Abstract
Molecular diagnosis of inborn errors of immunity (IEI) plays a critical role in determining patients’ long-term prognosis, treatment options, and genetic counseling. Over the past decade, the broader utilization of next-generation sequencing (NGS) techniques in both research and clinical settings has facilitated the evaluation of a significant proportion of patients for gene variants associated with IEI. In addition to its role in diagnosing known gene defects, the application of high-throughput techniques such as targeted, exome, and genome sequencing has led to the identification of novel disease-causing genes. However, the results obtained from these different methods can vary depending on disease phenotypes or patient characteristics. In this study, we conducted whole-exome sequencing (WES) in a sizable cohort of IEI patients, consisting of 303 individuals from 21 different clinical immunology centers in Türkiye. Our analysis resulted in likely genetic diagnoses for 41.1% of the patients (122 out of 297), revealing 52 novel variants and uncovering potential new IEI genes in six patients. The significance of understanding outcomes across various IEI cohorts cannot be overstated, and we believe that our findings will make a valuable contribution to the existing literature and foster collaborative research between clinicians and basic science researchers.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10875-024-01759-w.
Keywords: Inborn errors of immunity, next generation sequencing, whole exome sequencing, genetic diagnosis
Introduction
Inborn errors of immunity or primary immunodeficiencies (PIDs) represent a diverse group of disorders characterized by increased susceptibility to infections, malignancy, allergy, and immune dysregulation [1]. While these diseases occur at a frequency of approximately 1 in 10,000 in the general population, their prevalence is higher in societies with elevated rates of consanguinity, such as Türkiye [2–4]. The genetic pleiotropy and heterogeneity observed in IEI contribute to the broad range of clinical manifestations associated with these disorders [5]. The majority of IEI cases are monogenic diseases with autosomal recessive inheritance patterns [5]. Therefore, comprehensive genetic diagnosis is vital for effective management of patients with IEI. In the past decade, NGS methods have revolutionized genetic screening, greatly enhancing the diagnostic capabilities for IEI [6]. This progress has led to an unprecedented increase in the identification of genes causing immunodeficiencies, with approximately 500 genetic defects associated with immunodeficiency currently recognized [7].
Founded in 2018 in memory of Can Sucak, who suffered from ZAP70 deficiency, the Candan Bişeyler Foundation (CSCBF) actively supports research in the field of IEI and raises awareness in Türkiye. The “Hacettepe University Can Sucak Research Laboratory for Translational Immunology” is dedicated to providing genetic diagnosis for immunodeficiency patients and conducting advanced functional research in a comprehensive manner throughout the country. This study presents the results of a comprehensive investigation into the genetic diagnosis of an extensive cohort of IEI patients from a specialized immune deficiency research center in Türkiye.
Methods
Study Participants
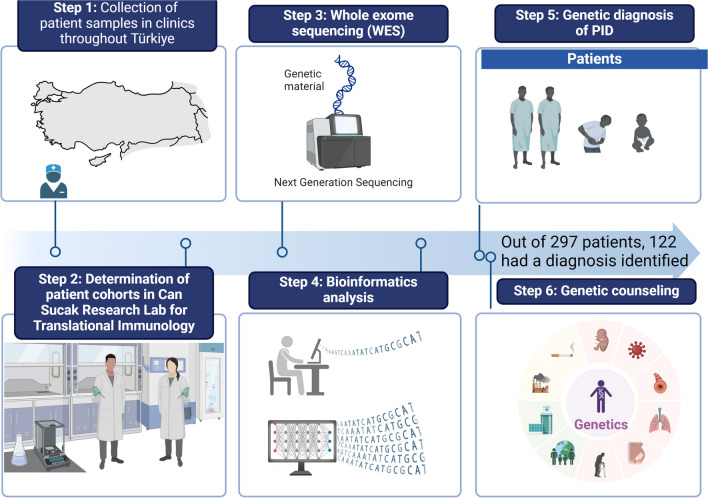
Patients diagnosed with IEI based on clinical and laboratory characteristics between 2020 and 2023 were included in the study. These patients were recruited from multiple clinical immunology centers in Türkiye. Blood samples were collected from the patients following the guidelines and approval of the local Ethics Committee of Hacettepe University. Informed consent forms were obtained from the participants or their parents. The study's workflow is illustrated in Fig. 1.
Fig. 1.
Schematic workflow of the study
Whole Exome Sequencing and Variant Analysis
Genomic DNA was isolated from peripheral blood samples using a DNA isolation kit (GeneAll). The NGS exome library was prepared utilizing the Illumina Nextera DNA Prep with Enrichment Kit. Sequencing was carried out on the Illumina NextSeq 550 platform, generating 150-bp paired-end reads. Mapping, variant calling, and annotation were performed using SEQ Platform v8 (Genomize). Copy number variation (CNV) analysis was conducted using SEQ Platform as well.
To identify causative variants, we employed a filtering strategy that involved screening all variants identified from the WES data. Our focus was on exonic and splice site variants, excluding synonymous variants, and we specifically looked for rare variants with a minor allele frequency of less than 1% in different strategic gene groups. Initially, we examined rare variants in known IEI genes (approximately 500), followed by potential candidate genes predicted by the human gene connectome [8]. Finally, we assessed variants across the entire set of genes (Supplementary Figure 2A).
Sanger Sequencing
To validate the identified variants, we conducted Sanger sequencing using standard protocols [9].
RT-qPCR
RT-qPCR was utilized to validate the effects of structural variants. Total RNA was isolated from peripheral blood mononuclear cells (PBMCs) obtained from both patients and healthy controls using the NucleoSpin RNA Plus Kit (Macherey-Nagel). Subsequently, cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). RT-qPCR was carried out on the CFX Connect System (Bio-Rad) using the iTaq Universal SYBR Green Supermix (Bio-Rad) [10].
Results
Technical Output of the Sequencing Data
The results of the WES data showed a total number of reads ranging from 21.7 to 77.6 million (median: 46.1) (Supplementary Figure 2B). The average depth of coverage varied between 24.5 and 134.2 (median: 64.1) (Supplementary Figure 2C). The target regions (exons and splice regions) were covered at a depth of 20X from 89.02% to 99.91%, and at a depth of 50X from 68.13% to 99.65% (Supplementary Figure 2D).
Patients
Our study involved a total of 303 individuals who were clinically diagnosed with IEI. These participants were recruited from 21 separate clinical immunology centers and they were selected after assessments with their clinicians. Especially, patients truly exhibited severe phenotypes of immunodeficiency were admitted to the study. However, six patients were excluded from the current analysis as they exhibited potential novel IEI-associated genes, pending further investigation through functional studies. Therefore, the analysis in this study includes 297 patients.
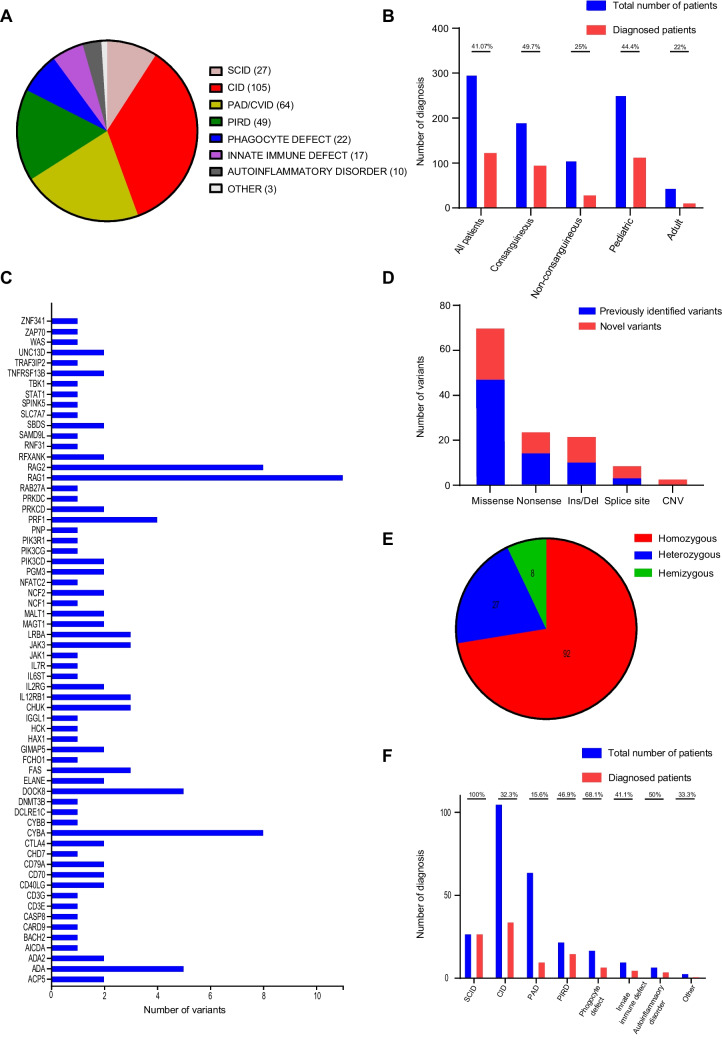
Among the included patients, there were 145 males and 152 females, representing a relatively balanced gender distribution. The age range of the participants varied from three months to 42 years, with a median age of nine years. The majority of the cohort consisted of pediatric patients (n=252), while a smaller subset comprised adult patients (n=45). A notable observation in our study was the high consanguinity rate, with 64.6% (192 out of 297 cases) of patients demonstrating consanguineous relationships within their families. The distribution of clinical diagnoses, classified according to the International Union of Immunological Societies (IUIS) classification, included 27 cases of Severe Combined Immunodeficiency (SCID), 105 cases of Combined Immunodeficiency (CID), 64 cases of Primary Antibody Deficiency (PAD), 49 cases of Primary Immune Regulatory Disorder (PIRD), 22 cases of congenital anomalies affecting phagocyte number/function, 17 cases of disorders of intrinsic and innate immunity, 10 cases of autoinflammatory disorders, and 3 cases of other classified IEI. These other cases potentially involve bone marrow failure or complement deficiencies, as illustrated in Fig. 2A.
Fig. 2.
