Abstract
Enigmatic mechanisms restore the resting state in activated lymphocytes following human immunodeficiency virus type 1 (HIV-1) infection, rarely allowing persistent nonproductive infection. We detail a mechanism whereby cellular factors could establish virological latency. The transcription factors YY1 and LSF cooperate in repression of transcription from the HIV-1 long terminal repeat (LTR). LSF recruits YY1 to the LTR via the zinc fingers of YY1. The first two zinc fingers were observed to be sufficient for this interaction in vitro. A mutant of LSF incapable of binding DNA blocked repression. Like other transcriptional repressors, YY1 can function via recruitment of histone deacetylase (HDAC). We find that HDAC1 copurifies with the LTR-binding YY1-LSF repressor complex, the domain of YY1 that interacts with HDAC1 is required to repress the HIV-1 promoter, expression of HDAC1 augments repression of the LTR by YY1, and the deacetylase inhibitor trichostatin A blocks repression mediated by YY1. This novel link between HDAC recruitment and inhibition of HIV-1 expression by YY1 and LSF, in the natural context of a viral promoter integrated into chromosomal DNA, is the first demonstration of a molecular mechanism of repression of HIV-1. YY1 and LSF may establish transcriptional and virological latency of HIV, a state that has recently been recognized in vivo and has significant implications for the long-term treatment of AIDS.
A subpopulation of stably infected CD4+ T lymphocytes containing integrated proviral DNA capable of producing virus upon stimulation has been identified in human immunodeficiency virus (HIV)-positive individuals (6, 7, 8, 15, 69). As antiretroviral therapy now allows significant inhibition of active HIV type 1 (HIV-1) replication, an understanding of factors that establish or maintain the integrated proviral state takes on new relevance. Potent repression of long terminal repeat (LTR) transcription could allow an activated, infected cell to return to the resting state and establish a stable nonproductive infection. This may occur via changes in local chromatin architecture surrounding the HIV promoter. While activation of the HIV LTR has been shown to be associated with changes in chromatin structure (13, 46, 51, 61–64), factors that result in durable repression of LTR expression are less well known.
We have identified two cellular factors, YY1 (δ, NF-E1, UCRBP, or CF1 [45, 52, 56, 40, 70]) and LSF (CP-2, LBP-1c, or UBP-1 [22, 26, 38, 40, 70]), that cooperate uniquely in recognition of the region −10 to +27 of the HIV-1 LTR (referred to as the RCS [repressor complex sequence]). These have been shown to specifically and synergistically repress HIV LTR expression and viral production (41, 49). Antibodies to either YY1 or LSF inhibit RCS complex formation, and mutations within the LTR that eliminate LSF binding and RCS complex formation ablate repression mediated by YY1 and/or LSF (41).
YY1, a zinc finger-containing transcriptional regulator with homology to the GLI-Krüppel family of proteins, is a ubiquitous cellular factor with the ability both to activate and repress gene expression (16, 32, 52, 56). YY1 has two N-terminal transactivation domains, while the C-terminal domain is required for direct DNA binding and for repression of some promoters (2, 4, 17). This broad spectrum of activity has been attributed to bending of DNA, interactions with other factors, or posttranscriptional modification of YY1 (52). However, activity depends on the promoter context and specific protein-protein interactions that YY1 establishes with other regulatory proteins (23, 32–34, 49, 50, 53, 71–73, 77) and with general transcription factors (5, 61).
LSF is the predominantly expressed member of a family of proteins (also termed LBP-1a, -1b, -1c, and -1d) that are produced from the differential splicing from two related genes (55, 74). All bind DNA except for LSF-ID (LBP1-d), which lacks a central encoding exon. LSF can bind the HIV LTR, and binding is associated with direct repression of transcription in vitro (18, 29, 44). However, this effect has not been observed in vivo, as transient expression of LSF alone had no observable effect on expression from the HIV LTR (49, 74, 76).
Genetic and biochemical studies have established that chromatin in living cells critically affect the transcriptional competence of a promoter sequence (3, 14, 36, 58, 68). A number of recent reports have documented the importance of histone deacetylases (HDACs) as the effector molecules of transcriptional downregulation in many genes (11, 20, 25, 39, 47). In addition, several transcriptional repressors that tether HDACs to the promoter have been described (2, 3, 21, 28, 31, 42, 43, 72, 73, 75).
To determine the domains of YY1 and LSF that participate in complex formation and regulation of the HIV promoter, we mapped the interactions of LSF and YY1 by using a number of chimeric YY1 and truncated LSF constructs. Our findings suggested a novel molecular mechanism of repression of an integrated HIV provirus in vivo, wherein LSF is required for recruitment of YY1 to the HIV LTR, and repression is mediated by YY1 via the action of HDAC.
We find that the YY1-LSF complex copurifies with HDAC1, identified by both Western blot analysis and enzymatic activity assay. Deletion of a glycine/alanine-rich domain of YY1, previously shown to specifically direct the interaction between YY1 and HDAC (72), ablates the ability of YY1 to repress the HIV-1 LTR. Further YY1-mediated repression of the LTR is ablated by the deacetylase inhibitor trichostatin A. This is the first time that the molecular mechanism by which the YY1-LSF complex represses HIV-1 transcription has been described and represents another important biological circumstance whereby the action of a transcriptional repressor is mediated by an HDAC.
MATERIALS AND METHODS
Nuclear extracts.
