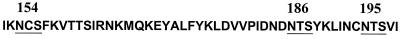Abstract
We examined the role of asparagine-linked glycosylation of the V2 loop of the human immunodeficiency virus (HIV) SF162 envelope on viral replication potential and neutralization susceptibility. We report that the asparagines located at the amino- and carboxy-terminal sites (at positions 154 and 195, respectively), as well as within the V2 loop of the SF162 envelope (at position 186), are glycosylated during in vitro replication of this virus in human peripheral blood mononuclear cells. Our studies indicate that glycosylation of the V2 loop, in particular at its base, facilitates the interaction of the HIV envelope with the CD4 and CCR5 receptor molecules present on the surface of target cells and affects viral replication kinetics in a cell type-dependent manner. In cells expressing high numbers of receptor molecules on their surfaces, the SF162-derived V2 loop-deglycosylated mutant viruses replicate as efficiently as the parental SF162 virus, while in cells expressing small numbers of receptor molecules, the mutant viruses replicate with markedly reduced efficiency. In addition to expanding the viral tropism, V2 loop glycosylation at the three sites examined prevents neutralization by anti-CD4 binding site antibodies. In contrast, glycosylation at the amino- and carboxy-terminal sites of the V2 loop but not within the loop itself offers protection against anti-V3 loop antibodies. Thus, the epitopes masked by the sugar molecules present on the three glycosylation sites examined are not identical but overlap.
Approximately 50% of the mass of the human and simian immunodeficiency virus (HIV and SIV, respectively) envelope glycoproteins is attributed to N- and O-linked sugar molecules (2, 28). Envelope glycosylation affects not only the intracellular transport of these viral proteins, but also their structure, and therefore influences the viral phenotype (6, 10, 15, 20, 23, 29, 31, 39, 45, 64).
Moreover, envelope glycosylation plays an important role in the interaction of these viruses with the immune system of the infected host by reducing the recognition of specific epitopes by T lymphocytes (3) and by decreasing epitope immunogenicity (44). During infection of macaques by SIV and simian/human immunodeficiency virus (SHIV), the emergence of neutralization-resistant viruses in the blood circulation of the infected host, which usually precedes or coincides with the development of disease, is associated with changes in the envelope glycosylation pattern (5, 36–38, 47). This resistance is partially due to the repositioning of glycosylation sites within the viral envelope, which results in the “masking” of neutralization epitopes (1, 41, 44, 47–49).
Removal of glycosylation sites located in the first hypervariable region (V1 loop) of the SIVmac239 envelope enhances the exposure of as yet unknown neutralization epitopes and increases their immunogenicity (44). During infection of macaques with mutant SIVmac239 viruses lacking asparagine-linked glycosylation sites in the V1 loop, high titers of antibodies that recognize the newly exposed epitopes are generated. Important for vaccine development is the observation that sera collected from macaques infected with such partially deglycosylated SIVmac239 mutant viruses have a higher neutralization potential against the parental fully glycosylated SIVmac239 virus than sera collected from macaques infected with the parental virus itself (44).
The above observations strongly suggest that investigation of the mechanisms by which HIV or SIV envelope glycosylation affects the viral phenotype will not only provide us with information regarding the biology of this pathogen, but may also assist us in the development of more effective anti-HIV (or anti-SIV) envelope-based immunogens.
We previously reported that on the background of the neutralization-resistant HIV-1 isolate SF162, the elimination of 30 amino acids (from T160 to Y189) from the central region of the second hypervariable envelope region (V2 loop) renders the virus (termed SF162ΔV2) highly susceptible to neutralization by antibodies present in sera from HIV-infected patients (54). It appears that on the background of the SF162 virus, this partial V2 loop deletion increases the exposure of neutralization epitopes whose structure is conserved among heterologous HIV-1 isolates. The above deletion results in the loss of one potential asparagine-linked glycosylation site located in the V2 loop (at position 186 of the SF162 envelope). Therefore, it is possible that the switch in viral phenotype observed upon V2 loop deletion is not due to the elimination of the 30 amino acids per se, but rather to the elimination of the sugar moieties present on the asparagine at position 186. In the studies presented here, we investigated the role of this particular potential glycosylation site, as well as the roles of all other potential N-linked glycosylation sites present in the V2 loop of the SF162 envelope, in viral replication, tropism, and susceptibility to antibody-mediated neutralization. This investigation expands our understanding of the relationship between HIV envelope structure and viral phenotype.
MATERIALS AND METHODS
Elimination by mutagenesis of potential N-linked glycosylation sites.
Elimination of the potential glycosylation sites was performed as previously described (43, 44) by replacing the asparagines (N) with glutamines (Q) at positions 154, 186, and 195 of the SF612 envelope, using the QuickChange site-directed mutagenesis kit (Stratagene). We designate the glycosylation mutant (GM) envelopes as follows: N 154 replaced by Q is GM154; N 186 replaced by Q is GM186; and N 195 replaced by Q is GM195.
The primers were 5′-AGAGGAGAAATAAAACAGTGCTCTTTCAAGGTC-3′ and 5′-GACCTTGAAAGAGCACTGTTTTATTTCTCCTCT-3′ for generating GM154; 5′-CCAATAGATAATGATCAGACAAGCTATAAATTG-3′ and 5′-CAATTTATAGCTTGTCTGATCATTATCTATTGG-3′ for generating GM186; and 5′-CTGTGTAATGACTGAGGTCTGACAATTTATCAATTT-3′ and 5′-AAATTGATAAATTGTCAGACCTCAGTCATTACACAG-3′ for generating GM195.
The introduction of mutations was verified by sequencing. Two clones of each mutant envelope were amplified and used for generation of infectious virions. The replication potential and neutralization susceptibility of all clones were evaluated and compared.
Generation of infectious virions with a partially deglycosylated V2 loop.