Patient and variant characteristics. A Distribution of the patients based on their clinical diagnosis. B Diagnostic yield of the patients. C Number of the detected variants and their distribution across different IEI genes. D Types of detected variants and their novelty. E Distribution of zygosity. F Number of diagnosis in patient groups
Results of Genetic Diagnosis and the Profile of Disease-Causing Variants
In our cohort, a genetic diagnosis was established in 122 out of the 297 patients examined, with a total of 127 potential genetic variants identified. This yielded a diagnostic rate of 41.1%. Among the 193 patients with consanguineous parents, causative genetic defects were identified in 95 individuals, resulting in a diagnostic rate of 49.7%. On the other hand, among the 106 patients from non-consanguineous parents, 28 individuals (25.7%) received a genetic diagnosis. The diagnostic rate was higher in pediatric patients, with 44.4% (112 out of 252) receiving a genetic diagnosis, compared to the adult group, which had a lower rate of 22% (10 out of 45) (Fig. 2B). Details of all identified genetic variants and their associated clinical features are presented in Table 1, Table 2 and Supplementary Table 3. In addition, variant characteristics including American College of Medical Genetics (ACMG) criteria and pathogenicity prediction scores were given in Supplementary Table 1). Overall, a total of 127 likely causative genetic anomalies were identified across 64 known IEI genes, as depicted in Fig. 2C. Among these genetic variants, 75 had been previously reported in public databases, while 52 were novel findings reported in this study (Fig. 2D). The variants consisted of 92 homozygous, 27 heterozygous, and 8 hemizygous mutations (Fig. 2E). The spectrum of variant types included 69 missense mutations, 24 nonsense mutations, 22 insertion/deletions (indels), 9 essential splice site variations, and 3 copy number variations (Figure 2D). CNV analysis was performed on 57 subjects using a strategy that incorporated samples with comparable mean read depths. The implications of the CNVs were validated through capillary sequencing or quantitative PCR (qPCR). The causality of monoallelic variants was evaluated based on clinical and laboratory features of the patients, literature associations, or different functional analyses (Supplementary Table 2). The diagnostic rates across different disease categories were as follows: Severe Combined Immunodeficiency (SCID) had a diagnostic rate of 100%, congenital anomalies affecting phagocyte number/function at 68.1%, autoinflammatory disorders at 50%, Primary Immune Regulatory Disorder (PIRD) at 46.9%, intrinsic and innate immunity defects at 41.1%, Combined Immunodeficiency (CID) at 32.3%, other forms of IEI at 33.3%, and Primary Antibody Deficiency (PAD) at 15.6%, and (Fig. 2F).
Table 1.
Details of the variants detected in the study
| Patient no | Clinical diagnosis (IUIS) | Age | Gender | Consan. | Gene | Variant | Transcript ID | Zygosity | Consequence | Novelty |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 [11] | Innate immune defect | 9 | M | + | CARD9 | c.883C>T p.Gln295Ter | NM_052813.4 | Hom | Nonsense | rs1833232307 |
| P2 [12] | CID | 6 | M | + | RFXANK | c.634C>T p.Arg212Ter | NM_003721.3 | Hom | Nonsense | rs747402973 |
| P3 [13] | SCID | 7 | M | + | CD3E | c.176G>A p.Trp59Ter | NM_000733.3 | Hom | Nonsense | rs121918659 |
| P4 | CID | 12 | F | + | NFATC2 | c.340_345delGAGATC p.Glu114_Ile115del | NM_173091.3 | Hom | Inframe Deletion | Novel |
| P5 [12] | SCID | 6 m | F | + | JAK3 | c.2134G>A p.Gly712Ser | NM_000215.4 | Hom | Missense | rs1178958564 |
| P6 [12] | SCID | 8 m | F | + | RAG2 | c.581C>A p.Ser194Ter | NM_000536.3 | Hom | Nonsense | Novel |
| P7 [12] | SCID | 2 | M | + | RAG1 | c.2005G>A p.Glu669Lys c.1307C>A p.Thr436Asn |
Comp. Het |
Missense Missense |
rs878853004 Novel |
|
| P8 [12] | SCID | 1 | M | + | RAG1 | c.2005G>A p.Glu669Lys c.1307C>A p.Thr436Asn | NM_000448.2 |
Comp. Het |
Missense Missense |
rs878853004 Novel |
| P9 [14] | PIRD | 6 | M | + | CD70 | c.332C>T p.Thr111Met | NM_001252.3 | Hom | Missense | rs1378830614 |
| P10 [14] | PIRD | 4 | M | + | CD70 | c.332C>T p.Thr111Met | NM_001252.3 | Hom | Missense | rs1378830614 |
| P11 [12] | Phagocyte defect | 9 | F | + | CYBA | c.58+4_58+7delAGTG | NM_000101.4 | Hom | Splice site/Deletion | rs771926427 |
| P12 [15] | CID | 6 | M | + | ZNF341 | c.1626C>G p.Tyr542Ter | NM_001282933.2 | Hom | Nonsense | rs376598954 |
| P13 | CID | 7 | F | + | ZAP70 | c.1010T>G p.Leu337Ala | NM_001079.4 | Hom | Missense | rs1254428002 |
| P14 [16, 17] | SCID | 3 m | M | + | RAG2 | c.105G>C p.Gly35Ala | NM_000536.4 | Hom | Missense | rs148508754 |
| P15 [16, 17] | SCID | 1 | M | + | RAG2 | c.105G>C p.Gly35Ala | NM_000536.4 | Hom | Missense | rs148508754 |
| P16 [18, 19] | PAD/CVID | 40 | F | + | TNFRSF13B | c.310C>T p.Cys104Arg | NM_012452.3 | Hom | Missense | rs34557412 |
| P17 | PAD/CVID | 3 | F | + | PIK3R1 | c.837-1G>A | NM_181523.2 | Hom | Splice site/Missense | Novel |
| P18 | CID | 20 | F | + | PGM3 | c.214G>A p.Gly72Ser | NM_001199919.1 | Hom | Missense | Novel |
| P19 | Other | 2 | F | - | SAMD9L | c.2639A>C p.His880Pro | NM_001350083 | Het | Missense | Novel |
| P20 [18] | PAD/CVID | 17 | M | + | TNFRSF13B | c.204dupA p.Leu69Tfs*11 | NM_012452.3 | Hom | Out of frame/Insertion | rs72553875 |
| P21 | PAD/CVID | 24 | F | + | CD79A | c.380-2A>G | NM_001783 | Hom | Splice Site/Missense | Novel |
| P22 | CID | 34 | F | + | DNMT3B | c.2029G>A p.Val677Met | NM_006892.4 | Hom | Missense | rs866792483 |
| P23 | PAD/CVID | 34 | F | + | AICDA | c.A100T p.Lys34Ter | NM_001330343 | Hom | Nonsense | Novel |
| P24 [20, 21] | Phagocyte defect | 2 | F | + | CYBA | c.G70A p.Gly24Arg | NM_000101.4 | Hom | Missense | rs28941476 |
| P25 [22] | CID | 13 | M | + | MALT1 | c.1318_1321delTGTC p.L440Valfs*6 | NM_006785.4 | Hom | Out of frame/Deletion | rs140664950 |
| P26 | Phagocyte defect | 10 | F | - | SBDS |
c.578T>C p.Lys193Pro c.184A>T p.Lys62Ter |
NM_016038.4 | Comp. Het |
Missense Nonsense |
rs120074160 rs1195681400 |
| P27 | CID | 10 | M | + | RFXANK | Exon 2-6 Deletion | NM_003721.3 | Hom | CNV | Novel |
| P28 | PIRD | 11 | F | - | MAGT1 | c.199-16A>G | NM_032121.5 | Hem | Splice Site/Missense | Novel |
| P29 | SCID | 6 m | F | + | ADA | c.551_555del p.Glu184Glyfs*2 c.241G>A p.Gly81Arg |
Comp. Het |
Out of frame/Deletion Missense |
Novel rs2065384316 |
|
| P30 | SCID | 1 | F | + | RAG1 | c.1767C>G p.Tyr589Ter | NM_000448.2 | Hom | Nonsense | Novel |
| P31 | SCID | 8 m | F | + | JAK3 | c.932delC p.Pro311Argfs*17 | NM_000215 | Hom | Out of frame/Deletion | Novel |
| P32 | Innate immune defect | 2 | M | + | TRAF3IP2 | c.559C>T p.Arg187Ter | NM_147686.3 | Hom | Nonsense | rs762395569 |
| P33 | SCID | 9 m | M | + | RAG1 | c.2126G>A p.Gly709Asp | NM_000448.2 | Hom | Missense | Novel |
| P34 | SCID | 1 | M | + | ADA | c.779A>G p.Glu260Gly | NM_000022.4 | Hom | Missense | rs1354071013 |
| P35 | Phagocyte defect | 10 | M | + | NCF2 | c.233G>A p.Gly78Glu | NM_000433.4 | Hom | Missense | rs137854519 |
| P36 | Phagocyte defect | 1 | F | + | CYBA | c.166dupC p.Arg56Profs*156 | NM_000101 | Hom | Out of frame/Insertion | rs1555550793 |
| P37 | PIRD | 9 | M | + | LRBA | c.646-1G>A | NM_006726.4 | Hom | Splice site/Missense | rs1741243666 |
| P38 | SCID | 10 m | F | + | JAK3 | c.2080G>T p.Glu694Ter | NM_000215.3 | Hom | Nonsense | Novel |
| P39 | SCID | 4 | M | - | IL2RG | c.437T>A p.Leu146Gln | NM_000206.2 | Hem | Missense | Novel |
| P40 | PIRD | 19 | M | + | PRKCD | c.1097G>A p.Gly366Glu | NM_001354680.2 | Hom | Missense | Novel |
| P41 | SCID | 1 | M | + | RAG2 | c.623T>A p.Val208Asp | NM_001243786.1 | Hom | Missense | Novel |
| P42 [23–26] | PIRD | 15 | M | - | CTLA4 | c.118G>A p.Val40Met | NM_005214.5 | Het | Missense | rs1553657378 |
| P43 | PIRD | 17 | M | - | JAK1 | c.2485A>G p.Asn829Asp | NM_001321853.2 | Het | Missense | Novel |
| P44 | SCID | 9 m | F | + | RAG1 | c.1767C>G p.Tyr589Ter | NM_000448.2 | Hom | Nonsense | Novel |
| P45 | PIRD | 18 | M | + | PRKCD | c.1097G>A p.Gly366Glu | NM_001354680.2 | Hom | Missense | Novel |
| P46 | Phagocyte defect | 32 | M | - | CYBB | c.770G>A p.Cys257Tyr | NM_000397.4 | Hem | Missense | Novel |
| P47 | CID | 8 | F | + | CHUK | c.499G>A p.Gly167Arg | NM_001278.5 | Hom | Missense | Novel |
| P48 | CID | 4 | F | + | CHUK | c.499G>A p.Gly167Arg | NM_001278.5 | Hom | Missense | Novel |
| P49 | SCID | 1 | F | + | RAG1 | c.742C>T p.Gln248Ter | NM_000448.2 | Hom | Nonsense | Novel |
