Abstract
The mutualistic symbiotic relationship between insects and bacteria greatly influences the growth and development of host insects. Tessaratoma javanica (Thunberg) (Hemiptera: Tessaratomidae), also referred to as the litchi stink bug, has recently been established as an important insect pest of Litchi chinensis Sonn. and causes substantial yield loss in India. To design effective and environmentally safe management strategies, an understanding of the diversity and functions of microbiota harbored across the development stages is very important. The assessment of the diversity of development-associated bacteria in T. javanica and their predicted functions was conducted using 16S rRNA gene sequences obtained by the Illumina MiSeq technology. The result showed that taxonomic analysis of associated bacteria in different developmental stages includes a total of 46 phyla, encompassing 139 classes, 271 orders, 474 families, and 893 genera of bacteria. All developmental stages of T. javanica shared a total of 42.82 percent of operational taxonomic units (OTUs), with a 97 % similarity threshold. Alpha diversity indices showed maximum species richness in the egg and adult stages. The phyla Proteobacteria followed by Firmicutes, Bacteriodetes, and Actinobacteria, exhibited the highest levels of abundance across all the developmental stages of T. javanica. Microbiota were most different between the egg and the 4th nymphal stage (χ2 = 711.67) and least different between the 2nd and 4th nymphal instars (χ2 = 44.45). The predicted functions of the microbiota associated with T. javanica are mainly involved in amino acid metabolism, cell motility, cellular processes and signaling, glycan biosynthesis and metabolism, lipid metabolism, and membrane transport. The present study documentation and information on symbiotic bacteria across T. javanica life stages will prompt the development of novel biological management strategies.
Keywords: Tessaratoma, 16s rRNA, Developmental stages, Litchi, Bacterial diversity
Graphical abstract
Highlights
-
•
Significant changes in bacterial diversity associated with developmental stages of Tessaratoma javanica, referred to as the litchi stink bug, cause substantial yield loss in litchi crops in India.
-
•
Less number of operational taxonomic units (OTUs) were found to be shared across all developmental stages of T. javanica.
-
•
The phyla Proteobacteria followed by Firmicutes, Bacteriodetes, and Actinobacteria exhibited the highest levels of abundance across all the developmental stages.
-
•
Proteobacteria dominate in the nymphal stages while Firmicutes in the egg stage of T. javanica.
1. Introduction
Arthropods, including insects, are one of the most diverse groups of animals, which belong to the class Insecta. It consists of millions of species found in almost all habitats and has a wide range of adaptations. Insects harbor diverse microorganisms in their gut called microbiota [1]. The microbial communities residing within insects constitute a significant proportion, ranging from 1 to 10 %, of their total biomass [2]. The intricate relationship between insects and their symbiotic microorganisms, encompassing bacterial, fungal, viral, protozoan, and archaea, plays a crucial role in shaping the diversity and adaptability of these organisms [3]. The insect and symbiotic bacteria relationship is a fascinating and complex interaction that has vital implications for both the insect host and the bacteria involved. These symbiotic bacteria are ubiquitous in insects and play a vital role in numerous aspects of insect biology which include immunity, development, reproduction, stress resistance, pesticide susceptibility, digestion, adaptability, absorption of nutrients, and degradation of toxic substances [[4], [5], [6], [7], [8], [9], [10], [11]]. A lot of insects have symbiotic bacteria that play crucial roles in several aspects of the host's life cycle, including development, nutritional needs, and evolutionary progress [[12], [13], [14], [15]]. The significance of bacteria in facilitating interactions between pests and host plants is widely accepted [16]. They also offer protection against invaders, allowing their hosts to withstand extreme temperatures and defend against pathogens [17]. In addition, these microbes are very important for getting rid of harmful secondary metabolites in plants, making semiochemicals, and sending important signals and cues [12,18]. Developmental stages of insects, host plant associations, and associated ecological factors can all have an impact on the diversity and abundance of microorganisms [6,[19], [20], [21], [22]]. Ultimately, relationships might continue over the necessary evolutionary time to permit the selection and establishment of true gut residents, which would not be possible due to the considerable diversity in the microbial community caused by host plants [23].
Litchi chinensis Sonn., commonly known as litchi, is a species of evergreen trees native to tropical and sub-tropical regions. It belongs to the family Sapindaceae and is susceptible to multiple kinds of pests [24]. Tessaratoma javanica (Thunberg) (Hemiptera: Tessaratomidae), commonly known as the litchi stink bug, holds significant economic importance as a major sucking pest of Litchi in India and various other countries such as India, Thailand, Malaysia, China, Vietnam, Australia, and the Philippines [[25], [26], [27], [28], [29], [30], [31], [32]]. Stink bugs also attack plants like Rambutan, longan, Pomegranate, Castor and Eucalyptus, Pummelo, Mahua, Mulberry, Kusum, Loquat, and Rose [25,27,28,33]. Other co-species, like Tessaratoma papillosa Drury, are common bug species that are also damaging to litchi. Several species of Tessaratoma have been documented as inflicting significant economic losses in terms of both quantity and quality of litchi, longan, and citrus in different parts of the world, including China, Australia, Myanmar, and Thailand [29,[34], [35], [36]]. Among them, T. javanica is one of them, which is very destructive to litchi plants. Earlier in India, T. javanica was considered a minor pest with a sporadic incidence of litchi [27]. In recent times, this particular insect pest has emerged as a significant threat to litchi cultivation in India. It has been observed that this pest is responsible for causing substantial economic losses to litchi crops, with numerous outbreaks being documented in India and its neighboring country, Bangladesh [24,25,37].
The pest generally inflicts damage upon litchi trees starting in the last week of February, coinciding with the emergence of flower panicles, and continuing until the fruits are ready for harvesting. Subsequently, insects consume newly emerging vegetative growth and enter a state of hibernation during the adult phase in the eastern region of India during the months characterized by harsh winter conditions [25]. The gregarious nymphs and adults of stink bugs pierce and suck from flower buds, flower stalks, young fruit stalks, and tender shoots. The act of feeding by this insect results in the subsequent decline and detachment of flowers and fruits, hence directly diminishing the overall yield of litchi as well as longan fruits [38]. T. javanica has an incomplete metamorphosis where eggs, five instar nymphs, and adult stages follow [39].
The studies of T. javanica have been restricted to its biology, morphology, DNA barcodes, population dynamics, seasonal occurrence, bioefficacy of insecticides, potential parasitoids, and their natural control [26,27,40,41]. The scientific study regarding the constitution and biomechanics of associated bacteria across all the stages of growth of T. javanica, a significant pest, is strikingly absent. We conducted comprehensive 16s rRNA amplicon sequence investigations to gain a fundamental understanding of the overall diversity and potential metabolic functions of the microbiota communities associated with T. javanica. Therefore, the present study was initiated to evaluate the diversity and variation of microbiota and their predicted functions among different developmental stages of the litchi stink bug, T. javanica using 16s rRNA amplicon (V3–V4 region) gene sequencing. The present findings enable us to understand the status of developmental stage-associated bacteria, which will lay the basis for understanding the role of associated bacteria in their growth, development, reproduction, immunity, digestion, and adaptability.
2. Material and methods
2.1. Sample collection
We collected specimens from litchi trees at the ICAR Research Complex for Eastern Region, Farming System Research Centre for Hill and Plateau Region, (ICAR RCER FSRCHPR) Ranchi, India, during the year 2021, representing different stages of development of T. javanica, including the egg stage, 2nd instar nymph, 4th instar nymph, and adult male and female. The geographical coordinates of the collection site are approximately 23°45′ N latitude and 85°30′ E longitude, with an elevation of 620 m above mean sea level (AMSL). We brought the insect specimens alive to the laboratory and stored them at a temperature of −20 °C, except for the eggs. The identification of these specimens was conducted using morphological features as defined by Parveen et al. [39]. A total of 25 samples (N = 5 for each developmental stage) the whole eggs and gut of the 2nd instar nymph, 4th instar nymph, adult males, and females were used for DNA extraction to compare the bacterial communities of different stages of developmental T. javanica. A total of 100 eggs (whole) and guts of 5 insects of the 2nd instar nymph, 4th instar nymph, adultmales, and females were taken in each replicate in a sterile environment for further processing.