Large-scale preparation of nuclear extract from CEM cells for chromatographic purification of the RCS complex were prepared as described previously (12), with the following minor modifications: buffers A and C were supplemented with 1 mM NaF, 1 mM Na2VO4, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml, and 1 μg of pepstatin A per ml. Chymostatin (1 μg/ml) was also added to buffer A, and 50 mM β-glycerophosphate was added to buffer C.
Ion-exchange chromatography.
Activated P11 phosphocellulose (Whatman, Clifton, N.J.) was equilibrated with 50 mM NaCl–50 mM HEPES (pH 7.9)–10% glycerol–0.2 mM EDTA–0.5 mM phenylmethylsulfonyl fluoride (PMSF)–0.5 mM dithiothreitol (DTT). CEM cell nuclear extract was loaded at 0.4 ml/min, washed, and eluted in a linear gradient of 50 mM to 1 M NaCl. Fractions shown by Western blotting with anti-YY1 antibody (α-YY1; C-20; Santa Cruz Biotechnology, Santa Cruz, Calif.) to contain YY1, and shown by electrophoretic mobility shift assay (EMSA) to contain RCS-binding activity, were pooled and dialyzed against 20 mM Tris-HCl (pH 7.9)–10% glycerol–0.2 mM EDTA–0.5 mM PMSF–0.5 mM DTT–50 mM NaCl before DEAE-cellulose chromatography. A DEAE-cellulose DE52 column (Whatman) was loaded with pooled fractions at 0.2 ml/min. The column was washed and eluted, and fractions were analyzed as described above. Fractions positive both in Western blot and gel shift analyses were subjected to further purification by DNA affinity chromatography.
DNA affinity chromatography.
A double-stranded oligonucleotide spanning positions −10 to +27 of the HIV-1 LTR was ligated and coupled to cyanogen bromide-activated Sepharose CL-4B (Pharmacia, Piscataway, N.J.) as previously described (27). Active fractions from DEAE-cellulose chromatography were equilibrated in buffer Z (25 mM HEPES [pH 7.6], 0.1 M NaCl, 20% glycerol, 12.5 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 0.1% Nonidet P-40). Affinity resin was washed extensively with buffer Z without glycerol and Nonidet P-40. Fractions were incubated for 10 min at 4°C with 10 μg of dI-dC per ml, loaded by gravity, washed, and eluted with a step gradient of 0.1 to 1 M NaCl. Western blot analysis for detection of HDAC was performed using a rabbit polyclonal antibody raised against a peptide corresponding to the C-terminal amino acids 319 to 334 of the molecule (60) and against LSF using rabbit polyclonal anti-CP2 antiserum (LSF, LBP-1c; gift from M. Sheffery). HDAC assays were performed as previously described (31).
Cell lines, transfections, and assays.
Transfections of HeLa cells were performed as previously described (49). HeLa-CD4-LTR (9) cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and transfected with 20 μg of plasmid DNA (prepared using an EndoFree plasmid kit [Qiagen, Valencia, Calif.]) by calcium phosphate coprecipitation as instructed by the manufacturer (ProFection system; Promega, Madison, Wis.). After 30 min at room temperature, the solution was added to the cells (2.5 × 105 to 4 × 105 cells/plate). Twelve hours after transfection, the cells were washed with phosphate-buffered saline (PBS) and fed fresh medium. Forty-eight hours later, the cells were harvested, cellular extracts were prepared, and chloramphenicol acetyltransferase (CAT) assays performed as previously described (49). To control for the effect of transcription factor overexpression on general cellular promoters, a reporter construct driven by the β-actin promoter, pHβ-actin-luciferase (67), was used in cotransfection and CAT expression normalized for luciferase activity. Other plasmids used have been previously described (2, 49). Luciferase assays were performed at 48 h as suggested by the manufacturer of the luciferase assay system (Promega), but cells were resuspended in 200 μl of lysis buffer, and one freeze-thaw step was performed. Up to 30 μl of cellular extract (normalized for protein concentration) in a final volume of 130 μl was used for luciferase reactions.
For virus production experiments, 2 × 104 HeLa cells were transfected with 10 μl of Superfect (Qiagen) and 2.5 to 3.0 μg of DNA in a volume of 0.6 ml for 3 h, washed with PBS, and then grown in 2 ml. Aliquots of culture medium were sampled for detection of HIV-1 p24gag protein by antigen capture enzyme-linked immunosorbent assay as instructed by the manufacturer (Coulter Corporation, Hialeah, Fla.).
Immunoprecipitation and EMSA.
Immunoprecipitation was performed with nuclear extracts prepared from a Jurkat T-cell line (49). Twenty-microliter samples of extract were mixed with antibody (rabbit immunoglobulin G; α-YY1-C20 [Upstate Biotechnology, Lake Placid, N.Y.] or α-LSF [gift from M. Sheffery and S. Swendenmann]) at 4°C for 1 h; 5 μl α-rabbit IgG agarose-conjugated antibody (Sigma, St. Louis, Mo.) was added, and incubation was continued for 1 h. The complex was precipitated by centrifugation at 3,000 rpm and 4°C for 5 min. The pellet was washed three times with PBS and resuspended in 30 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer before separation by SDS-PAGE and Western analysis with α-YY1 or α-LSF. Bands were visualized using horseradish peroxidase-conjugated α-rabbit IgG (Sigma).