| P50 | CID | 10 | M | - | CD40L | c.15C>A p.Tyr5Ter | NM_000074.3 | Hem | Nonsense | Novel |
| P51 | PIRD | 1 | M | + | UNC13D | c.2346_2349delGGAG p.Arg782SerfsTer12 | NM_199242.2 | Hom | Out of frame/Deletion | rs764196809 |
| P52 [27] | PAD/CVID | 2 | F | + | IGGL1 | c.425C>T p.Pro142Leu | NM_020070.4 | Hom | Missense | rs1064422 |
| P53 | Phagocyte defect | 1 | F | - | ELANE | c.703delG p.Val235TrpfsTer5 | NM_001972.4 | Het | Out of frame/Deletion | Novel |
| P54 | Autoinflammatory disorder | 42 | F | - | HCK | c.135_136delinsTG p.Pro46Ala | NM_002110.4 | Het | Indel | Novel |
| P55 | Phagocyte defect | 11 | M | + | CYBA | c.385G>A p.Glu129Lys | NM_000101.4 | Hom | Missense | rs1246768740 |
| P56 | PIRD | 17 | M | + | SLC7A7 | c.1417C>T p.Arg473Ter | NM_001126106.2 | Hom | Nonsense | rs386833808 |
| P57 [28, 29] | Phagocyte defect | 4 | M | + | NCF2 | c.196C>T p.Arg66Ter | NM_000433.3 | Hom | Nonsense | rs750782115 |
| P58 | SCID | 2 | M | + | DCLRE1C | c.1633del p.Glu545Asnfs*58 | NM_001350965.2 | Hom | Out of frame/Deletion | Novel |
| P59 | SCID | 8 m | F | + | RAG2 | c.712delC p.Val238LeufsTer10 | NM_001243786.1 | Hom | Out of frame/Deletion | Novel |
| P60 [30, 31] | Innate immune defect | 18 | F | + | IL12RB1 | c.523C>T p.Arg175Trp | NM_005535.3 | Hom | Missense | rs750667928 |
| P61 | CID | 12 | M | + | CD40L | c.15C>A p.Tyr5Ter | NM_000074.3 | Hom | Nonsense | Novel |
| P62 [32–34] | Autoinflammatory disorder | 15 | M | + | ADA2 | c.1072G>A p.Gly358Arg | NM_001282225.2 | Hom | Missense | rs45511697 |
| P63 [35, 36] | Innate immune defect | 2 | M | + | IL12RB1 | c.1456C>T p.Arg486Ter | NM_005535.3 | Hom | Nonsense | rs576374797 |
| P64 | CID | 2 | F | + | CHUK | c.499G>A p.Gly167Arg | NM_000074.3 | Hom | Missense | Novel |
| P65 | Phagocyte defect | 6 | M | + | CYBA | c.371C>T p.Ala124Val | NM_000101.4 | Hom | Missense | rs179363894 |
| P66 [37] | CID | 17 | M | + | GIMAP5 | c.667C>T p.Leu223Phe | NM_018384.5 | Hom | Missense | rs2116581086 |
| P67 [37] | CID | 12 | F | + | GIMAP5 | c.667C>T p.Leu223Phe | NM_018384.5 | Hom | Missense | rs2116581086 |
| P68 | PAD/CVID | 7 | F | + | CD79A | c.177dup p.Asn60GlnfsTer20 | NM_001783.4 | Hom | Out of frame/Insertion | Novel |
| P69 | PIRD | 1 | F | + | UNC13D | c.1082del p.Tyr361SerfsTer43 | NM_199242.2 | Hom | Out of frame/Deletion | Novel |
| P70 [38] | PIRD | 19 | M | - | FAS | c.361C>T p.Arg121Trp | NM_000043.6 | Het | Missense | rs121913078 |
| P71 [39, 40] | PIRD | 1 | F | + | PRF1 | c.1122G>A p.Trp374Ter | NM_005041.5 | Hom | Nonsense | rs104894176 |
| P72 | CID | 6 | M | + | DOCK8 | c.5831C>T p.Pro1944Leu | NM_203447.3 | Hom | Missense | rs775779897 |
| P73 | CID | 4 | F | + | DOCK8 | c.5831C>T p.Pro1944Leu | NM_203447.3 | Hom | Missense | rs775779897 |
| P74 [26, 41] | PIRD | 14 | F | - | CTLA4 | c.151C>T p.Arg51Ter | NM_005214.5 | Het | Nonsense | rs606231417 |
| P75 [42, 43] | Phagocyte defect | 5 | M | + | HAX1 | c.130_131insA p.Trp44Ter | NM_006118.4 | Hom | Out of frame | rs1572018284 |
| P76 | CID | 6 | F | + | PIK3CG | c.2159A>G p.Tyr720Cys | NM_002649.3 | Hom | Missense | rs199590448 |
| P77 | CID | 7 | M | + | MALT1 | c.1133T>G p.Phe378Cys | NM_006785.4 | Hom | Missense | novel |
| P78 | PIRD | 12 | M | - | MAGT1 | c.628-4T>C | NM_032121.5 | Hem | Splice site/Missense | novel |
| P79 [44] | Autoinflammatory disorder | 17 | M | + | ACP5 |
c.772_790del p.Ser258WTrpfs*39 |
NM_001322023.2 | Hom | Out of frame/Deletion | rs878853218 |
| P80 [45] | CID | 1 | F | + | PGM3 | c.821A>G p.Asn274Ser | NM_001199917.2 | Hom | Missense | rs587777562 |
| P81 [46] | CID | 9 | F | + | CD3G | c.80-1G>C | NM_000073.2 | Hom | Splice site/Missense | rs775848095 |
| P82 | Phagocyte defect | 2 | M | - | ELANE | c.367-8C>A | NM_001972.4 | Het | Splice site/Missense | novel |
| P83 [20, 47] | Phagocyte defect | 16 | F | - | CYBA | c.70G>A p.Gly24Arg c.373G>A p.Ala125Thr | Comp. Het |
Missense Missense |
rs28941476 rs119103269 | |
| P84 | Autoinflammatory disorder | 16 | M | + | ADA2 | c.319A>C p.Lys107Gln | NM_001282225.2 | Hom | Missense | novel |
| P85 | PIRD | 6 | M | - | FAS | c.761T>A p.Val254Asp | NM_000043.6 | Het | Missense | novel |
| P86 | CID | 3 | F | + | PNP | c.461+1G>A | NM_000270.3 | Hom | Splice site/Missense | novel |
| P87 [48–51] | PIRD | 9 | M | + | RAB27A | c.514_518del p.Gln172AsnfsTer2 | NM_004580.5 | Hom | Out of frame/Deletion | rs767481076 |
| P88 | CID | 16 | M | - | BACH2 | c.745del p.Ser249ValfsTer93 | NM_021813.2 | Het | Out of frame/Deletion | novel |
| P89 | CID | 6 | M | + | RNF31 | c.2846A>C p.Asn949Thr | NM_017999.5 | Hom | Missense | rs766565788 |
| P90 [44] | Autoinflammatory disorder | 2 | F | + | ACP5 | c.772_790del Ser258Trpfs*39 | NM_001322023.2 | Hom | Out of frame/Deletion | rs878853218 |
| P91 [39, 40] | PIRD | 2 | F | + | PRF1 | c.1122G>A p.Trp374Ter | NM_005041.5 | Hom | Nonsense | rs104894176 |
| P92 | Phagocyte defect | 14 | F | + | NCF1 | Exon 5-6 Dup | NM_000265 | Hom | CNV | novel |
| P93 | CID | 2 | M | - | CHD7 | c.1904A>T p.Asp635Val | NM_017780.4 | Het | Missense | rs752468864 |
| P94 | CID | 17 | M | + | FCHO1 | c.2183A>C p.Asn728Thr | NM_001161357.1 | Hom | Missense | novel |
| P95 [52–54] | PIRD | 4 | M | + | LRBA | c.2836_2839del p.Glu946Ter | NM_006726.4 | Hom | Out of frame/Deletion | rs777413769 |
| P96 | Innate immune defect | 8 | M | - | TBK1 | c.1055T>C p.Leu352Pro | NM_013254.4 | Het | Missense | novel |
| P97 | SCID | 1 | M | + | IL7R | c.337G>T p.Glu113Ter | NM_002185.5 | Hom | Nonsense | novel |
| P98 [52–54] | PIRD | 20 | F | + | LRBA | c.2836_2839del p.Glu946Ter | NM_006726.4 | Hom | Out of frame/Deletion | rs777413769 |
| P99 [55–57] | SCID | 9 m | F | + | PRKDC | c.9182T>G p.Leu3061Arg | NM_006904.7 | Hom | Missense | rs587777685 |
| P100 [58, 59] | SCID | 16 | F | + | RAG2 | c.104G>C p.Gly35Ala | NM_001243786.1 | Hom | Missense | rs148508754 |
| P101 [60, 61] | PAD/CVID | 6 | F | - | PIK3CD | c.1573G>A p.Glu525Lys | NM_005026.5 | Het | Missense | rs587777389 |
| P102 | PIRD | 14 | M | - | FAS | c.340G>A p.Glu114Lys | NM_000043.6 | Het | Missense | rs773565107 |
| P103 | Innate immune defect | 11 | F | - | STAT1 | c.1192G>A p.Gly397Ser | NM_007315.3 | Het | Missense | novel |
| P104 | CID | 12 | F | - | IL6ST | c.2093C>A p.Ala698Glu | NM_002184.4 | Het | Missense | rs745818447 |
| P105 [62, 63] | Innate immune defect | 10 | M | + | IL12RB1 | c.637C>T p.Arg213Trp | NM_005535.3 | Hom | Missense | rs121434494 |
| P106 | CID | 2 | F | + | DOCK8 | c.5766G>A p.Met1922Ile | NM_203447.4 | Hom | Missense | rs2057267200 |
| P107 | CID | 1 | F | + | DOCK8 | Exon 1-10 Deletion | NM_203447.4 | Hom | CNV | novel |
| P108 | CID | 5 | M | + | SPINK5 | c.2658_2662dupGAGCA p.Ile888ArgfsTer56 | NM_001127698.1 | Hom | Out of frame/Dup | novel |
| P109 [64] | SCID | 6 m | M | + | ADA | c.556G>A p.Glu186Lys | NM_000022.4 | Hom | Missense | rs1555844416 |
| P110 [65–67] | CID | 2 | M | + | RAG1 | c.2095C>T p.Arg699Trp | NM_000448.3 | Hom | Missense | rs199474676 |
| P111 | PIRD | 3 m | M | + | PRF1 | c.1267delC p.Gln423LysfsX17 | NM_005041.5 | Hom | Out of frame/Deletion | novel |
| P112 | SCID | 3 m | M | + | IL2RG | c.511G>T p.Glu171Ter | NM_000206.2 | Hem | Nonsense | novel |
| P113 [68, 69] | PAD/CVID | 7 | F | + | CASP8 | c.919C>T p.Arg307Trp | be NM_001080125.1 | Hom | Missense | rs17860424 |
| P114 | CID | 18 | F | + | DOCK8 | c.5831C>T p.Pro1944Leu | NM_203447.4 | Hom | Missense | rs775779897 |
| P115 [64] | SCID | 9 m | M | + | ADA | c.556G>A p.Glu186Lys | NM_000022.4 | Hom | Missense | rs1555844416 |
| P116 | SCID | 1 | F | + | RAG1 | c.1307C>A p.Thr436Asn | NM_000448.2 | Hom | Missense | novel |
| P117 [29, 70, 71] | SCID | 1 | F | + | RAG1 | c.2210G>A p.Arg737His | NM_000448.3 | Hom | Missense | rs104894286 |
| P118 [20] | Phagocyte defect | 5 | F | + | CYBA | c.70G>A p.Gly24Arg | NM_000101.4 | Hom | Missense | rs28941476 |
| P119 | PIRD | 3 | F | + | PRF1 | c.1385C>A p.Ser462Ter | NM_005041.5 | Hom | Nonsense | rs1564723653 |
| P120 | CID | 4 | M | - | WAS | c.37C>T p.Arg13Ter | NM_000377.3 | Hem | Nonsense | rs193922415 |
| P121 | CID | 5 | M | - | WAS | c.91G>A p.Glu31Lys | NM_000377.3 | Hem | Missense | rs1557006239 |
| P122 | PAD/CVID | 9 | M | - | PIK3CD | c.1573G>A p.Glu525Lys | NM_005026.5 | Het | Missense | rs587777389 |
SCID Severe combined immunodeficiency, CID Combined immunodeficiency, PAD Primary antibody deficiency, CVID Common variable immunodeficiency, PIRD Primary immune regulation disorder, m months, M Male, F Female, Consan Consanguinity, Hom Homozygous, Het Heterozygous, Hem Hemizygous, CNV Copy number variation
Table 2.