2.2. DNA extraction and amplicon sequencing of 16S rRNA gene
The method used to perform the extraction of genomic DNA from each sample was based on the protocol described by Naaz et al. [42], with slight modifications. Each sample was subjected to a disinfection process involving the application of 70 % ethanol for a duration of 30 s. This was followed by the application of 0.1 % mercuric chloride for a duration of 1 min. Subsequently, the samples were washed three times with double distilled water for a duration of 60 s to eliminate any exterior contamination. The 4th instar nymph and adult male and female stinkbug wings and backplates were first removed and then dissected aseptically to remove the midgut. We then homogenized the guts in an Eppendorf tube using a sterilized micropestle. 700 μl of CTAB buffer was added to the Eppendorf tube with homogenized gut samples and incubated for 1 h in a water bath at 65 °C. Equal volumes of phenol, chloroform, and isoamyl alcohol (in a ratio of 25:24:1 v/v) were added and well mixed after the duration of 1 h. Tubes were then spun at 10,000 rpm in a centrifuge (REMI India Ltd.) machine for 12 min. The aqueous phase was shifted to a new tube, and the liquid above it was discarded. Then the prechilled isopropanol (450 μl) was added and kept at −20 °C for 1 h to precipitate the DNA. The tubes were spun again in a centrifuge machine at 10,000 rpm for 12 min and discarded the supernatant. The DNA pellet was sedimented at the bottom of the Eppendorf tube, followed by three washes with 70 % ethanol and subsequent air drying of the tube. After that, 100 μl of TE buffer (Tris EDTA) was added to dissolve the pellet. Subsequently, RNAse was added at a concentration of 10 μl/ml and incubated at a temperature of 37 °C for a duration of 30 min. The DNA samples were subsequently subjected to gel electrophoresis and analyzed using a 0.8 % agarose gel in 1X TAE buffer. Following this, the samples were stored at a temperature of −20 °C to facilitate subsequent amplification procedures.
2.3. PCR amplification
DNA concentration and purity were checked by the fluorescent (Qubit fluorometer 4.0). The stage-associated metagenomic sequencing of bacterial communities was done for V3–V4 hypervariable region of 16S rRNA amplicon while employing the primer with added Illumina adaptor overhang sequences (16S rRNA gene-specific sequences are underlined) 314F (5′- TCGTCGGCAGGGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) and 805R (5′- GTCTCGTGGGCTCGGAGATGTGTATAAGAGCTACTAGGGTATCTAATCC-3′) [43]. The polymerase chain reaction (PCR) was conducted under the specified conditions: an initial denaturation phase at 98 °C for 30 s, followed by 27 cycles consisting of a 10 s denaturation at 98 °C, annealing at 55 °C for 30 s, extension at 72 °C for 30 s, and a final extension step for 10 min at 72 °C. The sequencing library was created using the KAPA HiFi Hotstart Ready Mix kit (Roche, Basel, CHE) according to the manufacturer protocols. Nuclease - free water was used for the false-positive PCR product. The Nextera XT Index Kit (Illumina, San Diego, CA, USA) was utilized to attach the dual index adapters on the Illumina MiSeq platform. The sequencing procedure employed for the paired-end run, with a read length of 300 base pairs, was conducted using the MiSeq Reagent kit V3, which provides 600 cycles. Sequence read archives (SRA) for the present study have been placed in the NCBI SRA database with the accession number: PRJNA980004 (https://www.ncbi.nlm.nih.gov/sra/.PRJNA980004).
2.4. Sequencing and statistical analysis of data
The raw sequences passed quality assessments for base quality parameters, base composition distribution, and GC dispersion using FastQC software version 0.11.28 [44]. Adapter sequences at the 3'end of the reads, primer sequences, and low-quality bases were removed through TrimGlore tools [45]. The raw sequences that has been trimmed were subjected to processing and analysis utilizing the Mothur software (version 1.30.2). The criterion of overlap of more than 10 bp and mismatch rate of 0.02 were used to reassign paired-end reads. Contigs that could not be aligned were removed, and duplicates were merged. Chimeric regions were also removed by using the UCHIME algorithm [46]. The non-bacterial i.e., chloroplast, mitochondria, archea, and eukaryota sequences were removed using commands of taxonomic classifications and undesirables implemented in the Mothur pipeline. The clean reads were clustered together using UPARSE software [47]. The Uclust program, version 1.2.22 [48], was used to pool and cluster the processed sequences into Operational Taxonomic Units (OTUs) based on 97 % similarity. The sequences of each OTU were screened with features –classifier-classify-sklearn against the 16S rRNA database on GREENGENES database v.13.8–99 by PyNAST [49] tools using a threshold value of 80 % which targeted the V3–V4 region of 16S rRNA.
The downstream analysis of biome files was done by using an online Microbiome Analyst platform (https://www.microbiomeanalyst.ca/) and R package. The analysis comprised the study of different alpha diversity indexes, mainly Ace, Chao1, Shannon, and Simpson, utilizing the Bray-Curtis dissimilarity index. The significant differences in alpha diversity were analyzed using the Kruskal-Wallis H test performed in SPSS (version 22.0). The box plots of different alpha diversity indices were constructed by using a vegan package in R. The beta diversity across different developmental stages was calculated based on the Bray-Curtis dissimilarities index using principal coordinate analysis (PCoA). The Venn Diagram package in R was used to plot the Venn diagram to represent unique and shared OTUs among developmental stages of T. javanica. The bar plot was constructed to represent the relative abundance of top phyla across developmental stages. The average dissimilarities among developmental stages of T. javanica using SIMPER analysis and similarity percentage between phyla were analyzed in PAST 4.03 [50]. The metabolic functional pathways of bacterial communities were predicted using PICRUSt v1.1.4 [51]. Comparative analysis of functional pathways was done using Welch's t-test in STAMP [52]. The differences in pathways were observed with FDR-adjusted p value < 0.05. The significant differences among bacterial communities' composition of developmental stages were determined by using linear discriminate analysis (LDA) effect size (LEfSe).
3. Result
3.1. Sequencing profile of bacterial communities
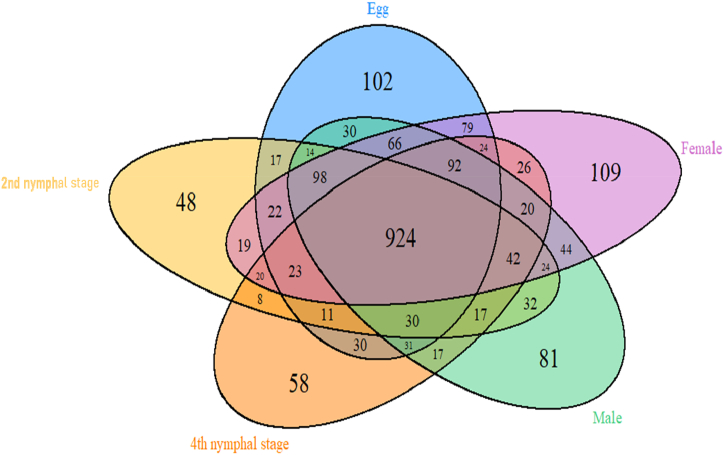
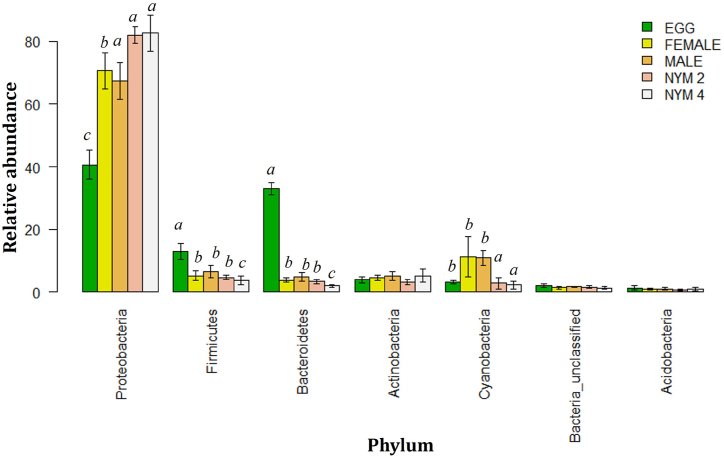
The rarefaction curve reached the plateau region which suggests a sufficient number of samples were taken for analysis (Supplementary Fig. 1). The 16s rRNA amplicon gene sequences of the 25 samples were obtained using the Illumina MiSeq platform. A total of 2158 filtered OTUs were identified from studying the developmental stages of T. javanica. The highest number of OTUs (Operational Taxonomic Units) was observed from the adult female stage (female: 1632), while the lowest number of OTUs was observed from the 2nd instar nymphal stage (II: 1349). The Venn diagram depicted an overall total of 924 operational taxonomic units (OTUs) that were found to be common across all developmental stages of T. javanica, using a similarity threshold of 97 % (Fig. 1). The observed differences in the number of OTUs across distinct developmental phases serve as an indicator of the intricate nature of microbial communities. The major phyla of bacteria associated with T. javanica were Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Cyanobacteria (Fig. 2). Among the top 5 classified genera, three genera such as Enterobacteriaceae_unclassified, Streptophyta_unclassified, and Bacteria unclassified were unidentified.
