Abstract
Previous studies have indicated the efficacy of momordin Ic (MIc), a plant-derived triterpenoid, against several types of cancers, implying its potential for further development. However, comprehensive insights into the molecular mechanisms and targets of MIc in cholangiocarcinoma (CCA) are lacking. This study aimed to investigate the actions of MIc against CCA at the molecular level. Network pharmacology analysis was first employed to predict the mechanisms and targets of MIc. The results unveiled the potential involvement of MIc in apoptosis and cell migration, pinpointing Src and FAK as key targets. Subsequently, cell-based assays, in accordance with FAK/Src-associated metastasis, were conducted, demonstrating the ability of MIc to attenuate the metastatic behaviours of KKU-452 cells. The in vitro results further indicated the capability of MIc to suppress the epithelial-mesenchymal transition (EMT) process, notably by downregulating EMT regulators, including N-cadherin, vimentin, ZEB2 and FOXC1/2 expression. Furthermore, MIc suppressed the activation of the FAK/Src signalling pathway, influencing critical downstream factors such as MMP-9, VEGF, ICAM-1, and c-Myc. Molecular docking simulations also suggested that MIc could interact with FAK and Src domains and restrain kinases from being activated by hindering ATP binding. In conclusion, this study employs a comprehensive approach encompassing network pharmacology analysis, in vitro assays, and molecular docking to unveil the mechanisms and targets of MIc in CCA. MIc mitigates metastatic behaviours and suppresses key pathways, offering a promising avenue for future therapeutic strategies against this aggressive cancer.
Keywords: Momordin Ic, Cholangiocarcinoma, Network pharmacology, Antimetastasis, FAK, Src
1. Introduction
Cholangiocarcinoma (CCA) or bile duct cancer, is a cancer that threatens human life, especially in individuals residing in Southeast Asia, due to its high incidence and mortality in the region [1]. Infection of the liver flukes by Opisthorchis viverrini and Clonorchis sinensis by ingestion of raw or fermented fish is the most important risk factor for CCA in this area [2,3]. The mechanisms of liver fluke-induced CCA are caused by infection-related inflammation and the resulting oxidative stress [3]. While surgical resection is effective for early-stage CCA, advanced stage or metastasis cases often lack this option and rely on chemotherapy. The current guidelines regarding the regimen of advanced biliary tract cancers are cisplatin plus gemcitabine, showing an advantage in overall survival from this combination compared to gemcitabine alone [4]. Moreover, second-line treatments such as 5-fluorouracil and oxaliplatin are recommended when gemcitabine-based treatments are not successful [5]. Nonetheless, resistance to current chemotherapeutic agents in CCA is a major problem in the failure of therapy [[4], [5], [6]]. Hence, counteractions to improve CCA treatment are important. Several strategies have been applied to enhance treatment effectiveness, including the use of bioactive phytochemicals.
Medicinal plants have a long history of yielding bioactive components for drug development. Cumulative evidence has highlighted the importance of plant-derived phytochemicals as valuable sources of effective therapeutic agents [7,8]. Among these, momordin Ic (MIc), abundantly present in edible plants such as Momordica charantia and Kochia scoparia, has gained attention. These plants have been used in traditional Chinese medicines [9,10]. Previous studies demonstrated an excellent spectrum of pharmacological activities of MIc, such as anti-inflammatory [11], anti-rheumatic [10] and hepatoprotective effects [12]. The antitumour activities of MIc have been reported in several types of cancers, such as hepatocellular carcinoma and colon cancer [13,14]. MIc also exhibits potent apoptosis-inducing effects in CCA cells [15]. Nevertheless, the anticancer activities of MIc against CCA and the biological molecular mechanisms accounting for these effects remain challenging to investigate.
Network pharmacology is an approach in drug development that integrates systems biology and network analysis. Instead of focusing on a single target, network pharmacology works on multiple targets, in which a drug works against several diseases and yields overall interactions of possible drug targets [16]. The approach thus represents a powerful tool to investigate the molecular mechanism(s) and target(s) of an agent of interest [17,18]. In this study, a network of possible targets of MIc against CCA was initially constructed using network pharmacology to predict the relevant mechanisms and targets. Based on network pharmacology analysis, a cell-based assessment was then performed to elucidate the actions of MIc against CCA. KKU-452, a CCA cell line with high metastatic potential [19], was used as a cancer cell model since it represents advanced-stage CCA characteristics. Consequently, molecular docking analysis was applied as supporting evidence of network pharmacology and cell-based assays regarding the molecular interaction between MIc and its targets. Accordingly, by utilizing network pharmacology, in vitro investigation, and molecular docking, our results demonstrated not only the effectiveness of MIc but also mechanistic insight into the anticancer effect of MIc against CCA, implying its potential for further development.
2. Material and methods
2.1. Materials
Reagents and chemicals for cell culture were purchased from Gibco BRL Life Technologies (NY, USA). MIc (purity ≥98 %, Cat. No. CFN99726) was obtained from ChemFaces Biochemical (Wuhan, China). StemXVivo® EMT Inducing Media Supplement (Cat. No. CCM017) was purchased from R&D Systems (MN, USA). Antibodies against Src (#BF0357), N-cadherin (#AF5239), ZEB2 (#AF5278) and FOXC1/2 (#DF3252) were purchased from Affinity Biosciences (Ohio, USA). Antibodies against c-Myc (#5605), p-FAK (Tyr925, #3284) and p-Src (Tyr416, #2101) were purchased from Cell Signaling Technology (MA, USA). Antibodies against VEGF (sc-7269), ICAM-1 (sc-8439), vimentin (sc-6260), MMP-9 (sc-12759), FAK (sc-271195), β-actin (sc-47778), goat anti-mouse IgG-HRP (sc-2031), goat anti-rabbit IgG-HRP (sc-2030) and mouse anti-goat anti-rabbit IgG-HRP (sc-2354) were purchased from Santa Cruz Biotechnology (CA, USA). Goat polyclonal anti-mouse IgG (Alexa Fluor 488) (ab150113) and donkey polyclonal anti-rabbit IgG (Alexa Fluor 488) (ab150073) were purchased from Abcam (MA, USA).
2.2. Identification of MIc targets in CCA
MIc targets were retrieved by uploading MIc Canonical SMILES into the Swiss Target Prediction database (http://www.swisstargetprediction.ch). CCA-associated targets were retrieved using the keyword “cholangiocarcinoma” in the OMIM (https://www.omim.org), DisGeNET (https://www.disgenet.org), and GeneCards (https://www.genecards.org) databases. The acquired sets of MIc and CCA targets were then unioned and intersected using VENNY 2.1 (https://bioinfogp.cnb.csic.es/tools/venny) and displayed as a Venn diagram.
2.3. Gene Ontology (GO) and Kyoto Encyclopedia of genes and Genomes (KEGG) enrichment analysis
KEGG and GO analyses were conducted to pinpoint the cellular pathways and processes associated with the MIc targets. KEGG maps genes onto established pathways, offering details on specific biological pathways. In contrast, GO categorizes genes based on their functional characteristics across defined aspects. In brief, KEGG addresses pathways, while GO categorizes genes functionally. KEGG enrichment analysis was performed using Shiny GO 0.77 (http://bioinformatics.sdstate.edu/go) [20] with human species selected and a p value (FDR: false discovery rate) cut-off of 0.05. For GO enrichment analysis, Cytoscape (version 3.10.0) with ClueGo (version 2.5.10) was applied for functional annotation clustering—according to the “Homo sapiens” model—based on 4 aspects, including biological process (GO:0008150), cellular component (GO:0005575), immune system process (GO:0002376), and molecular function (GO:0003674), with a p value (two-sided hypergeometric test) cut-off of 0.05.