Clinical features of the patients associated with detected gene defects
| Patient no | Clinical diagnosis (IUIS classification) | Gene | Variant | Associated features of the patients |
|---|---|---|---|---|
| P1 | Innate immune defect | CARD9 | c.883C>T p.Gln295Ter | Invasive fungal infection, HSM, dermatitis, elevated IgG and IgE |
| P2 | CID | RFXANK | c.634C>T p.Arg212Ter | Failure to thrive, respiratory and gastrointestinal infections, low CD4+ T cells |
| P3 | SCID | CD3E | c.176G>A p.Trp59Ter | T - B+ NK+ |
| P4 | CID | NFATC2 | c.340_345delGAGATC p.Glu114_Ile115del |
EBV-associated lymphoproliferation, recurrent pulmonary infections, hypogammaglobulinemia |
| P5 | SCID | JAK3 | c.2134G>A p.Gly712Ser | T - B+ NK+ |
| P6 | SCID | RAG2 | c.581C>A p.Ser194Ter | T - B- NK+ |
| P7 | SCID | RAG1 | c.2005G>A p.Glu669Lys c.1307C>A p.Thr436Asn | T - B- NK+ |
| P8 | SCID | RAG1 | c.2005G>A p.Glu669Lys c.1307C>A p.Thr436Asn | T - B- NK+ |
| P9 | PIRD | CD70 | c.332C>T p.Thr111Met | Burkitt lymphoma, hypogammaglobulinemia, reduced memory B cells |
| P10 | PIRD | CD70 | c.332C>T p.Thr111Met | Recurrent pulmonary infections, non-Hodgkin lymphoma, hypogammaglobulinemia |
| P11 | Phagocyte defect | CYBA | c.58+4_58+7delAGTG | Pulmonary Aspergillus infections, lymphadenitis, defective oxidative burst |
| P12 | CID | ZNF341 | c.1626C>G p.Tyr542Ter | Early onset eczema, recurrent skin and pulmonary infections, eosinophilia, elevated IgE |
| P13 | CID | ZAP70 | c.1010T>G p.Leu337Ala | CMV infection, chronic diarrhea, recurrent bacterial infections, low CD8+ T cells |
| P14 | SCID | RAG2 | c.105G>C p.Gly35Ala | T - B- NK+ |
| P15 | SCID | RAG2 | c.105G>C p.Gly35Ala | T - B- NK+ |
| P16 | PAD/CVID | TNFRSF13B | c.T310C p.Cys104Arg | Recurrent pulmonary infections, ITP, panhypogammaglobulinemia, reduced switched memory B cells |
| P17 | PAD/CVID | PIK3R1 | c.837-1G>A | Recurrent pulmonary infections, septic arthritis, agammaglobulinemia |
| P18 | CID | PGM3 | c.G214A p.Gly72Ser | Severe atopy, bacterial and viral infections, scoliosis, achondroplasia, dysgerminoma, reduced B and memory B cells, elevated IgE |
| P19 | Other | SAMD9L | c.A2639C p.His880Pro | Aplastic anemia, recurrent bacterial infections, agammaglobulinemia, reduced NK cells |
| P20 | PAD/CVID | TNFRSF13B | c.204dupA p.Leu69Tfs*11 | Lichen planus, panhypogammaglobulinemia |
| P21 | PAD/CVID | CD79A | c.380-2A>G | IBD, recurrent diarrhea, agammaglobulinemia, undetectable CD19+ B cells |
| P22 | CID | DNMT3B | c.G2029A p.Val677Met | Recurrent pulmonary infections, osteoporosis, agammaglobulinemia, reduced T and B cells |
| P23 | PAD/CVID | AICDA | c.A100T p.Lys34Ter | Rheumatoid arthritis, bacterial infections, elevated IgM |
| P24 | Phagocyte defect | CYBA | c.G70A p.Gly24Arg | BCGitis, anal and liver abscess, defective oxidative burst |
| P25 | CID | MALT1 | c.1318_1321delTGTC p.L440Valfs*6 | Bacterial, viral, fungal infections, defective T cell proliferation |
| P26 | Phagocyte defect | SBDS |
c.T578C p.Lys193Pro c.A184T p.Lys62Ter |
Recurrent sinopulmonary infections, gingivitis, neutropenia |
| P27 | CID | RFXANK | Exon 2-6 Deletion | Failure to thrive, recurrent sinopulmonary and gastrointestinal infections, warts, low CD4+ T cells |
| P28 | PIRD | MAGT1 | c.199-16A>G | EBV infection, lymphoma, hypogammaglobulinemia, decreased memory B cells |
| P29 | SCID | ADA | c.551_555del p.Glu184Glyfs*2 c.G241A p.Gly81Arg | T - B- NK- |
| P30 | SCID | RAG1 | c.C1767G p.Tyr589Ter | T - B- NK+ |
| P31 | SCID | JAK3 | c.932delC p.Pro311Argfs*17 | T - B+ NK- |
| P32 | Innate immune defect | TRAF3IP2 | c.C559T p.Arg187Ter | CMC, alopecia areata, skin rashes |
| P33 | SCID | RAG1 | c.G2126A p.Gly709Asp | T - B- NK+ |
| P34 | SCID | ADA | c.A779G p.Glu260Gly | T - B- NK- |
| P35 | Phagocyte defect | NCF2 | c.G233A p.Gly78Glu | Recurrent infections, aphthous stomatitis, cervical lymphadenitis, occasional skin infections, defective oxidative burst |
| P36 | Phagocyte defect | CYBA | c.166dupC p.Arg56Profs*156 | Recurrent infections, cervical lymphadenitis, defective oxidative burst |
| P37 | PIRD | LRBA | c.646-1G>A | AIHA, HSM, hypogammaglobulinemia, slightly decreased CD4+ T cells |
| P38 | SCID | JAK3 | c.G2080T p.Glu694Ter | T - B+ NK- |
| P39 | SCID | IL2RG | c.437T>A p.Leu146Gln | T - B+ NK- |
| P40 | PIRD | PRKCD | c.1097G>A p.Gly366Glu | BCGosis, meningitis, lymphoproliferation, CGD-like presentation |
| P41 | SCID | RAG2 | c.623T>A p.Val208Asp | T - B- NK+ |
| P42 | PIRD | CTLA4 | c.118G>A p.Val40Met | AIHA, enteropathy, reduced T and B cells |
| P43 | PIRD | JAK1 | c.2485A>G p.Asn829Asp | IBD, lymphopenia, vitiligo, recurrent diarrhea, lymphopenia |
| P44 | SCID | RAG1 | c.C1767G p.Tyr589Ter | T - B- NK+ |
| P45 | PIRD | PRKCD | c.1097G>A p.Gly366Glu | SLE, thrombocytopenia, failure to thrive, skin rashes, mental retardation, hypogammaglobulinemia |
| P46 | Phagocyte defect | CYBB | c.770G>A p.Cys257Tyr | Lymphoproliferation, granulomatous hepatitis, cytopenia, defective oxidative burst |
| P47 | CID | CHUK | c.499G>A p.Gly167Arg | Recurrent bacterial, viral, fungal infections, chronic diarrhea, failure to thrive, hepatic fibrosis, absent secondary lymphoid tissues, hypogammaglobulinemia, reduced switched memory B cells |
| P48 | CID | CHUK | c.499G>A p.Gly167Arg | Recurrent bacterial, viral, fungal infections, chronic diarrhea, failure to thrive, absent secondary lymphoid tissues, hypogammaglobulinemia, reduced switched memory B cells |
| P49 | SCID | RAG1 | c.742C>T p.Gln248Ter | T - B- NK+ |
| P50 | CID | CD40L | c.15C>A p.Tyr5Ter | Recurrent sinopulmonary infections, hypereosinophilia, eosinophilic gastroenteritis, memory B cells absent |
| P51 | PIRD | UNC13D | c.2346_2349delGGAG p.Arg782SerfsTer12 | HLH, pancytopenia, reduced naive T and RTE cells |
| P52 | PAD/CVID | IGGL1 | c.425C>T p.Pro142Leu | Recurrent bacterial, viral, fungal infections, panhypogammaglobulinemia |
| P53 | Phagocyte defect | ELANE | c.703delG p.Val235TrpfsTer5 | Recurrent bacterial infections, severe congenital neutropenia |
| P54 | Autoinflammatory disorder | HCK | c.135_136delinsTG p.Pro46Ala | Nodulocystic acnes, cutaneous vasculitis, HSM |
| P55 | Phagocyte defect | CYBA | c.385G>A p.Glu129Lys | Lung granulomas, chronic diarrhea, defective oxidative burst |
| P56 | PIRD | SLC7A7 | c.1417C>T p.Arg473Ter | Mental motor retardation, failure to thrive, skeletal anomalies, acanthosis nigricans, AIHA, lymphopenia |
| P57 | Phagocyte defect | NCF2 | c.196C>T p.Arg66Ter | Recurrent bacterial, fungal infections, lung granulomas, defective oxidative burst |
| P58 | SCID | DCLRE1C | c.1633delT p.Glu545AsnfsTer | T - B- NK+ |
| P59 | SCID | RAG1 | c.712delC p.Val238LeufsTer10 | T - B- NK+ |
| P60 | Innate immune defect | IL12RB1 | c.523C>T p.Arg175Trp | BCGitis |
| P61 | CID | CD40L | c.15C>A p.Tyr5Ter | Asymptomatic, reduced switched memory B cells |
| P62 | Autoinflammatory disorder | ADA2 | c.1072G>A p.Gly358Arg | Recurrent pulmonary infections, reduced switched memory B and marginal zone B cells |
| P63 | Innate immune defect | IL12RB1 | c.1456C>T p.Arg486Ter | BCGitis, BCG lymphadenitis |
| P64 | CID | CHUK | c.499G>A p.Gly167Arg | Recurrent pulmonary infections, absent secondary lymphoid tissues, hypogammaglobulinemia, reduced switched memory B cells |
| P65 | Phagocyte defect | CYBA | c.371C>T p.Ala124Val | Recurrent sinopulmonary infections, recurrent fungal infections, deafness, defective oxidative burst |
| P66 | CID | GIMAP5 | c.667C>T p.Leu223Phe | Hodgkin lymphoma |
| P67 | CID | GIMAP5 | c.667C>T p.Leu223Phe | Hodgkin lymphoma |
| P68 | PAD/CVID | CD79A | c.177dup p.Asn60GlnfsTer20 | Chronic diarrhea, elevated hepatic transaminases, failure to thrive, agammaglobulinemia |