Fig. 1.
Venn diagram showing unique and shared OTUs (924 OTUs shared among all developmental stages) among developmental stages of Tessaratoma javanica.
Fig. 2.
Bar plot showing the relative abundance at phylum level in different developmental stages of Tessaratoma javanica. Upper different alphabets represent significant differences (p < 0.05) among developmental stages. NYM 2 refers to 2nd nymphal stage and NYM 4 – 4th nymphal stage.
3.2. Bacterial diversity of T. javanica
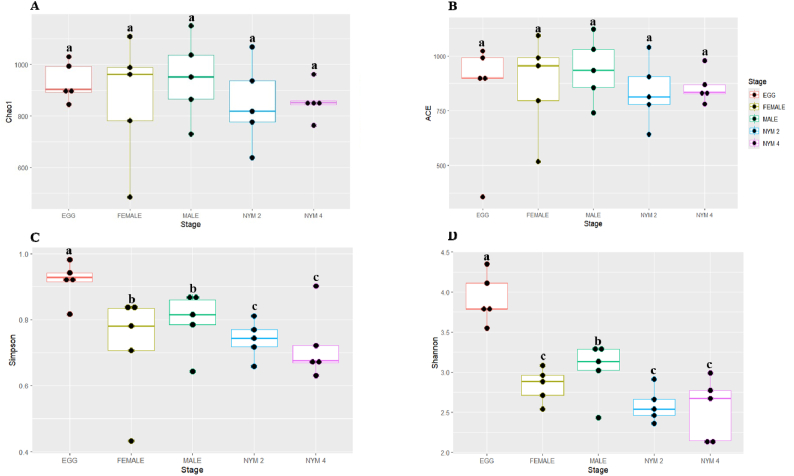
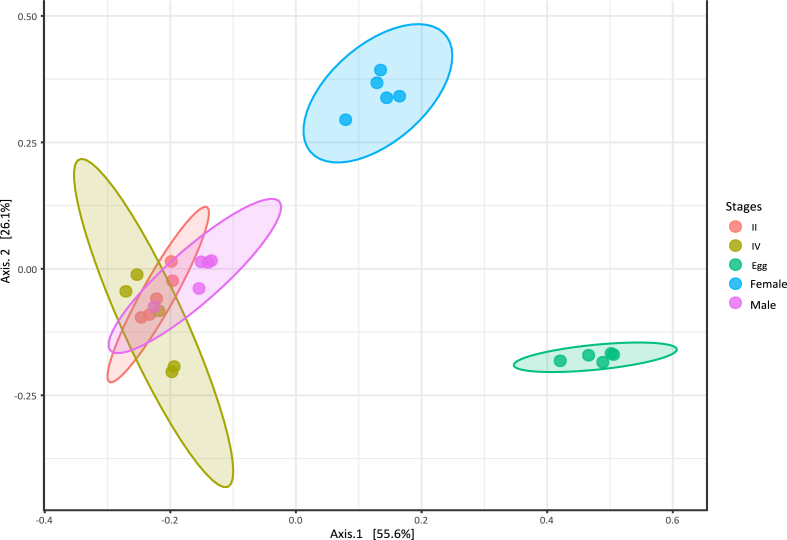
The alpha diversity indices show the species abundance, species richness, and distribution of samples. Different alpha indices Ace, Chao1, Shannon, and Simpson of the bacterial communities of T. javanica across the developmental stage were analyzed, and results were compared using the Kruskal-Wallis H test (P < 0.05) (Table 1). The Chao1 indices were observed to be highest in the adult male stage (946.82 %) and the lowest in the egg stages (833.23 %) (Fig. 3A). The highest Ace was observed in the adult male stage (936.25 %) and lowest in the egg stage (833.57 %) (Fig. 3B). The Simpson and Shannon species diversity indices were highest in the egg stage (0.95 ± 0.00 %) and (3.92 ± 0.14 %), respectively, and the lowest was found in the 4th nymphal stage, i.e., Simpson (0.72 ± 0.01 %) and Shannon (2.54 ± 0.10 %) (Fig. 3C-D). Beta diversity of bacterial communities in different developmental stages was analyzed by the Bray - Curtis dissimilarity method (Fig. 4). The result indicated that bacterial communities of the 2nd nymphal stage, 4th nymphal stages, and male adult stage overlapped together, which suggests that bacterial communities shared among these developmental stages. The egg stage and female adult stage were clustered separately, indicating distinct bacterial communities associated with them (ANOSIM, R = 0.85354, P < 0.001).
Table 1.
Species richness and alpha diversity indices of the bacterial community during the developmental stages of Tessaratoma javanica.
| Stage | Chao1 | Ace | Shannon | Simpson | Fisher | Observed |
|---|---|---|---|---|---|---|
| EGG | 833.23 ± 124.52 | 833.57 ± 121.85 | 3.92 ± 0.14a | 0.95 ± 0.00a | 108.10 ± 17.91 | 689.20 ± 102.01 |
| LSB 2 | 848.25 ± 73.07 | 835.79 ± 66.10 | 2.59 ± 0.10c | 0.74 ± 0.02c | 108.41 ± 9.67 | 703.20 ± 53.68 |
| LSB4 | 856.02 ± 31.56 | 857.90 ± 33.22 | 2.54 ± 0.18c | 0.72 ± 0.01c | 112.07 ± 5.48 | 726.40 ± 24.70 |
| Male | 946.82 ± 71.91 | 936.25 ± 66.55 | 3.03 ± 0.16b | 0.82 ± 0.03b | 121.71 ± 11.47 | 778.60 ± 60.28 |
| Female | 865.09 ± 108.66 | 870.62 ± 100.28 | 2.834 ± 0.01c | 0.81 ± 0.02b | 109.73 ± 16.44 | 708.60 ± 88.48 |
| P value | <0.5 (NS) | <0.5 (NS) | <0.001 | <0.001 | <0.5 (NS) | <0.5 (NS) |
| F value (4,16) | 0.24 | 0.24 | 15.74 | 28.31 | 0.17 | 0.23 |
Data in the tables show diversity indices as mean ± standard error of the mean of different developmental stages and abbreviation represents developmental stage LSB 2 referred to as Litchi stink bug 2nd instar nymph, LSB 4 = 4th nymphal stage.
Fig. 3.
Box plot of different alpha diversity indices A) Chao 1, B) Ace, C) Simpson, and D) Shannon across the developmental stages of Tessaratoma javanica. Different alphabetical letters indicate the significant differences among developmental stages (p < 0.05). NYM 2 refers to 2nd Nymphal stage and NYM 4 refers to 4th nymphal stage.
Fig. 4.
Principal coordinate analysis (PCoA) plot visualizes the beta diversity based on the Bray-Curtis dissimilarity index among developmental stages of Tessaratoma javanica (ANOSIM, R = 0.85354, P ≤ 0.001). Abbreviations of II refer to the 2nd nymphal stage, IV – 4th nymphal stage.
3.3. Relative abundance of bacterial communities
The taxonomic investigation of bacteria linked with different stages of development, ranging from phyla to genus level, encompasses a total of 46 bacterial phyla, which includes unclassified bacteria. Furthermore, this research indicates the presence of 139 classes, 271 orders, 474 families, and 893 genera, showing a substantial abundance of bacterial communities related to the subject matter. The most prevalent phyla of bacteria observed across several developmental stages are Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Cyanobacteria. Phyla and genera exhibit varying percentages of abundance (Table 2, Table 3). The phylum Proteobacteria was dominant in each developmental stage in egg (40.70 ± 2.01 %), 2nd nymphal stage (82.02 ± 1.23 %), 4th nymphal stage (82.66 ± 2.57 %), adult male (67.44 ± 2.65 %), and female adult stage (70.63 ± 2.57 %).