Histidine-tagged YY1 (His-YY1) and His-LSF were expressed and harvested in Escherichia coli as previously described (53, 65). The RCS oligonucleotide (−10 to +27 of HIV-1 LTR [49]) was end labeled, and 104 cpm was incubated with various amounts of His-LSF for 20 min at 25°C. Glutathione S-transferase (GST)–YY1 or His-YY1 was added, total protein content was normalized by the addition of bovine serum albumin, and the reaction continued for 30 min. EMSA was then performed as previously described (49). Supershifts were performed by addition of either concentrated α-YY1 (C20-X; Santa Cruz Biotechnology, Santa Cruz, Calif.) or α-LSF (65) to the reaction mixture.
In vitro protein interaction mapping.
GST-YY1/GFI-1 chimeras (17) and GST-LSF deletion (54, 55) constructs were expressed and harvested in E. coli as previously described. Proteins were visualized by Coomassie brilliant blue staining, and protein content was normalized by densitometric analysis. LSF was transcribed and translated in vitro by T7 RNA polymerase in rabbit reticulocyte (Promega) in the presence of [35S]methionine as instructed by the manufacturer (54). Following capture of GST-YY1/GFI-1 chimera proteins on glutathione-agarose beads, equal volumes of beads were incubated with [35S]methionine-labeled LSF in incubation buffer (17) for 1 h at 4°C. The beads were washed in incubation buffer–100 mM KCl and resuspended in 20 μl of 2× SDS loading buffer. Retained LSF was separated by SDS-PAGE (10% gel) dried, and visualized by autoradiography. Similarly, YY1 was transcribed and translated in vitro, and then incubated with captured GST-LSF constructs, and retained YY1 was visualized by autoradiography following electrophoresis.
RESULTS
YY1 and LSF interact in vivo.
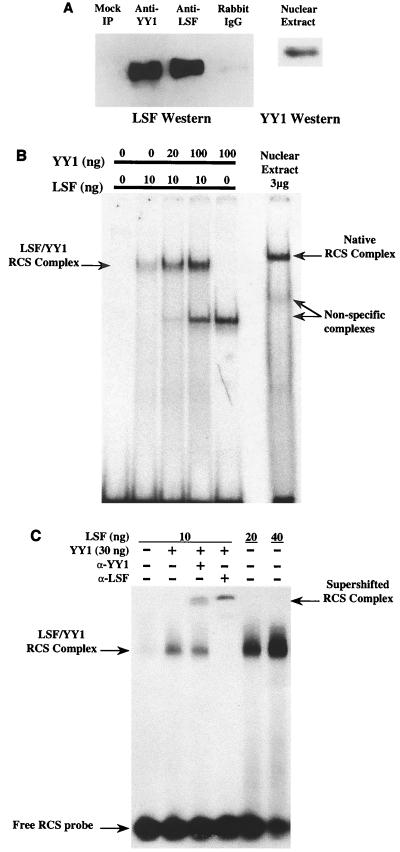
Complex formation at the RCS can be ablated by either α-YY1 or α-LSF, suggesting that both YY1 and LSF are necessary to form this regulatory complex (49). We sought evidence that these factors interacted directly in the absence of a DNA-binding sequence. Jurkat CD4+ T-cell nuclear extracts were incubated with α-YY1, α-LSF, or a nonspecific rabbit polyclonal antiserum. Antibody-protein complexes were precipitated by addition of α-IgG-agarose beads and centrifugation. Precipitates were then assayed for the presence of LSF by Western blot analysis. Immunoprecipitation was specific, as only trace amounts of LSF were recovered by the nonspecific antiserum. α-YY1 precipitated approximately 75% of the LSF activity that could be recovered by α-LSF (Fig. 1A). This indicates that LSF interacts with YY1 in vivo in the absence of the HIV RCS-binding site and suggests that direct protein-protein interaction between YY1 and LSF is necessary for complex formation at the HIV LTR.
FIG. 1.
YY1 and LSF associate in vivo and in vitro, in the absence of a DNA-binding site or other factors. (A) Immunoprecipitations of Jurkat nuclear extracts using either α-YY1, α-LSF, or a nonspecific rabbit polyclonal antiserum. Mock immunoprecipitations (IP) were performed in the absence of antibody. Precipitates were assayed by Western blot using α-LSF. Approximately 75% of the LSF protein recovered by α-LSF is also immunoprecipitated by α-YY1. To demonstrate the recognition of YY1, a Western blot of input nuclear extract is displayed at the right. (B) EMSA was performed using the RCS-binding site and the indicated amounts of LSF and YY1; total amount of protein was normalized by the addition of bovine serum albumin. The mobility of the native RCS complex formed by nuclear extract is displayed at the right. Nonspecific interactions with the RCS are indicated. (C) Complexes supershifted by the addition of either α-YY1 or α-LSF. Addition of α-YY1 had no effect on the LSF complexes in the absence of YY1 protein (not shown).
LSF and YY1 associate without cofactors in vitro.
Romerio et al. (49) showed that both LSF and YY1 form a complex at the RCS site of the HIV LTR. It was not known whether LSF and YY1 were sufficient to form this complex. We sought to reconstitute this complex in vitro, in the absence of other factors. Histidine-tagged recombinant LSF and YY1 proteins were expressed and harvested from E. coli and allowed to interact with an oligonucleotide encoding the HIV LTR RCS. Up to 100 ng of His-YY1 alone did not form a detectable specific complex with the RCS (Fig. 1B), although a low-molecular-weight band was observed when some protein or nuclear extract preparations were used. This band became more intense with the age of the preparation, suggesting it was the result of protein degradation. Ten nanograms of LSF, however, formed a specific EMSA band (Fig. 1B and C). The addition of increasing amounts (20 to 100 ng) of YY1 resulted in increasing levels of complex formation in the presence of 10 ng of LSF (Fig. 1B). As little as 20 ng of His-LSF induced a very prominent protein-DNA complex (Fig. 1C), similar to previous studies (18, 26). Under these conditions, no additional effect of YY1 was discernible. YY1 specifically enhanced RCS complex formation in the presence of low amounts of LSF, as all reactions were normalized for total protein content by the addition of BSA.