| P69 | PIRD | UNC13D | c.1082del p.Tyr361SerfsTer43 | HLH, pancytopenia |
| P70 | PIRD | FAS | c.361C>T p.Arg121Trp | Splenomegaly, lymphadenopathy, ITP |
| P71 | PIRD | PRF1 | c.1122G>A p.Trp374Ter | HLH, HSM, reduced NK cells |
| P72 | CID | DOCK8 | c.5831C>T p.Pro1944Leu | Human papillomavirus (HPV) infections, recurrent sinopulmonary and gastrointestinal infections, elevated IgE, reduced naive and increased memory CD8+ T cells |
| P73 | CID | DOCK8 | c.5831C>T p.Pro1944Leu | Recurrent sinopulmonary and gastrointestinal infections, severe atopy, eosinophilia, elevated IgE, reduced naive and increased memory CD8+ T cells |
| P74 | PIRD | CTLA4 | c.151C>T p.Arg51Ter | Lymphadenopathy, lymphopenia, hypogammaglobulinemia, reduced switched memory B cells |
| P75 | Phagocyte defect | HAX1 | c.130_131insA p.Trp44Ter | Recurrent perianal abscess, neutropenia |
| P76 | CID | PIK3CG | c.2159A>G p.Tyr720Cys | Severe atopic dermatitis, multiple food allergies, eosinophilia, hypogammaglobulinemia |
| P77 | CID | MALT1 | c.1133T>G p.Phe378Cys | Failure to thrive, moniliasis, necrotizing skin lesions, lymphoproliferation |
| P78 | PIRD | MAGT1 | c.628-4T>C | Recurrent sinopulmonary infections, wet cough, panhypogammaglobulinemia |
| P79 | Autoinflammatory disorder | ACP5 |
c.772_790del p.Ser258WTrpfs*39 |
B-ALL, failure to thrive, spondyloenchondrodysplasia, intracranial calcification, mild MR |
| P80 | CID | PGM3 | c.821A>G p.Asn274Ser | Facial dysmorphic features, pancytopenia, T cell lymphopenia, reduced T lymphocyte activation |
| P81 | CID | CD3G | c.80-1G>C | Recurrent sinopulmonary infections, AIHA, panhypogammaglobulinemia, reduced memory and switched memory B cells |
| P82 | Phagocyte defect | ELANE | c.367-8C>A | Early onset IBD, oral aphtosis, recurrent gastrointestinal infections, severe congenital neutropenia |
| P83 | Phagocyte defect | CYBA | c.70G>A p.Gly24Arg c.373G>A p.Ala125Thr | Colitis, perianal abscess, defective oxidative burst |
| P84 | Autoinflammatory disorder | ADA2 | c.319A>C p.Lys107Gln | EBV associated Hodgkin lymphoma, splenomegaly, anemia, hypogammaglobulinemia |
| P85 | PIRD | FAS | c.761T>A p.Val254Asp | Lymphoproliferation, elevated DNT |
| P86 | CID | PNP | c.461+1G>A | Autoimmune hemolytic anemia, neurological impairment, osteomyelitis, lymphopenia |
| P87 | PIRD | RAB27A | c.514_518del p.Gln172AsnfsTer2 | Preseptal cellulitis, partial albinism, cytopenia |
| P88 | CID | BACH2 | c.745del p.Ser249ValfsTer93 | IBD, pancreatitis, hypogammaglobulinemia |
| P89 | CID | RNF31 | c.2846A>C p.Asn949Thr | Chronic diarrhea, hypoalbunemia, lymphoplasmacytic inflammation |
| P90 | Autoinflammatory disorder | ACP5 | c.772_790del Ser258Trpfs*39 | Recurrent viral infections, thrombocytopenia, AIHA |
| P91 | PIRD | PRF1 | c.1122G>A p.Trp374Ter | Sepsis, HSM, cytopenia, recurrent moniliasis, HLH |
| P92 | Phagocyte defect | NCF1 | Exon 5-6 Dup | Necrotizing pneumonia, lymphopenia, neutropenia |
| P93 | CID | CHD7 | c.1904A>T p.Asp635Val | Facial dysmorphic features, recurrent pulmonary infections, chronic severe diarrhea, reduced CD3 lymphocytes |
| P94 | CID | FCHO1 | c.2183A>C p.Asn728Thr | BCG lymphadenitis, abdominal pain, hepatitis, elevated IgE, eosinophilia |
| P95 | PIRD | LRBA | c.2836_2839del p.Glu946Ter | Recurrent pulmonary infections, IBD, panhypogammaglobulinemia, reduced switched memory B cells |
| P96 | Innate immune defect | TBK1 | c.1055T>C p.Leu352Pro | Enteroviral meningitis, recurrent sinopulmonary infections, failure to thrive |
| P97 | SCID | IL7R | c.337G>T p.Glu113Ter | T- B+ NK+ |
| P98 | PIRD | LRBA | c.2836_2839del p.Glu946Ter | Recurrent sinopulmonary infections, CMV colitis, EBV, arthritis, deafness, hyper IgM phenotype, absent B lymphocytes |
| P99 | SCID | PRKDC | c.9182T>G p.Leu3061Arg | T- B- NK+ |
| P100 | SCID | RAG2 | c.104G>C p.Gly35Ala | T- B- NK+ |
| P101 | PAD/CVID | PIK3CD | c.1573G>A p.Glu525Lys | Lichen planus, fulminant hepatic failure, granuloma, ITP, lymphoproliferation, reduced switched memory B cells |
| P102 | PIRD | FAS | c.340G>A p.Glu114Lys | AIHA, cytopenia, HSM, lymphoproliferation, crescentic GLN, agammaglobulinemia, elevated DNT, reduced Treg cells |
| P103 | Innate immune defect | STAT1 | c.1189A>G p.Asn3Asp | Recurrent pulmonary infections, bronchiectasis, CMC, nail dystrophia, severe growth retardation, hypothyroidism, hypergammaglobulinemia, CD4+ T cel lymphopenia |
| P104 | CID | IL6ST | c.2093C>A p.Ala698Glu | Recurrent pulmonary infections, bronchiectasis, severe eczema, hypogammaglobulinemia, elevated IgE, lymphopenia |
| P105 | Innate immune defect | IL12RB1 | c.637C>T p.Arg213Trp | Severe pulmonary tuberculosis, vasculitis, recurrent arthritis |
| P106 | CID | DOCK8 | c.5766G>A p.Met1922Ile | Severe eczema, multiple food allergies, recurrent infections, elevated IgE, lymphopenia |
| P107 | CID | DOCK8 | Exon 1-10 Deletion | Recurrent infections, growth retardation, failure to thrive, food allergies, elevated IgE, hypogammaglobulinemia, lymphopenia |
| P108 | CID | SPINK5 | c.2658_2662dupGAGCA p.Ile888ArgfsTer56 | Recurrent bacterial infections, failure to thrive, reduced memory B cells, elevated IgE, |
| P109 | SCID | ADA | c.556G>A p.Glu186Lys | T- B- NK- |
| P110 | CID | RAG1 | c.2095C>T p.Arg699Trp | Erythroderma, severe recurrent infections, T cell lymphopenia |
| P111 | PIRD | PRF1 | c.1267delC p.Gln423LysfsX17 | Sepsis, pancytopenia, HLH |
| P112 | SCID | IL2RG | c.511G>T p.Glu171Ter | T- B+ NK- |
| P113 | PAD/CVID | CASP8 | c.919C>T p.Arg307Trp | Recurrent bacterial infections, HSM, hypogammaglobulinemia, low B cells, increased DNT cells |
| P114 | CID | DOCK8 | c.5831C>T p.Pro1944Leu | Recurrent pulmonary and cutaneous infections, bronchiectasis, T cell lymphopenia, high IgE |
| P115 | SCID | ADA | c.556G>A p.Glu186Lys | T- B- NK- |
| P116 | SCID | RAG1 | c.1307C>A p.Thr436Asn | T- B- NK+ |
| P117 | SCID | RAG1 | c.2322G>A p.Arg737His | T- B- NK+ |
| P118 | Phagocyte defect | CYBA | c.G70A p.Gly24Arg | Recurrent infections, lung granulomas, defective oxidative burst |
| P119 | PIRD | PRF1 | c.1385C>A p.Ser462Ter | Hemophagocytic lymphohistiocytosis HLH, HSM, low NK cells |
| P120 | CID | WAS | c.37C>T p.Arg13Ter | Thrombocytopenia, eczema, recurrent bacterial infections, poor polysaccharide vaccine response |
| P121 | CID | WAS | c.91G>A p.Glu31Lys | Thrombocytopenia, eczema, recurrent bacterial infections, low T cells |
| P122 | PAD/CVID | PIK3CD | c.1573G>A p.Glu525Lys | EBV infection, lymphadenopathy, reduced IgA and IgG |
HSM Hepatosplenomegaly, ITP Immune thrombocytopenic purpura, IBD Inflammatory bowel disease, CMC Chronic mucocutaneous candidiasis, AIHA Autoimmune hemolytic anemia, SLE Systemic lupus erythematosus, HLH Hemophagocytic lymphohistiocytosis, RTE recent thymic emigrant, B-ALL B-cell acute lymphoblastic leukemia, MR mental retardation, DNT Double negative T cells, GLN Glomerulonephritis
Discussion
Advancements in NGS, with WES at the forefront, have been instrumental in the diagnostic processes of IEI by pinpointing causative genetic aberrations [72]. Genetic diagnosis now routinely assists in the delineation of IEI, underscoring its significance in the strategic management of patient treatments. Literature suggests a wide-ranging diagnostic yield for targeted and exome sequencing, from 10% to 70%, across various IEI patient groups [23, 58, 68, 73–79] . In this study, out of the 127 causative genetic defects in 122 patients, we identified 52 novel IEI-causing variants. We also discovered novel and very rare gene variants in NFATC2, CHUK, and PIK3CG genes, which have limited reported cases in the literature [80–83].