Table 2.
Relative abundance of the top five bacterial genus of Tessaratoma javanica.
| Genus | Egg | LSB 2 | LSB 4 | Male | Female | F value | P value |
|---|---|---|---|---|---|---|---|
| Erwinia | 6.91 ± 0.59c | 41.16 ± 4.09a | 44.6 ± 2.58a | 37.34 ± 3.67a | 29.97 ± 4.78b | 26.07 | <0.001 |
| Enterobacteriaceae_unclassified | 4.61 ± 1.27c | 27.50 ± 2.81a | 25.26 ± 6.23a | 14.96 ± 1.01b | 25.37 ± 3.59a | 9.84 | <0.001 |
| Streptophyta_unclassified | 3.09 ± 0.31b | 2.84 ± 0.81b | 2.21 ± 0.54b | 10.83 ± 1.09a | 11.22 ± 2.92a | 12.05 | <0.001 |
| Stenotrophomonas | 0.32 ± 0.06b | 1.76 ± 0.13a | 0.39 ± 0.06b | 0.27 ± 0.07b | 0.31 ± 0.07b | 61.25 | <0.001 |
| Bacteria_unclassified | 2.11 ± 0.20a | 1.61 ± 0.19a | 1.39 ± 0.16a | 1.68 ± 0.12a | 1.42 ± 0.20a | 2.60 | <0.05 |
Data are shown as mean ± SE. The abbreviation represents different developmental stages: LSB2- 2nd instar nymph, LSB4 – 4th instar nymph. Significant differences among developmental stages were represented by small letters alphabet using Turkey's test with p < 0.05.
Table 3.
Relative abundance of bacterial communities at the phylum level in Tessaratoma javanica.
| Phylum | Egg | LSB 2 | LSB 4 | Male | Female | F value | P value |
|---|---|---|---|---|---|---|---|
| Proteobacteria | 40.70 ± 2.01c | 82.02 ± 1.23a | 82.66 ± 2.57a | 67.44 ± 2.65b | 70.63 ± 2.57b | 67.28 | <0.05 |
| Firmicutes | 12.96 ± 1.08a | 4.68 ± 0.28b | 3.73 ± 0.66c | 6.58 ± 0.82b | 5.29 ± 0.68b | 25.76 | <0.05 |
| Bacteroidetes | 33.05 ± 0.88a | 3.46 ± 0.31b | 2.00 ± 0.17c | 4.97 ± 0.61b | 3.88 ± 0.32b | 582.44 | <0.05 |
| Actinobacteria | 3.93 ± 0.47a | 3.21 ± 0.36a | 5.20 ± 0.93a | 5.19 ± 0.60a | 4.56 ± 0.35a | 2.184 | <0.05 |
| Cyanobacteria | 3.27 ± 0.24b | 2.87 ± 0.81b | 2.27 ± 0.54b | 10.91 ± 1.10a | 11.31 ± 2.90a | 12.16 | <0.05 |
| Bacteria_unclassified | 2.11 ± 0.20a | 1.61 ± 0.18a | 1.39 ± 0.16a | 1.68 ± 0.12a | 1.42 ± 0.20a | 2.61 | <0.05 |
| Acidobacteria | 1.29 ± 0.30a | 0.80 ± 0.12a | 0.97 ± 0.23a | 1.07 ± 0.21a | 0.99 ± 0.17a | 0.66 | <0.05 |
Data in the table are expressed as the mean ± SE. Significant variations among stages of growth were identified using Turkey's test with a significance level of p ≤ 0.05, where different letters (a, b, c) were used to represent these differences.
The 4th nymph stage exhibited the largest relative abundance of Proteobacteria (82.66 ± 2.57 %), followed by the 2nd nymph stage (82.02 ± 1.23 %). The female stage displayed a relatively lower abundance (70.63 ± 2.57 %), while the egg stage had the lowest abundance (40.70 ± 2.01 %) of Proteobacteria (Fig. 2). The prevalence of Firmicutes was found to be notably greater during the egg stage (12.96 ± 1.08 %) and attained the lowest level during the 4th nymphal stage (3.73 ± 0.66 %). The phylum Actinobacteria was highest in the 4th nymph stage (5.20 ± 0.93 %) and lowest in the 2nd nymph stage (3.21 ± 0.36 %). The Cyanobacteria was highest in the female stage (11.31 ± 2.90 %) and lowest in the 4th nymphal stage (2.27 ± 0.54 %). Other remaining bacterial phyla Acidobacteria, Chloroflexi, Nitrospirae, Verrucomicrobia, Gemmatimonadetes, Euryarchaeota, Synergistetes, Thermi, Tenericutes, Armatimonadetes, TM7, Spirochaetes, Fusobacteria, Chlorobi, Planctomycetes, Crenarchaeota, WS3, Elusimicrobia, Deferribacteres, NC10 and Bacteria_unclassified were found in very low abundance. The predominant genera Erwinia and Enterobacteriaceae_unclassified were observed (Table 2). The investigation performed employing SIMPER (Similarity Percentage) technique revealed that the dissimilarities noticed across the developmental stages of T. javanica were mostly attributed to the presence of shared taxa, namely Proteobacteria and Bacteroidetes, throughout distinct stages (Supplementary Table 1). Maximum dissimilarities were observed in the egg and 4th nymphal stage with the χ2 value of 711.67, and minimum dissimilarities were observed between the nymphal stage 2nd and 4th nymphal stage with χ2 value of 44.45. Erwinia, Enterobacteriaceae_unclassified, Streptophyta_unclassified, Stenotrophomonas, and Bacteria_unclassified were the most dominant genera among developmental stages (Table 2). The relative abundance of Erwinia was significantly increased in the larval stage, (44.64 ± 2.58 %) in the 4th nymphal stage and (41.16 ± 4.09 %) in 2nd nymphal stage followed by the adult stage (37.34 ± 3.67 %) in the male and (2.97 ± 4.78) in female and lowest were found in egg stage (6.91 ± 0.59 %). Enterobacteriaceae_unclassified was observed highest in 2nd nymphal stage (27.50 ± 2.81 %) followed by 25.26 ± 6.23 % in the female adult stage and then in 4th nymphal stage (25.26 ± 6.23 %). The lowest Enterobacteriaceae_unclassified was observed in the egg stage (4.61 ± 1.27 %). Streptophyta_unclassified were found highest in the adult female stage (11.22 ± 2.92 %) and (10.83 ± 1.09 %) in the male adult stage followed by the egg stage (3.09 ± 0.31 %) and lowest was found in the nymphal stage, (2.84 ± 0.81 %) in 2nd nymphal stage and (2.21 ± 0.54) in 4th nymphal stage.
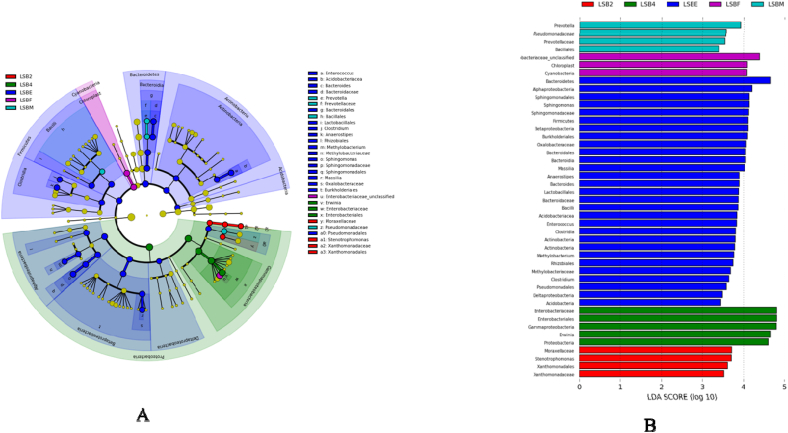
LEfSe analysis revealed that thirty clades were continuously present in the developmental stages of T. javanica (Fig. 5A). Overall, the phylum Proteobacteria was enriched in all the developmental stages. We found that significant differences at bacterial phylum to genus level and LDA score were greater than 2 (Fig. 5B). Each developmental stage of T. javanica had a unique, significantly different bacterial taxa from phylum to genus level.