RCS protein-DNA complexes were supershifted by either α-YY1 or α-LSF (Fig. 1C), confirming that these complexes contained both LSF and YY1. α-LSF completely shifted the RCS complex, whereas under these conditions only a fraction of complexes were supershifted by the addition of excess α-YY1. The quantity of α-YY1 (5 μl) completely supershifted YY1 complexes formed on a canonical adeno-associated virus P5-binding site (data not shown). One interpretation of these results is that all complexes bound to DNA in these conditions contain LSF multimers (54, 55), but all complexes do not contain YY1 accessible to antibody. The effect of the antibody was specific, as α-YY1 had no effect on the mobility of the complex in the absence of YY1 protein (data not shown). Although other factors may be present in the RCS complex in vivo, YY1 and LSF are sufficient to form a complex at the RCS.
The addition of YY1 to LSF bound to the RCS was not associated with a further change in the mobility of the DNA-protein complex. As LSF (64 kDa) binds other sites as a tetramer (54), it is possible that YY1 (apparent molecular mass of 68 kDa) replaces LSF molecules within the RCS complex. Alternatively, the addition of YY1 to the multimeric LSF might not significantly affect complex mobility in the native electrophoresis conditions used.
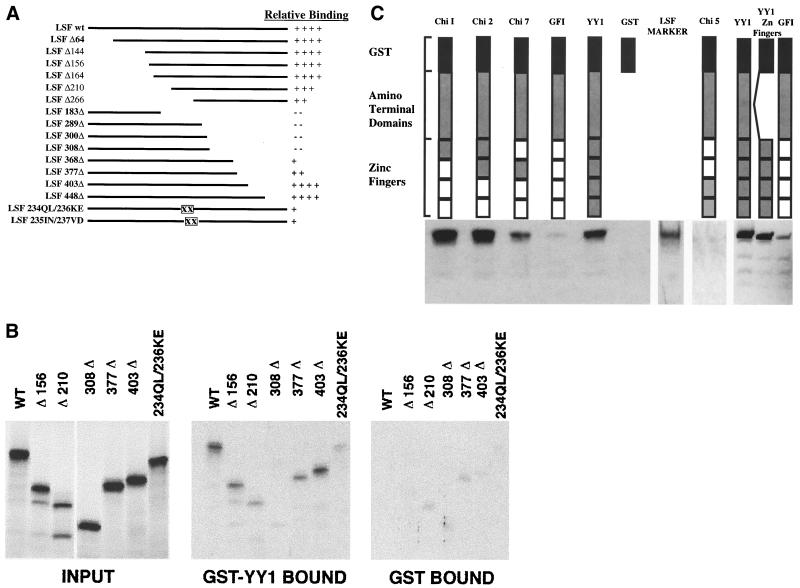
Interaction domains of YY1 and LSF.
The interaction of LSF with YY1 was mapped using constructs that contained a series of nested deletions within the coding region of LSF (Fig. 2A). Neither the carboxyl nor amino terminus of LSF was required for interaction with YY1. However, the central region of the protein was required for interaction with YY1, as amino-terminal deletions beyond amino acid 164 and carboxy-terminal deletions prior to amino acid 403 resulted in marked diminution of the ability of LSF to bind YY1 (Fig. 2B). Binding was lost altogether when C-terminal sequences between amino acid residues 308 and 368 were removed. Further amino acid substitutions within this region, which impair multimerization (55), markedly decreased the ability of the mutant LSF to bind full-length YY1. A likely possibility is that LSF recognizes YY1 only in its multimeric conformation.
FIG. 2.
Mapping of the YY1-LSF interaction domains. (A) Representation of wild-type (wt) LSF and LSF deletion mutants used to identify the region of interaction between LSF with YY1. ΔX, deletion up to codon X; XΔ, deletion after codon X; XX, mutated single codons. The amount of LSF bound to GST-YY1 varied from 2.5 to 7% of the input, depending on the experiment. All values were normalized to the amount of wild-type LSF bound to GST-YY1 in the experiment. (B) Representative autoradiographs showing input LSF constructs and LSF constructs retained by GST-YY1 and by GST. (C) Graphical representation of the YY1 chimeras, all of which contained the GST tag. All constructs also contained the N-terminal region of YY1 (amino acids 1 to 294) except YY1 Zn Fingers, which lacked this region. YY1 is the full-length wild-type YY1 molecule. Nonshaded regions represent GFI-1 zinc fingers (related to Krüppel zinc finger proteins). GFI contains only GFI-1 zinc fingers, Chi 1 contained the first YY1 zinc finger, Chi 2 contained the first two YY1 zinc fingers, Chi 5 contained the last two YY1 zinc fingers, Chi 7 contained the second YY1 zinc finger only, and YY1 Zn Fingers contained all four YY1 zinc fingers without the YY1 amino-terminal region. The first two zinc fingers of YY1 are required for optimal binding of LSF. Chi 1, Chi 2, Chi 7, and YY1 bound LSF, whereas Chi 5, GFI-1, and GST exhibited background levels of LSF binding. A lane containing only a diluted aliquot of labeled LSF serves as a marker. When normalized for protein concentration, a construct expressing only the YY1 zinc fingers fused to GST binds LSF as avidly as intact GST-YY1. Background levels of binding varied between experiments, as shown.