293T cells were cotransfected with pUC DNA vectors expressing the 5′ and 3′ (containing the parental SF162 or the mutagenized envelope gene) halves of the viral genome using Lipofectamine, as previously described (54). At 72 h posttransfection, the cell supernatants were collected, subjected to centrifugation (10 min at room temperature at 2,000 rpm), filtered through 0.45-μm-pore membranes, and used to inoculate human peripheral blood mononuclear cells (PBMC) that had been stimulated for 3 days with 3 μg of phytohemagglutinin (PHA; Sigma) per ml. Following a 3-h incubation at 37°C, the viral inoculum was removed, and the cells were cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS) (BioWhittaker), penicillin (100 U/ml), streptomycin (100 μg/ml), glutamine (2 mM) and interleukin-2 (IL-2; 20 U/ml) (Hoffmann-La Roche, Inc.). Virus production was monitored by determining the concentration of p24 antigen in the cell supernatant every 3 to 4 days using an in-house p24 detection enzyme-linked immunosorbent assay (ELISA). Supernatants with high p24 content were collected, filtered, and stored as 0.5-ml aliquots at −80°C. These viral stocks were titrated in PBMC and used for all subsequent experiments.
Electrophoretic mobility analysis of virion-associated gp120 proteins.
Intact infectious viral particles produced in PBMC were first separated from free gp120 molecules by centrifugation at 13,500 × g for 2 h at 4°C. The viral pellet was resuspended in 40 μl of lysis buffer (50 mM Tris-HCl, 100 mM 2-mercaptoethanol, 2% sodium dodecyl sulfate [SDS], 0.1% bromophenol blue, 10% glycerol) and subjected to boiling for 5 min. The denatured viral proteins were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 5% gels, then transferred to Immobilon P membranes (Millipore), and incubated overnight at 4°C with goat-anti-SF2 gp120 serum antibodies (Chiron; 1:1,000 dilution) and for 2 h at room temperature with protein G-horseradish peroxidase (Bio-Rad; 1:1,000 dilution). Visualization of the gp120 envelope molecules was performed with the use of enhanced chemiluminescence reagents (Amersham).
Infectivity assays. (i) PBMC and macrophages.
PHA-activated human PBMC were generated as previously described (9) from the blood of HIV-negative donors (New York Blood Center) and cultured in the above-mentioned RPMI medium. Macrophages were prepared by the plastic adherence method as previously described (9) and cultured in RPMI medium supplemented with 5% homologous human serum, 10% FBS, penicillin, streptomycin, and glutamine. PBMC (2 × 106) and macrophages (105 per well of a 12-well plate) were inoculated in triplicate with 200 or 400 50% tissue culture infective doses (TCID50), respectively, of each isolate for 3 h at 37°C. The viral inoculum was removed, and the cells were cultured in the appropriate medium (2 ml for PBMC and 1 ml for macrophages). The p24 antigen concentration in the cell supernatants was determined every 3 to 4 days, during which time the medium was replaced. The replication of all isolates (two clones for each mutant) was tested simultaneously. Experiments were repeated at least three times using target cells from different donors to eliminate potential donor-specific effects on viral infectivity.
(ii) HeLa cell lines.
Infection of HeLa cells expressing various numbers of CD4 and CCR5 receptor molecules on their surface by the various viruses was analyzed as previously described (25, 42). Briefly, 2.5 × 105 cells in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 μg/ml), and glutamine (2 mM) were added per well of a 12-well plate (Corning). The next day, the medium was aspirated and the cells were inoculated with 500 TCID50 of each isolate and incubated for 3 h at 37°C. Triplicate wells for each isolate were used. The inoculum was removed, and fresh DMEM was added (1 ml per well). After 72 h, the medium was aspirated and the cells were fixed for 10 min with acetone-methanol (1:1) at room temperature. Thereafter, the cells were incubated (1 h at room temperature) with serum (1:150 dilution) from HIV-positive patients, then with a sheep anti-human immunoglobulin G (IgG)-horseradish peroxidase-coupled antibody (Amersham; 1:100 dilution), and finally with a diaminobenzidine peroxidase substrate solution (Sigma). The number of foci of infected cells present in each well was determined.
Neutralization assays.
The neutralization assays were performed as previously reported (54, 58). Briefly, each virus (100 TCID50 in 50 μl of the above RPMI medium) was mixed with an equal volume of serially diluted heat-inactivated HIV-positive serum or monoclonal antibodies (MAbs) for 1 h at 37°C. For each serum or MAb dilution, triplicate wells of a U-bottomed 96-well plate were used. PBMC (4 × 105 in 0.1 ml of medium) were added to each well. Virus was also incubated with cells in the absence of HIV-positive serum or MAb. These wells served as positive controls (0% neutralization). Approximately 24 h later, half the cell supernatant of each well (containing virus and serum or MAbs) was replaced with fresh medium. This procedure was repeated twice. The percent neutralization in the presence of each serum or MAb dilution was determined as previously described (58). Neutralization experiments were performed two to three times using pooled PBMC from two donors in order to eliminate potential effects on viral infectivity that may be associated with the cells of a particular donor.
Determination of the relative envelope content of wild-type and mutant virions.
We used well-established ELISAs to determine envelope content. Briefly, intact infectious virions from the wild-type and mutant viruses produced in PBMC were separated from free gp120 molecules by centrifugation as mentioned above. The virion pellet was lysed with Tris-buffered saline (TBS)–2% Empigen detergent by an overnight incubation at room temperature. Equal volumes (200 μl per well) from the viral lysates were added to ELISA wells coated with either 6203 (5 μg/ml) or 6205 (5 μg/ml) sheep antibodies (International Enzymes, Inc.), and twofold serial dilutions were made in TBS–2% Empigen. The first polyclonal antibody recognizes various regions of the HIV-1 p24gag protein, while the second recognizes the carboxy-terminal 15 amino acids of the HIV-1 gp120 subunit. The relative amount of the captured viral proteins was determined as previously described (55, 58). The ratio of optical density at 490 nm signals from the 6205 and 6203 wells reports on the relative gp120 to p24 protein ratio in these virions.
RESULTS
Glycosylation of asparagines at positions 154, 186, and 195 of the SF162 envelope.