2.4. PPI (protein-protein interaction) construction
PPI network construction was performed using the STRING database (version 11.0) (https://string-db.org) in accordance with the “Homo sapiens” model. The interaction confidence was set at 0.9. The network nodes and edges represent proteins (targets) and interactions, respectively. The core target proteins were identified and sorted using Cytoscape—cytoHubba plug-in (version 0.1)—based on the degree topological algorithm.
2.5. Cell line and cell culture
The human CCA cell line KKU-452 was kindly provided by the Cholangiocarcinoma Research Institute, Khon Kaen University and was cultivated in Ham's F12 media (Gibco-BRL Life Technologies, Grand Island, New York) containing 10 % foetal calf serum (HiMedia Laboratories, Mumbai, India) supplemented with 10 mM HEPES (pH 7.3), sodium bicarbonate, 100 μg/mL gentamicin and 100 U/mL penicillin at 37 °C in a humidified incubator with 5 % CO2.
2.6. Cytotoxicity assay
The sulforhodamine B (SRB) assay was carried out to assess the effect of MIc on KKU-452 cell viability. Briefly, the cells were seeded in 96-well plates and treated with MIc at various concentrations (0.1–10 μM) for 24 h. Following treatment, the cells were fixed with 10 % trichloroacetic acid and stained with 0.4 % SRB dye. Thereafter, the excess dye was washed with 1 % acetic acid, and the protein-bound dye was solubilized in 10 mM Tris solution. A microplate reader (Sunrise™, TECAN Austria GmbH, Austria) was used to determine the colour intensity at 540 nm.
2.7. Clonogenic assay
KKU-452 cells were cultured onto 6-well plates at a density of 2.5 × 105 cells/well and treated with vehicle at various concentrations of MIc (0.5, 1, 2.5 μM) for 48 h. After complete treatment, the cells were trypsinized and plated onto a new 6-well plate at a density of 600 viable cells per well in fresh media for further incubation for 7 days with fresh media replacement every 2 days. Thereafter, the cultured cells were rinsed, fixed with absolute methanol at 4 °C for 15 min and stained with 0.5 % crystal violet in 2 % ethanol for 20 min. The plates were then washed, air-dried, and photographed using a ChemiDoc™ MP imaging system (Bio-Rad Laboratories, USA). The number of colonies in each well was counted using Image-Pro Plus (Media Cybernetics, Inc.)
2.8. Migration assay
The wound healing method was used to determine the ability of cells to migrate. KKU-452 cells were cultured in a 24-well plate and allowed to attach overnight. The bottom of the well was scratched using a sterile pipette tip to make a wound, and the detached cells were removed by washing with PBS. The cultured cells were then treated with vehicle or various concentrations of MIc (0.5, 1 and 2.5 μM) for 48 h. A series of wound images were photographed at 0 h as baseline and after complete treatment using a Nikon ECLIPSE TS100 inverted microscope (Nikon Instruments Inc., Japan). The closing of the scratched wound, representing the migration process, was determined by the capture of the denuded area along the scratch using Image-Pro Plus software (Media Cybernetics, LP, USA). The wound distance was determined by dividing the area by the length of the scratch. The migration ability expressed as the percentage of wound gap closure of the control and each treatment condition was calculated by comparing the wound distance with its baseline value (at 0 h) as follows:
2.9. Invasion assay
The Transwell invasion assay was carried out using Matrigel-coated 8 μm pore size polycarbonate membrane Transwell inserts (BD Biosciences, Bedford, MA). KKU-452 cells treated with vehicle or MIc (0.5, 1 or 2.5 μM) were seeded in the top compartment of the Transwell chamber. Medium containing 10 % FBS was added to the bottom chamber as a chemoattractant. To allow invasion, the cells were further incubated for 48 h. After complete incubation, the invading cells were fixed using methanol and stained using crystal violet. The cells were then photographed under a microscope (ECLIPSE Ni–U upright Microscope, Nikon Instruments Inc., Japan) and analysed by ImagePro Plus software (Media Cybernetics, LP, USA).
2.10. Adhesion assay
The cell adhesion assay was performed using a 96-well plate coated with 100 μL of 0.64 μg/mL fibronectin per well. KKU-452 cells were exposed to vehicle or different concentrations of MIc (0.5, 1, 2.5 μM) for 48 h. Thereafter, control cells or MIc-treated cells were trypsinized, counted and placed on fibronectin-precoated plates, and allowed to attach for 30 min. The unattached cells were then removed and the attached cells were fixed with methanol, stained with crystal violet and photographed using a Nikon ECLIPSE TS100 inverted microscope (Nikon Instruments Inc., Japan). After that, 5 % acetic acid in 10 % methanol was added to solubilize the dye and the absorbance was measured at 540 nm using a microplate reader (Sunrise™, TECAN Austria GmbH, Austria).
2.11. EMT induction
StemXVivo® EMT-inducing media supplement (R&D Systems) was used to induce the EMT process in KKU-452 cells, following the manufacturer's instructions. Briefly, the cells were cultured in their original media that were supplemented with the induction reagent. Three days after plating, the media were removed from the plates and fresh media containing EMT-inducing media supplement were added. The cultured cells were additionally grown for 2 days. Thereafter, the cells were ready for analysis. To test the effect of MIc on the EMT process, KKU-452 cells were cultured in media containing StemXVivo® EMT-inducing media supplement in the absence or presence of the compound at 1 or 2.5 μM. Five days after plating, the cells were used for determining protein expression using Western blotting and immunocytofluorescence analyses.
2.12. Western blot analysis
KKU-452 cells were treated with vehicle or MIc (0.5, 1, 2.5 μM) for 24 or 48 h. After treatment, the cells were harvested, and whole cell lysates were collected using RIPA lysis buffer containing 1 % Halt™ proteinase and phosphatase inhibitor cocktail (78441, Thermo Scientific, Rockford, USA). Protein samples (30 μg) were resolved by 10 % SDS-PAGE and blotted onto a polyvinylidene difluoride membrane. The membranes were incubated with 5 % bovine serum albumin in TBS buffer with Tween-20 (TBST) to block nonspecific binding sites. Subsequently, the blots were probed with specific antibodies at 4 °C overnight against N-cadherin (1:500), vimentin (1:1000), ZEB2 (1:1000), FOXC1/2 (1:1000), p-FAK (1:1000), p-Src (1:1000), FAK (1:1000), Src (1:1000), VEGF (1:1000), ICAM-1 (1:1000), MMP-9 (1:1000), c-Myc (1:1000) and β-actin (1:5000). After complete incubation, the membranes were intensively washed with TBST and further incubated with goat anti-mouse IgG-HRP (1:2000), goat anti-rabbit IgG-HRP (1:2000) or mouse anti-goat anti-rabbit IgG-HRP (1:2000) secondary antibodies for 2 h at room temperature. The target protein bands were identified using an enhanced chemiluminescence detection system (Luminata™ Forte Western HRP Substrate, Merck Millipore, Watford, UK) and imaged by a ChemiDoc™ MP Imaging system.
