proliferation, increases Lgr5+ ISC numbers | Promotes proliferation via alpha-linolenic acid production and IL-22 | Hua et al. (2023) | |
| ATCC334 | Normal mouse and porcine epithelial cells | Stimulates epithelial cell proliferation | Induces epithelial expression of Reg3a to inhibit pathogens and stimulate damage repair | Bai et al. (2021) | |
| Lactobacillus plantarum | |||||
| Normal Drosophila | Stimulates ISC proliferation | Nox1-dependant reactive oxygen species generation | Jones et al. (2013) | ||
| Lactobacillus reuteri | |||||
| D8 | C. rodentium-infected mouse, DSS-induced mouse colitis model, TNF-damaged mouse organoids | Stimulates proliferation, induces differentiation of Paneth cells, increases Lgr5+ ISC numbers | Induces R-spondin expression and activates Wnt/β-catenin pathway; | Wu et al. (2020) | |
| D8 | Stimulates lamina propria lymphocyte secretion of IL-22 and STAT3 signaling pathway | Hou et al. (2018) | |||
| 17938 | Normal neonatal mice | Stimulates proliferation | Increases microbial phylogenetic diversity | Preidis et al. (2012) | |
| Lactobacillus rhamnosus | |||||
| GG | Normal mouse colonic organoids, normal mouse | Stimulates colonic epithelial proliferation | Microbe-induced leptin expression triggers JAK-STAT signaling in a nox1 and leptin receptor-dependent manner | Darby et al. (2020), Jones et al. (2013) | |
| GG | Normal mouse | Stimulates ISC proliferation | Nox1-dependant reactive oxygen species generation | Lyu et al. (2022) | |
| GG | Intrauterine growth retardation rat pups | Stimulates proliferation and goblet cell differentiation | Stimulates WNT/β-catenin signaling | Banasaz et al. (2002) | |
Many species and strains of intestinal bacteria have been implicated in affecting ISC proliferation and differentiation (Table 1). Due to the amount of genetic variability between strains of a bacterial species – especially in those that were isolated from different hosts (Frese et al., 2011) – it is probable that many of the observed effects of a given microbe are strain-specific. For example, in mice, Lactobacillus reuteri strain 17938 increases murine small intestinal crypt cell proliferation, but strain 6475 does not (Preidis et al., 2012). Comparative genomics reveals that although these strains are members of the same species, they share only 70% of their genes (Saulnier et al., 2011). As these strains differ in their ability to induce crypt cell proliferation, these genetic differences may code for factors that modulate ISCs, potentially providing key targets for further exploring the relationship between ISCs and the microbiome. Strain-specific differences are particularly relevant when drawing conclusions from studies linking specific microbial metabolites or factors from a microbial species to ISC regulation, as not all strains of a species may produce the same factor.
Many different mechanisms have been proposed to explain how microbes regulate ISCs (Table 1 and Table 2), suggesting that microbial regulation of ISCs is most likely complex and multi-factorial, involving multiple pathways and mechanisms. Of the major niche pathways, the mechanism often proposed for microbial stimulation of ISC proliferation is the activation of WNT/β-catenin signaling (Kim et al., 2021, Wu et al., 2020, Lyu et al., 2022), which is unsurprising, as this pathway is the major proliferation-stimulating pathway in LGR5+ ISCs, and without proper WNT/β-catenin signaling, the intestinal epithelium is unable to self-renew (Korinek et al., 1998, Pinto et al., 2003, Kuhnert et al., 2004). Whether microbes induce these pathways directly, with microbial cells themselves interacting with the epithelial surface, or indirectly, via microbial metabolites, secreted proteins, or outer membrane vesicles remains poorly understood. Due to the presence of the mucus barrier that overlays the intestinal epithelium in the absence of injury or disease, many studies have concluded that microbes are unable to physically interact with the epithelium, hypothesizing that the ISC niche is a sterile environment, free from microbes themselves (Hansson and Johansson, 2010, Johansson et al., 2008, Johansson et al., 2014). However, the crypt location would be ideal for microbial modulation of ISC pathways through direct microbial communication with the various ISC niche components—potentially through interaction of microbial cell wall components with the epithelium or through the release of outer membrane vesicles or metabolites that can locally interact with the niche—yet whether these direct interactions occur remains unknown.
Table 2.
Summary of the known effects of microbial metabolites and factors on ISC regulation.
| Microbial factor | Description | Effects on ISCs | Mechanisms | Reference |
|---|---|---|---|---|
| Butyrate | SCFA | Stimulates proliferation | Stimulates MEK-ERK signaling | Park et al. (2016) |
| Stimulates proliferation | Unspecified | Boffa et al. (1992) | ||
| Inhibits proliferation | Unspecified | Kaiko et al. (2016) | ||
| Deoxycholic acid | Secondary bile acid | Inhibits proliferation | Possible FXR inactivation of EGFR signaling | Dossa et al. (2016) |
| Flagellin | Flagella component | Stimulates proliferation | Upregulation of NOX1 and stimulation of EGFR signaling | van der Post et al. (2021) |
| Indole-3-aldehyde | Tryptophan metabolite | Promotes proliferation and Paneth cell differentiation | Stimulation of IL-22 secretion and STAT3-dependent proliferation stimulation | Hou et al. (2018) |
| Promotes proliferation and goblet cell differentiation | Promotes IL-10 signaling | Powell et al. (2020) | ||
| Indoleacetic acid | Tryptophan metabolite | Inhibits proliferation | Aryl hydrocarbon receptor (AhR) E3 ubiquitin ligase degradation of β-catenin | Kawajiri et al. (2009) |
| Lactate | Organic acid | Stimulates proliferation, enhances survival of Lgr5+ ISCs | Gpr81-dependent Wnt3 expression and WNT/β-catenin pathway stimulation | Lee et al. (2018) |
| Lipopolysaccharide | Gram-negative bacterial cell membrane | Inhibits proliferation, promotes cell death, promotes goblet cell differentiation | TLR4-induced apoptosis | Naito et al. (2017) |
| Muramyl-dipeptide | Peptidoglycan (cell wall) motif | Stimulates proliferation, enhances survival of Lgr5+ ISCs | Nod2-mediated cytoprotection | Levy et al. (2020), Nigro et al. (2014) |
| Propionate | SCFA | Stimulates proliferation, increases ISC stemness, increased LGR5+ cells | Stimulates MEK-ERK signaling and WNT signaling | Park et al. (2016), Duan et al. (2023) |
| Succinic acid | Organic acid | Inhibits proliferation | Unspecified | Inagaki et al. (2007) |
| Valproic acid | SCFA | Stimulates proliferation, suppresses secretory lineage differentiation | Stimulates Notch pathway | Yin et al. (2014) |
| Stimulates proliferation | Stimulates MEK-ERK signaling | Park et al. (2016) |
In addition to WNT/β-catenin signaling, other pathways of microbial regulation of ISCs have also been suggested. ISCs robustly express the pattern recognition receptor Nod2, which recognizes the peptidoglycan motif muramyl dipeptide (MDP) that is a cell wall component in all bacteria (Nigro et al., 2014, Ogura et al., 2003). This finding serves as a clue that microbial cells may be able to physically interact directly with ISCs. MDP has been linked to beneficial effects in epithelial repair and has recently emerged as a microbial-responsive mechanism that aids in ISC survival in murine and organoid studies. Nod2 has been shown to trigger a pathway of ISC cytoprotection when stimulated by MDP; a mechanism thought to enhance the ability of ISCs to regenerate crypts upon exposure to cytotoxic stressors such as reactive oxygen species, radiation damage, or the chemotherapeutic agent doxorubicin (Nigro et al., 2014, Levy et al., 2020, Lee et al., 2019).