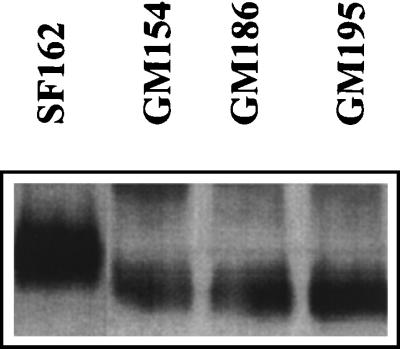
The positions of the three potential asparagine-linked glycosylation sites in the V2 loop of the SF162 gp120 envelope subunit are shown in Fig. 1. It is expected that the individual elimination by mutagenesis of a utilized N-linked glycosylation site on the HIV-1 gp120 will reduce the molecular size of this protein by approximately 2.5 kDa (24). This in turn will increase the relative electrophoretic mobility of the mutated envelope gp120 protein compared to the fully glycosylated parental protein. In contrast, if the site is not utilized, no change in electrophoretic mobility will be recorded. To examine whether the asparagines at the positions of interest are glycosylated during the in vitro replication of SF162 in human PBMC, we compared the electrophoretic mobility of virion-associated gp120 molecules from the GM154, GM186, and GM195 virions to that of gp120 molecules derived from the parental SF162 virus (Fig. 2). The gp120 molecules derived from all three mutant viruses migrated faster than those derived from the SF162 virus. This indicates that the asparagines at all three positions are normally glycosylated when SF162 is propagated in human PBMC in vitro.
FIG. 1.
Amino acid sequence of the V2 loop of the SF162 envelope. The amino acid sequence of the second hypervariable region of the SF162 envelope from isoleucine 152 to isoleucine 199 is shown. Underlined are the positions of potential N-linked glycosylation sites. The numbers indicate the location of asparagines (N) at positions 154, 186, and 195 of the SF162 envelope.
FIG. 2.
Electrophoretic mobility of virion-associated gp120 molecules during SDS-PAGE. The preparation of virion-associated gp120 molecules from the parental SF162 and the three glycosylation mutant viruses and SDS-PAGE were performed as described in Materials and Methods. No attempt was made to correct for differences in the gp120 content of the samples.
Viral replication in PBMC and macrophages.
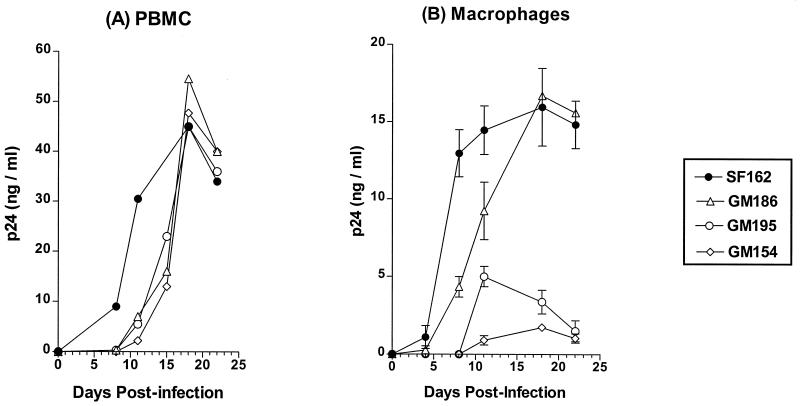
To examine whether envelope glycosylation at positions 154, 186, and 195 of the SF162 envelope affects the replication potential of this virus, we compared the replication kinetics of the three glycosylation mutant viruses to that of the parental SF162 virus using as target cells PHA-activated human PBMC and primary macrophages (Fig. 3). All three mutant viruses replicated to titers as high as those of the parental SF162 virus in PBMC, with only slight delays in their replication kinetics (Fig. 3A). Although minor variations in the relative replication kinetics of the three mutant isolates were recorded from experiment to experiment, the GM154 virus consistently replicated with slower kinetics than the GM186 and GM195 viruses.
FIG. 3.
Replication in activated human PBMC (A) and primary human macrophages (B). The replication of the mutant viruses was compared to that of the parental SF162 isolate, as described in Materials and Methods. Values indicate the mean p24 concentration from triplicate wells, and the bars indicate the standard deviation from the mean (in the case of PBMC, the standard deviation has been omitted for clarity). One out of three independent experiments is shown.
Similar to our observation in the case of PBMC, the elimination of the glycosylation site at position 186 delays viral replication in macrophages, although the virus (GM186) grows to titers as high as those of the parental SF162 virus (Fig. 3B). In contrast, elimination of the glycosylation sites located at the base of the V2 loop had a more profound effect on viral replication potential in primary macrophages, such that the GM154 and GM195 isolates replicate with a marked delay and to very low titers in these cells.
Infection of HeLa cell lines expressing various numbers of CD4 and CCR5 molecules on their surface.
Differentiated human macrophages express fewer CD4 and CCR5 molecules than activated PBMC on their surfaces (16, 35, 63). The decreased replication ability of GM154 and GM195 viruses in macrophages could be due to a less efficient utilization of the CD4 and/or CCR5 molecules by their partially deglycosylated gp120 envelope molecules. To examine this, we compared the efficiency with which the various isolates infect HeLa cell lines expressing various amounts of CD4 or CCR5 molecules on their surfaces (25, 42).
Our results (Table 1) are in agreement with previous reports indicating that the infectious potential of SF162 is dependent on the presence of both CD4 and CCR5 molecules on the surface of target cells (8, 42). The sole presence of CD4 molecules (at both low and high numbers; cells HI-R and HI-J, respectively) on the target membranes in the absence of CCR5 molecules does not allow efficient entry. However, this may be dependent on the viral genetic background, because it has been shown that on the background of the ADA virus, deglycosylation of the V2 loop at positions very similar to those examined here alters the viral tropism such that the virus infects CCR5-expressing cells in a CD4-independent fashion (23). When CD4 numbers are kept low, an increase in the number of CCR5 molecules (compare the results obtained with HI-R cells to those obtained with the RC-12 cells) results in a 10-fold increase in entry of SF162 and GM186, but only a 2-fold increase in entry of GM154 and GM195. When the CCR5 numbers are kept low, an increase in the number of CD4 molecules (compare the results obtained with the RC-12 cells to those obtained with the JC-10 cells) results in an increase in entry of SF162 and GM186 by 13- and 12-fold, respectively, that of GM154 by 5-fold, and that of GM195 by 7.5-fold. When the CD4 numbers are high and the number of CCR5 molecules increases (compare the results obtained with the JC-10 cells to those obtained with the JC-53 cells), no further increase in the infectivity of SF162 and GM186 takes place, but in contrast a 3-fold increase in the case of GM186 and a 4.5-fold increase in the case of GM195 is recorded.