YY1 DNA-binding activity and many YY1-protein interactions map to the carboxyl-terminal zinc fingers of the molecule. Therefore, YY1 interaction domains were mapped using chimeric YY1 recombinants (Fig. 2C). These chimeras expressed GST fused to the N-terminal domain of the protein and had various numbers of YY1 zinc finger domains replaced by the structurally homologous GFI-1 zinc fingers (17). No deleterious effect on function or stability of YY1 was observed (17). Figure 2C shows that the interaction with LSF did not require the third and fourth zinc fingers of YY1. LSF bound in vitro to constructs that contained YY1 zinc finger 1 or 2, or both. In these assays, chimera 2, which expresses zinc fingers 1 and 2, bound LSF more avidly than intact YY1. Chimera 1 bound LSF nearly as well as YY1, while chimera 7 could bind LSF in vitro but less avidly. Thus, either zinc finger allowed binding to LSF, but binding was optimal when both fingers were present. Chimera 5, containing only the last two zinc fingers of YY1, did not bind LSF. The GST-Y/GFI-1 construct containing the entire YY1 amino-terminal domain fused to the zinc finger domain of the GFI-1 protein retained minimal amounts of LSF. Finally, a chimera expressing only the YY1 zinc finger domains, and lacking the entire amino-terminal YY1 region, bound LSF at least as well as intact YY1. Therefore, YY1 requires only zinc fingers 1 and 2 to recognize LSF.
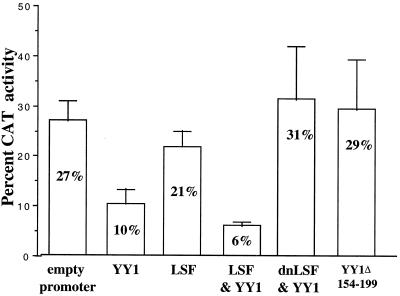
LSF competent to bind DNA is required for repression of HIV LTR expression.
Shirra and Hansen (54) and Shirra et al. (55) demonstrated that LSF binds a canonical simian virus 40 late promoter site via the formation of homotetramers. Further, binding can be blocked by the expression of a dominant negative mutant defective in DNA binding but with remaining ability to multimerize (LSF 234QL/236KE or dnLSF). We performed experiments to test the effect of dnLSF using the HeLa-CD4-LTR cell line (9). The LTR reporter carried by this cell line exists within the native chromatin structure of the genome. Transfection of YY1 inhibited Tat-activated CAT activity in these cells (Fig. 3), in agreement with previous studies using plasmid-based reporters (49). In the setting of a chromosomally integrated reporter gene, the provision of LSF augmented repression mediated by YY1, confirming the effect of YY1 and LSF on an integrated HIV-1 promoter. Significantly, dnLSF abolished the ability of YY1 to repress CAT expression, confirming that LSF capable of binding DNA is required to allow YY1 to repress HIV-1 LTR expression (Fig. 3). As in previous studies (49), these effects were specific to the HIV LTR; results are normalized to the expression of a cotransfected β-actin–luciferase reporter gene, whose expression was not significantly affected by YY1, LSF, or dnLSF (not shown).
FIG. 3.
Repression by YY1 and LSF requires functional LSF and HDAC interaction-competent YY1. Expression of an integrated LTR-CAT reporter in HeLa-CD4-LTR cells, when activated by 200 ng of pAR-Tat, was inhibited by 2.5 μg of CMV-YY1 or 2.5 μg of both CMV-YY1 and CMV-LSF; 2.5 μg of CMV-LSF had no effect on expression of CAT; 2.5 μg of dnLSF (pCMV-LSF 234QL/236KE), incapable of binding DNA but capable of forming inactive multimers, blocked inhibition of Tat-activated LTR expression by 2.5 μg of YY1; 2.5 μg of CMV-YY1Δ154-199, incapable of interacting with HDAC, was unable to inhibit Tat-activated expression. All transfections received a total of 5 μg of CMV promoter-driven plasmid. Data are from at least four independent transfections, normalized for expression of cotransfected β-actin–luciferase.
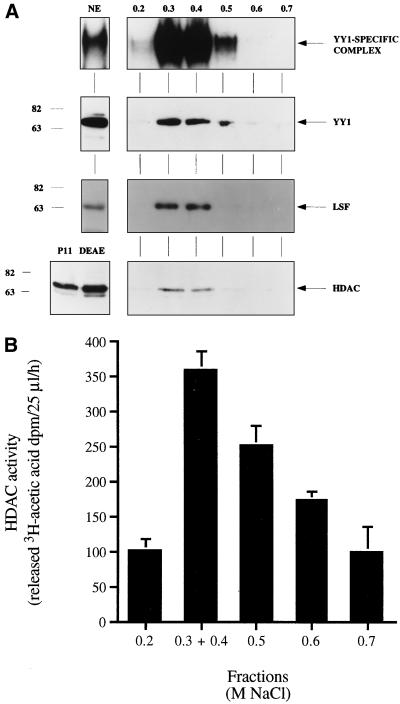
Copurification of HDAC with the YY1-LSF complex.