Many microbially derived metabolites and secreted products are thought to provide important components to the ISC niche (Table 2). Recognition that small intestinal microbial products such as MDP and lactate can maintain the proliferative capacity of the ISC niche in the context of damage highlights the importance of maintaining a diverse, healthy microbiome. Several strains promote proliferation and facilitate epithelial renewal following damage, as seen in DSS-induced mouse colitis models, radiation-induced rat injury models, and during pathogenic infection (Wu et al., 2020, Zhang et al., 2020, Zhang et al., 2023, Lu et al., 2020, Hua et al., 2023) (Table 1). These findings demonstrate that microbes play an important role in repairing the small intestinal epithelium following damage. Commensal microbes, such as Lactobacillus reuteri D8, can stimulate epithelial proliferation and repair to reduce intestinal pro-inflammatory cytokine secretion and serum LPS concentrations (Wu et al., 2020). Additionally, in the context of damage due to inflammation or radiation injury, commensal microbes – especially lactobacilli – have been documented to play a very important role in facilitating small intestinal epithelial renewal by stimulating ISC proliferation via IL-22 secretion from immune cells (Zhang et al., 2023, Hua et al., 2023, Qiu et al., 2017, Ge et al., 2022, Hamade et al., 2022, Hou et al., 2018). In contrast, other evidence suggests that certain microbes might enhance damage. The microbial endotoxin lipopolysaccharide (LPS), which is often present in higher amounts during pathogenic infection, has been shown to inhibit cellular proliferation and increase apoptosis in bot the small intestine upon binding to the toll-like receptor 4 (TLR4) (Naito et al., 2017, Neal et al., 2012). Further work is necessary to understand both the positive and detrimental effects of the small intestinal microbial composition on ISC regulation.
Ways to Study Microbial Regulation of Intestinal Stem Cells:
Most of the research studying the pro-proliferative effects of various microbes on the ISC has been conducted in murine models. However, it has not yet been ascertained how applicable these discoveries in murine systems are to human biology. The expanded use of human intestinal organoid model systems provides the opportunity to overcome the hurdles of host and species-specific differences. In 2011, Sato et al. pioneered the development of ex vivo tissue-derived human small intestinal organoids (HIOs), which allow the direct cultivation of human ISCs in vitro (Sato et al., 2011). Unlike transformed cell lines, HIOs are genetically stable and closely model the cellular makeup of the in vivo intestinal epithelium (Blutt et al., 2018, Foulke-Abel et al., 2014, Sato et al., 2011, Sato et al., 2009), which presents many advantages. HIOs can be grown in multiple formats, facilitating the study of many scientific questions involving microbe-epithelial interactions (Figure 2). In a 3D format, bacteria can be microinjected into the lumen of the HIO to assess how live microbes affect the rate of organoid growth and ISC proliferation (Poletti et al., 2021). Williamson et al. recently developed a high-throughput method of 3D organoid microinjection to evaluate how various microbes influenced gastrointestinal physiology (Williamson et al., 2018). Additional studies in 3D HIOs have analyzed the impacts of microbes or their secreted products on various aspects of epithelial biology (Co et al., 2019, Dheer and Young, 2021). HIOs can also be grown in a 2D monolayer or transwell format, which allows easy access to the epithelial apical surface for the application of microbes or their products and more closely mimics the physiologic contact of the epithelium and the intestinal microbiome (VanDussen et al., 2015, Wilke et al., 2020). Transwells also facilitate access to the basolateral side of the epithelium to address microbial-cell interactions that may occur in this region and model systemic infections (Wang et al., 2018). Work in organoid models has allowed researchers to determine the colonization patterns of microbes and how microbial factors affect the apical surface of the epithelial cells (Rajan et al., 2018, Zhang et al., 2020). As HIOs have the advantage of preserving the host’s genetic makeup in vitro, microbial-HIO studies can encompass multiple individuals with a variety of genetic backgrounds, permitting the assessment of demographic factors such as sex, ethnicity, and age to be studied in the context of the microbiome. The advent of HIOs presents a powerful tool that can be used to gain a comprehensive understanding of the intricate interplay between the human small intestinal microbiome and ISCs. To circumvent disparities in the microbiota between mice and humans, future research endeavors should supplement rodent models with the integration of microbiome components into HIOs.
Figure 2: Formats of Human Intestinal Organoids.
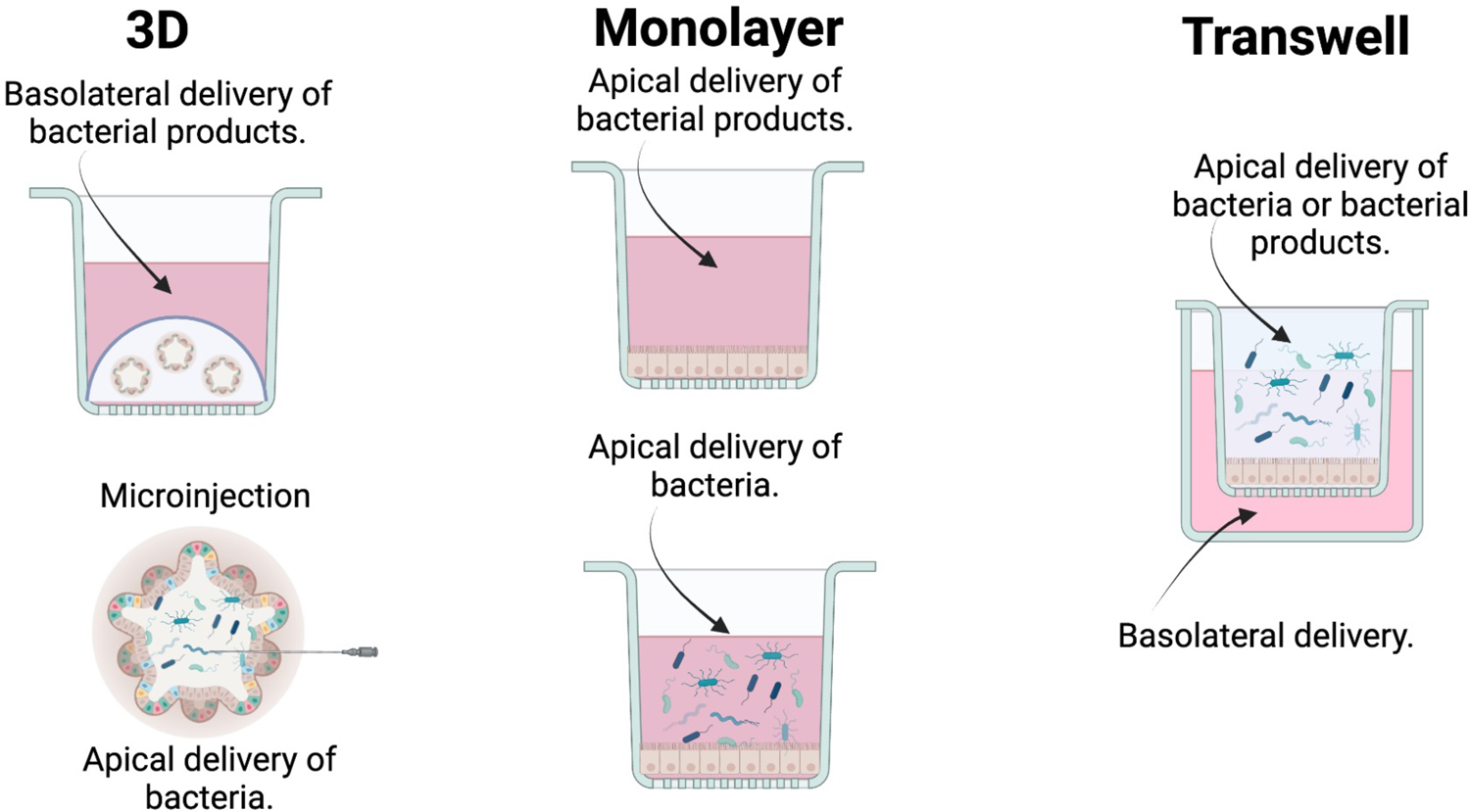
Human intestinal organoids (HIOs) can be used in a variety of formats depending on the scientific questions to be explored. Created in BioRender.