TABLE 1.
Infectivity in HeLa cells expressing various numbers of CD4 and CCR5 molecules on their surfacea
| Cell line | CD4 level | CCR5 level | No. of foci/wellb
|
|||
|---|---|---|---|---|---|---|
| SF162 | GM186 | GM154 (Nt) | GM195 (Ct) | |||
| HI-R | Low | Absent | 5 | 7 | 5 | 5 |
| HI-J | High | Absent | 8 | 9 | 5 | 5 |
| RC-12 | Low | Intermediate | 50 | 50 | 10 | 10 |
| JC-10 | High | Low | 650 | 600 | 50 | 75 |
| JC-53 | High | High | 700 | 750 | 150 | 350 |
CD4 and CCR5 expression levels were reported previously (42). The values in this table represent the number of foci per well (average of three wells). One representative out of three independent experiments is shown.
Nt and Ct, amino- and carboxy-terminal, respectively, sides of the V2 loop.
Neutralizations.
Several reports have highlighted the important role of envelope glycosylation on the susceptibility of HIV and SIV to antibody-mediated neutralization (1, 5, 36, 44, 47–49). To evaluate whether glycosylation of the asparagines located at positions 154, 186, and 195 of the V2 loop of the SF162 envelope offers protection against antibody-mediated neutralization, we compared the neutralization susceptibility of the GM154, GM186, and GM195 mutant viruses to that of the parental SF162 virus using sera collected from HIV-infected patients (Table 2). These studies also allowed us to compare the relative protective role of each of these three glycosylation sites.
TABLE 2.
Neutralization by HIV-positive seraa
| Serum | SF162 | GM186 | GM154 (Nt) | GM195 (Ct) |
|---|---|---|---|---|
| Pooledb | 100 | 200 | 10,000 | 10,000 |
| A | 50 | 200 | 2,000 | 1,000 |
| B | 100 | 100 | 100 | 200 |
| C | 150 | 700 | 1,000 | 400 |
| D | NDc | 700 | 10,000 | 10,000 |
The values indicate the serum dilution that results in 90% inhibition of infection. Also see Table 1, footnote b.
The sera were mixed at a 1:1 ratio.
ND, 90% inhibition was not achieved with a serum dilution of 1:10.
All three mutant viruses were more susceptible to neutralization by pooled HIV-positive sera than the parental SF162 virus. The GM154 and GM195 viruses were more susceptible to neutralization than GM186. Neutralization studies conducted with individual sera indicated that a given serum neutralizes the three mutant viruses to different extents. For example, serum A neutralizes the GM154 and GM195 isolates 10- and 5-fold, respectively, more efficiently than the GM186 virus. Our studies also indicate that each mutant virus is not equally susceptible to neutralization by the various sera tested. For instance, isolate GM154 was highly susceptible to neutralization by serum D, less susceptible to neutralization by serum A, and even less susceptible to neutralization by serum C.
To examine whether glycosylation of the V2 loop masks neutralization epitopes located within the V2 loop itself or other envelope regions, we used MAbs with known epitope specificity (Table 3). The three mutant viruses, like the parental SF162 virus, were resistant to neutralization by the two anti-V2 loop MAbs tested, G3.4 and G3.136. Therefore, the increased susceptibility to neutralization of the mutant viruses by HIV-positive sera is most likely not due to an increase in the exposure of epitopes located within the V2 loop, but rather in regions located outside the V2 loop. To examine this possibility, we used MAbs that recognize epitopes located outside the V2 loop. All four viruses were resistant to neutralization by MAbs 17b and 48d, which recognize CD4-induced epitopes (59, 60), which may be involved in the binding of the HIV envelope to chemokine receptor molecules (62, 65). In contrast, all three mutant viruses were more susceptible, to various degrees, than SF162 to neutralization by the anti-CD4 binding site MAb IgGCD4. GM154 was approximately 1 log more susceptible to neutralization than either GM186 or GM195, which in turn were 2- to 3-fold more susceptible than SF162. Similarly, all three mutant viruses were more susceptible to neutralization by MAb IgG1b12, which recognizes a complex epitope overlapping the CD4 binding site. In contrast to what we observed for the IgGCD4 MAb, both GM154 and GM195 were approximately 1 log more susceptible to neutralization than GM186, which in turn was 1 log more susceptible to neutralization than SF162. The two anti-V3 loop MAbs examined, 447D and 391-95D, efficiently neutralized both GM154 and GM195 but not GM186, which was as resistant to neutralization as SF162. MAb 447D recognizes the GPxR motif at the tip of the V3 loop and was reported to neutralize several primary isolates (14, 19), while MAb 391-95D recognizes a conformational epitope flanking the tip of the V3 loop (50).
TABLE 3.
Neutralization by MAbsa
| MAb | Epitope specificity | Inhibitory MAb concn (μg/ml)
|
|||
|---|---|---|---|---|---|
| SF162 | GM186 | GM154 (Nt) | GM195 (Ct) | ||
| G3.4 | V2 loop | ND | ND | ND | ND |
| G3.136 | V2 loop | ND | ND | ND | ND |
| 17b | Complex | ND | ND | ND | ND |
| 48d | Complex | ND | ND | ND | ND |
| IgGCD4 | CD4-b.s | 7.5 | 3 | 0.2 | 2.4 |
| IgG1b12 | CD4-b.s | 5 | 0.4 | 0.05 | 0.05 |
| 447D | V3 loop | ND | ND | 0.3 | 0.7 |
| 391-95D | V3 loop | ND | ND | 0.05 | 0.4 |
Values indicate the antibody concentration that results in 90% inhibition of infection. Average values from three independent experiments are shown in each case. CD4-b.s, CD4 binding site; ND, 90% neutralization was not achieved at the highest MAb concentration used in these experiments (10 μg/ml). See also Table 1, footnote b.
Relative envelope content of wild-type and mutant viruses.