Fig. 5.
A) LEfSe Bar diagram showing significant differences in different life stages from phylum to genus level shows enriched bacterial diversity. B) LDA (Linear discriminate analysis) score was greater than 2. The lowercase alphabet represented significant differences. Differences in the colour of nodes indicate diversity in bacterial communities across developmental stages of Tessaratoma javanica. Abbreviations of LSBE refer to egg stage, LSB2 - 2nd instar nymph stage, LSB4-4th nymphal stage, LSBF-Female stage, and LSBM-Male stage.
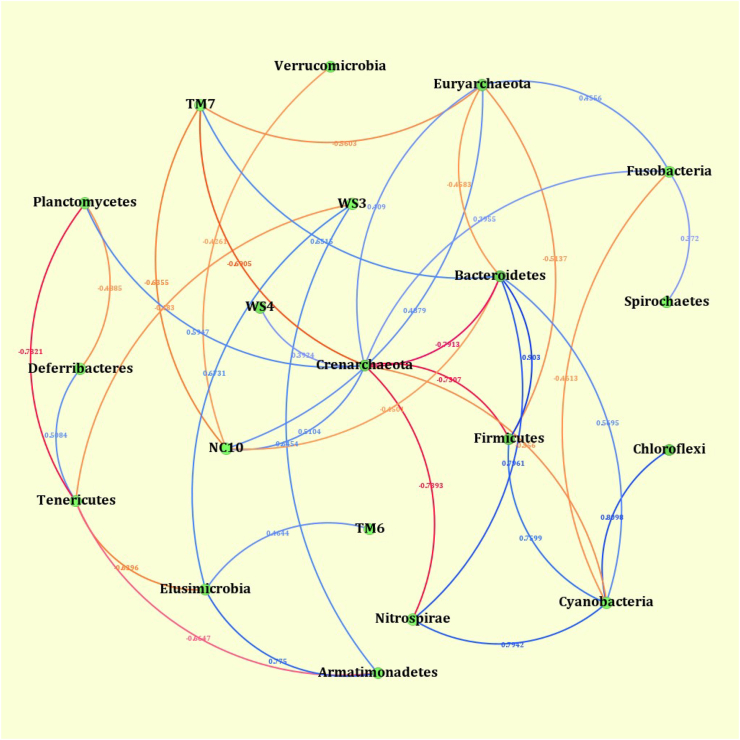
Correlation analysis was used to untangle the microbial co-occurrence pattern among the different phyla of bacteria (Fig. 6). These network relationships show the positive and negative relationships among the phyla of bacteria. The Firmicutes were positively and strongly correlated with Bacteroidetes and Cyanobacteria.
Fig. 6.
Correlation analysis shows the relationship between major phyla. Each node represents a taxon. The blue bars represent the positive correlation and the red bars represent the negative correlation. The deeper the color of the bar (darker blue or darker red), represent the stronger the correlation.
3.4. Functional prediction and their differences among developmental stages of T. javanica
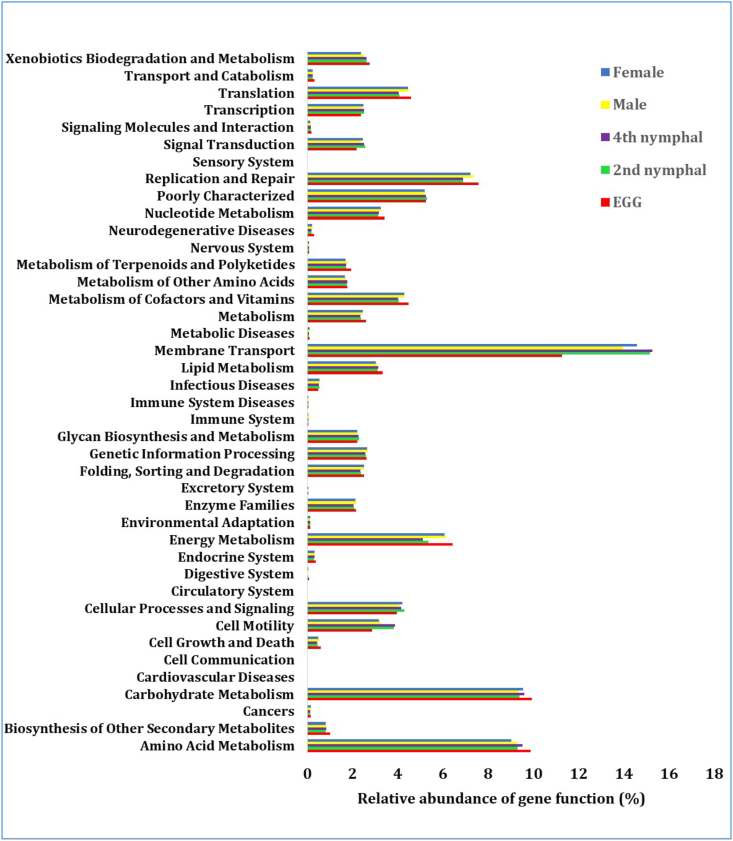
The application of PICRUSt analysis was employed to get insights into the functional characteristics of bacterial communities throughout the various developmental phases of T. javanica, utilizing 16s rRNA gene sequences. A total of 6910 KEGG Orthology (KO) groupings were recorded (Fig. 7). The predicted pathway using the MetaCys database was compared between the egg and other stages. The majority of bacterial genes play crucial roles in many metabolic functional pathways, including amino acid metabolism, glucose and energy metabolism, membrane transport, replication and repair, translation, and transcription. (Supplementary Fig. 2g). Comparative analysis was observed between the egg and 2nd instar nymph, where membrane transport function was more active in the 2nd instar nymph than in the egg stage (Supplementary Fig. 2a). The membrane transport function shows a considerable increase from the egg stage to the 4th instar nymph stage (Supplementary Fig. 2b). In the egg and adult female stages, the functional gene involved in membrane transport was more prevalent in the adult female stage than egg stage, whereas amino acid metabolism was higher in the egg stage than the adult female stage (Supplementary Fig. 2c). A comparison of functional pathways between the 2nd instar nymph and adult female stage revealed a considerable increase in cell motility function in the 2nd instar nymph relative to the adult female stage (Supplementary Fig. 2d). Metabolic functional pathways between 2nd instar nymph and adult male, only membrane transport was higher in 2nd instar nymph than adult male (Supplementary Fig. 2e). In the 4th instar nymph and adult female stages, cell motility was higher in the 4th instar nymph than adult female stage but translation function was higher in the adult female than 4th instar nymph (Supplementary Fig. 2f). The comparison was conducted among the egg stage and other developmental stages, revealing that the membrane transport function was comparatively reduced during the egg stage in comparison to the other stages of development (Supplementary Fig. 2g).
Fig. 7.
KEGG- Level 2 metabolic functions of bacterial communities associated with the different developmental stages of Tessaratoma javanica.
4. Discussion
This study is the first to use Illumina sequencing and functional pathway annotation to look at the bacterial communities that inhabit the gut of the litchi stink bug T. javanica at different stages of development. Symbiotic bacterial communities are of utmost importance in facilitating the growth and development of host organisms [12,18,53]. Insects undergo both incomplete and complete metamorphosis, wherein they experience significant structural transformations that result in alterations in microbial diversity from the egg stage to adults [[54], [55], [56]]. The present study results showed that the predominant bacterial phylum was Proteobacteria in all developmental stages, which was similarly reported in the cospecies T. papillosa [57]. Each developmental stage of T. javanica was dominated by the phylum Proteobacteria, followed by Firmicutes, which follows the pattern of bacteria reported from Hemipteran and other insects as well [[58], [59], [60], [61], [62], [63]]. Bacteria Erwinia and Enterobacteriaceae can degrade polysaccharides and aid in nitrogen fixation which may help in the development, growth, and reproduction of associated hosts. Diverse bacterial communities play a crucial role in the assimilation of nutrients from delicate plant tissues, such as plant parts, flowers, and fruits. These bacterial communities contribute to the growth and development of host plants, including litchi and longan trees, by participating in the metabolism of secondary metabolites [61,64] providing evidence that the host plant can affect the gut microbial communities of an insect. As the sap suckers of T. javanica, proteobacteria bacteria play a vital role in the metabolism of nutrients and secondary metabolites, such as the degradation of plant cell walls which enhance the nutrition of the nymphal stage [65,66]. The other phyla, Bacteroidetes, Actinobacteria, and Cyanobacteria were the remaining identified dominant phyla.