Additional methods to study complex communities of bacteria are being developed that, when combined with organoid technology, can be used to assess host-microbe and ISC-microbe interactions. Robert Britton’s laboratory pioneered the development of mini-bioreactor arrays (MBRAs) that can be utilized to cultivate complex human small intestinal microbial communities and metabolites (Auchtung et al., 2015). Using the MBRA system, stable microbial communities can be reliably and reproducibly cultured within eight days, with approximately 94% of the microbes present from the original sample represented in the MBRA community (Auchtung et al., 2015). Supernatants from these communities can be collected and their biological effects on ISCs assessed using several different assays. Other laboratories are actively expanding the repertoire of culturable bacteria; an advancement that, when paired with HIO studies, will substantially contribute to the scientific community’s ability to uncover the mechanistic relationship between complex communities of microbes and ISCs (Wang et al., 2018, Rettedal et al., 2014, Kim et al., 2016, Kim et al., 2022, Afrizal et al., 2022).
Microbial Regulation of the Large Intestine Stem Cell:
Unlike the small intestinal epithelial landscape, which has a crypt and villus architecture, the colonic epithelial landscape is much different, and only consists of deep glandular crypt regions. The colon is home to the greatest and most diverse population of microbes in the body; therefore, it is also important to understand how the resident bacteria in the colon may be impacting the regeneration processes of the colonic stem cell. Due to the ease of sampling and characterizing the microbiome in the large intestine, there is a plethora of research examining whether large intestinal microbial communities modulate ISCs. Driving this work is the association of colorectal cancer and the microbial composition in the large intestine. Large intestinal microbial dysbiosis strongly correlates with the development of cancer in the large intestine(Kim and Lee, 2021) and there is much focus on understanding the effect of the large intestinal microbiome on epithelial proliferation.
Several groups have found that various large intestinal microbes and microbial products can influence ISC proliferation in the colon including Bacteroides fragilis, and Lactobacillus rhamnosus GG (Wu et al., 2003, Zhang et al., 2023, Darby et al., 2020). Colonic organisms have been postulated to interact with the colonic ISC via several different mechanisms, including a toxin-mediated destruction of epithelial E-cadherin which subsequently triggers an epithelial repair response characterized by β-catenin translocation to the nucleus, a hallmark of ISC activation(Wu et al., 2003). Other mechanisms include activation of STAT2 signaling via the induction of IL-22 (Zhang et al., 2023), leptin mediated induction of epithelial proliferation, and proliferation via pathways that involve Nox (Darby et al., 2020). Most of the work linking specific large intestinal microbial metabolites to ISC regulation involve short-chain fatty acids (SCFAs), such as butyrate, acetate, propionate, and valproic acid, which are produced by microbial fermentation of dietary polysaccharides and are found primarily in the colon. The most well-studied SCFA is butyrate, which serves as a direct energy source for colonocytes and has been shown to modulate inflammation (Lee et al., 2017, Donohoe et al., 2011, Dou et al., 2020, Chang et al., 2014). Butyrate can stimulate proliferation in healthy colonic epithelium but inhibits proliferation in tumor cell lines (Whitehead et al., 1986, Sakata, 1987, Kien et al., 2007, Frankel et al., 1994), a phenomenon known as the “butyrate paradox” (Mariadason et al., 2001, Comalada et al., 2006). Although detailed mechanisms explaining how colonic microbial metabolites regulate ISCs have not been fully elucidated, evidence from the literature suggests that many microbial metabolites can regulate epithelial proliferation. With the high prevalence of colonic cancers worldwide (Boustany et al., 2023), it is important to understand the relationship between resident colonic microbiota and their influences on epithelial proliferation. Recent work has identified a crypt-specific microbiota that resides deep within murine colonic crypts (Pedron et al., 2012, Saffarian et al., 2019) which implies local interactions between the two entities. Further research is needed to fully understand and appreciate whether organisms that live in the large intestinal crypt region provide local signals that regulate ISCs.
Conclusion and Discussion:
The intestinal lumen is a highly dynamic and changing environment that is home to trillions of bacteria that constitute the intestinal microbiome, a complex ecosystem of organisms that live in symbiosis with the intestinal epithelium. Epithelial maintenance is critical for intestinal health and originates from ISCs. The relationship between the intestinal microbiota and ISCs is highly dynamic and understanding its complexity is still in its infancy. Rodent, zebrafish, and Drosophila model systems provide solid evidence that the microbiome plays an important regulatory role in ISC functioning. In addition to functioning under homeostasis, the microbiome has also been linked to enhanced intestinal repair mechanisms that occur following injury. A deep focus on the role of the microbiome in regeneration and renewal following intestinal damage will begin to shed light on the utilization and manipulation of the microbiome to treat intestinal disease. Collectively, this area of research implies that the intestinal microbiome holds tremendous potential as a therapeutic target. In addition, the microbiome can be regulated by many external and internal factors including dietary factors (Perler et al., 2023) such as fiber (Myhrstad et al., 2020) and sugars (Di Rienzi and Britton, 2020), prebiotic (Bedu-Ferrari et al., 2022), probiotic (Hemarajata and Versalovic, 2013) and antibiotic use (Fishbein et al., 2023), stress (Segerberg-Konttinen, 1988), physical activity (Holzhausen et al., 2022), sleep (Klimashina et al., 1989), aging (Badal et al., 2020), genetics (Hall et al., 2017), and disease (Durack and Lynch, 2019). Understanding how these variables indirectly affect ISC dynamics and whether they have the potential to modulate intestinal regeneration and repair via their effects on the microbiome is an emerging area of research. It is tantalizing to speculate as to whether simple interventions such a modulation of diet might modulate ISC renewal in humans, as this has been demonstrated in mice (Hou et al., 2021). Deeper knowledge of microbiome-ISC interactions will provide an understanding of the potential application of the microbiome to human health. We predict that the field will continue to advance with the use of new model systems to fully understand how the microbiome “vibes” with ISCs in humans and further define the human ISC niche.
Acknowledgments:
This work was supported by the National Institute of Health grants U01- DK103168, and U19- AI116497, and the Translational Research Institute for Space Health grant -. NNX16AO69A. We are grateful for the support and guidance from Dr. Mary K. Estes in the completion of this manuscript for providing feedback and editing the manuscript. Images were created using BioRender.