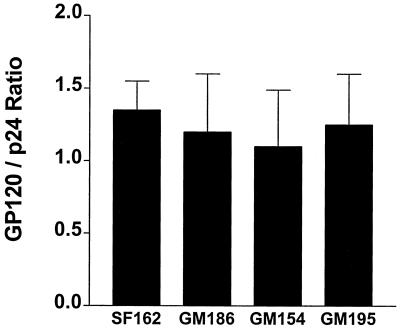
The observed differences in the replication potential as well as the susceptibility to neutralization between the SF162 virus and the three mutant viruses could be due to less efficient incorporation of the modified envelope proteins into the virion membrane. It is therefore possible that the mutant viruses express fewer envelope molecules on their surface. Our results indicate, however, that the relative ratio of gp120 to p24 proteins in the SF162 and mutant virions is similar (Fig. 4), suggesting that similar numbers of gp120 molecules are present on the surface of all the viruses examined.
FIG. 4.
Relative ratio of gp120 to p24 molecules in parental and mutant virions. The quantitation of gp120 and p24 proteins present in intact virions produced in PBMC was performed with the use of ELISA methodologies as described in Materials and Methods. The mean gp120-to-p24 ratio and the standard deviation from the mean from three independent experiments is shown.
DISCUSSION
In the studies presented here, we evaluated the role of asparagine-linked glycosylation sites located in the V2 loop of the HIV-1 envelope on the replication potential and neutralization susceptibility of the R5-using, macrophage-tropic, neutralization-resistant HIV-1 SF162 isolate (7, 8, 51, 52, 58). Three asparagine-linked glycosylation sites exist in this region of the SF162 envelope (Fig. 1); one is located within the V2 loop itself (position 186), and the other two are located at the amino and carboxy terminus of the base of this loop (positions 154 and 195, respectively).
During the in vitro replication of the SF162 virus in human PBMC, all three positions are glycosylated (Fig. 2). However, elimination of any one of these positions does not abrogate the ability of the virus to replicate in these cells (Fig. 3). These results are in agreement with those indicating that the pattern of HIV, SIV, or SHIV envelope glycosylation does not significantly affect viral replication ability in PBMC (5, 43, 47). In connection with the role of envelope glycosylation in viral tropism, previous studies have suggested that changes in envelope glycosylation could expand the tropism of the virus (10, 46). However, these studies were primarily conducted with established T-cell lines as target cells, and it is unclear whether an expansion in replication in established T-cell lines would have any relevance in vivo. Our current studies indicate that V2 loop glycosylation profoundly affects the ability of HIV to replicate in primary macrophages. In particular, glycosylation at the base of the V2 loop (at both the amino- and carboxy-terminal sides) appears to be crucial for efficient replication in these cells (Fig. 3).
The fact that the effect of the introduced modifications on viral replication is more pronounced when the target cells are macrophages than PBMC is most likely explained by the smaller number of CD4 and CCR5 receptor molecules on the surface of the former than the latter cells (16, 35, 63). Our studies conducted with HeLa cell lines (Table 1) expressing different numbers of CD4 and CCR5 molecules on their surfaces (42) suggest that the introduced modifications affect envelope interaction with both the CD4 and CCR5 receptor molecules on the surface of target cells. While glycosylation at position 186 appears to play a more important role during the interaction of the envelope with CD4 than with CCR5, glycosylation at positions 154 and 195 appears to play a role during both envelope-CD4 and envelope-CCR5 interaction (Table 1). Alternatively, the more efficient replication of the mutant viruses in PBMC than in macrophages could be related to potential differences in the oligomerization, glycosylation, and/or CD4 association of CCR5 molecules in these two cell types (17, 27). In this case, the fact that the SF162 virus replicates efficiently in both activated PBMC and macrophages, while the mutant viruses replicate less efficiently in macrophages, would suggest that the partially deglycosylated envelopes but not the wild-type envelope interacts less efficiently with the CCR5 molecules present on the surface of macrophages than those present on the surface of activated PBMC. Future studies will address this possibility.
We previously reported that CD4 binding to the SF162 gp120 envelope subunit is influenced by an envelope region comprising the V2 loop (56). Our current studies suggest that in addition to the amino acid sequence of the V2 loop, the extent of glycosylation of this region also influences the binding of HIV to the CD4 receptor. In addition to affecting envelope-CD4 binding, V2 loop glycosylation may affect the type and/or the extent of CD4-mediated envelope conformational changes (55) that are required for envelope-CCR5 binding. This may explain the observation that deglycosylation of the base of the V2 loop reduces the efficiency of interaction of virion-associated envelope molecules with CCR5 molecules present on the surface of target cells. Alternatively, the sugar molecules present on the V2 loop of SF162 interact directly with CCR5 molecules on the target cell surface. A third possible explanation for our observations could be that V2 loop deglycosylation alters the conformation of currently unknown envelope epitopes that interact with the CCR5 molecules.
As fully V2 loop-glycosylated envelopes appear to require less CCR5 for cell entry than partially deglycosylated envelopes (Table 1), our studies suggest that glycosylation of the V2 loop of the HIV envelope may offer CCR5-using HIV-1 isolates an initial advantage over CCR5-using isolates with a partially deglycosylated V2 loop in establishing infection in a new host by permitting a more efficient interaction of their envelope with the surface of naive CD4+ T lymphocytes, which express small numbers of CCR5 molecules.
In addition to allowing HIV to infect macrophages, glycosylation of the V2 loop offers protection from antibody-mediated neutralization (Tables 1 and 2). Removal of any one of the three asparagine-linked glycosylation sites examined renders the virus more susceptible to neutralization by antibodies produced during natural HIV infection. The recorded differences in neutralization susceptibility are not due to differences in the numbers of virion-associated envelope molecules between SF162 and the mutant viruses (Fig. 4). Most likely, V2 loop deglycosylation increases the exposure of neutralization epitopes. We previously reported that the elimination of 30 amino acids (from T160 to Y189), including the asparagine at position 186, from the central region of the V2 loop from the SF162 isolate renders the virus, termed SF162ΔV2, highly susceptible to neutralization by the same serum antibodies examined here (54). The studies conducted here reveal that the elimination of the glycosylation site at position 186 from the SF162 background renders the virus (GM186) more susceptible to neutralization than the parental SF162 virus. Even so, the GM186 virus is severalfold less susceptible to neutralization than the SF162ΔV2 virus with the sera tested (see reference 54 for comparison). Therefore, although the carbohydrate side chains present at position 186 of the SF162 envelope mask neutralization epitopes and contribute to the neutralization-resistant phenotype of this virus, this masking is not as extensive as that offered by the 30 amino acids of the V2 loop themselves. These amino acids may either mask additional epitopes or more efficiently mask the same epitopes as those masked by the sugar molecules present on the asparagine at position 186.