YY1 can act via the recruitment of HDACs (72, 73). As a nucleosome is present near the RCS when the HIV-1 LTR is integrated (46, 51, 62), we examined whether a histone deacetylase was present in the RCS DNA affinity chromatography fractions. The YY1-LSF complex was purified by phosphocellulose P11, DEAE-cellulose column, and DNA affinity chromatography as previously described (49). YY1, LSF, and the RCS-binding activity copurified in the 0.3 and 0.4 M NaCl fractions of the final step of purification (Fig. 4A). RCS-binding activity was enriched about 10,000-fold by this procedure. As shown in Fig. 4A, a rabbit polyclonal antibody raised against amino acids 319 to 334 of HDAC1 was able to detect a protein with apparent molecular mass of 66 kDa in the 0.3 and 0.4 M NaCl pooled fractions. HDAC1, a 55-kDa protein, migrates at this apparent molecular mass in our gel system (59).
FIG. 4.
YY1, LSF, and HDAC1 copurify with RCS-binding activity. (A) Activities of crude nuclear extract and elution fractions from an RCS DNA affinity chromatography column. Shown are results of EMSA using the RCS probe (top panel), and of Western blotting using rabbit polyclonal α-YY1 (second panel), rabbit polyclonal α-CP2 (LSF) (third panel), and rabbit polyclonal α-HDAC1/2 (bottom panel). EMSA was performed with 4 μg of nuclear extract (NE) and 20 ng of DNA affinity column eluate. Western blotting was performed with 20 μg of nuclear extract and 200 ng of DNA affinity column eluate. An arrow indicates the YY1-specific complex, as validated by α-YY1 interference in EMSA. Positions of molecular weight markers are indicated in kilodaltons. (B) HDAC activity of DNA affinity chromatography fractions correlates with the presence of the YY1-LSF complex.
To rule out the possibility that these fractions contained a protein immunologically similar but enzymatically unrelated to HDAC, we assayed the HDAC activity of the DNA affinity chromatography fractions. As expected, the 0.3 and 0.4 NaCl fractions showed strong HDAC activity, as measured by release of 3H-acetic acid (Fig. 4B). These results indicate that active HDAC1 copurifies with the YY1-LSF complex and suggest that the YY1-LSF complex represses HIV-1 transcription via the recruitment of HDAC.
Repression of the HIV-1 LTR by YY1 requires interaction with an HDAC.
A recent report has demonstrated that the Gly/Ala-rich domain of YY1 mediates the interaction with the HDAC and is required for repression of the adeno-associated virus P5 promoter by YY1 (72). To test whether the mechanism of repression of the HIV-1 LTR by YY1 is mediated by an HDAC, we performed a series of transient transfection experiments using a mutant YY1 deleted of the Gly/Ala-rich domain (YY1Δ154-199 [2]) required for interaction with HDAC.
Initial experiments were performed as previously (41, 49), demonstrating that cotransfection of YY1 inhibited Tat-activated, LTR-driven CAT expression. However, YY1Δ154-199 was unable to inhibit CAT activity, indicating the absolute requirement of the Gly/Ala-rich domain of YY1 for efficient repression of the HIV-1 LTR (data not shown). Chromatin remodeling effects on gene activity have often been imputed in studies using transfected, plasmid-encoded reporter genes that may not reflect the activity of genes contained in native chromatin. However the LTR reporter carried by the HeLa-CD4-LTR cell line (9) exists within the native chromatin structure of the genome. Significantly, YY1Δ154-199 failed to repress CAT expression, confirming that the Gly/Ala-rich HDAC interaction domain is required for repression of the HIV-1 LTR by YY1 (Fig. 3). Although YY1Δ154-199 activated CAT expression, this effect was not significant when normalized for modest activation observed of a cotransfected β-actin–luciferase reporter. Repression was also blocked by addition of the specific HDAC inhibitor trichostatin A to the culture medium (data not shown). This is the first demonstration of both cooperative repression by YY1 and LSF and the lack of repression by YY1Δ154-199 in the context of a chromosomally integrated HIV-1 promoter. It is also the first demonstration in this context that YY1 requires its HDAC interaction domain to mediate repression.
Repression of the HIV-1 virion production by YY1 requires interaction with an HDAC.
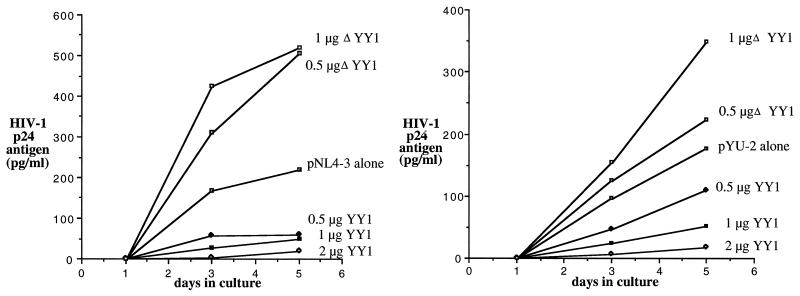
Support for the role of HDAC in repression of HIV-1 virion production was demonstrated by cotransfection of HeLa cells with the infectious molecular clone pNL4-3 (1) or pYU-2 (37) and empty cytomegalovirus (CMV) vector, CMV-YY1, or CMV-YY1Δ154-199. As these cells support HIV replication but cannot be infected, a measurement of the effect of YY1 on a single round of viral replication can be made. The influence of YY1 on viral production was assayed by testing of culture supernatant for the presence of the viral protein p24gag. Cotransfection with a vector expressing YY1 produced dose-dependent inhibition of either CXCR4 (pNL4-3)- or CCR5 (pYU-2)-tropic virus, whereas cotransfection of YY1Δ154-199 failed to inhibit HIV production (Fig. 5). Again, YY1Δ154-199 activated HIV expression above normal levels. Similar results were seen in the CD4+ T-cell line CEM when transfected with pNL4-3 or pYU-2 and empty CMV vector, CMV-YY1, or CMV-YY1Δ154-199 (data not shown). These findings suggest the possibility of ongoing competition between constitutive cellular YY1 and HIV LTR activating factors. However, as CMV-YY1Δ154-199 weakly activated a β-actin–luciferase reporter, secondary activating effect of YY1Δ154-199 on the HIV-1 LTR cannot be excluded.