Footnotes
Conflict of Interest:
The authors certify they have no conflicts of interest to declare. All authors agree with the content of this manuscript.
References:
- AFRIZAL A, HITCH TCA, VIEHOF A, TREICHEL N, RIEDEL T, ABT B, BUHL EM, KOHLHEYER D, OVERMANN J & CLAVEL T 2022. Anaerobic single-cell dispensing facilitates the cultivation of human gut bacteria. Environ Microbiol, 24, 3861–3881. [DOI] [PubMed] [Google Scholar]
- ANDERSSON-ROLF A, ZILBAUER M, KOO BK & CLEVERS H 2017. Stem Cells in Repair of Gastrointestinal Epithelia. Physiology (Bethesda), 32, 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARRIETA MC, WALTER J & FINLAY BB 2016. Human Microbiota-Associated Mice: A Model with Challenges. Cell Host Microbe, 19, 575–8. [DOI] [PubMed] [Google Scholar]
- AUCHTUNG JM, ROBINSON CD & BRITTON RA 2015. Cultivation of stable, reproducible microbial communities from different fecal donors using minibioreactor arrays (MBRAs). Microbiome, 3, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BADAL VD, VACCARIELLO ED, MURRAY ER, YU KE, KNIGHT R, JESTE DV & NGUYEN TT 2020. The Gut Microbiome, Aging, and Longevity: A Systematic Review. Nutrients, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAI Y, HUANG Y, LI Y, ZHANG B, XIAO C, HOU X & YU L 2021. The Murine Reg3a Stimulated by Lactobacillus casei Promotes Intestinal Cell Proliferation and Inhibits the Multiplication of Porcine Diarrhea Causative Agent in vitro. Front Microbiol, 12, 675263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANASAZ M, NORIN E, HOLMA R & MIDTVEDT T 2002. Increased enterocyte production in gnotobiotic rats mono-associated with Lactobacillus rhamnosus GG. Appl Environ Microbiol, 68, 3031–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARKER N, VAN DE WETERING M & CLEVERS H 2008. The intestinal stem cell. Genes Dev, 22, 1856–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARKER N, VAN ES JH, KUIPERS J, KUJALA P, VAN DEN BORN M, COZIJNSEN M, HAEGEBARTH A, KORVING J, BEGTHEL H, PETERS PJ & CLEVERS H 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature, 449, 1003–7. [DOI] [PubMed] [Google Scholar]
- BARKER N, VAN OUDENAARDEN A & CLEVERS H 2012. Identifying the stem cell of the intestinal crypt: strategies and pitfalls. Cell Stem Cell, 11, 452–60. [DOI] [PubMed] [Google Scholar]
- BEDU-FERRARI C, BISCARRAT P, LANGELLA P & CHERBUY C 2022. Prebiotics and the Human Gut Microbiota: From Breakdown Mechanisms to the Impact on Metabolic Health. Nutrients, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEVINS CL & SALZMAN NH 2011. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol, 9, 356–68. [DOI] [PubMed] [Google Scholar]
- BIECHELE TL & MOON RT 2008. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods Mol Biol, 468, 99–110. [DOI] [PubMed] [Google Scholar]
- BLUTT SE, CRAWFORD SE, RAMANI S, ZOU WY & ESTES MK 2018. Engineered Human Gastrointestinal Cultures to Study the Microbiome and Infectious Diseases. Cell Mol Gastroenterol Hepatol, 5, 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOFFA LC, LUPTON JR, MARIANI MR, CEPPI M, NEWMARK HL, SCALMATI A & LIPKIN M 1992. Modulation of colonic epithelial cell proliferation, histone acetylation, and luminal short chain fatty acids by variation of dietary fiber (wheat bran) in rats. Cancer Res, 52, 5906–12. [PubMed] [Google Scholar]
- BOUSTANY A, ONWUZO S, ALMOMANI A & ASAAD I 2023. Epidemiology and risk of colorectal cancer in patients with a history of Helicobacter pylori infection: a population-based study. Ann Gastroenterol, 36, 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUCHON N, BRODERICK NA, CHAKRABARTI S & LEMAITRE B 2009a. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev, 23, 2333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUCHON N, BRODERICK NA, POIDEVIN M, PRADERVAND S & LEMAITRE B 2009b. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe, 5, 200–11. [DOI] [PubMed] [Google Scholar]
- BURCLAFF J, BLITON RJ, BREAU KA, OK MT, GOMEZ-MARTINEZ I, RANEK JS, BHATT AP, PURVIS JE, WOOSLEY JT & MAGNESS ST 2022. A Proximal-to-Distal Survey of Healthy Adult Human Small Intestine and Colon Epithelium by Single-Cell Transcriptomics. Cell Mol Gastroenterol Hepatol, 13, 1554–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANG PV, HAO L, OFFERMANNS S & MEDZHITOV R 2014. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A, 111, 2247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLEVERS H 2013. The intestinal crypt, a prototype stem cell compartment. Cell, 154, 274–84. [DOI] [PubMed] [Google Scholar]
- CLEVERS HC & BEVINS CL 2013. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol, 75, 289–311. [DOI] [PubMed] [Google Scholar]
- CO JY, MARGALEF-CATALA M, LI X, MAH AT, KUO CJ, MONACK DM & AMIEVA MR 2019. Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions. Cell Rep, 26, 2509–2520 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COMALADA M, BAILON E, DE HARO O, LARA-VILLOSLADA F, XAUS J, ZARZUELO A & GALVEZ J 2006. The effects of short-chain fatty acids on colon epithelial proliferation and survival depend on the cellular phenotype. J Cancer Res Clin Oncol, 132, 487–97. [DOI] [PubMed] [Google Scholar]
- DARBY TM, NAUDIN CR, LUO L & JONES RM 2020. Lactobacillus rhamnosus GG-induced Expression of Leptin in the Intestine Orchestrates Epithelial Cell Proliferation. Cell Mol Gastroenterol Hepatol, 9, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVE M, HIGGINS PD, MIDDHA S & RIOUX KP 2012. The human gut microbiome: current knowledge, challenges, and future directions. Transl Res, 160, 246–57. [DOI] [PubMed] [Google Scholar]
- DHEER R & YOUNG VB 2021. Stem-cell-derived models: tools for studying role of microbiota in intestinal homeostasis and disease. Curr Opin Gastroenterol, 37, 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DI RIENZI SC & BRITTON RA 2020. Adaptation of the Gut Microbiota to Modern Dietary Sugars and Sweeteners. Adv Nutr, 11, 616–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONOHOE DR, GARGE N, ZHANG X, SUN W, O’CONNELL TM, BUNGER MK & BULTMAN SJ 2011. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab, 13, 517–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOSSA AY, ESCOBAR O, GOLDEN J, FREY MR, FORD HR & GAYER CP 2016. Bile acids regulate intestinal cell proliferation by modulating EGFR and FXR signaling. Am J Physiol Gastrointest Liver Physiol, 310, G81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOU X, GAO N, YAN D & SHAN A 2020. Sodium Butyrate Alleviates Mouse Colitis by Regulating Gut Microbiota Dysbiosis. Animals (Basel), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUAN C, WU J, WANG Z, TAN C, HOU L, QIAN W, HAN C & HOU X 2023. Fucose promotes intestinal stem cell-mediated intestinal epithelial development through promoting Akkermansia-related propanoate metabolism. Gut Microbes, 15, 2233149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUCKWORTH CA 2021. Identifying key regulators of the intestinal stem cell niche. Biochem Soc Trans, 49, 2163–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DURACK J & LYNCH SV 2019. The gut microbiome: Relationships with disease and opportunities for therapy. J Exp Med, 216, 20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FISHBEIN SRS, MAHMUD B & DANTAS G 2023. Antibiotic perturbations to the gut microbiome. Nat Rev Microbiol. [DOI] [PubMed] [Google Scholar]