With the exception of serum B, which neutralized the GM154 and GM186 mutant viruses to the same extent as the parental SF162 virus, all the other HIV-positive human sera tested neutralized all three mutant viruses more efficiently than the SF162 virus. The fact that the three mutant viruses are not equally susceptible to neutralization by any given HIV-positive serum (Table 2) suggests that the sugar molecules present at positions 154, 186, and 195 of the SF162 envelope occlude neutralizing epitopes that are not identical but most likely overlap. It appears that glycosylation of the two asparagines located at the base of the V2 loop provides better protection from antibody-mediated neutralization to the SF162 isolate than glycosylation of the asparagine located within the V2 loop itself (position 186). Regardless, since the sera tested here were collected from patients infected with viruses heterologous to SF162, the epitopes that become exposed upon V2 loop deglycosylation of the SF162 envelope have common structural features among heterologous primary isolates. The fact that a given mutant virus is differentially susceptible to neutralization by the various HIV-positive sera tested suggests that the titers of antibodies recognizing the epitopes that are normally occluded by V2 loop glycosylation differ among the sera tested. Overall, the above results suggest that during HIV (or SIV) infection, even though the anti-envelope antibody responses evolve to neutralize the virus more efficiently (4, 12, 13, 32, 33), the virus itself can easily adapt and escape neutralization by altering the pattern of envelope glycosylation of the V2 loop (and most likely other regions) without affecting its replication ability in activated PBMC.
The sugar molecules present at the glycosylation sites examined appear to occlude neutralization epitopes located outside the V2 loop itself but within the CD4 binding site and the V3 loop (Table 3). These results are in agreement with conclusions from numerous studies reporting on a structural and functional interaction between the V1V2 region, the CD4 binding site, and the V3 loop of the HIV envelope (18, 21, 30, 34, 57, 66–69), as well as with results obtained following elucidation of the crystal structure of the gp120 core, which suggest that the V1V2 region of the HIV envelope folds over elements of the CD4 binding site and the V3 loop (26, 66–69). The three glycosylation sites examined, however, do not contribute equally to protection from neutralization by anti-CD4 binding site and anti-V3 loop antibodies. Glycosylation at the amino-terminal side (GM154) of the base of the V2 loop is more effective in preventing the binding of antibodies to these two envelope regions than glycosylation of the carboxy-terminal side (GM195). Also, in contrast to the protection from neutralization by anti-V3 loop MAbs offered by the glycosylation of the base of the V2 loop, glycosylation within the loop itself (GM186) does not appear to offer protection by such antibodies.
We previously reported that several V3 loop epitopes, including that recognized by MAb 391-95D, are occluded within the functional, virion-associated envelope (55). Our current studies indicate that this occlusion may be due (at least partially) to the sugar molecules present at the base of the V2 loop. Several groups, including ours, have reported that the V2 loop is implicated in determining viral tropism, either directly or in collaboration with the V3 loop (21, 22, 40, 61), which may interact with elements of the CCR5 (or CXCR4) coreceptor (11, 53, 70). Our present results suggest that V2 loop glycosylation, by masking specific V3 loop epitopes, can indirectly affect the interaction of the V3 loop with the coreceptor molecules and thus influence viral tropism.
In contrast to the observations made with the anti-CD4 binding site and anti-V3 loop MAbs, glycosylation of the three sites examined does not increase susceptibility to neutralization by MAbs 17b and 48d. These two MAbs recognize overlapping conformational epitopes that are CD4 induced and involved in envelope-chemokine receptor binding (60, 62, 65). Since elimination of the two glycosylation sites at the base of the V2 loop results in a less efficient interaction of the HIV envelope with the CCR5 chemokine receptor molecules on the surface of susceptible cells (Table 1), the sugar molecules present at these sites may be affecting an envelope-CCR5 binding step which follows that involving the epitopes recognized by MAbs 17b and 48d.
In summary, our current studies indicate that glycosylation of the V2 loop of HIV not only expands its cellular tropism by allowing it to more efficiently replicate in primary macrophages, but also protects the virus from neutralization by cross-reactive neutralizing antibodies. It would be interesting to compare the immunogenicity of envelope proteins derived from the GM154, GM186, and GM195 viruses to that of the SF162 envelope to determine whether the mutant envelopes elicit the generation of higher titers of neutralizing antibodies not only against SF162, but also against heterologous HIV-1 isolates.
ACKNOWLEDGMENTS
This work was supported partially by a Jonathan S. Canno Research Fund award from AMFAR (02572-23-RGV) and by the NIH (AI 44309-01).
We thank C. Cheng-Mayer for many helpful comments and suggestions. We acknowledge D. Kabat for providing the HeLa cell lines and S. Zolla-Pazner, M. Gorny, J. Robinson, and D. Burton for providing the MAbs used in this study.