FIG. 5.
YY1 directly affects production of HIV in vitro. Production of HIV-1 is inhibited by YY1 but not by YY1Δ154-199 lacking the HDAC interaction domain following transfection of HeLa cells with 0.5 μg of the CXCR4 prototypic clone pNL4-3 (left) or 1 μg of the CCR5 prototypic clone pYU-2 (right). Data are representative of three transfections.
DISCUSSION
YY1 and LSF interact and cooperate in repression of HIV-1 LTR expression.
We show that LSF and YY1 interact with one another both in vitro and in vivo. Interaction was observed in the absence of (i) a DNA-binding site, (ii) other cellular factors, and (iii) YY1 C-terminal zinc fingers required for DNA binding to canonical YY1 sites (Fig. 1 and 2 and reference 17). As a majority of the LSF that can be recovered by immunoprecipitation can also be recovered in association with YY1, this complex is likely to be formed in the cell prior to binding to viral regulatory elements. Further evidence of YY1 and LSF interaction is provided by the observation that YY1 alone does not bind the RCS site in EMSA, but α-YY1 supershifted a significant fraction of the RCS-protein complex.
Overexpression of LSF alone does not repress transcription from the HIV LTR (49, 76). However, an LTR reporter gene is inhibited by YY1 expression, and this effect is augmented by coexpression of LSF. This occurs in the context of both plasmid-based (49) and chromatin-based reporter genes (Fig. 3). Further, both YY1 and LSF function are required for inhibition of the HIV LTR. While expression of LSF does not significantly inhibit LTR expression, LSF synergizes with YY1 in repression, and dominant negative LSF prevents repression by YY1 (Fig. 3). Consistent with this model, preliminary studies show that the replication of a provirus containing TAR mutations that block RCS formation but allow Tat function (48) is unaffected by YY1 (data not shown).
YY1 is known to interact with a number of cellular factors via a Gly/Ala-rich region within residues 154 to 199 (2). LSF, however, interacts with the zinc finger domain of YY1. The Gly/Ala domain was present on all chimeric YY1 constructs, but only YY1, chimeras 1, 2, and 7, and the YY1 zinc fingers alone were able to specifically bind LSF in vitro (Fig. 2C). These results indicate that the first and, to a lesser extent, second zinc finger domains of YY1 participate in interaction with LSF. While zinc fingers often mediate DNA binding, examples of protein-protein interactions mediated by zinc fingers have been documented (3, 19, 30, 66). The interaction of YY1 with LSF via its carboxy-terminal zinc fingers creates an attractive model. Within the RCS complex, the amino terminus of YY1 might be accessible for interacting with other factors, such as HDAC, as well as the nearby basal transcription complex, the complex of proteins that binds TAR, or nearby nucleosomes.
Taken together, our findings show that LSF is required for recruitment of YY1 to the HIV promoter. LSF may facilitate YY1 recognition of the LTR, guiding YY1 onto a site that was inaccessible or of low affinity in its absence. Alternatively, YY1 may enter the RCS complex solely through protein-protein interaction with LSF. Given the sensitivity of this interaction to mutations within the core of LSF that impair multimerization, it is likely that YY1 recognizes a structure displayed by LSF multimers. The location of the RCS within the HIV LTR may position YY1 to directly inhibit the basal transcription complex or activators of the LTR, as well as recruit mediators of repression such as HDACs.
Interaction of HDAC with the YY1-LSF repressive complex.
The multiprotein complex containing the human factors YY1 and LSF, previously isolated from CEM cells, detected in primary T cells, and shown to repress the HIV-1 LTR, copurifies with a 65-kDa protein which we have identified as HDAC1 (Fig. 4A). The anti-HDAC antibody used to perform the Western blot analysis was reactive to both HDAC1 and HDAC2 but not to HDAC3 (73). However, based on the molecular weight found in Western blot analysis (60, 72), HDAC1 is likely the protein copurified with YY1 and LSF.
We have demonstrated that the Gly/Ala-rich domain of YY1, which mediates the interaction with HDAC (72), is absolutely required for efficient repression of the HIV-1 LTR by the YY1-LSF complex (Fig. 3). This was not an obvious result, given the many molecular mechanisms of repression that have been attributed to YY1. This suggests that recruitment of an HDAC is a necessary event in the mechanism of repression of HIV-1 gene expression by YY1. Indeed, the fact that transfection of YY1Δ154-199 resulted in modest upregulation of LTR expression and HIV production suggests that LTR expression may in part reflect the competing influences of cellular HDAC and histone acetyltransferase activity.
The Gly/Ala-rich domain has also been shown to mediate interaction between YY1 and the general transcription factors TFIIB, TATA binding protein, and TAFII55 and with the transcription factor p300/CBP (2). These interactions have been proposed to be relevant for repression by YY1 through a mechanism of quenching or direct repression (2, 10, 24, 35). Although our studies provide no direct evidence for this speculation, YY1 might utilize multiple mechanisms to repress the HIV-1 LTR.