2.13. Immunocytofluorescence analysis
After treatment with vehicle or MIc (0.5, 1, 2.5 μM) for 24 or 48 h, KKU-452 cells were seeded on a cell culture slide, fixed with 4 % paraformaldehyde, permeabilized with 0.2 % Triton X-100 and blocked with 5 % bovine serum albumin. The cells were then immunolabelled with specific primary antibodies at 4 °C overnight against N-cadherin (1:200), vimentin (1:500), ZEB2 (1:500), FOXC1/2 (1:500), p-FAK (1:500), p-Src (1:500), VEGF (1:500), ICAM-1 (1:500), MMP-9 (1:500) and c-Myc (1:500). Thereafter, the cells were washed with PBS and incubated with goat polyclonal anti-mouse IgG (Alexa Fluor 488) (1:1000) or donkey polyclonal anti-rabbit IgG (Alexa Fluor 488) (1:1000) at room temperature. Subsequently, the cells were mounted with VECTASHIELD® PLUS Antifade Mounting Medium with DAPI (Vector Laboratories, CA, USA, H-1200-10), visualized and photographed under an upright fluorescence microscope (Nikon Eclipse TS2-FL, Nikon Instruments Inc., Japan).
2.14. Molecular docking
Molecular docking was performed according to a previous report [21] with minor modifications. The 3D structure of the ligand MIc (PubChem ID: 176596) was drawn using ChemDraw 3D (version 12.0). Ligand energy minimization was performed using PyRx (Pyrx-Python Prescription 0.8) with the Merck molecular force field parameter (MMFF94). The 3D crystal structures of human Src and FAK proteins were obtained from the Protein Data Bank and refined by removing water, solvent, and ligands using PyMOL software (version 2.4.1) (PyMOL Molecular Graphics System, Schrödinger). Molecular docking between the ligand and proteins was then performed using PyRx. The complexes were visually analysed by PyMOL, and 2D interactions were examined using Discovery Studio Visualizer (version 21.1.0.20298) (BIOVIA, San Diego, CA, USA).
2.15. Statistical analysis
Differences between the control and treatment groups in in vitro assays were analysed using analysis of variance with a post hoc Student-Newman-Keuls (SNK) multiple comparison test, where p < 0.05 was considered statistically significant. Statistical analysis was carried out using GraphPad Prism 8.0 software.
3. Results
3.1. MIc targets in CCA
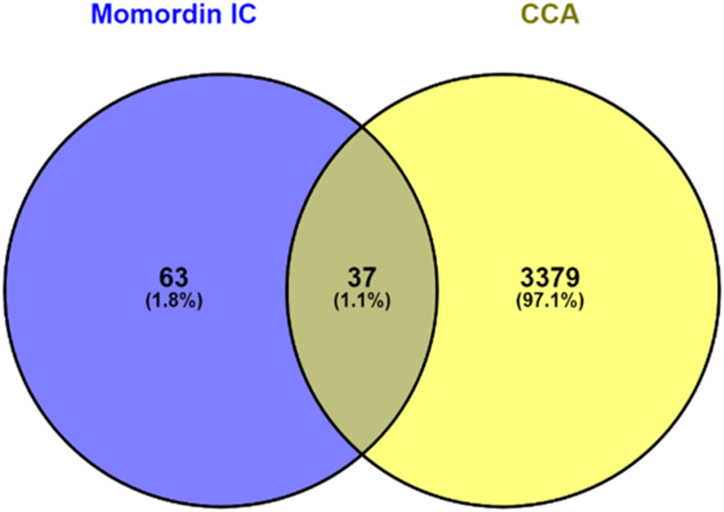
For MIc, 100 targets were predicted based on the Swiss Target Prediction database. For CCA, a total of 4016 targets were acquired from the GeneCards, DisGeNET, and OMIM databases. The 37 possible targets of MIc against CCA were then sorted out using a Venn diagram (Fig. 1) and are listed in Table 1.
Fig. 1.
Venn diagram displaying the intersection of MIc targets and CCA targets.
Table 1.
Possible targets of MIc in CCA.
| No. | Targets | Description |
|---|---|---|
| 1 | CYP2D6 | Cytochrome P450 2D6 |
| 2 | TPH1 | Tryptophan 5-hydroxylase 1 |
| 3 | SIRT1 | NAD-dependent deacetylase sirtuin 1 |
| 4 | PTGER1 | Prostanoid EP1 receptor |
| 5 | AGTR1 | Type-1 angiotensin II receptor |
| 6 | CASR | Calcium sensing receptor |
| 7 | SRC | Tyrosine-protein kinase SRC |
| 8 | LCK | Tyrosine-protein kinase LCK |
| 9 | MAPK14 | MAP kinase p38 alpha |
| 10 | KDR | Vascular endothelial growth factor receptor 2 |
| 11 | ITGA4 | Integrin alpha-4 |
| 12 | VDR | Vitamin D receptor |
| 13 | RORC | Nuclear receptor ROR-gamma |
| 14 | NR3C1 | Glucocorticoid receptor |
| 15 | GRB2 | Growth factor receptor-bound protein 2 |
| 16 | BCL2L1 | Apoptosis regulator Bcl-X |
| 17 | RRM1 | Ribonucleoside-diphosphate reductase M1 chain |
| 18 | PPM1A | Protein phosphatase 2C alpha |
| 19 | PTPA | Protein phosphatase 2A regulatory subunit B′ |
| 20 | PTPN6 | Protein-tyrosine phosphatase 1C |
| 21 | METAP2 | Methionine aminopeptidase 2 |
| 22 | CASP3 | Caspase-3 |
| 23 | F10 | Thrombin and coagulation Factor X |
| 24 | MMP9 | Matrix metalloproteinase 9 |
| 25 | CASP7 | Caspase-7 |
| 26 | CASP8 | Caspase-8 |
| 27 | DPP4 | Dipeptidyl peptidase IV |
| 28 | MME | Neprilysin |
| 29 | F2 | Thrombin |
| 30 | ACE | Angiotensin-converting enzyme |
| 31 | DNMT3B | DNA (cytosine-5)-methyltransferase 3B |
| 32 | IL2 | Interleukin-2 |
| 33 | TLR9 | Toll-like receptor (TLR7/TLR9) |
| 34 | GLI1 | Zinc finger protein GLI1 |
| 35 | JUN | Proto-oncogene c-JUN |
| 36 | STAT3 | Signal transducer and activator of transcription 3 |
| 37 | TYMS | Thymidylate synthase |
3.2. GO and KEGG enrichment analysis
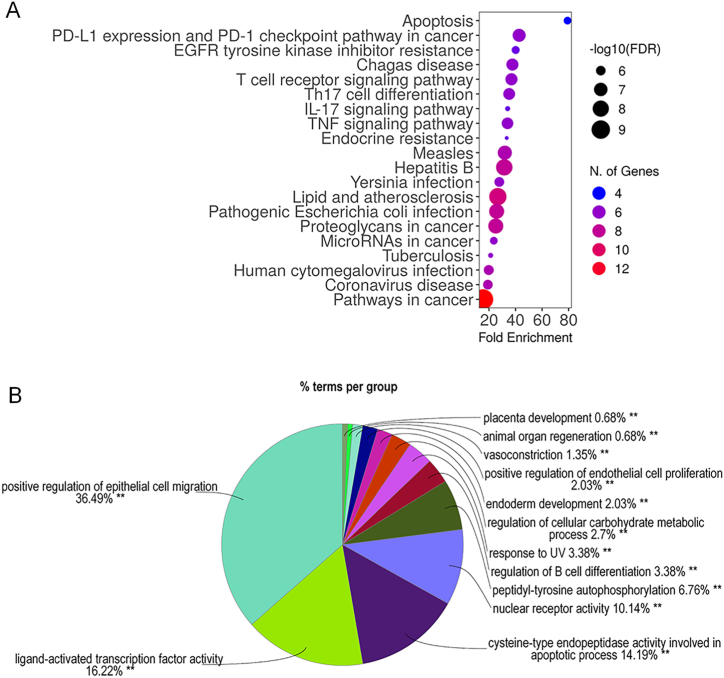
KEGG enrichment analysis was used to retrieve information from the candidate targets regarding their involvement in the relevant pathways. In this study, KEGG enrichment analysis implied that the candidate targets were components of the pathways involved in apoptosis, PD-1 and PDL-1 signalling, and EGFR inhibitor resistance, with fold enrichment percentages of 79.2 %, 42.7 %, and 40.1 %, respectively (Fig. 2A). The data suggested that MIc may have an action in these pathways, especially apoptosis (79.2 %), according to the highest percentage of enrichment. GO enrichment analysis was further carried out to gain additional information on the relevant gene functions. Regarding four aspects (biological process, cellular component, immune system process, and molecular function), GO enrichment analysis indicated that MIc may be active against CCA due to its attribution to signalling molecules involved mainly in the regulation of epithelial cell migration (36.5 %) and ligand-activated transcription factors (16.2 %) (Fig. 2B).