- FOULKE-ABEL J, IN J, KOVBASNJUK O, ZACHOS NC, ETTAYEBI K, BLUTT SE, HYSER JM, ZENG XL, CRAWFORD SE, BROUGHMAN JR, ESTES MK & DONOWITZ M 2014. Human enteroids as an ex-vivo model of host-pathogen interactions in the gastrointestinal tract. Exp Biol Med (Maywood), 239, 1124–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKEL WL, ZHANG W, SINGH A, KLURFELD DM, DON S, SAKATA T, MODLIN I & ROMBEAU JL 1994. Mediation of the trophic effects of short-chain fatty acids on the rat jejunum and colon. Gastroenterology, 106, 375–80. [DOI] [PubMed] [Google Scholar]
- FRESE SA, BENSON AK, TANNOCK GW, LOACH DM, KIM J, ZHANG M, OH PL, HENG NC, PATIL PB, JUGE N, MACKENZIE DA, PEARSON BM, LAPIDUS A, DALIN E, TICE H, GOLTSMAN E, LAND M, HAUSER L, IVANOVA N, KYRPIDES NC & WALTER J 2011. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet, 7, e1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GE Y, SUN H, XU L, ZHANG W, LV J & CHEN Y 2022. The amelioration of alcohol-induced liver and intestinal barrier injury by Lactobacillus rhamnosus Gorbach-Goldin (LGG) is dependent on Interleukin 22 (IL-22) expression. Bioengineered, 13, 12650–12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEHART H & CLEVERS H 2019. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol, 16, 19–34. [DOI] [PubMed] [Google Scholar]
- GORDON HA & BRUCKNER-KARDOSS E 1961. Effect of the normal microbial flora on various tissue elements of the small intestine. Acta Anat (Basel), 44, 210–25. [DOI] [PubMed] [Google Scholar]
- GREGORIEFF A, PINTO D, BEGTHEL H, DESTREE O, KIELMAN M & CLEVERS H 2005. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology, 129, 626–38. [DOI] [PubMed] [Google Scholar]
- GREICIUS G, KABIRI Z, SIGMUNDSSON K, LIANG C, BUNTE R, SINGH MK & VIRSHUP DM 2018. PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc Natl Acad Sci U S A, 115, E3173–E3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREICIUS G & VIRSHUP DM 2019. Stromal control of intestinal development and the stem cell niche. Differentiation, 108, 8–16. [DOI] [PubMed] [Google Scholar]
- GREIG CJ, ALPER A, GOODMAN AL & COWLES RA 2018. Mucosal homeostasis is altered in the ileum of gnotobiotic mice. J Surg Res, 231, 331–337. [DOI] [PubMed] [Google Scholar]
- HALL AB, TOLONEN AC & XAVIER RJ 2017. Human genetic variation and the gut microbiome in disease. Nat Rev Genet, 18, 690–699. [DOI] [PubMed] [Google Scholar]
- HAMADE DF, ESPINAL A, YU J, LEIBOWITZ BJ, FISHER R, HOU W, SHIELDS D, VAN PIJKEREN JP, MUKHERJEE A, EPPERLY MW, VLAD AM, COFFMAN L, WANG H, SAIFUL HUQ M, PATEL R, HUANG J & GREENBERGER JS 2022. Lactobacillus reuteri Releasing IL-22 (LR-IL-22) Facilitates Intestinal Radioprotection for Whole-Abdomen Irradiation (WAI) of Ovarian Cancer. Radiat Res, 198, 89–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANSSON GC & JOHANSSON ME 2010. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes, 1, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HASAN N & YANG H 2019. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ, 7, e7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEMARAJATA P & VERSALOVIC J 2013. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap Adv Gastroenterol, 6, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLZHAUSEN EA, MALECKI KC, SETHI AK, GANGNON R, CADMUS-BERTRAM L, DEBLOIS CL, SUEN G, SAFDAR N & PEPPARD PE 2022. Assessing the relationship between physical activity and the gut microbiome in a large, population-based sample of Wisconsin adults. PLoS One, 17, e0276684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU Q, YE L, LIU H, HUANG L, YANG Q, TURNER JR & YU Q 2018. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ, 25, 1657–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU Y, WEI W, GUAN X, LIU Y, BIAN G, HE D, FAN Q, CAI X, ZHANG Y, WANG G, ZHENG X & HAO H 2021. A diet-microbial metabolism feedforward loop modulates intestinal stem cell renewal in the stressed gut. Nat Commun, 12, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUA Q, ZHANG H, XU R, TIAN C, GAO T, YUAN Y, HAN Y, LI Y, QI C, ZHONG F & MA A 2023. Lacticaseibacillus casei ATCC334 Ameliorates Radiation-Induced Intestinal Injury in Rats by Targeting Microbes and Metabolites. Mol Nutr Food Res, 67, e2200337. [DOI] [PubMed] [Google Scholar]
- HUGENHOLTZ F & DE VOS WM 2018. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci, 75, 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUMAN MICROBIOME PROJECT C. 2012. Structure, function and diversity of the healthy human microbiome. Nature, 486, 207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICHIKAWA H & SAKATA T 1997. Effect of L-lactic acid, short-chain fatty acids, and pH in cecal infusate on morphometric and cell kinetic parameters of rat cecum. Dig Dis Sci, 42, 1598–610. [DOI] [PubMed] [Google Scholar]
- INAGAKI A, ICHIKAWA H & SAKATA T 2007. Inhibitory effect of succinic acid on epithelial cell proliferation of colonic mucosa in rats. J Nutr Sci Vitaminol (Tokyo), 53, 377–9. [DOI] [PubMed] [Google Scholar]
- JOHANSSON ME, GUSTAFSSON JK, HOLMEN-LARSSON J, JABBAR KS, XIA L, XU H, GHISHAN FK, CARVALHO FA, GEWIRTZ AT, SJOVALL H & HANSSON GC 2014. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut, 63, 281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHANSSON ME, PHILLIPSON M, PETERSSON J, VELCICH A, HOLM L & HANSSON GC 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A, 105, 15064–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JONES RM, LUO L, ARDITA CS, RICHARDSON AN, KWON YM, MERCANTE JW, ALAM A, GATES CL, WU H, SWANSON PA, LAMBETH JD, DENNING PW & NEISH AS 2013. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J, 32, 3017–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JUGE N 2012. Microbial adhesins to gastrointestinal mucus. Trends Microbiol, 20, 30–9. [DOI] [PubMed] [Google Scholar]