Genus Erwinia was dominant across developmental stages of T. javanica, which is a similar result as observed in A. suturalis [67]. Genus Erwinia metabolizes nitrogen, sulphur, and phosphorus [68] and helps in the digestion and absorption of nutrients [67]. Stink bugs frequently exhibit a symbiotic association with the family Enterobacteriaceae, belonging to the class Gammaproteobacteria and the order Enterobacteriales, with a higher abundance during the 4th nymphal stage [57]. The presence of Enterobacteriaceae bacteria in fruit flies during their larval or adult stage has been observed to potentially contribute to sugar metabolism and exert a significant influence on courting and reproductive behaviours (Zhao et al., 2018), and gammaproteobacteria is an essential symbiont that helps Hemiptera in overall host fitness [69].
The egg stage exhibited a higher level of bacterial diversity, aligning with findings from field-collected samples of T. papillosa and other hemipteran insects [57]. A significant amount of microbial variety was identified in the embryonic phase of insects when examining the bacterial makeup across several stages of development [[70], [71], [72]]. The eggs that were grown in the laboratory for extended durations had a limited bacterial diversity [73,74]. Hence, it is plausible that a substantial bacterial variety during the egg stage is a prevalent occurrence in direct field-collected samples, as observed in the present investigation. The complex and high diversity of eggs may also be environmentally dependent compared to host feeding stages because the egg stage directly contacts the environment of surroundings like soil and plant nectar [15]. The vertical spread of high egg stage variety across females on the egg surface is necessary to compensate for the dietary needs of newly emerged larvae and facilitate their complete growth [75].
The bacterial communities in the 2nd and 4th nymphal stages were almost similar, and the adult stages had higher bacterial diversity in comparison with the nymphal stage. The nymphal stages and male adult bacterial communities overlap and cluster together in PCoA results, which is also reported in other species of litchi stink bug, i.e., T. papillosa [57]. Several studies have reported variations in bacterial diversity among different life stages of bugs. For instance, Liu et al. [57], Guo et al. [76], Xue et al. [77], and Xue et al. [67] observed higher bacterial diversity in the nymphs of certain bug species. Conversely, Gao et al. [73], Ali et al. [70], Andongma et al. [71], Zhao et al. [72], Hu et al. [78], and Noman et al. [79] found higher bacterial diversity in the adult stage of other insect species. The diversity of bacterial communities in various developmental stages is contingent upon the insect species and the environmental conditions in which they reside and obtain nourishment. The gut bacteria between nymphs and adults of incomplete metamorphosis insects may have smaller changes [67]. Differences in their living environment, feeding habits, life histories, and sources of nutrition can account for the variation in bacterial diversity between nymphs and adults. These factors contribute to notable changes in the composition of the intestinal bacterial community [3,73,[80], [81], [82]]. The studies conducted by Choudhary et al. [25] and Xue et al. [67] are relevant to the subject matter in discussion. The species richness in female adults was higher than that of males; the PCoA result, therefore, shows slight differences among them. The present study hypothesis was that females harbor an abundant and diverse gut microbiota compared to males due to their wide adaptability and high reproduction capability. Therefore, dietary factors significantly influence the dynamic alterations observed in the bacterial community composition of insects.
The ability to predict the function of gut-associated bacteria in stink bugs or other insects offers numerous benefits that inhance our comprehension of ecology, evolution, and potential future applications. Understanding the role and function of gut-associated bacteria sheds light on the complicated interaction that exists between insects and their microbial partners. It clarifies how these relationships affect an insect's ability to adapt to its surroundings, its capacity to interact with other animals in its ecosystem, and the advantages of a specific diet. Comparative research on different stink bug species might highlight recurring patterns or adaptations in the bacterial functional profile of these insect's guts. Microbial communities are involved in many metabolic functions in insects. Numerous genes were found to be involved in many metabolic processes, such as the breakdown of amino acids, the creation of secondary metabolites, the metabolism of carbohydrates and, the breakdown of cofactors and vitamins, and the metabolism of lipids. The findings are consistent with previous studies [6,83,84]. The functional analysis revealed that there was a higher prevalence of amino acid metabolism, biosynthesis of other secondary metabolites, and carbohydrate metabolism in the egg stage compared to other stages. This finding confirms that metabolic function plays a crucial role in the survival and development of an insect from the egg to the larva stage. However, all host-associated bacteria are not always beneficial. Diet-altered variations of gut microbiota may be unfavorable to host insects [81]. This study information can inspire scientific curiosity and deepen our understanding of microbial life and its relationship with the host and other organisms. This work will also help the creation of new ways to get rid of pests that depend on microbes, like symbiont-mediated RNAi and paratrangenesis [18]. However, there were some limitations in this present study. The sequences generated by the next-gen sequencing platform in the present study cannot be assigned to specific bacterial species levels, which should be addressed in future studies using high-resolution profiling of gut bacteria using more sophisticated bioinformatics and sequencing platforms such as PacBio SMRT sequencing systems [85]. Predicted functions of the bacterial community by PICRUSt were annotated in the classification at the genus level due to the limitations of the sequencing platform. Furthermore, the information provided by PICRUSt about the roles played by gut bacteria in host physiology is limited; hence, future research utilizing metagenomic and metatranscriptomics techniques will be necessary to identify potential microbial functions.
5. Conclusion
In conclusion, gut-associated bacteria are essential partners of stink bugs because they play a vital role in insect development, nutrition, health, ecological interaction, and evolution. The present investigation of the bacterial population associated with T. javanica in different developmental stages reveals fresh knowledge about the associations of symbiotic bacteria and their variation across the developmental stages. Therefore, more research on these studies will help to examine the specifics of these symbiotic relationships with the hosts. The current study will also facilitate the development of microbial-dependent emerging pest management strategies, such as symbiont-mediated RNAi and paratrangenesis methods against T. javanica.
Data availability
The raw datasets generated in the process of this investigation have been deposited and are accessible in the NCBI SRA database under the bio-project accession ID PRJNA980004.