Fig. 2.
GO and KEGG enrichment analysis of the potential targets of MIc in CCA. (A) KEGG enrichment analysis demonstrates the pathways involved. The y- and x-axes represent significant KEGG pathways and the percentage of fold enrichment, respectively. The gradient of colour represents target counts, and the size of nodes represents the false discovery rate. (B) Overview chart of GO enrichment analysis demonstrates the functions or signalling involved as the percentage of terms (function/signalling) per group.
3.3. PPI network construction
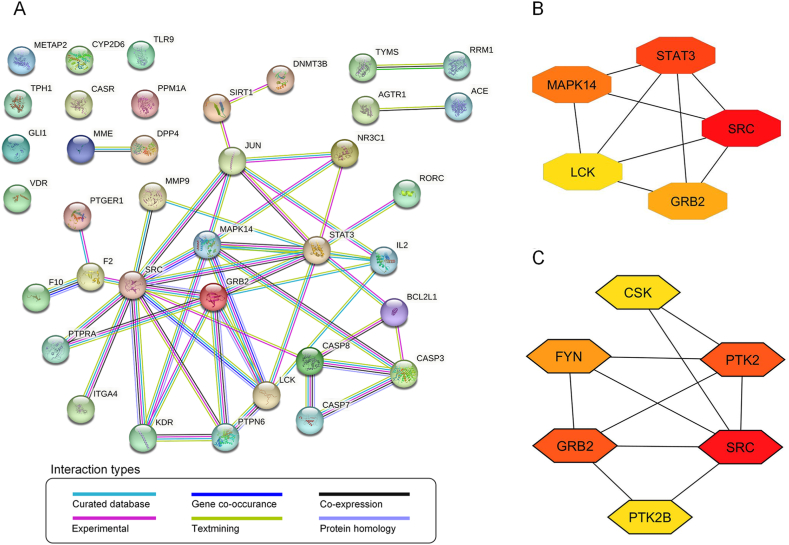
To gain a better understanding of the target interactions, PPIs of all intersected targets were constructed and visualized based on STRING analysis. The high confidence interaction between nodes (targets) analysed in the full string network is depicted in Fig. 3A, consisting of 37 nodes and 49 edges, with each edge representing the PPIs. The average node degree, an average number of interactions on a protein, was 1.77. The local clustering coefficient, corresponding to the wellness of the connected nodes, was 0.45. Out of 37 targets (nodes), major interactions (46 edges) link 23 targets together, forming the main cluster. The average node degree (1.77) and local clustering coefficient (0.45) indicate moderate connections and clustering among proteins in the network. However, 23 out of 37 proteins formed the main cluster (62 % of all possible targets), suggesting an important, tightly interconnected group of proteins. This might represent a specific biological pathway, a protein signalling pathway, or a functional module within the interaction network.
Fig. 3.
Protein-protein interaction (PPI) analysis. (A) PPI network of 37 potential MIc targets against CCA obtained from the STRING database. (B) The top 5 targets of potential MIc targets and (C) the top 5 targets of Src with only “physical interactions” were ranked using Cytoscape with cytoHubba plug-in. The higher degree value is represented by colours ranging from red to yellow.
To identify the most contributing targets inside the cluster, the PPI network was analysed using Cytoscape. The targets were then ranked according to the degree parameter. Since Degree relates to the number of connected nodes with individual nodes, a higher Degree represents a characteristic of a hub (highly connected target). Our results showed that the highest degree target was Src (Fig. 3B). STRING analysis was run again to identify any physical interaction of Src—analysed on the physical subnetwork—followed by degree ranking using Cytoscape. The results showed that the most Src-associated proteins were GRB2 and PTK2 (Fig. 3C). Since PTK2, encoding focal adhesion kinase (FAK), forms a complex with Src (FAK-Src complex) and GRB2 functions in downstream signalling, Src and FAK were then selected as the most likely targets of MIc for further in vitro investigation in this study.
3.4. MIc diminished metastasis-related behaviours of CCA cells
PPI analysis indicated that MIc may interact with Src and FAK (PTK2). Moreover, the interaction is involved in the process of apoptosis and cell migration according to enrichment analysis. Apoptosis is an important physiological process of cellular suicide that eliminates unwanted cells to maintain tissue homoeostasis. The process also plays a critical role in the metastatic process since malignant tumour cells must overcome this programmed cell death to metastasize [22]. We previously reported the effect of MIc on the apoptosis of CCA cells [15]. Therefore, in this study, we focused on cancer metastasis, comprising cell migration as a fundamental hallmark. The effect of MIc on metastasis potential, including the migration, invasion, and adhesion abilities of CCA cells, was then examined.
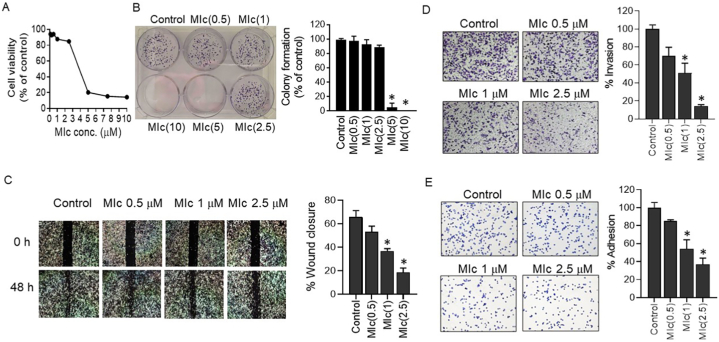
The effect of MIc on metastatic potential was initially investigated by determining its effect on cell migration and invasion in KKU-452 cells. To ascertain that the antimetastatic activity was not due to the cytotoxic and antiproliferation effects of the tested compound, SRB and clonogenic assays were carried out to determine the effect of MIc on cell viability and proliferation, respectively, prior to determining cell migration and invasion. As shown in Fig. 4A and B, MIc at concentrations ranging from 0.5 to 2.5 μM nonsignificant affected CCA cell viability and proliferation. Therefore, these concentrations were used for subsequent experiments. The effect of MIc on migration and invasion properties was assessed using wound-healing and Transwell invasion methods, respectively. The results showed that the wound field gap closure, which represents the migration ability, was 65.7 ± 5.6 % in the control group. For the treatment groups, the wound gap was closed by 53.2 ± 4.8 %, 36.8 ± 2.0 %, and 18.5 ± 3.8 % when treated with MIc at 0.5, 1 and 2.5 μM, respectively. The results indicated that treatment with MIc significantly reduced the motility of CCA cells (Fig. 4C). Similarly, the compound significantly inhibited the invasiveness of KKU-452 cells in a dose-dependent manner (Fig. 4D). These data indicated the antimigratory and anti-invasive efficacy of MIc in CCA cells. Apart from motility and invasive properties, the ability of tumour cells to adhere to the extracellular matrix has been considered essential for metastatic cancer cells to settle at the secondary target site [23]. In the present study, the effect of MIc on the adhesion of KKU-452 cells was also determined. The results showed that the compound decreased the cell-matrix interactions (Fig. 4E), suggesting the antiadhesive activity of MIc against CCA cells.