Among the 297 patients evaluated, a genetic etiology was confirmed in 122 individuals, resulting in a diagnostic yield of 41.1%. Diagnostic success exhibited pronounced variation among the different IEI subtypes: cases of SCID reached a 100% genetic identification rate, whereas CID and PID manifested lower diagnostic rates of 31% and 45%, respectively. Within the PAD cohort, genetic causality was determined in a mere 15.6% of cases (10 patients). This notably diminished diagnostic yield in Primary Antibody Deficiencies is in concordance with prior regional studies conducted by Fırtına S et al. [84]. In contrast, patients with probable Mendelian susceptibility to mycobacterial diseases and chronic granulomatous disease (CGD) demonstrated significantly higher diagnostic rates, with near-complete success in CGD patients.
The discrepancies in diagnostic success among IEI subtypes are primarily attributed to the complex nature of these disorders rather than limitations of WES. Factors such as the specific type of immunodeficiency, diverse clinical presentations, patient medical histories, and environmental influences affect the probability of achieving a genetic diagnosis [72]. Other factors include variable gene penetrance, the distinction between monogenic and polygenic influences, and various environmental considerations such as pathogenic exposures and age at presentation [85, 86]. Consanguinity plays a significant role in genetic diagnosis, as most IEI cases have autosomal recessive inheritance. Consanguineous populations or those from isolated regions with distinct phenotypes have reported higher diagnostic yields [87]. In our study, the consanguinity rate was 64.6%, and a diagnosis was made in 49.7% of those cases. We found 27 heterozygous variants in 21 unrelated patients, which can provide insights into the impact of heterozygous variants on protein function and aid in the search for novel IEI genes.
Currently, approximately 500 genetic etiologies leading to IEI are known [7]. Although the use of NGS, particularly WES, is increasing, it has limitations. Exome sequencing focuses on coding regions and essential splice sites, making it challenging to detect structural variations [72] and the use of short-read sequencing as in our study makes it difficult to map reads to repeated sequences, and pseudogenes [88]. Long-read sequencing (LRS) technologies both for exome or genome, have the capacity to enhance the detection of genetic variations and regions that are challenging to analyze with existing short-read NGS techniques [88–90]. However, the cost and complexity of analyzing large datasets pose challenges for WGS. In our study, we only identified three structural variants in 57 patients. Nevertheless, studies have shown the effectiveness of WGS in detecting both CNVs and coding variants [91, 92]. Reducing the cost of WGS and developing user-friendly bioinformatic tools may make it a routine diagnostic approach for IEI screening.
In conclusion, our findings highlight the limited success of WES in the genetic investigation of presumed IEI. The prospective adoption of WGS could enhance diagnostic yields, potentially surpassing WES in clinical examinations. With our substantial study cohort and diverse clinical presentations, the genetic variations we have identified will significantly contribute to the diagnosis of future IEI cases and guide the development of optimized NGS panels for these conditions.
Supplementary Information
(PDF 806 kb)
Acknowledgements
We express our gratitude to the “Can Sucak Candan Biseyler” Foundation (CSCBF) for their valuable support and contributions throughout this study. The CSCBF was established in 2018 to honor the memory of Can Sucak, who tragically passed away due to complications of primary immunodeficiency. The foundation actively supports research in the field of primary immunodeficiency and raises awareness about this condition. Additionally, we would like to acknowledge The Hospital Research Foundation (THRF) for their support of GC.
Author Contribution
B. E, U. A, C. I, D. P, B. O, C. B, S. T and M. K performed the experiments and analyzed data the with G. C. C. A, Ç. A, F. Ç, G. S, S. B. E, A. O, S. B, E. K. A, A. K, B. K, H. U, D. F. K, F. Ç, T. A, D. Ö, E. A, E. S. A, E. K, M. K, M. Y, Z. B, S. A, D.Ç.A, Ö. K, A. P. S, Ş. N. G, S. K, I. R, U. M, N. D. C, Ş. H, S. S. K, A. M, F. D, A. I and I. T provided clinical care of the patients, clinical data and patient materials. B. E, G. C, A. I and I. T wrote the manuscript. B. E, A. I, and I. T conceptualized and coordinated the study and provided laboratory resources. All authors critically reviewed the manuscript and agreed to its publication.
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK). The study received support from the “Sucak Candan Biseyler” Foundation and the Clinical Immunology Society, which provided the necessary Whole Exome Sequencing (WES) kits for the research.
Data Availability
No datasets were generated or analysed during the current study.
Declarations
Consent to Participate
Informed consent was obtained from all individual participants who were included in the study.
Consent for Publication
The manuscript does not contain any personal data of individual participants.
Conflict of Interests
The authors declare no competing interests.
Ethics Approval
This study was conducted in accordance with the principles outlined in the Declaration of Helsinki. Approval for the study was obtained from the local Ethics Committee of Hacettepe University (Approval number: GO 20/407).
Footnotes
The original version of this paper was updated due to several errors within the main Table 1 of the manuscript. Four variants were given with different transcript IDs of the same gene. There were also 2 nomenclature errors in the variants of P58 and P117.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Aydan Ikinciogulları and Ilhan Tezcan contributed equally to this work.
Change history
11/14/2024
The original version of this paper was updated due to several errors within the main Table 1 of the manuscript. Four variants were given with different transcript IDs of the same gene. There were also 2 nomenclature errors in the variants of P58 and P117.
Change history
11/14/2024
A Correction to this paper has been published: 10.1007/s10875-024-01841-3
References
- 1.Notarangelo LD, Bacchetta R, Casanova JL, Su HC. Human inborn errors of immunity: An expanding universe. Sci Immunol. 2020;5(49) [DOI] [PMC free article] [PubMed]
- 2.Eldeniz FC, Gul Y, Yorulmaz A, Guner SN, Keles S, Reisli I. Evaluation of the 10 Warning Signs in Primary and Secondary Immunodeficient Patients. Front Immunol. 2022;13:900055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyts I, Bousfiha A, Duff C, Singh S, Lau YL, Condino-Neto A, et al. Primary Immunodeficiencies: A Decade of Progress and a Promising Future. Front Immunol. 2020;11:625753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanal O, Tezcan I. Thirty years of primary immunodeficiencies in Turkey. Ann N Y Acad Sci. 2011;1238:15–23. [DOI] [PubMed] [Google Scholar]
- 5.Conley ME, Casanova JL. Discovery of single-gene inborn errors of immunity by next generation sequencing. Curr Opin Immunol. 2014;30:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vorsteveld EE, Hoischen A, van der Made CI. Next-Generation Sequencing in the Field of Primary Immunodeficiencies: Current Yield, Challenges, and Future Perspectives. Clin Rev Allergy Immunol. 2021;61(2):212–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2022;42(7):1473–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itan Y, Casanova JL. Novel primary immunodeficiency candidate genes predicted by the human gene connectome. Front Immunol. 2015;6:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halacli SO, Ayvaz DC, Sun-Tan C, Erman B, Uz E, Yilmaz DY, et al. STK4 (MST1) deficiency in two siblings with autoimmune cytopenias: A novel mutation. Clin Immunol. 2015;161(2):316–23. [DOI] [PubMed] [Google Scholar]
- 10.Aba Ü, Maslak IC, Ipsir C, Pehlivan D, Warnock NI, Tumes DJ, et al. A Novel Homozygous Germline Mutation in Transferrin Receptor 1 (TfR1) Leads to Combined Immunodeficiency and Provides New Insights into Iron-Immunity Axis. J Clin Immunol. 2024;44(2) [DOI] [PMC free article] [PubMed]
- 11.Erman B, Firtina S, Aksoy BA, Aydogdu S, Genc GE, Dogan O, et al. Invasive Saprochaete capitata Infection in a Patient with Autosomal Recessive CARD9 Deficiency and a Review of the Literature. J Clin Immunol. 2020;40(3):466–74. [DOI] [PubMed] [Google Scholar]
- 12.Erman B, Cipe F. Genetic Screening of the Patients with Primary Immunodeficiency by Whole-Exome Sequencing. Pediatr Allergy Immunol Pulmonol. 2020;33(1):19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erman B, Firtina S, Fisgin T, Bozkurt C, Cipe FE. Biallelic Form of a Known CD3E Mutation in a Patient with Severe Combined Immunodeficiency. J Clin Immunol. 2020;40(3):539–42. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh S, Kostel Bal S, Edwards ESJ, Pillay B, Jimenez Heredia R, Erol Cipe F, et al. Extended clinical and immunological phenotype and transplant outcome in CD27 and CD70 deficiency. Blood. 2020;136(23):2638–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Béziat V, Li J, Lin JX, Ma CS, Li P, Bousfiha A, et al. A recessive form of hyper-IgE syndrome by disruption of ZNF341-dependent STAT3 transcription and activity. Sci Immunol. 2018;3(24) [DOI] [PMC free article] [PubMed]