CRediT authorship contribution statement
Anita kumari: Writing – review & editing, Writing – original draft, Data curation. Jaipal Singh Choudhary: Writing – review & editing, Writing – original draft, Supervision, Project administration, Formal analysis, Conceptualization. Anand Kumar Thakur: Supervision, Project administration, Investigation. Sushmita Banra: Writing – review & editing, Writing – original draft, Formal analysis. Priti Kumari Oraon: Writing – review & editing, Writing – original draft, Formal analysis. Kanika Kumari: Writing – review & editing, Writing – original draft, Formal analysis. Subhash Kumar Sahu: Writing – review & editing, Writing – original draft, Formal analysis. Mohammed Fahad Albeshr: Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are acknowledging the Council of Scientific and Industrial Research (CSIR), New Delhi for granting a scholarship to the first author under the CSIR-UGC net program. We express our gratitude to the ICAR-Research Complex for Eastern Region, Farming System Research Centre for Hill and Plateau Region (ICAR RCER FSRCHPR), Plandu, Ranchi, for generously providing us with field and laboratory facilities during the duration of our research. The last author acknowledged the support through the Researchers Supporting Project number (RSP2024R436), King Saud University, Riyadh, Saudi Arabia. We authors are also very thankful to the editor and anonymous reviewers for their thoughtful comments to improve the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e32384.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Liu Y., et al. The gut microbiota diversity of five Orthoptera (Insecta, Polyneoptera) insects determined by DNA metabarcoding. Biodivers. Data J. 2023;11 doi: 10.3897/BDJ.11.e98162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douglas A.E. Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 2015;60:17–34. doi: 10.1146/annurev-ento-010814-020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engel P., Moran N.A. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 2013;37(5):699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 4.Dumra N., et al. Comparative evaluation of sublethal doses of different insecticides on the ovipositional behavior of whitefly (Bemisia tabaci) in Brinjal. J. King Saud Univ. Sci. 2024;36(2) [Google Scholar]
- 5.Choudhary J.S., et al. High taxonomic and functional diversity of bacterial communities associated with melon fly, Zeugodacus cucurbitae (Diptera: tephritidae) Curr. Microbiol. 2021;78(2):611–623. doi: 10.1007/s00284-020-02327-2. [DOI] [PubMed] [Google Scholar]
- 6.Naaz N., et al. Developmental stage-associated microbiota profile of the peach fruit fly, Bactrocera zonata (Diptera: tephritidae) and their functional prediction using 16S rRNA gene metabarcoding sequencing. 3 Biotech. 2020;10:1–13. doi: 10.1007/s13205-020-02381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia X., et al. Gut microbiota mediate insecticide resistance in the diamondback moth, Plutella xylostella (L.) Front. Microbiol. 2018;9:25. doi: 10.3389/fmicb.2018.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ankrah N.Y., Douglas A.E. Nutrient factories: metabolic function of beneficial microorganisms associated with insects. Environ. Microbiol. 2018;20(6):2002–2011. doi: 10.1111/1462-2920.14097. [DOI] [PubMed] [Google Scholar]
- 9.Douglas A.E. The microbial dimension in insect nutritional ecology. Funct. Ecol. 2009;23(1):38–47. [Google Scholar]
- 10.Brummel T., et al. Drosophila lifespan enhancement by exogenous bacteria. Proc. Natl. Acad. Sci. USA. 2004;101(35):12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosengaus R.B., et al. Disruption of the termite gut microbiota and its prolonged consequences for fitness. Appl. Environ. Microbiol. 2011;77(13):4303–4312. doi: 10.1128/AEM.01886-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillon R.J., Dillon V.M. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 2004;49(1):71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 13.Janson E.M., et al. Phytophagous insect–microbe mutualisms and adaptive evolutionary diversification. Evol. 2008;62(5):997–1012. doi: 10.1111/j.1558-5646.2008.00348.x. [DOI] [PubMed] [Google Scholar]
- 14.Kuechler S.M., et al. Repeated evolution of bacteriocytes in lygaeoid stinkbugs. Environ. Microbiol. 2019;21(11):4378–4394. doi: 10.1111/1462-2920.14804. [DOI] [PubMed] [Google Scholar]
- 15.Yong H.S., et al. High diversity of bacterial communities in developmental stages of Bactrocera carambolae (Insecta: tephritidae) revealed by Illumina MiSeq sequencing of 16S rRNA gene. Curr. Microbiol. 2017;74:1076–1082. doi: 10.1007/s00284-017-1287-x. [DOI] [PubMed] [Google Scholar]
- 16.Frago E., et al. Insect symbionts as hidden players in insect–plant interactions. TREE. 2012;27(12):705–711. doi: 10.1016/j.tree.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 17.Deng J., et al. Associated bacteria of a pine sawyer beetle confer resistance to entomopathogenic fungi via fungal growth inhibition. Environ. Microbiome. 2022;17(1):47. doi: 10.1186/s40793-022-00443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., et al. The pivotal roles of gut microbiota in insect plant interactions for sustainable pest management. NPJ BIOFILMS MICROBI. 2023;9(1):66. doi: 10.1038/s41522-023-00435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choudhary J.S., et al. Biointensive Integrated Pest Management for Horticultural Crops. 2021. Biointensive integrated pest management of litchi; pp. 23–37. [Google Scholar]
- 20.Kim J.M., et al. Effects of diet type, developmental stage, and gut compartment in the gut bacterial communities of two Cerambycidae species (Coleoptera) J. Microbiol. 2017;55:21–30. doi: 10.1007/s12275-017-6561-x. [DOI] [PubMed] [Google Scholar]
- 21.Li D.D., et al. Fall armyworm gut bacterial diversity associated with different developmental stages, environmental habitats, and diets. Insects. 2022;13(9):762. doi: 10.3390/insects13090762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raymann K., et al. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017;15(3) doi: 10.1371/journal.pbio.2001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira N.C., et al. Non-targeted metabolomics reveals differences in the gut metabolic profile of the fall armyworm strains when feeding different food sources. J. Insect Physiol. 2022;139 doi: 10.1016/j.jinsphys.2022.104400. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava K., Choudhary J.S. In: Trends in Horticultural Entomology. Mani M., editor. 2022. Pests and their management in litchi; pp. 719–734. [Google Scholar]
- 25.Choudhary J.S., et al. Litchi stink bug (Tessaratoma javanica) outbreak in Jharkhand, India, on litchi, Phytopara. 2013;41(1):73–77. [Google Scholar]
- 26.Hitendra K., Gajendra S. Biology of litchi bug, Tessaratoma javanica Thunberg (Hemiptera: pentatomidae) on litchi. Pantnagar J. Res. 2007;5(1):17–20. [Google Scholar]
- 27.Kumar H., et al. Population dynamics and seasonal occurrence of Tessaratoma javanica Thunberg in litchi orchards. Ann. Plant Prot. Sci. 2008;16(1):70–73. [Google Scholar]
- 28.Hassan M.E., et al. Report of litchi stink bug, Tessaratoma javanica (Hemiptera: Tessaratomidae) on mahua tree in Chhattisgarh. Records Zool. Sur. India. 2014;114(2):263–268. [Google Scholar]
- 29.Menzel C.M. Lychee Production in the Asia-Pacific Region. Food and Agricultural Organization of the United Nations; Bangkok, Thailand: 2002. Lychee production in Australia; pp. 14–27. [Google Scholar]
- 30.Papademetriou M.K., Dent F.J. vol. 4. 2002, Food and Agriculture Organization, RAP Publication; 2002. Lychee production in the Asia-Pacific region. (Regional Office for Asia and the Pacific). [Google Scholar]
- 31.Pu Z.L., et al. Selected Works of Pu Zhelong Sun Yat-Sen University Guangdong Scientech Association. 1992. Utilizing Eupelmid wasp Anastatus sp to control litchi stink bug Tessaratoma papillosa; pp. 135–169. [Google Scholar]
- 32.Schulte M.J., et al. Effects of azadirachtin injection in litchi trees (Litchi chinensis Sonn.) on the litchi stink bug (Tessaratoma papillosa Drury) in northern Thailand. J. Pest. Sci. 2006;79:241–250. [Google Scholar]
- 33.Mehra B.P., Kapur A.P. Bionomics and control of T. javanica (thunberg): a sporadic pest of Kusum, Schleichera oleosa in chota nagpur. Indian J. Entomol. 1955;17(1):76–88. [Google Scholar]
- 34.Fuping L., et al. Toxicity of neem seed extract to (Tessaratoma papillosa (Drury) relative to its allozyme genotypes, Kun Chong xue bao. Acta Entomol. Sin. 2006;49(2):241–246. [Google Scholar]
- 35.Leksawasdi P., Kumchu C. Mass rearing and releasing of the parasitoid anastatus sp. Agric. Nat. Resour. 1991;25(1):47–53. [Google Scholar]
- 36.Shichou H., et al. Mass releasing Anastatus japonicus to control Tessaratoma papillosa in HongKong, J. Biol. Control. 1999;15:54–56. [Google Scholar]
- 37.Mondal M.F., et al. First report of litchi stink bug (Tessaratoma javanica Thunberg) outbreak in Bangladesh. Int. J. Trop. Insect Sci. 2021;41:383–387. [Google Scholar]
- 38.Zheng Z.L. Overview on the biological and ecological characteristics of Tessaratoma papillosa Drury, South China. Fruits. 2014;43:25–33. [Google Scholar]