Fig. 4.
Effects of MIc on the metastatic behaviours of CCA cells. KKU-452 cells were exposed to different MIc concentrations. The initial evaluation included (A) cell viability and (B) colony forming ability. Subsequently, assessments consisting of (C) migration, (D) invasion, and (E) adhesion capabilities of MIc were performed. Representative figures and quantitative graphs are provided. Data are expressed as the mean ± SD from three separate experiments. *, p < 0.05 compared to the respective control.
3.5. MIc suppressed EMT in CCA cells
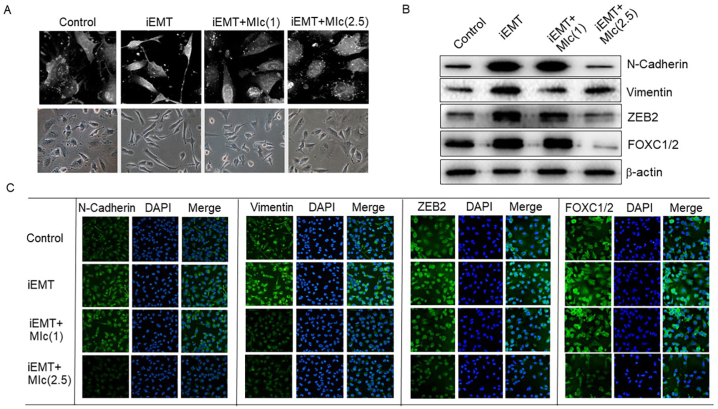
Since EMT is a crucial process that facilitates the metastatic capability of neoplastic cells [24], the effect of MIc on EMT was subsequently evaluated. In this study, EMT was chemically induced in KKU-452 cells using commercial StemXVivo® EMT-inducing media supplement. The results showed that after treatment with the inducing reagent, KKU-452 cells altered their morphology into spindle-shaped fibroblastic-like cells, as imaged with a holotomography microscope (Tomocube Inc., Daejeon, Korea) (Fig. 5A), which clearly confirmed the effectiveness of EMT induction. Upon the induction of EMT, the expression levels of EMT marker proteins, including N-cadherin, vimentin, ZEB2, and FOXC1/2, also increased, as determined by Western blot and immunofluorescence analyses (Fig. 5B and C). Interestingly, MIc suppressed morphological changes in EMT-induced KKU-452 cells (Fig. 5A). In addition, the expression of the mesenchymal markers N-cadherin and vimentin and the EMT-inducing transcription factors ZEB2 and FOXC1/2 was also downregulated after 2.5 μM MIc exposure for 48 h (Fig. 5B and C).
Fig. 5.
Effects of MIc on EMT in CCA cells. KKU-452 cells were chemically induced to undergo EMT (iEMT). (A) The effect of MIc on cell morphology at concentrations of 1 and 2.5 μM was determined. EMT marker expression, including N-cadherin, vimentin, ZEB2, and FOXC1/2, was further evaluated by (B) Western blot (Supplementary Fig. S1 Images of original blot 5B) and (C) immunocytofluorescence analyses. The data shown are representative of two reproducible experiments.
3.6. MIc inhibited FAK/Src signalling and its metastasis-associated downstream molecules
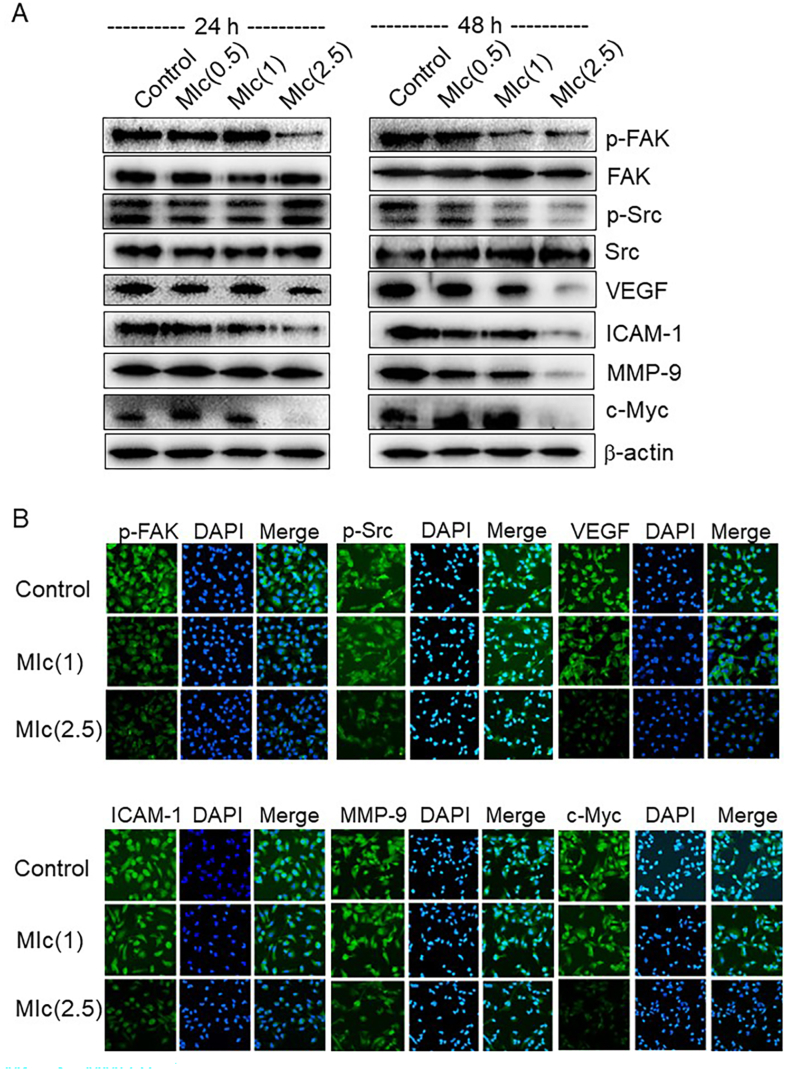
Since network pharmacology analysis revealed FAK and Src as key targets of MIc and the critical contribution of the FAK/Src pathway to the regulation of CCA metastasis has been reported [25], the effect of MIc on these signalling networks was determined in this study. The results showed that p-FAK and p-Src protein levels were markedly decreased after treatment with the compound at concentrations of 0.5, 1 and 2.5 μM for 48 h, although total FAK and Src protein levels were not significantly affected, as evidenced by immunoblotting analysis (Fig. 6A). These data suggested that MIc affected the phosphorylation of FAK and Src. Immunocytofluorescence analysis reflected a similar trend for p-FAK and p-Src proteins (Fig. 6B). To further examine the mechanistic role of MIc, the expression profile of metastasis-related proteins, which are downstream molecules under the control of the FAK/Src pathway, was investigated. As shown in Fig. 6A and B, treatment of KKU-452 cells with 2.5 μM MIc for 48 h, decreased the expression of VEGF, ICAM-1, MMP-9 and c-Myc.
Fig. 6.
Effects of MIc on p-FAK, p-Src, and their downstream effector molecules. KKU-452 cells were treated with MIc (0.5–2.5 μM) for 24 and 48 h. (A) The expression of p-FAK and p-Src, as well as their downstream effector molecules, including VEGF, ICAM-1, MMP-9, and c-Myc, was determined by Western blot analysis (Supplementary Fig. S2 Images of original blot 6A). (B) Immunocytofluorescence analysis was also performed in cells treated with MIc for 48 h. The data shown are representative of two reproducible experiments.