- 16.Meshaal SS, El Hawary RE, Abd Elaziz DS, Eldash A, Alkady R, Lotfy S, et al. Phenotypical heterogeneity in RAG-deficient patients from a highly consanguineous population. Clin Exp Immunol. 2019;195(2):202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabori U, Mark Z, Amariglio N, Etzioni A, Golan H, Biloray B, et al. Detection of RAG mutations and prenatal diagnosis in families presenting with either T-B severe combined immunodeficiency or Omenn's syndrome. Clinical Genetics. 2004;65(4):322–6. [DOI] [PubMed] [Google Scholar]
- 18.Castigli E, Wilson SA, Garibyan L, Rachid R, Bonilla F, Schneider L, Geha RS. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat Genet. 2005;37(8):829–34. [DOI] [PubMed] [Google Scholar]
- 19.Salzer U, Chapel HM, Webster ADB, Pan-Hammarström Q, Schmitt-Graeff A, Schlesier M, et al. Mutations in encoding TACI are associated with common variable immunodeficiency in humans. Nat Genet. 2005;37(8):820–8. [DOI] [PubMed] [Google Scholar]
- 20.Köker MY, van Leeuwen K, de Boer M, Çelmeli F, Metin A, Özgür TT, et al. Six different mutations including three novel mutations in ten families from Turkey, resulting in autosomal recessive chronic granulomatous disease. Eur J Clin Invest. 2009;39(4):311–9. [DOI] [PubMed] [Google Scholar]
- 21.Rae J, Noack D, Heyworth PG, Ellis BA, Curnutte JT, Cross AR. Molecular analysis of 9 new families with chronic granulomatous disease caused by mutations in, the gene encoding p22. Blood. 2000;96(3):1106–12. [PubMed] [Google Scholar]
- 22.Sefer AP, Abolhassani H, Ober F, Kayaoglu B, Bilgic Eltan S, Kara A, et al. Expanding the Clinical and Immunological Phenotypes and Natural History of MALT1 Deficiency. J Clin Immunol. 2022;42(3):634–52. [DOI] [PubMed] [Google Scholar]
- 23.Rae W, Ward D, Mattocks C, Pengelly RJ, Eren E, Patel SV, et al. Clinical efficacy of a next-generation sequencing gene panel for primary immunodeficiency diagnostics. Clin Genet. 2018;93(3):647–55. [DOI] [PubMed] [Google Scholar]
- 24.Egg D, Rump IC, Mitsuiki N, Rojas-Restrepo J, Maccari ME, Schwab C, et al. Therapeutic options for CTLA-4 insufficiency. J Allergy Clin Immunol. 2022;149(2):736–46. [DOI] [PubMed] [Google Scholar]
- 25.Hoshino A, Tanita K, Kanda K, Imadome KI, Shikama Y, Yasumi T, et al. High frequencies of asymptomatic Epstein-Barr virus viremia in affected and unaffected individuals with CTLA4 mutations. Clin Immunol. 2018;195:45–8. [DOI] [PubMed] [Google Scholar]
- 26.Schwab C, Gabrysch A, Olbrich P, Patino V, Warnatz K, Wolff D, et al. Phenotype, penetrance, and treatment of 133 cytotoxic T-lymphocyte antigen 4-insufficient subjects. J Allergy Clin Immunol. 2018;142(6):1932–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minegishi Y, Coustan-Smith E, Wang YH, Cooper MD, Campana D, Conley ME. Mutations in the human λ5/14.1 gene result in B cell deficiency and agammaglobulinemia. J Exp Med. 1998;187(1):71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulkarni M, Hule G, de Boer M, van Leeuwen K, Kambli P, Aluri J, et al. Approach to Molecular Diagnosis of Chronic Granulomatous Disease (CGD): an Experience from a Large Cohort of 90 Indian Patients. J Clin Immunol. 2018;38(8):898–916. [DOI] [PubMed] [Google Scholar]
- 29.Noack D, Rae J, Cross AR, Muñoz J, Salmen S, Mendoza JA, et al. Autosomal recessive chronic granulomatous disease caused by novel mutations in , the gene encoding the p67-component of phagocyte NADPH oxidase. Hum Genet. 1999;105(5):460–7. [DOI] [PubMed] [Google Scholar]
- 30.de Beaucoudrey L, Samarina A, Bustamante J, Cobat A, Boisson-Dupuis S, Feinberg J, et al. Revisiting human IL-12Rbeta1 deficiency: a survey of 141 patients from 30 countries. Medicine (Baltimore). 2010;89(6):381–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatipoglu N, Guvenc BH, Deswarte C, Koksalan K, Boisson-Dupuis S, Casanova JL, Bustamante J. Inherited IL-12Rbeta1 Deficiency in a Child With BCG Adenitis and Oral Candidiasis: A Case Report. Pediatrics. 2017;140(5) [DOI] [PMC free article] [PubMed]
- 32.Carmona-Rivera C, Khaznadar SS, Shwin KW, Irizarry-Caro JA, O'Neil LJ, Liu Y, et al. Deficiency of adenosine deaminase 2 triggers adenosine-mediated NETosis and TNF production in patients with DADA2. Blood. 2019;134(4):395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashem H, Kumar AR, Muller I, Babor F, Bredius R, Dalal J, et al. Hematopoietic stem cell transplantation rescues the hematological, immunological, and vascular phenotype in DADA2. Blood. 2017;130(24):2682–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashem H, Vatsayan A, Gupta A, Nagle K, Hershfield M, Dalal J. Successful reduced intensity hematopoietic cell transplant in a patient with deficiency of adenosine deaminase 2. Bone Marrow Transplant. 2017;52(11):1575–6. [DOI] [PubMed] [Google Scholar]
- 35.Asilsoy S, Bilgili G, Turul T, Dizdarer C, Kalkan S, Yasli H, et al. Interleukin-12/-23 receptor beta 1 deficiency in an infant with draining BCG lymphadenitis. Pediatr Int. 2009;51(2):310–2. [DOI] [PubMed] [Google Scholar]
- 36.Fieschi C, Dupuis S, Catherinot E, Feinberg J, Bustamante J, Breiman A, et al. Low penetrance, broad resistance, and favorable outcome of interleukin 12 receptor beta1 deficiency: medical and immunological implications. J Exp Med. 2003;197(4):527–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park AY, Leney-Greene M, Lynberg M, Gabrielski JQ, Xu X, Schwarz B, et al. GIMAP5 deficiency reveals a mammalian ceramide-driven longevity assurance pathway. Nat Immunol. 2024;25(2):282–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bettinardi A, Brugnoni D, Quiros-Roldan E, Malagoli A, La Grutta S, Correra A, Notarangelo LD. Missense mutations in the Fas gene resulting in autoimmune lymphoproliferative syndrome: a molecular and immunological analysis. Blood. 1997;89(3):902–9. [PubMed] [Google Scholar]
- 39.Balta G, Okur H, Unal S, Yarali N, Gunes AM, Unal S, et al. Assessment of clinical and laboratory presentations of familial hemophagocytic lymphohistiocytosis patients with homozygous W374X mutation. Leuk Res. 2010;34(8):1012–7. [DOI] [PubMed] [Google Scholar]
- 40.Zur Stadt U, Beutel K, Kolberg S, Schneppenheim R, Kabisch H, Janka G, Hennies HC. Mutation spectrum in children with primary hemophagocytic lymphohistiocytosis: molecular and functional analyses of PRF1, UNC13D, STX11, and RAB27A. Hum Mutat. 2006;27(1):62–8. [DOI] [PubMed] [Google Scholar]
- 41.Kuehn HS, Ouyang W, Lo B, Deenick EK, Niemela JE, Avery DT, et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science. 2014;345(6204):1623–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Germeshausen M, Grudzien M, Zeidler C, Abdollahpour H, Yetgin S, Rezaei N, et al. Novel HAX1 mutations in patients with severe congenital neutropenia reveal isoform-dependent genotype-phenotype associations. Blood. 2008;111(10):4954–7. [DOI] [PubMed] [Google Scholar]
- 43.Klein C, Grudzien M, Appaswamy G, Germeshausen M, Sandrock I, Schaffer AA, et al. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease). Nat Genet. 2007;39(1):86–92. [DOI] [PubMed] [Google Scholar]
- 44.Briggs TA, Rice GI, Daly S, Urquhart J, Gornall H, Bader-Meunier B, et al. Tartrate-resistant acid phosphatase deficiency causes a bone dysplasia with autoimmunity and a type I interferon expression signature. Nat Genet. 2011;43(2):127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stray-Pedersen A, Backe PH, Sorte HS, Morkrid L, Chokshi NY, Erichsen HC, et al. PGM3 mutations cause a congenital disorder of glycosylation with severe immunodeficiency and skeletal dysplasia. Am J Hum Genet. 2014;95(1):96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowe JH, Delmonte OM, Keles S, Stadinski BD, Dobbs AK, Henderson LA, et al. Patients with CD3G mutations reveal a role for human CD3gamma in T(reg) diversity and suppressive function. Blood. 2018;131(21):2335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teimourian S, Zomorodian E, Badalzadeh M, Pouya A, Kannengiesser C, Mansouri D, et al. Characterization of six novel mutations in CYBA: the gene causing autosomal recessive chronic granulomatous disease. Br J Haematol. 2008;141(6):848–51. [DOI] [PubMed] [Google Scholar]
- 48.Cetica V, Hackmann Y, Grieve S, Sieni E, Ciambotti B, Coniglio ML, et al. Patients with Griscelli syndrome and normal pigmentation identify RAB27A mutations that selectively disrupt MUNC13-4 binding. J Allergy Clin Immunol. 2015;135(5):1310–8 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mamishi S, Modarressi MH, Pourakbari B, Tamizifar B, Mahjoub F, Fahimzad A, et al. Analysis of RAB27A gene in griscelli syndrome type 2: novel mutations including a deletion hotspot. J Clin Immunol. 2008;28(4):384–9. [DOI] [PubMed] [Google Scholar]
- 50.Sarper N, Ipek IO, Ceran O, Karaman S, Bozaykut A, Inan S. A rare syndrome in the differential diagnosis of hepatosplenomegaly and pancytopenia: report of identical twins with Griscelli disease. Ann Trop Paediatr. 2003;23(1):69–73. [DOI] [PubMed] [Google Scholar]
- 51.Sepulveda FE, Debeurme F, Menasche G, Kurowska M, Cote M, Pachlopnik Schmid J, et al. Distinct severity of HLH in both human and murine mutants with complete loss of cytotoxic effector PRF1, RAB27A, and STX11. Blood. 2013;121(4):595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cagdas D, Halacli SO, Tan C, Lo B, Cetinkaya PG, Esenboga S, et al. A Spectrum of Clinical Findings from ALPS to CVID: Several Novel LRBA Defects. J Clin Immunol. 2019;39(7):726–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gamez-Diaz L, August D, Stepensky P, Revel-Vilk S, Seidel MG, Noriko M, et al. The extended phenotype of LPS-responsive beige-like anchor protein (LRBA) deficiency. J Allergy Clin Immunol. 2016;137(1):223–30. [DOI] [PubMed] [Google Scholar]