- 39.Parveen S., et al. Biology, morphology and DNA barcodes of Tessaratoma javanica (thunberg) (Hemiptera: Tessaratomidae) Zootaxa. 2015;3936(2):261–271. doi: 10.11646/zootaxa.3936.2.6. [DOI] [PubMed] [Google Scholar]
- 40.Choudhary J.S., et al. Determination bio-efficacy of insecticides against litchi stink bug, Tessaratoma javanica (Thunberg) (Hemiptera: Tessaratomidae): an emerging major pest of litchi. Litchi chinensis Sonn, The Bioscan. 2015;10(1):217–220. [Google Scholar]
- 41.Choudhary J.S., et al. New record of the litchi stink bug, Tessaratoma javanica (thunberg) egg parasitoids and their natural control effect in litchi orchards from India. Entomol. Gen. 2015;35(3):187–197. [Google Scholar]
- 42.Naaz N., et al. Identification and evaluation of cultivable gut bacteria associated with peach fruit fly, Bactrocera zonata (Diptera: tephritidae) Phytoparasitica. 2016;44(2):165–176. [Google Scholar]
- 43.Klindworth A., et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nuc. Aci. Res. 2013;41(1) doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc Available online at:
- 45.Krueger F., et al. FelixKrueger/TrimGalore: v0.6.7—DOI via zenodo. 2021. [DOI]
- 46.Rognes T., et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4 doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 48.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 49.Caporaso J.G., et al. PyNAST: a fexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26(2):266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammer O., Harper D.A. Past: paleontological statistics software package for education and data anlysis. Palaeont. Electr. 2001;4(1):1. [Google Scholar]
- 51.Langille M.G.I., et al. Predictive functional profing of microbial communities using 16S rRNA marker gene sequence. Nat. Biotechnol. 2013;31(9):814–882. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parks D.H., et al. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong W. Intestinal microbiota in various animals. Integ. Zool. 2022;17(3):331–332. doi: 10.1111/1749-4877.12633. [DOI] [PubMed] [Google Scholar]
- 54.Chen B., et al. Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 2016;6(1) doi: 10.1038/srep29505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammer T.J., et al. Caterpillars lack a resident gut microbiome. Proc. Natl. Acad. Sci. USA. 2017;114(36):9641–9646. doi: 10.1073/pnas.1707186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao X., et al. The divergence in bacterial components associated with Bactrocera dorsalis across developmental stages. Fron. Microbiol. 2018;9:114. doi: 10.3389/fmicb.2018.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z.H., et al. Stage correlation of symbiotic bacterial community and function in the development of litchi bugs (Hemiptera: Tessaratomidae) Ant. Leeu. 2022;115(1):125–139. doi: 10.1007/s10482-021-01685-6. [DOI] [PubMed] [Google Scholar]
- 58.Husseneder C., et al. Bacteria associated with Piezodorus guildinii (Hemiptera: pentatomidae), with special reference to those transmitted by feeding. Environ. Entomol. 2017;46(1):159–166. doi: 10.1093/ee/nvw112. [DOI] [PubMed] [Google Scholar]
- 59.Lim L., Ab Majid A.H. Metagenomic 16S rDNA amplicon data of microbial diversity of guts of fully fed tropical bed bugs. Cimex hemipterus (F.) (Hemiptera: Cimicidae), Data brief. 2020;30 doi: 10.1016/j.dib.2020.105575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chandler J.A., et al. Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Gen. 2011;7(9) doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colman D.R., et al. Do diet and taxonomy influence insect gut bacterial communities. Mol. Ecol. 2012;21(20):5124–5137. doi: 10.1111/j.1365-294X.2012.05752.x. [DOI] [PubMed] [Google Scholar]
- 62.Engel P., et al. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA. 2012;109(27):11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yun J.H., et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host, App. Environ. Microbiol. 2014;80(17):5254–5264. doi: 10.1128/AEM.01226-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dillon R., Charnley K. Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Res. Microbiol. 2002;153(8):503–509. doi: 10.1016/s0923-2508(02)01361-x. [DOI] [PubMed] [Google Scholar]
- 65.Ben-Yosef M., et al. Symbiotic bacteria enable olive fly larvae to overcome host defences. R. Soc. Open Sci. 2015;2(7):150–170. doi: 10.1098/rsos.150170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bozorov T.A., et al. Characterization of the gut microbiota of invasive Agrilus mali Matsumara (Coleoptera: buprestidae) using high-throughput sequencing: uncovering plant cell-wall degrading bacteria. Sci. Rep. 2019;9(1):4923. doi: 10.1038/s41598-019-41368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xue H., et al. Gut bacterial diversity in different life cycle stages of Adelphocoris suturalis (Hemiptera: Miridae) Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.670383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Friedl M.A., et al. Carbon source dependence and photostimulation of conidiation in Hypocrea atroviridis, App. Environ. Microbiol. 2008;74(1):245–250. doi: 10.1128/AEM.02068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kashkouli M., et al. Heritable gammaproteobacterial symbiont improves the fitness of Brachynema germari Kolenati (Hemiptera: pentatomidae) Environ. Entomol. 2019;48(5):1079–1087. doi: 10.1093/ee/nvz089. [DOI] [PubMed] [Google Scholar]
- 70.Ali H., et al. Pyrosequencing uncovers a shift in bacterial communities across the life stages of Octodonta nipae (Coleoptera: chrysomelidae) Front. Microbiol. 2019;10:466. doi: 10.3389/fmicb.2019.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andongma A.A., et al. Assessment of the bacteria community structure across life stages of the Chinese citrus fly, Bactrocera minax (Diptera: Tephritidae) BMC Microbiol. 2019;19:1–9. doi: 10.1186/s12866-019-1646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao C., et al. The developmental stage symbionts of the pea aphid-feeding Chrysoperla sinica (Tjeder) Front. Microbiol. 2019;10:2454. doi: 10.3389/fmicb.2019.02454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao X., et al. Biodiversity of the microbiota in Spodoptera exigua (Lepidoptera: Noctuidae) J. Appl. Microbiol. 2019;126(4):1199–1208. doi: 10.1111/jam.14190. [DOI] [PubMed] [Google Scholar]
- 74.Wang X., et al. Variability of gut microbiota across the life cycle of Grapholita molesta (Lepidoptera: Tortricidae) Front. Microbiol. 2020;11:1366. doi: 10.3389/fmicb.2020.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lam K., et al. Ovipositing female house flies provision offspring larvae with bacterial food. Entomol. Exp. Appl. 2009;133(3):292–295. [Google Scholar]
- 76.Guo D., et al. Dynamic gut microbiota of Apolygus lucorum across different life stages reveals potential pathogenic bacteria for facilitating the pest management, microbiol. Ecol. 2024;87:9. doi: 10.1007/s00248-023-02324-5. [DOI] [PubMed] [Google Scholar]
- 77.Xue H., et al. Dynamics and diversity of symbiotic bacteria in Apolygus lucorum at different developmental stages. J. Cotton Res. 2023;6:1–11. [Google Scholar]
- 78.Hu Y., et al. Dynamic of composition and diversity of gut microbiota in Triatoma rubrofasciata in different developmental stages and environmental conditions. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.587708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Noman M.S., et al. Diversity of bacteria in different life stages and their impact on the development and reproduction of Zeugodacus tau (Diptera: tephritidae) Insect Sci. 2021;28(2):363–376. doi: 10.1111/1744-7917.12768. [DOI] [PubMed] [Google Scholar]
- 80.Chouaia B., et al. Developmental stages and gut microenvironments influence gut microbiota dynamics in the invasive beetle Popillia japonica Newman (Coleoptera: scarabaeidae) Environ. Microbiol. 2019;21(11):4343–4359. doi: 10.1111/1462-2920.14797. [DOI] [PubMed] [Google Scholar]
- 81.Luo J., et al. Variation of gut microbiota caused by an imbalance diet is detrimental to bugs' survival. Sci. Total Environ. 2021;771 doi: 10.1016/j.scitotenv.2020.144880. [DOI] [PubMed] [Google Scholar]
- 82.Ma M., et al. Composition and diversity of gut bacterial community in different life stages of a leaf beetle Gastrolina depressa. Microbiol. Ecol. 2023;86:590–600. doi: 10.1007/s00248-022-02054-0. [DOI] [PubMed] [Google Scholar]
- 83.Li Y., et al. Bt GS57 interaction with gut microbiota accelerates Spodoptera exigua mortality. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.835227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y.P., et al. The adaptive evolution in the fall armyworm Spodoptera frugiperda (Lepidoptera: noctuidae) revealed by the diversity of larval gut bacteria. Genes. 2023;14(2):321. doi: 10.3390/genes14020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu L., et al. High-resolution profiling of gut bacterial communities in an invasive beetle using PacBio SMRT sequencing system. Insects. 2019;10(8):248. doi: 10.3390/insects10080248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw datasets generated in the process of this investigation have been deposited and are accessible in the NCBI SRA database under the bio-project accession ID PRJNA980004.