3.7. MIc interacted with FAK and Src in molecular docking analysis
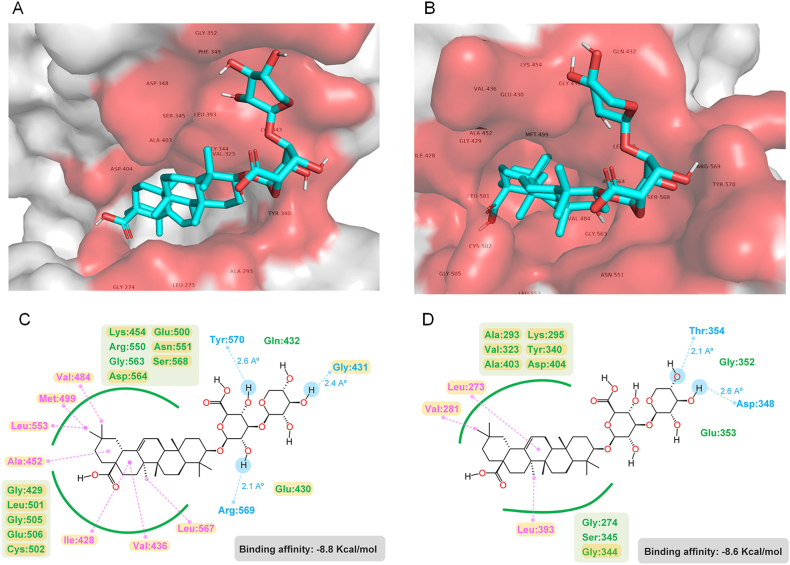
To further verify whether FAK and Src are molecular targets of MIc conferring its anticancer activities against CCA cells, the molecular interplay between MIc and these proteins was investigated. Molecular docking was applied to examine the possible interaction. Both FAK and Src are nonreceptor tyrosine kinases. Src's SH1 domain plays a pivotal role in catalytic activity, and the phosphorylation state of the domain (Tyr416) contributes to its active form. Active Src then phosphorylates FAK at the kinase domain, turning FAK into full catalytic activity. Phosphorylation also enables FAK in the active conformation to be available for sequential phosphorylation at the C-terminal region (Tyr925) [26]. Therefore, the Src SH1 (PDB ID: 4MXO) and FAK kinase (PDB ID: 3BZ3) domains were selected in the docking analysis since they correspond to the activities and phosphorylation stages of Src and FAK investigated. Our results showed that MIc bound with FAK and Src with binding affinities of −8.8 and −8.6 kcal/mol, respectively. The binding residues as well as ligand-receptor interactions are shown in Fig. 7A and B. The major interactions include hydrogen and van der Waals bonding. With the retrieved data from reported protein crystallography [27,28], our model showed that MIc shared binding pockets with the FAK and Src inhibitors, PF-562,271 and bosutinib, respectively (Fig. 7C and D).
Fig. 7.
Molecular interactions between MIc and FAK/Src. MIc binds to the kinase domain of (A) FAK (PDB ID: 3BZ3) and (B) Src (PDB ID: 4MOX) in a 3D representation. The amino acid residues occupied by MIc are highlighted in red on the surface. The 2D interactions of MIc with (C) FAK and (D) Src show hydrophobic interactions, including van der Waals (green) and alkyl to alkyl/pi (magenta), as well as a hydrophilic interaction involving hydrogen bonding (blue). The yellow box indicates residues shared by both MIc and FAK inhibitor (PF-562,271) or Src inhibitor (bosutinib).
4. Discussion
Identifying potent agents that can enhance the clinical efficacy of treatment outcomes is imperative for CCA therapy. For decades, active phytochemicals obtained from medicinal herbs and dietary plants have been of significant interest for drug development due to their health-promoting and therapeutic properties. Gaining insight into the mechanisms through which these compounds impact cancer is an essential prerequisite in the development of potent anticancer treatments. In the present study, the antimetastatic potential of MIc was demonstrated by using network pharmacology and cell-based assessments accompanied by molecular docking analysis. Thus, the compound could be an attractive agent for further development as a therapeutic drug for metastatic CCA treatment.
Network pharmacology integrates pharmacology, systems biology, and network analysis to comprehend intricate interactions among biological molecules (proteins, genes, metabolites) in diseases and drug actions [16]. In this analysis, 37 possible targets of MIc against CCA were initially revealed. The GO and KEGG enrichment analyses were then conducted based on these targets. The KEGG analysis suggested the likely involvement of MIc in the apoptotic pathway, while the GO analysis indicated that MIc is likely associated with cell migration function. Subsequently, protein-protein interactions demonstrated that Src, FAK, and GRB2 emerged as the most plausible intracellular targets. Bear in mind that network pharmacology is an in-silico analysis. Its primary function is to predict interactions and outcomes. However, due to its computational nature and reliance on existing data, experimental validation becomes crucial to verify and confirm these computational predictions for accuracy and reliability. Accumulating data reveal that the interplay among Src, FAK, and GRB2 is crucial to both apoptosis and cell motility [25,29]. FAK and Src function in tandem as FAK/Src complex, leading to the activation of GRB2, which consequently activates the Ras/MAPK signalling pathway and eventually facilitates cell proliferation and migration [30]. Since we previously reported the apoptotic-inducing activity of MIc against CCA [15], in this study, cell-based assessments were emphasized on the effect of MIc on the downstream FAK/Src pathway relevant to CCA metastasis.
Considering that increased motility and invasive capabilities are fundamental phenomena of metastatic malignant cells [23], the antimetastatic potential of MIc was initially evaluated by examining its inhibitory impact on these two behaviours in CCA KKU-452 cells. The results revealed that MIc exhibited antimigratory and anti-invasive efficacy against the cells. Furthermore, the compound also diminished the adhesive ability of cancer cells to adhere to the extracellular matrix, indicating its potential to suppress the settling of metastatic CCA cells at target organ sites. Consistently, previous studies have reported the ability of MIc to attenuate the migratory and invasive capacities of hepatocellular carcinoma cells [13,31].
The critical mechanism to enhance cellular migratory and invasive capacities is mediated via EMT. This process is considered a key mechanism that drives the metastasis of several cancers, including CCA [24]. At present, a number of EMT inhibitors have been under clinical trials at different phases of studies for the treatment of various cancers [32]. During EMT, cells undergo morphological changes in which cells transition from epithelial to mesenchymal phenotypes through intricate molecular interactions [33]. To simulate EMT in an in vitro model, several experimental strategies have been established [34,35]. In this study, commercial EMT induction medium was used. Upon induction of EMT, KKU-452 cells changed their phenotypic trait into a spindle-like morphology, which was accompanied by a substantial increase in the molecular signature for mesenchymal cell types, including N-cadherin, vimentin, ZEB2, and FOXC1/2. N-cadherin serves as an important marker of the ongoing EMT process [33]. Vimentin plays key roles in several fundamental cellular processes, such as cell integrity, migratory machinery, and extracellular attachment, emerging as a master EMT regulator [36]. Its overexpression during tumour metastasis, which is associated with poor patient prognosis, underscores its importance [37]. During EMT, vimentin modulates the expression of EMT-activating transcription factors such as snail, slug, ZEB2 and FOXC1/2, which subsequently results in an increased production of effector molecules to facilitate cancer spread [36]. This study demonstrated the inhibitory effect of MIc on EMT and EMT-related molecules in CCA cells. Similarly, a previous study reported that the compound negatively impacts the EMT process of liver cancer cells [13,31]. These findings suggest the potential of MIc to be developed as an EMT-based therapy for the treatment of metastatic malignancy.