- KABIRI Z, GREICIUS G, MADAN B, BIECHELE S, ZHONG Z, ZARIBAFZADEH H, EDISON ALIYEV,J, WU Y, BUNTE R, WILLIAMS BO, ROSSANT J & VIRSHUP DM 2014. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development, 141, 2206–15. [DOI] [PubMed] [Google Scholar]
- KAIKO GE, RYU SH, KOUES OI, COLLINS PL, SOLNICA-KREZEL L, PEARCE EJ, PEARCE EL, OLTZ EM & STAPPENBECK TS 2016. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell, 165, 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KASTL AJ JR., TERRY NA, WU GD & ALBENBERG LG 2020. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell Mol Gastroenterol Hepatol, 9, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATOH M 2007. Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev, 3, 30–8. [DOI] [PubMed] [Google Scholar]
- KATOH M & KATOH M 2007. WNT signaling pathway and stem cell signaling network. Clin Cancer Res, 13, 4042–5. [DOI] [PubMed] [Google Scholar]
- KAWAJIRI K, KOBAYASHI Y, OHTAKE F, IKUTA T, MATSUSHIMA Y, MIMURA J, PETTERSSON S, POLLENZ RS, SAKAKI T, HIROKAWA T, AKIYAMA T, KUROSUMI M, POELLINGER L, KATO S & FUJII-KURIYAMA Y 2009. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci U S A, 106, 13481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHOURY KA, FLOCH MH & HERSH T 1969. Small intestinal mucosal cell proliferation and bacterial flora in the conventionalization of the germfree mouse. J Exp Med, 130, 659–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIEN CL, BLAUWIEKEL R, BUNN JY, JETTON TL, FRANKEL WL & HOLST JJ 2007. Cecal infusion of butyrate increases intestinal cell proliferation in piglets. J Nutr, 137, 916–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM HJ, LEE J, CHOI JH, BAHINSKI A & INGBER DE 2016. Co-culture of Living Microbiome with Microengineered Human Intestinal Villi in a Gut-on-a-Chip Microfluidic Device. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM J & LEE HK 2021. Potential Role of the Gut Microbiome In Colorectal Cancer Progression. Front Immunol, 12, 807648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM MB, HWANGBO S, JANG S & JO YK 2022. Bioengineered Co-culture of organoids to recapitulate host-microbe interactions. Mater Today Bio, 16, 100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM S, SHIN YC, KIM TY, KIM Y, LEE YS, LEE SH, KIM MN, O E, KIM KS & KWEON MN 2021. Mucin degrader Akkermansia muciniphila accelerates intestinal stem cell-mediated epithelial development. Gut Microbes, 13, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLIMASHINA MM, VIKTOROVSKII IV, MIKHAILOVA NP & V’IUNOV K,A 1989. [Substrate specificity of the biotransformation enzymes in a Nocardia erythropolis culture]. Izv Akad Nauk SSSR Biol, 429–34. [PubMed] [Google Scholar]
- KORINEK V, BARKER N, MOERER P, VAN DONSELAAR E, HULS G, PETERS PJ & CLEVERS H 1998. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet, 19, 379–83. [DOI] [PubMed] [Google Scholar]
- KOZONI V, TSIOULIAS G, SHIFF S & RIGAS B 2000. The effect of lithocholic acid on proliferation and apoptosis during the early stages of colon carcinogenesis: differential effect on apoptosis in the presence of a colon carcinogen. Carcinogenesis, 21, 999–1005. [DOI] [PubMed] [Google Scholar]
- KUHNERT F, DAVIS CR, WANG HT, CHU P, LEE M, YUAN J, NUSSE R & KUO CJ 2004. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A, 101, 266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KURILSHIKOV A, MEDINA-GOMEZ C, BACIGALUPE R, RADJABZADEH D, WANG J, DEMIRKAN A, LE ROY CI, RAYGOZA GARAY JA, FINNICUM CT, LIU X, ZHERNAKOVA DV, BONDER MJ, HANSEN TH, FROST F, RUHLEMANN MC, TURPIN W, MOON JY, KIM HN, LULL K, BARKAN E, SHAH SA, FORNAGE M, SZOPINSKA-TOKOV J, WALLEN ZD, BORISEVICH D, AGREUS L, ANDREASSON A, BANG C, BEDRANI L, BELL JT, BISGAARD H, BOEHNKE M, BOOMSMA DI, BURK RD, CLARINGBOULD A, CROITORU K, DAVIES GE, VAN DUIJN CM, DUIJTS L, FALONY G, FU J, VAN DER GRAAF A, HANSEN T, HOMUTH G, HUGHES DA, IJZERMAN RG, JACKSON MA, JADDOE VWV, JOOSSENS M, JORGENSEN T, KESZTHELYI D, KNIGHT R, LAAKSO M, LAUDES M, LAUNER LJ, LIEB W, LUSIS AJ, MASCLEE AAM, MOLL HA, MUJAGIC Z, QIBIN Q, ROTHSCHILD D, SHIN H, SORENSEN SJ, STEVES CJ, THORSEN J, TIMPSON NJ, TITO RY, VIEIRA-SILVA S, VOLKER U, VOLZKE H, VOSA U, WADE KH, WALTER S, WATANABE K, WEISS S, WEISS FU, WEISSBROD O, WESTRA HJ, WILLEMSEN G, PAYAMI H, JONKERS D, ARIAS VASQUEZ A, DE GEUS EJC, MEYER KA, STOKHOLM J, SEGAL E, ORG E, WIJMENGA C, KIM HL, KAPLAN RC, SPECTOR TD, UITTERLINDEN AG, RIVADENEIRA F, FRANKE A, LERCH MM, FRANKE L, SANNA S, D’AMATO M, PEDERSEN O, et al. 2021. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet, 53, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI Y, MASATOSHI H, MA Y, GUO Y & ZHANG B 2021. Role of Vitamin K in Intestinal Health. Front Immunol, 12, 791565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE C, CHOI C, KANG HS, SHIN SW, KIM SY, PARK HC & HONG SN 2019. NOD2 Supports Crypt Survival and Epithelial Regeneration after Radiation-Induced Injury. Int J Mol Sci, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE C, KIM BG, KIM JH, CHUN J, IM JP & KIM JS 2017. Sodium butyrate inhibits the NF-kappa B signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int Immunopharmacol, 51, 47–56. [DOI] [PubMed] [Google Scholar]
- LEE YS, KIM TY, KIM Y, LEE SH, KIM S, KANG SW, YANG JY, BAEK IJ, SUNG YH, PARK YY, HWANG SW, O E, KIM KS, LIU S, KAMADA N, GAO N & KWEON MN 2018. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe, 24, 833–846 e6. [DOI] [PubMed] [Google Scholar]
- LESHER S, WALBURG HE JR. & SACHER GA JR. 1964. Generation Cycle in the Duodenal Crypt Cells of Germ-Free and Conventional Mice. Nature, 202, 884–6. [DOI] [PubMed] [Google Scholar]
- LEVY A, STEDMAN A, DEUTSCH E, DONNADIEU F, VIRGIN HW, SANSONETTI PJ & NIGRO G 2020. Innate immune receptor NOD2 mediates LGR5(+) intestinal stem cell protection against ROS cytotoxicity via mitophagy stimulation. Proc Natl Acad Sci U S A, 117, 1994–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU X, NAGY P, BONFINI A, HOUTZ P, BING XL, YANG X & BUCHON N 2022. Microbes affect gut epithelial cell composition through immune-dependent regulation of intestinal stem cell differentiation. Cell Rep, 38, 110572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOGAN CY & NUSSE R 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol, 20, 781–810. [DOI] [PubMed] [Google Scholar]
- LU X, XIE S, YE L, ZHU L & YU Q 2020. Lactobacillus Protects Against S. Typhimurium-Induced Intestinal Inflammation by Determining the Fate of Epithelial Proliferation and Differentiation. Mol Nutr Food Res, 64, e1900655. [DOI] [PubMed] [Google Scholar]