REFERENCES
- 1.Back N K T, Smit L, De Jong J-J, Keulen W, Schutten M, Goudsmit J, Tersmette M. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology. 1994;199:431–438. doi: 10.1006/viro.1994.1141. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein H B, Tucker S P, Hunter E, Schutzbach J S, Compans R W. Human immunodeficiency virus type 1 envelope glycoprotein is modified by O-linked oligosaccharides. J Virol. 1994;68:463–468. doi: 10.1128/jvi.68.1.463-468.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botarelli P, Houlden B A, Haigwood N L, Servis C, Montagna D, Abrignani S. N-glycosylation of HIV-gp120 may constrain recognition by T lymphocytes. J Immunol. 1991;147:3128–3132. [PubMed] [Google Scholar]
- 4.Burns D P, Collignon C, Desrosiers R C. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J Virol. 1993;67:4104–4113. doi: 10.1128/jvi.67.7.4104-4113.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng-Mayer C, Brown A, Harouse J, Luciw P A, Mayer A J. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J Virol. 1999;73:5294–5300. doi: 10.1128/jvi.73.7.5294-5300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng-Mayer C, D. S, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 7.Cheng-Mayer C, Homsy J, Evans L A, Levy J A. Identification of human immunodeficiency virus subtypes with distinct patterns of sensitivity to serum neutralization. Proc Natl Acad Sci USA. 1988;85:2815–2819. doi: 10.1073/pnas.85.8.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of HIV-1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J A. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng-Mayer C, Seto D, Levy J A. Altered host range of HIV-1 after passage through various human cell types. Virology. 1991;181:288–294. doi: 10.1016/0042-6822(91)90494-v. [DOI] [PubMed] [Google Scholar]
- 11.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 12.Cole K S, Murphey-Corb M, Narayan O, Joag S V, Shaw G M, Montelaro R C. Common themes of antibody maturation to simian immunodeficiency virus, simian-human immunodeficiency virus, and human immunodeficiency virus type 1 infections. J Virol. 1998;72:7852–7859. doi: 10.1128/jvi.72.10.7852-7859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole K S, Rowles J L, Jagerski B A, Murphey-Corb M, Unangst T, Clements J E, Robinson J, Wyand M S, Desrosiers R C, Montelaro R C. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J Virol. 1997;71:5069–5079. doi: 10.1128/jvi.71.7.5069-5079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conley A J, Gorny M K, Kessler II J A, Boots L J, Ossorio-Castro M, Koenig S, Lineberger D W, Emini E A, Williams C, Zolla-Pazner S. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody 447-52D. J Virol. 1994;68:6994–7000. doi: 10.1128/jvi.68.11.6994-7000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewar R L, Vasudevachais M B, Natarajan V, Salzman N P. Biosynthesis and processing of human immunodeficiency virus type 1 envelope glycoproteins: effects of monensin on glycosylation and transport. J Virol. 1988;63:2452–2456. doi: 10.1128/jvi.63.6.2452-2456.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Marzio P, Tse J, Landau N R. Chemokine receptor regulation and HIV type 1 tropism in monocyte-macrophages. AIDS Res Hum Retroviruses. 1998;14:129–138. doi: 10.1089/aid.1998.14.129. [DOI] [PubMed] [Google Scholar]
- 17.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N P, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 18.Fox D G, Balfe P, Palmer C P, May J C, Arnold C, McKeating J A. Length polymorphism within the second variable region of the human immunodeficiency virus type 1 envelope glycoprotein affects accessibility of the receptor binding site. J Virol. 1997;71:759–765. doi: 10.1128/jvi.71.1.759-765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorny M K, Conley A J, Karwowska S, Buchbinder A, Xu J-Y, Emini E A, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruters R A, Neefjes J J, Tersmette M, De Golde R E Y, Tulp A, Huisman H G, Miedema F, Ploegh H L. Interference with HIV-induced syncytium formation and viral infectivity by inhibitors of trimming glycosidase. Nature (London) 1987;330:74–77. doi: 10.1038/330074a0. [DOI] [PubMed] [Google Scholar]
- 21.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koito A, Stamatatos L, Cheng-Mayer C. Small amino acid sequence changes within the V2 domain can affect the function of a T-cell line-tropic human immunodeficiency virus type 1 envelope gp120. Virology. 1995;206:878–884. doi: 10.1006/viro.1995.1010. [DOI] [PubMed] [Google Scholar]
- 23.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L J, Choe H, Sodroski J. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 25.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature (London) 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lapham C K, Zaitseva M B, Lee S, Romanstseva T, Golding H. Fusion of monocytes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nat Med. 1999;5:303–308. doi: 10.1038/6523. [DOI] [PubMed] [Google Scholar]
- 28.Leonard C K, Spellman M W, Riddle L, Harris R J, Thomas J N, Gregory T J. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 29.Mathews T J, Weinhold K J, Lyerly H K, Langois A J, Wigzell H, Bolognesi D P. Interaction between the human T-cell lymphotropic virus type IIIB envelope glycoprotein gp120 and the surface antigen CD4: role of carbohydrate in binding and cell fusion. Proc Natl Acad Sci USA. 1987;84:5424–5428. doi: 10.1073/pnas.84.15.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo H, Stamatatos L, Ip J E, Barbas C F, Parren P W H I, Burton D R, Moore J P, Ho D D. Human immunodeficiency virus type 1 mutants that escape neutralization by human monoclonal antibody IgG1b12. J Virol. 1997;71:6869–6874. doi: 10.1128/jvi.71.9.6869-6874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montefiori D C, Robinson W E, Mitchell W M. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1988;85:9248–9252. doi: 10.1073/pnas.85.23.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montelaro R C, Cole K S, Hammond S A. Maturation of immune responses to lentivirus infection: implications for AIDS vaccine development. AIDS Res Hum Retroviruses. 1998;14(Suppl. 3):S255–S259. [PubMed] [Google Scholar]
- 33.Moog C, Fleury H J, Pellegrin I, Kirn A, Aubertin A M. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J Virol. 1997;71:3734–3741. doi: 10.1128/jvi.71.5.3734-3741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore J P, Sattentau Q J, Yoshiyama H, Thali M, Charles M, Sullivan N, Poon S-W, Fung M S, Traincard F, Pinkus M, Robey G, Robinson J E, Ho D D, Sodroski J. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J Virol. 1993;67:6136–6151. doi: 10.1128/jvi.67.10.6136-6151.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naif H M, Li S, Alali M, Sloane A, Wu L, Kelly M, Lynch G, Lloyd A, Cunningham A L. CCR5 expression correlates with susceptibility of maturing monocytes to human immunodeficiency virus type 1 infection. J Virol. 1998;72:830–836. doi: 10.1128/jvi.72.1.830-836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narayan S V, Mukherjee S, Jia F, Li Z, Wang C, Foresman L, McCormick-Davis C, Stephens E B, Joag S V, Narayan O. Characterization of a neutralization-escape variant of SHIVKU-1, a virus that causes acquired immune deficiency syndrome in pig-tailed macaques. Virology. 1999;256:54–63. doi: 10.1006/viro.1999.9605. [DOI] [PubMed] [Google Scholar]