Numerous intracellular signal transduction cascades have been implicated in the metastatic process of cancer cells. As mentioned above, the data from network pharmacology analysis indicated that FAK and Src are potential targets of MIc against CCA. Moreover, accumulating evidence supports the pivotal role of the FAK/Src pathway in the regulation of EMT and other metastasis-related events [25,38]. Notably, dysregulated activation of the FAK/Src cascade has been observed in CCA [39], making it an attractive target for therapeutic intervention against CCA metastasis. The effect of MIc on the FAK/Src signalling pathway was then investigated in this study. As expected, the compound exhibited a suppressive effect on FAK/Src activation, as evidenced by substantial reductions in p-FAK and p-Src protein levels, as assessed through immunoblotting and immunofluorescence analyses. Subsequently, various metastasis-related downstream effectors, including MMP-9, VEGF, ICAM-1, and c-Myc, were significantly decreased in MIc-exposed cells. MMP-9, known for promoting extracellular matrix proteolysis, facilitates the mobility and invasiveness of metastatic cancer, and its activation is also implicated during EMT [40]. VEGF is a key regulator of tumour angiogenesis and vascular permeability, while ICAM-1 promotes cancer cell adhesion to endothelial walls, facilitating colonization at secondary sites [41]. In addition to CCA, MIc has also been demonstrated to be effective in downregulating VEGF, ICAM-1, and MMP-9 in hepatocellular carcinoma cells [31]. According to c-Myc, the dysregulated expression of this oncogenic transcription factor is frequently found in cancers, and this phenomenon is associated with aggressive behaviours of cancer cells [42]. Prior studies demonstrated that inhibition of c-Myc underlies the antimetastatic effect of several natural compounds, including caffeine [43] and triterpene acid [44]. In this study, the suppressive effect of MIc on c-Myc expression in CCA was clearly demonstrated. Collectively, our findings indicated that reducing the metastatic potential of KKU-452 CCA cells by MIc can be achieved by inhibiting of the FAK/Src pathway and its downstream effector molecules.
Additionally, molecular docking was conducted to obtain corroborating evidence for the impact of MIc on FAK and Src. These two proteins are nonreceptor tyrosine kinases that play a pivotal role upstream of metastasis regulation [25]. Both kinases consist of several functional domains that contribute to their activity and regulation [26]. The catalytic domains of both FAK and Src were selected for docking in this study since their function affects the activity of FAK and Src as well as the phosphorylation state investigated in our study. Docking results revealed MIc binding to FAK and Src at pocket sites coinciding with those of the competitive ATP inhibitors PF-562,271 (FAK inhibitor) and bosutinib (Src inhibitor) [27,28]. Hence, it is presumed that MIc obstructs FAK and Src by binding to their catalytic domains, thwarting ATP binding. This interference hampers ATP binding, consequently maintaining the kinase structures in an inactive state, leading to diminished phosphorylation and catalytic function. Notably, our docking result could not refer to the specificity of MIc with FAK and Src. Since MIc binds with FAK and Src mainly via hydrophobic interactions, particularly those lacking binding specificity, other molecular targets might contribute to the MIc action observed. To confirm both affinity and selectivity, a definitive binding study, such as X-ray crystallography, between MIc and FAK/Src is needed. Furthermore, the broader range of molecular targets and signalling pathways contributing to the antimetastatic effect of MIc in CCA necessitates further investigation.
In conclusion, MIc might have a beneficial effect for the treatment of metastatic CCA. The compound suppresses EMT and metastatic behaviours of CCA cells by inhibiting the activation of the oncogenic FAK/Src pathway and its metastasis-related downstream molecular targets. Additional pharmacological studies are needed for the future development of MIc as an antimetastatic agent.
Funding
This research was supported by NSRF under the Fundamental Fund of Khon Kaen University (66).
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request.
Code availability
Not applicable.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
CRediT authorship contribution statement
Piman Pocasap: Writing – original draft, Methodology, Formal analysis, Conceptualization. Auemduan Prawan: Validation, Supervision, Resources. Sarinya Kongpetch: Visualization, Validation, Resources, Methodology. Laddawan Senggunprai: Writing – review & editing, Methodology, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e32352.
Contributor Information
Piman Pocasap, Email: pimapo@kku.ac.th.
Auemduan Prawan, Email: peuamd@kku.ac.th.
Sarinya Kongpetch, Email: sarinyako@kku.ac.th.
Laddawan Senggunprai, Email: laddas@kku.ac.th.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Khan A., Tavolari S., Brandi G. Cholangiocarcinoma: epidemiology and risk factors. Liver Int. 2019;39:19–31. doi: 10.1111/liv.14095. [DOI] [PubMed] [Google Scholar]
- 2.Sithithaworn P., Andrews R.H., Nguyen V.D., et al. The current status of opisthorchiasis and clonorchiasis in the Mekong Basin. Parasitol. Int. 2012;61:10–16. doi: 10.1016/j.parint.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sripa B., Brindley P.J., Mulvenna J., et al. The tumorigenic liver fluke Opisthorchis viverrini--multiple pathways to cancer. Trends Parasitol. 2012;28:395–407. doi: 10.1016/j.pt.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Najran P., Lamarca A., Mullan D., et al. Update on treatment options for advanced bile duct tumours: radioembolisation for advanced cholangiocarcinoma. Curr. Oncol. Rep. 2017;19:50. doi: 10.1007/s11912-017-0603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banales J.M., Cardinale V., Carpino G., et al. Expert consensus document: cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA) Nat. Rev. Gastroenterol. Hepatol. 2016;13:261–280. doi: 10.1038/nrgastro.2016.51. [DOI] [PubMed] [Google Scholar]
- 6.O'Hagan K. Updates in cholangiocarcinoma. J. Adv. Pract. Oncol. 2022;13:320–323. doi: 10.6004/jadpro.2022.13.3.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerekes D., Horvath A., Kusz N., et al. Coumarins, furocoumarins and limonoids of Citrus trifoliata and their effects on human colon adenocarcinoma cell lines. Heliyon. 2022;8 doi: 10.1016/j.heliyon.2022.e10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swainson N.M., Pengoan T., Khonsap R., et al. In vitro inhibitory effects on free radicals, pigmentation, and skin cancer cell proliferation from Dendrobium hybrid extract: a new plant source of active compounds. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e20197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo M., Zeng B., Wang H., et al. Kochia scoparia saponin momordin ic modulates hacat cell proliferation and apoptosis via the wnt/β-catenin pathway. Evid. Based Complement. Alternat. Med. 2021 doi: 10.1155/2021/5522164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi J., Lee K.-T., Jung H.-J., Park H.-S., Park H.-J. Anti-rheumatoid arthritis effect of theKochia scoparia fruits and activity comparison of momordin ic, its prosapogenin and sapogenin. Arch Pharm. Res. (Seoul) 2002;25:336–342. doi: 10.1007/BF02976636. [DOI] [PubMed] [Google Scholar]