- LYU L, ZHOU X, ZHANG M, LIU L, LIU T, NIU H, WU Y, LIANG C, HAN X & ZHANG L 2022. Lactobacillus derived from breast milk facilitates intestinal development in IUGR rats. J Appl Microbiol, 133, 503–514. [DOI] [PubMed] [Google Scholar]
- MARIADASON JM, VELCICH A, WILSON AJ, AUGENLICHT LH & GIBSON PR 2001. Resistance to butyrate-induced cell differentiation and apoptosis during spontaneous Caco-2 cell differentiation. Gastroenterology, 120, 889–99. [DOI] [PubMed] [Google Scholar]
- MARTINEZ-GURYN K, LEONE V & CHANG EB 2019. Regional Diversity of the Gastrointestinal Microbiome. Cell Host Microbe, 26, 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- METCALFE C, KLJAVIN NM, YBARRA R & DE SAUVAGE FJ 2014. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell, 14, 149–59. [DOI] [PubMed] [Google Scholar]
- MORRIS O & JASPER H 2021. Reactive Oxygen Species in intestinal stem cell metabolism, fate and function. Free Radic Biol Med, 166, 140–146. [DOI] [PubMed] [Google Scholar]
- MYHRSTAD MCW, TUNSJO H, CHARNOCK C & TELLE-HANSEN VH 2020. Dietary Fiber, Gut Microbiota, and Metabolic Regulation-Current Status in Human Randomized Trials. Nutrients, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAITO T, MULET C, DE CASTRO C, MOLINARO A, SAFFARIAN A, NIGRO G, BERARD M, CLERC M, PEDERSEN AB, SANSONETTI PJ & PEDRON T 2017. Lipopolysaccharide from Crypt-Specific Core Microbiota Modulates the Colonic Epithelial Proliferation-to-Differentiation Balance. mBio, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEAL MD, SODHI CP, JIA H, DYER M, EGAN CE, YAZJI I, GOOD M, AFRAZI A, MARINO R, SLAGLE D, MA C, BRANCA MF, PRINDLE T JR., GRANT Z, OZOLEK J & HACKAM DJ 2012. Toll-like receptor 4 is expressed on intestinal stem cells and regulates their proliferation and apoptosis via the p53 up-regulated modulator of apoptosis. J Biol Chem, 287, 37296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NELSON DP & MATA LJ 1970. Bacterial flora associated with the human gastrointestinal mucosa. Gastroenterology, 58, 56–61. [PubMed] [Google Scholar]
- NIGRO G, ROSSI R, COMMERE PH, JAY P & SANSONETTI PJ 2014. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe, 15, 792–8. [DOI] [PubMed] [Google Scholar]
- OGURA Y, LALA S, XIN W, SMITH E, DOWDS TA, CHEN FF, ZIMMERMANN E, TRETIAKOVA M, CHO JH, HART J, GREENSON JK, KESHAV S & NUNEZ G 2003. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut, 52, 1591–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAI R, TARNAWSKI AS & TRAN T 2004. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell, 15, 2156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK JH, KOTANI T, KONNO T, SETIAWAN J, KITAMURA Y, IMADA S, USUI Y, HATANO N, SHINOHARA M, SAITO Y, MURATA Y & MATOZAKI T 2016. Promotion of Intestinal Epithelial Cell Turnover by Commensal Bacteria: Role of Short-Chain Fatty Acids. PLoS One, 11, e0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PECK BCE, SHANAHAN MT, SINGH AP & SETHUPATHY P 2017. Gut Microbial Influences on the Mammalian Intestinal Stem Cell Niche. Stem Cells Int, 2017, 5604727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEDRON T, MULET C, DAUGA C, FRANGEUL L, CHERVAUX C, GROMPONE G & SANSONETTI PJ 2012. A crypt-specific core microbiota resides in the mouse colon. mBio, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PERLER BK, FRIEDMAN ES & WU GD 2023. The Role of the Gut Microbiota in the Relationship Between Diet and Human Health. Annu Rev Physiol, 85, 449–468. [DOI] [PubMed] [Google Scholar]
- PINTO D, GREGORIEFF A, BEGTHEL H & CLEVERS H 2003. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev, 17, 1709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POLETTI M, ARNAUTS K, FERRANTE M & KORCSMAROS T 2021. Organoid-based Models to Study the Role of Host-microbiota Interactions in IBD. J Crohns Colitis, 15, 1222–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POTTEN CS 1998. Stem cells in gastrointestinal epithelium: numbers, characteristics and death. Philos Trans R Soc Lond B Biol Sci, 353, 821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POWELL DN, SWIMM A, SONOWAL R, BRETIN A, GEWIRTZ AT, JONES RM & KALMAN D 2020. Indoles from the commensal microbiota act via the AHR and IL-10 to tune the cellular composition of the colonic epithelium during aging. Proc Natl Acad Sci U S A, 117, 21519–21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PREIDIS GA, SAULNIER DM, BLUTT SE, MISTRETTA TA, RIEHLE KP, MAJOR AM, VENABLE SF, FINEGOLD MJ, PETROSINO JF, CONNER ME & VERSALOVIC J 2012. Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine. FASEB J, 26, 1960–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QIU Y, JIANG Z, HU S, WANG L, MA X & YANG X 2017. Lactobacillus plantarum Enhanced IL-22 Production in Natural Killer (NK) Cells That Protect the Integrity of Intestinal Epithelial Cell Barrier Damaged by Enterotoxigenic Escherichia coli. Int J Mol Sci, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAJAN A, VELA L, ZENG XL, YU X, SHROYER N, BLUTT SE, POOLE NM, CARLIN LG, NATARO JP, ESTES MK, OKHUYSEN PC & MARESSO AW 2018. Novel Segment- and Host-Specific Patterns of Enteroaggregative Escherichia coli Adherence to Human Intestinal Enteroids. mBio, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RETTEDAL EA, GUMPERT H & SOMMER MO 2014. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat Commun, 5, 4714. [DOI] [PubMed] [Google Scholar]
- ROAGER HM & LICHT TR 2018. Microbial tryptophan catabolites in health and disease. Nat Commun, 9, 3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBBE C, CAPON C, CODDEVILLE B & MICHALSKI JC 2004. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J, 384, 307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAFFARIAN A, MULET C, REGNAULT B, AMIOT A, TRAN-VAN-NHIEU J, RAVEL J, SOBHANI I, SANSONETTI PJ & PEDRON T 2019. Crypt- and Mucosa-Associated Core Microbiotas in Humans and Their Alteration in Colon Cancer Patients. mBio, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAKATA T 1987. Stimulatory effect of short-chain fatty acids on epithelial cell proliferation in the rat intestine: a possible explanation for trophic effects of fermentable fibre, gut microbes and luminal trophic factors. Br J Nutr, 58, 95–103. [DOI] [PubMed] [Google Scholar]
- SANTOS AJM, LO YH, MAH AT & KUO CJ 2018. The Intestinal Stem Cell Niche: Homeostasis and Adaptations. Trends Cell Biol, 28, 1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARTOR RB 2015. Gut microbiota: Optimal sampling of the intestinal microbiota for research. Nat Rev Gastroenterol Hepatol, 12, 253–4. [DOI] [PubMed] [Google Scholar]