- 37.Overbaugh J, Rudensey L M. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J Virol. 1992;66:5937–5948. doi: 10.1128/jvi.66.10.5937-5948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R E, Morton W R. Variation in simian immunodeficiency virus Env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pal R, Hoke G M, Sarngadharan M G. Role of oligosaccharides in the processing and maturation of envelope glycoproteins of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1988;86:3384–3388. doi: 10.1073/pnas.86.9.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer C, Balfe P, Fox D, May J C, Frederiksson R, Fenyo E M, McKeating J A. Functional characterization of the V1V2 region of human immunodeficiency virus type 1. Virology. 1996;220:436–449. doi: 10.1006/viro.1996.0331. [DOI] [PubMed] [Google Scholar]
- 41.Papandreou M-J, Fenouillet E. Effect of changes in the glycosylation of the human immunodeficiency virus type 1 envelope on the immunoreactivity and sensitivity to thrombin of its third variable domain. Virology. 1998;241:163–167. doi: 10.1006/viro.1997.8930. [DOI] [PubMed] [Google Scholar]
- 42.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reitter J N, Desrosiers R C. Identification of replication-competent strains of simian immunodeficiency virus lacking multiple attachment sites for N-linked carbohydrates in variable regions 1 and 2 of the surface envelope protein. J Virol. 1998;72:5399–5407. doi: 10.1128/jvi.72.7.5399-5407.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 45.Robinson W E, Montefiori D C, Mitchell W M. Evidence that mannosyl residues are involved in human immunodeficiency virus type 1 (HIV-1) pathogenesis. AIDS Res Hum Retroviruses. 1987;3:265–282. doi: 10.1089/aid.1987.3.265. [DOI] [PubMed] [Google Scholar]
- 46.Rudensey L M, Kimata J T, Benveniste R E, Overbaugh J. Progression to AIDS in macaques is associated with changes in the replication, tropism, and cytopathic properties of the simian immunodeficiency virus variant population. Virology. 1995;207:528–542. doi: 10.1006/viro.1995.1113. [DOI] [PubMed] [Google Scholar]
- 47.Rudensey L M, Kimata J T, Long E M, Chackerian B, Overbaugh J. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR5-coreceptor recognition. J Virol. 1998;72:209–217. doi: 10.1128/jvi.72.1.209-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schonning K, Bolmstedt A, Novotny J, Lund O S, Olofsson S, Hansen J-E S. Induction of antibodies against epitopes inaccessible on the HIV type 1 envelope oligomer by immunization with recombinant monomeric glycoprotein 120. AIDS Res Hum Retroviruses. 1998;14:1451–1456. doi: 10.1089/aid.1998.14.1451. [DOI] [PubMed] [Google Scholar]
- 49.Schonning K, Jansson B, Olofsson S, Nielsen J O, Hansen J-E S. Resistance to V3-directed neutralization caused by an N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology. 1996;218:134–140. doi: 10.1006/viro.1996.0173. [DOI] [PubMed] [Google Scholar]
- 50.Seligman S J, Binley J M, Gorny M K, Burton D R, Zolla-Pazner S, Sokolowski K A. Characterization by serial competition ELISAs of HIV-1 V3 loop epitopes recognized by monoclonal antibodies. Mol Immunol. 1996;33:737–745. doi: 10.1016/0161-5890(96)00044-2. [DOI] [PubMed] [Google Scholar]
- 51.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature (London) 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 52.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamatatos L, Cheng-Mayer C. An envelope modification that renders a primary, neutralization-resistant, clade B HIV-1 isolate highly susceptible to neutralization by sera from other clades. J Virol. 1998;72:7840–7845. doi: 10.1128/jvi.72.10.7840-7845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stamatatos L, Cheng-Mayer C. Structural modulations of the envelope gp120 glycoprotein of human immunodeficiency virus type 1 upon oligomerization and differential V3 loop epitope exposure of isolates displaying distinct tropism upon virion-soluble receptor binding. J Virol. 1995;69:6191–6198. doi: 10.1128/jvi.69.10.6191-6198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stamatatos L, Werner A, Cheng-Mayer C. Differential regulation of cellular tropism and sensitivity to soluble CD4 neutralization by the envelope gp120 of human immunodeficiency virus type 1. J Virol. 1994;68:4973–4979. doi: 10.1128/jvi.68.8.4973-4979.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stamatatos L, Wiskerchen M, Cheng-Mayer C. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV-1 isolate on viral envelope structure, cell-entry and replication. AIDS Res Hum Retroviruses. 1998;14:1129–1139. doi: 10.1089/aid.1998.14.1129. [DOI] [PubMed] [Google Scholar]
- 58.Stamatatos L, Zolla-Pazner S, Gorny M, Cheng-Mayer C. Binding of antibodies to virion-associated gp120 molecules of primary-like human immunodeficiency virus type 1 (HIV-1) isolates: effect on HIV-1 infection of macrophages and peripheral blood mononuclear cells. Virology. 1997;229:360–369. doi: 10.1006/viro.1997.8443. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thali M, Moore J P, Furma C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toohey K, Wehrly K, Nishio J, Perryman S, Chesebro B. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology. 1995;213:70–79. doi: 10.1006/viro.1995.1547. [DOI] [PubMed] [Google Scholar]
- 62.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR5. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 63.Tuttle D L, Harrison J K, Anders C, Sleasman J W, Goodenow M M. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J Virol. 1998;72:4962–4969. doi: 10.1128/jvi.72.6.4962-4969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker B D, Kowalski M, Goh W C, Kozarsky K, Krieger M, Rosen C, Rohrschneider L, Haseltine W A, Sodroski J. Inhibition of human immunodeficiency virus syncytium formation and virus replication by castanosperine. Proc Natl Acad Sci USA. 1987;84:8120–8124. doi: 10.1073/pnas.84.22.8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR5. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 66.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature (London) 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 67.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 69.Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao L, Owen S M, Goldman I, Lal A A, deJong J J, Goudsmit J, Lal R B. CCR5 coreceptor usage of non-syncytium-inducing primary HIV-1 is independent of phylogenetically distinct global HIV-1 isolates: delineation of consensus motif in the V3 domain that predicts CCR-5 usage. Virology. 1998;240:83–92. doi: 10.1006/viro.1997.8924. [DOI] [PubMed] [Google Scholar]