- 11.Yoo S.R., Jeong S.J., Lee N.R., Shin H.K., Seo C.S. Quantification analysis and in vitro anti-inflammatory effects of 20-hydroxyecdysone, momordin ic, and oleanolic acid from the fructus of kochia scoparia. Pharmacogn Mag. 2017;13:339–344. doi: 10.4103/0973-1296.211023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Na-Young K., Mi-Kyung L., Myoung-Ju P., et al. Momordin ic and oleanolic acid from kochiae fructus reduce carbon tetrachloride-induced hepatotoxicity in rats. J. Med. Food. 2005;8:177–183. doi: 10.1089/jmf.2005.8.177/. [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Han Y., Wang M., Zhao Q., Chen X., Liu X. Natural triterpenoid saponin momordin Ic suppresses HepG2 cell invasion via COX-2 inhibition and PPARγ activation. Toxicol. Vitro. 2020;65 doi: 10.1016/j.tiv.2020.104784. [DOI] [PubMed] [Google Scholar]
- 14.Xianjun F., Xirui X., Jie T., et al. Momordin Ic induces G0/1 phase arrest and apoptosis in colon cancer cells by suppressing SENP1/c-MYC signaling pathway. J. Pharmacol. Sci. 2021;146:249–258. doi: 10.1016/j.jphs.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Malikrong P., Prawan A., Kongpetch S., Senggunprai L. Momordin ic triggers mitochondria-mediated apoptosis of cholangiocarcinoma cells and enhances the efficacy of conventional chemotherapeutic drugs. FARMACIA. 2023;71 doi: 10.31925/farmacia.2023.3.9. [DOI] [Google Scholar]
- 16.Hopkins A.L. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 17.Iksen I., Witayateeraporn W., Wirojwongchai T., et al. Identifying molecular targets of Aspiletrein-derived steroidal saponins in lung cancer using network pharmacology and molecular docking-based assessments. Sci. Rep. 2023;13:1545. doi: 10.1038/s41598-023-28821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakle N.S., More S.A., Mokale S.N. A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: an updated prototype in drug discovery. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-74251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saensa-Ard S., Leuangwattanawanit S., Senggunprai L., et al. Establishment of cholangiocarcinoma cell lines from patients in the endemic area of liver fluke infection in Thailand. Tumour Biol. 2017;39 doi: 10.1177/1010428317725925. [DOI] [PubMed] [Google Scholar]
- 20.Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2019;36:2628–2629. doi: 10.1093/bioinformatics/btz931%JBioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaewmeesri P., Pocasap P., Kukongviriyapan V., Prawan A., Kongpetch S., Senggunprai L. Anti-metastatic potential of natural triterpenoid cucurbitacin B against cholangiocarcinoma cells by targeting Src protein. Integr. Cancer Ther. 2022;21 doi: 10.1177/15347354221124861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su Z., Yang Z., Xu Y., Chen Y., Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol. Cancer. 2015;14:48. doi: 10.1186/s12943-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tahtamouni L., Ahram M., Koblinski J., Rolfo C. Molecular regulation of cancer cell migration, invasion, and metastasis. Anal. Cell Pathol. 2019;2019 doi: 10.1155/2019/1356508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaquero J., Guedj N., Clapéron A., Nguyen Ho-Bouldoires T.H., Paradis V., Fouassier L. Epithelial-mesenchymal transition in cholangiocarcinoma: from clinical evidence to regulatory networks. J. Hepatol. 2017;66:424–441. doi: 10.1016/j.jhep.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J., Yi Q., Tang L. The roles of nuclear focal adhesion kinase (FAK) on Cancer: a focused review. J. Exp. Clin. Cancer Res. 2019;38:250. doi: 10.1186/s13046-019-1265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tapial Martínez P., López Navajas P., Lietha D. Fak structure and regulation by membrane interactions and force in focal adhesions. Biomolecules. 2020;10 doi: 10.3390/biom10020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levinson N.M., Boxer S.G. A conserved water-mediated hydrogen bond network defines bosutinib's kinase selectivity. Nat. Chem. Biol. 2014;10:127–132. doi: 10.1038/nchembio.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts W.G., Ung E., Whalen P., et al. Antitumor activity and pharmacology of a selective focal adhesion kinase inhibitor, pf-562,271. Cancer Res. 2008;68:1935–1944. doi: 10.1158/0008-5472.CAN-07-5155. [DOI] [PubMed] [Google Scholar]
- 29.Chuang H.H., Zhen Y.Y., Tsai Y.C., et al. FAK in Cancer: from mechanisms to therapeutic strategies. Int. J. Mol. Sci. 2022;23 doi: 10.18632/oncotarget.10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng S.Y.S., Sun G., Schlaepfer D.D., Pallen C.J. Grb2 promotes integrin-induced focal adhesion kinase (FAK) autophosphorylation and directs the phosphorylation of protein tyrosine phosphatase α by the Src-FAK kinase complex. Mol. Cell Biol. 2014;34:348–361. doi: 10.1128/MCB.00825-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J., Liu Q., Xiao H., Luo X., Liu X. Suppressive effects of momordin Ic on HepG2 cell migration and invasion by regulating MMP-9 and adhesion molecules: involvement of p38 and JNK pathways. Toxicol. Vitro. 2019;56:75–83. doi: 10.1016/j.tiv.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Santamaria P.G., Moreno-Bueno G., Portillo F., Cano A. EMT: present and future in clinical oncology. Mol. Oncol. 2017;11:718–738. doi: 10.1002/1878-0261.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/jci39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J., Hahm E.R., Marcus A.I., Singh S.V. Withaferin A inhibits experimental epithelial-mesenchymal transition in MCF-10A cells and suppresses vimentin protein level in vivo in breast tumors. Mol. Carcinog. 2015;54:417–429. doi: 10.1002/mc.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y.L., Chou C.K., Kim M., et al. Assessing metastatic potential of breast cancer cells based on EGFR dynamics. Sci. Rep. 2019;9:3395. doi: 10.1038/s41598-018-37625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Usman S., Waseem N.H., Nguyen T.K.N., et al. Vimentin is at the heart of epithelial mesenchymal transition (emt) mediated metastasis. Cancers. 2021;13 doi: 10.3390/cancers13194985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du L., Li J., Lei L., He H., Chen E., Dong J., Yang J. High vimentin expression predicts a poor prognosis and progression in colorectal cancer: a study with meta-analysis and TCGA Database. BioMed Res. Int. 2018;2018 doi: 10.1155/2018/6387810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigiracciolo D.C., Cirillo F., Talia M., et al. Lappano, Focal adhesion kinase fine tunes multifaced signals toward breast cancer progression. Cancers. 2021;13 doi: 10.3390/cancers13040645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong L., Lu D., Chen R., et al. Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma. Cancer Cell. 2022;40:70–87.e15. doi: 10.1016/j.ccell.2021.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Scheau C., Badarau I.A., Costache R., et al. The role of matrix metalloproteinases in the epithelial-mesenchymal transition of hepatocellular carcinoma. Anal. Cell Pathol. 2019;2019 doi: 10.1155/2019/9423907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim E.J., Kang J.H., Kim Y.J., Kim S., Lee S.J. ICAM-1 promotes cancer progression by regulating SRC activity as an adapter protein in colorectal cancer. Cell Death Dis. 2022;13:417. doi: 10.1038/s41419-022-04862-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ala M. Target c-Myc to treat pancreatic cancer. Cancer Biol. Ther. 2022;23:34–50. doi: 10.1080/15384047.2021.2017223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meisaprow P., Aksorn N., Vinayanuwattikun C., Chanvorachote P., Sukprasansap M. Caffeine induces g0/g1 cell cycle arrest and inhibits migration through integrin αv, β3, and fak/akt/c-myc signaling pathway. Molecules. 2021;26:7659. doi: 10.3390/molecules26247659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi H.S., Kim S.L., Kim J.H., Deng H.Y., Yun B.S., Lee D.S. Triterpene acid (3-O-p-coumaroyltormentic acid) isolated from Aronia extracts inhibits breast cancer stem cell formation through downregulation of c-Myc protein. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19092528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request.