- SATO T, VAN ES JH, SNIPPERT HJ, STANGE DE, VRIES RG, VAN DEN BORN M, BARKER N, SHROYER NF, VAN DE WETERING M & CLEVERS H 2011. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature, 469, 415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATO T, VRIES RG, SNIPPERT HJ, VAN DE WETERING M, BARKER N, STANGE DE, VAN ES JH, ABO A, KUJALA P, PETERS PJ & CLEVERS H 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature, 459, 262–5. [DOI] [PubMed] [Google Scholar]
- SAULNIER DM, SANTOS F, ROOS S, MISTRETTA TA, SPINLER JK, MOLENAAR D, TEUSINK B & VERSALOVIC J 2011. Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS One, 6, e18783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAVAGE DC & BLUMERSHINE RV 1974. Surface-surface associations in microbial communities populating epithelial habitats in the murine gastrointestinal ecosystem: scanning electron microscopy. Infect Immun, 10, 240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEGERBERG-KONTTINEN M 1988. Malignant tumors found during medicolegal postmortem examinations. Am J Forensic Med Pathol, 9, 357. [DOI] [PubMed] [Google Scholar]
- STEARNS JC, LYNCH MD, SENADHEERA DB, TENENBAUM HC, GOLDBERG MB, CVITKOVITCH DG, CROITORU K, MORENO-HAGELSIEB G & NEUFELD JD 2011. Bacterial biogeography of the human digestive tract. Sci Rep, 1, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANG Q, JIN G, WANG G, LIU T, LIU X, WANG B & CAO H 2020. Current Sampling Methods for Gut Microbiota: A Call for More Precise Devices. Front Cell Infect Microbiol, 10, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THEODOSIOU NA & TABIN CJ 2003. Wnt signaling during development of the gastrointestinal tract. Dev Biol, 259, 258–71. [DOI] [PubMed] [Google Scholar]
- TIAN H, BIEHS B, WARMING S, LEONG KG, RANGELL L, KLEIN OD & DE SAUVAGE FJ 2011. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature, 478, 255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN DER FLIER LG & CLEVERS H 2009. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol, 71, 241–60. [DOI] [PubMed] [Google Scholar]
- VAN DER POST S, BIRCHENOUGH GMH & HELD JM 2021. NOX1-dependent redox signaling potentiates colonic stem cell proliferation to adapt to the intestinal microbiota by linking EGFR and TLR activation. Cell Rep, 35, 108949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANDUSSEN KL, MARINSHAW JM, SHAIKH N, MIYOSHI H, MOON C, TARR PI, CIORBA MA & STAPPENBECK TS 2015. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut, 64, 911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANUYTSEL T, SENGER S, FASANO A & SHEA-DONOHUE T 2013. Major signaling pathways in intestinal stem cells. Biochim Biophys Acta, 1830, 2410–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VASAPOLLI R, SCHUTTE K, SCHULZ C, VITAL M, SCHOMBURG D, PIEPER DH, VILCHEZ-VARGAS R & MALFERTHEINER P 2019. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology, 157, 1081–1092 e3. [DOI] [PubMed] [Google Scholar]
- VUIK F, DICKSVED J, LAM SY, FUHLER GM, VAN DER LAAN L, VAN DE WINKEL A, KONSTANTINOV SR, SPAANDER M, PEPPELENBOSCH MP, ENGSTRAND L & KUIPERS EJ 2019. Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United European Gastroenterol J, 7, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG JY, MCCORMACK SA, VIAR MJ & JOHNSON LR 1991. Stimulation of proximal small intestinal mucosal growth by luminal polyamines. Am J Physiol, 261, G504–11. [DOI] [PubMed] [Google Scholar]
- WANG M, AHRNE S, JEPPSSON B & MOLIN G 2005. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol, 54, 219–31. [DOI] [PubMed] [Google Scholar]
- WANG Y, KIM R, GUNASEKARA DB, REED MI, DISALVO M, NGUYEN DL, BULTMAN SJ, SIMS CE, MAGNESS ST & ALLBRITTON NL 2018. Formation of Human Colonic Crypt Array by Application of Chemical Gradients Across a Shaped Epithelial Monolayer. Cell Mol Gastroenterol Hepatol, 5, 113–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITEHEAD RH, YOUNG GP & BHATHAL PS 1986. Effects of short chain fatty acids on a new human colon carcinoma cell line (LIM1215). Gut, 27, 1457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILKE G, WANG Y, RAVINDRAN S, STAPPENBECK T, WITOLA WH & SIBLEY LD 2020. In Vitro Culture of Cryptosporidium parvum Using Stem Cell-Derived Intestinal Epithelial Monolayers. Methods Mol Biol, 2052, 351–372. [DOI] [PubMed] [Google Scholar]
- WILLIAMSON IA, ARNOLD JW, SAMSA LA, GAYNOR L, DISALVO M, COCCHIARO JL, CARROLL I, AZCARATE-PERIL MA, RAWLS JF, ALLBRITTON NL & MAGNESS ST 2018. A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology. Cell Mol Gastroenterol Hepatol, 6, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU H, XIE S, MIAO J, LI Y, WANG Z, WANG M & YU Q 2020. Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa. Gut Microbes, 11, 997–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU S, MORIN PJ, MAOUYO D & SEARS CL 2003. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology, 124, 392–400. [DOI] [PubMed] [Google Scholar]
- XING PY, PETTERSSON S & KUNDU P 2020. Microbial Metabolites and Intestinal Stem Cells Tune Intestinal Homeostasis. Proteomics, 20, e1800419. [DOI] [PubMed] [Google Scholar]
- YAN KS, CHIA LA, LI X, OOTANI A, SU J, LEE JY, SU N, LUO Y, HEILSHORN SC, AMIEVA MR, SANGIORGI E, CAPECCHI MR & KUO CJ 2012. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A, 109, 466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YIN X, FARIN HF, VAN ES JH, CLEVERS H, LANGER R & KARP JM 2014. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods, 11, 106–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZACHOS NC, KOVBASNJUK O, FOULKE-ABEL J, IN J, BLUTT SE, DE JONGE HR, ESTES MK & DONOWITZ M 2016. Human Enteroids/Colonoids and Intestinal Organoids Functionally Recapitulate Normal Intestinal Physiology and Pathophysiology. J Biol Chem, 291, 3759–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG W, ZHOU Q, LIU H, XU J, HUANG R, SHEN B, GUO Y, AI X, XU J, ZHAO X, LIU Y, WANG Y & ZHI F 2023. Bacteroides fragilis strain ZY-312 facilitates colonic mucosa regeneration in colitis via motivating STAT3 signaling pathway induced by IL-22 from ILC3 secretion. Front Immunol, 14, 1156762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG X, TONG Y, LYU X, WANG J, WANG Y & YANG R 2020. Prevention and Alleviation of Dextran Sulfate Sodium Salt-Induced Inflammatory Bowel Disease in Mice With Bacillus subtilis-Fermented Milk via Inhibition of the Inflammatory Responses and Regulation of the Intestinal Flora. Front Microbiol, 11, 622354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Z, GENG J, TANG X, FAN H, XU J, WEN X, MA ZS & SHI P 2014. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J, 8, 881–93. [DOI] [PMC free article] [PubMed] [Google Scholar]