- 54.Lo B, Zhang K, Lu W, Zheng L, Zhang Q, Kanellopoulou C, et al. AUTOIMMUNE DISEASE. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science. 2015;349(6246):436–40. [DOI] [PubMed] [Google Scholar]
- 55.Esenboga S, Akal C, Karaatmaca B, Erman B, Dogan S, Orhan D, et al. Two siblings with PRKDC defect who presented with cutaneous granulomas and review of the literature. Clin Immunol. 2018;197:1–5. [DOI] [PubMed] [Google Scholar]
- 56.Mathieu AL, Verronese E, Rice GI, Fouyssac F, Bertrand Y, Picard C, et al. PRKDC mutations associated with immunodeficiency, granuloma, and autoimmune regulator-dependent autoimmunity. J Allergy Clin Immunol. 2015;135(6):1578–88 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Burg M, Ijspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119(1):91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu H, Zhang VW, Stray-Pedersen A, Hanson IC, Forbes LR, de la Morena MT, et al. Rapid molecular diagnostics of severe primary immunodeficiency determined by using targeted next-generation sequencing. J Allergy Clin Immunol. 2016;138(4):1142–51 e2. [DOI] [PubMed] [Google Scholar]
- 59.Walter JE, Rosen LB, Csomos K, Rosenberg JM, Mathew D, Keszei M, et al. Broad-spectrum antibodies against self-antigens and cytokines in RAG deficiency. J Clin Invest. 2015;125(11):4135–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coulter TI, Chandra A, Bacon CM, Babar J, Curtis J, Screaton N, et al. Clinical spectrum and features of activated phosphoinositide 3-kinase delta syndrome: A large patient cohort study. J Allergy Clin Immunol. 2017;139(2):597–606 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lucas CL, Kuehn HS, Zhao F, Niemela JE, Deenick EK, Palendira U, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110delta result in T cell senescence and human immunodeficiency. Nat Immunol. 2014;15(1):88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altare F, Ensser A, Breiman A, Reichenbach J, Baghdadi JE, Fischer A, et al. Interleukin-12 receptor beta1 deficiency in a patient with abdominal tuberculosis. J Infect Dis. 2001;184(2):231–6. [DOI] [PubMed] [Google Scholar]
- 63.Sakai T, Matsuoka M, Aoki M, Nosaka K, Mitsuya H. Missense mutation of the interleukin-12 receptor beta1 chain-encoding gene is associated with impaired immunity against Mycobacterium avium complex infection. Blood. 2001;97(9):2688–94. [DOI] [PubMed] [Google Scholar]
- 64.Adams SP, Wilson M, Harb E, Fairbanks L, Xu-Bayford J, Brown L, et al. Spectrum of mutations in a cohort of UK patients with ADA deficient SCID: Segregation of genotypes with specific ethnicities. Clin Immunol. 2015;161(2):174–9. [DOI] [PubMed] [Google Scholar]
- 65.Lee PP, Chan KW, Chen TX, Jiang LP, Wang XC, Zeng HS, et al. Molecular diagnosis of severe combined immunodeficiency--identification of IL2RG, JAK3, IL7R, DCLRE1C, RAG1, and RAG2 mutations in a cohort of Chinese and Southeast Asian children. J Clin Immunol. 2011;31(2):281–96. [DOI] [PubMed] [Google Scholar]
- 66.Reiff A, Bassuk AG, Church JA, Campbell E, Bing X, Ferguson PJ. Exome sequencing reveals RAG1 mutations in a child with autoimmunity and sterile chronic multifocal osteomyelitis evolving into disseminated granulomatous disease. J Clin Immunol. 2013;33(8):1289–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang ZY, Zhao XD, Jiang LP, Liu EM, Cui YX, Wang M, et al. Clinical characteristics and molecular analysis of three Chinese children with Omenn syndrome. Pediatr Allergy Immunol. 2011;22(5):482–7. [DOI] [PubMed] [Google Scholar]
- 68.Simon AJ, Golan AC, Lev A, Stauber T, Barel O, Somekh I, et al. Whole exome sequencing (WES) approach for diagnosing primary immunodeficiencies (PIDs) in a highly consanguineous community. Clin Immunol. 2020;214:108376. [DOI] [PubMed] [Google Scholar]
- 69.Niemela J, Kuehn HS, Kelly C, Zhang M, Davies J, Melendez J, et al. Caspase-8 Deficiency Presenting as Late-Onset Multi-Organ Lymphocytic Infiltration with Granulomas in two Adult Siblings. J Clin Immunol. 2015;35(4):348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schuetz C, Huck K, Gudowius S, Megahed M, Feyen O, Hubner B, et al. An immunodeficiency disease with RAG mutations and granulomas. N Engl J Med. 2008;358(19):2030–8. [DOI] [PubMed] [Google Scholar]
- 71.Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, et al. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93(5):885–96. [DOI] [PubMed] [Google Scholar]
- 72.Picard C, Fischer A. Contribution of high-throughput DNA sequencing to the study of primary immunodeficiencies. Eur J Immunol. 2014;44(10):2854–61. [DOI] [PubMed] [Google Scholar]
- 73.Al-Mousa H, Abouelhoda M, Monies DM, Al-Tassan N, Al-Ghonaium A, Al-Saud B, et al. Unbiased targeted next-generation sequencing molecular approach for primary immunodeficiency diseases. J Allergy Clin Immunol. 2016;137(6):1780–7. [DOI] [PubMed] [Google Scholar]
- 74.Bisgin A, Boga I, Yilmaz M, Bingol G, Altintas D. The Utility of Next-Generation Sequencing for Primary Immunodeficiency Disorders: Experience from a Clinical Diagnostic Laboratory. Biomed Res Int. 2018;2018:9647253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Erman B, Bilic I, Hirschmugl T, Salzer E, Boztug H, Sanal O, et al. Investigation of Genetic Defects in Severe Combined Immunodeficiency Patients from Turkey by Targeted Sequencing. Scand J Immunol. 2017;85(3):227–34. [DOI] [PubMed] [Google Scholar]
- 76.Kojima D, Wang X, Muramatsu H, Okuno Y, Nishio N, Hama A, et al. Application of extensively targeted next-generation sequencing for the diagnosis of primary immunodeficiencies. J Allergy Clin Immunol. 2016;138(1):303–5 e3. [DOI] [PubMed] [Google Scholar]
- 77.Moens LN, Falk-Sorqvist E, Asplund AC, Bernatowska E, Smith CI, Nilsson M. Diagnostics of primary immunodeficiency diseases: a sequencing capture approach. PLoS One. 2014;9(12):e114901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nijman IJ, van Montfrans JM, Hoogstraat M, Boes ML, van de Corput L, Renner ED, et al. Targeted next-generation sequencing: a novel diagnostic tool for primary immunodeficiencies. J Allergy Clin Immunol. 2014;133(2):529–34. [DOI] [PubMed] [Google Scholar]
- 79.Okano T, Imai K, Naruto T, Okada S, Yamashita M, Yeh TW, et al. Whole-Exome Sequencing-Based Approach for Germline Mutations in Patients with Inborn Errors of Immunity. J Clin Immunol. 2020;40(5):729–40. [DOI] [PubMed] [Google Scholar]
- 80.Bainter W, Lougaris V, Wallace JG, Badran Y, Hoyos-Bachiloglu R, Peters Z, et al. Combined immunodeficiency with autoimmunity caused by a homozygous missense mutation in inhibitor of nuclear factor ?B kinase alpha (IKKalpha). Sci Immunol. 2021;6(63):eabf6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharma M, Fu MP, Lu HY, Sharma AA, Modi BP, Michalski C, et al. Human complete NFAT1 deficiency causes a triad of joint contractures, osteochondromas, and B-cell malignancy. Blood. 2022;140(17):1858–74. [DOI] [PubMed] [Google Scholar]
- 82.Takeda AJ, Maher TJ, Zhang Y, Lanahan SM, Bucklin ML, Compton SR, et al. Human PI3Kgamma deficiency and its microbiota-dependent mouse model reveal immunodeficiency and tissue immunopathology. Nat Commun. 2019;10(1):4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thian M, Hoeger B, Kamnev A, Poyer F, Kostel Bal S, Caldera M, et al. Germline biallelic PIK3CG mutations in a multifaceted immunodeficiency with immune dysregulation. Haematologica. 2020;105(10):e488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Firtina S, Ng YY, Ng OH, Kiykim A, Ozek EY, Kara M, et al. Primary antibody deficiencies in Turkey: molecular and clinical aspects. Immunol Res. 2022;70(1):44–55. [DOI] [PubMed] [Google Scholar]
- 85.Edwards ESJ, Bosco JJ, Ojaimi S, O'Hehir RE, van Zelm MC. Beyond monogenetic rare variants: tackling the low rate of genetic diagnoses in predominantly antibody deficiency. Cell Mol Immunol. 2021;18(3):588–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rojas-Restrepo J, Caballero-Oteyza A, Huebscher K, Haberstroh H, Fliegauf M, Keller B, et al. Establishing the Molecular Diagnoses in a Cohort of 291 Patients With Predominantly Antibody Deficiency by Targeted Next-Generation Sequencing: Experience From a Monocentric Study. Front Immunol. 2021;12:786516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abolhassani H, Chou J, Bainter W, Platt CD, Tavassoli M, Momen T, et al. Clinical, immunologic, and genetic spectrum of 696 patients with combined immunodeficiency. J Allergy Clin Immunol. 2018;141(4):1450–8. [DOI] [PubMed] [Google Scholar]
- 88.Mantere T, Kersten S, Hoischen A. Long-Read Sequencing Emerging in Medical Genetics. Front Genet. 2019;10:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanford Kobayashi E, Batalov S, Wenger AM, Lambert C, Dhillon H, Hall RJ, et al. Approaches to long-read sequencing in a clinical setting to improve diagnostic rate. Sci Rep. 2022;12(1):16945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Troskie RL, Jafrani Y, Mercer TR, Ewing AD, Faulkner GJ, Cheetham SW. Long-read cDNA sequencing identifies functional pseudogenes in the human transcriptome. Genome Biol. 2021;22(1):146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Belkadi A, Bolze A, Itan Y, Cobat A, Vincent QB, Antipenko A, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci U S A. 2015;112(17):5473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thaventhiran JED, Lango Allen H, Burren OS, Rae W, Greene D, Staples E, et al. Whole-genome sequencing of a sporadic primary immunodeficiency cohort. Nature. 2020;583(7814):90–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 806 kb)
Data Availability Statement
No datasets were generated or analysed during the current study.