YY1-LSF-HDAC may alter the chromatin structure of the HIV-1 LTR.
Studies of the chromatin structure of the integrated HIV-1 provirus in chronically and acutely infected cells lines have detected the presence of a large nucleosome-free, DNase I-hypersensitive region spanning nucleotides 223 to 450 of the HIV-1 genome. This corresponds to the portion of the LTR including the enhancer and the promoter regions, up to the transcription start site. Upon treatment with tetradecanoyl phorbol acetate or tumor necrosis factor alpha, the 3′ boundary of the nucleosome-free region was extended a further 140 nucleotides, indicating the alteration of the nucleosome, termed nuc-1 (51, 62–64). Additional DNase I-hypersensitive sites and nucleosome-protected regions have been identified all along the integrated HIV-1 genome (62, 62).
More recently, Pazin et al. (46) have shown that binding of both Sp1 and the p50 subunit of NF-κB to the HIV-1 LTR alters the local nucleosomal array in vicinity of the HIV-1 promoter and produces the DNase I-hypersensitive region between nucleotides 223 and 450. However, it is the p65 subunit of NF-κB that induces changes in the nucleosome nuc-1, perhaps through the recruitment of a histone acetyltransferase (51), and enhances transcriptional activity (46).
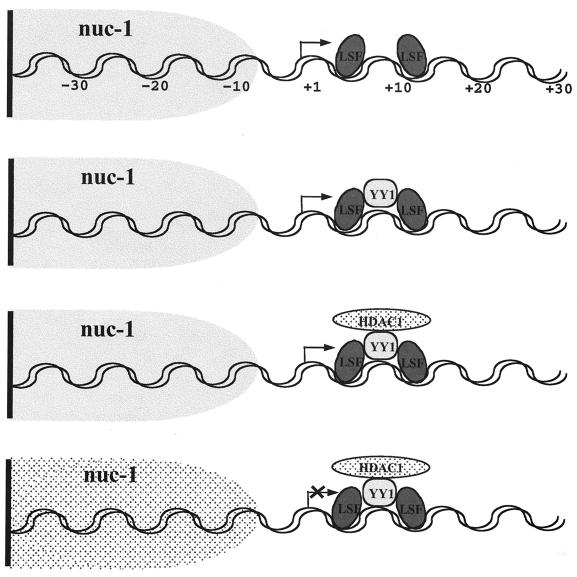
Our previous results suggested that LSF allows YY1 to recognize a site on the LTR that YY1 could not bind by itself (49). Therefore, LSF might primarily act as a docking molecule for YY1, which in turn acts by tethering HDAC (Fig. 6). In this model, YY1 may be a limiting factor for repression of the LTR, required for the recruitment of the HDAC to the HIV-1 promoter. The finding that overexpression of the mutant YY1Δ154-199 results in activation of HIV expression is consistent with such a model. El Kharroubi et al. (13) have shown that activation of the integrated proviral HIV genome requires alteration of the local chromatin via acetylation of the nucleosome adjacent to the start site. The recruitment of HDAC by YY1 might prevent such changes in the local chromatin, maintaining the nucleosome in a deacetylated state, preserving higher-order nucleosome structure, and thereby inhibiting gene expression (Fig. 6).
FIG. 6.
Model of recruitment by LSF of YY1 and then HDAC to the HIV promoter.
In vitro assays have shown that assembly of nuc-1 on naked HIV-1 DNA can be inhibited by the presence of LSF (51). Further, one previous report suggested that LSF is important for efficient activation of the HIV-1 LTR (26), although this has been disputed (76). However, we find no evidence that wild-type LSF activates LTR expression. While previous evidence that LSF is an activator of HIV conflicts with our findings, these studies were performed in very different experimental systems. Indeed, through interaction with other factors, in the absence of YY1, or in other cellular milieus, LSF might direct LTR activation.
Regulation of HIV expression within resting CD4 lymphocytes.
The HIV-1 enhancer and promoter possess a multiplicity of sequences recognized by cellular and viral regulatory factors. The roles of cellular enhancers such as Sp1 and NF-κB and of the viral activator Tat in active HIV gene expression have been extensively studied. As discussed above, changes in chromatin structure about an integrated HIV promoter during activation have been documented. However, mechanisms that downregulate HIV expression are largely unknown. We have shown that YY1 and LSF are capable of cooperating to inhibit HIV transcription. Of the many possible mechanisms through which YY1 might downregulate transcription, we can now link this function to the recruitment of an HDAC; our studies strongly suggest that this enzyme is HDAC1. Thus, the YY1-LSF repressor complex recruits factors capable of potent and durable inhibition of HIV-1 LTR promoter expression.
A large nucleosome-free, DNase I-hypersensitive region spanning nucleotides 223 to 450 of the HIV-1 has been observed in the chromatin structure of the integrated HIV-1 provirus in chronically and acutely infected cells lines. Activation of LTR expression extends the 3′ boundary of this nucleosome-free region a further 140 nucleotides. Each nucleosome is entwined by 1.65 turns of a left-handed superhelix of DNA that corresponds to 147 bp. This indicates that the DNA protected by one nucleosome has been exposed, presumably by remodeling of the nucleosome structure. The binding of both Sp1 and the p50 or p65 subunits of NF-κB to the HIV-1 LTR alters the local nucleosomal array in vicinity of the HIV-1 promoter and perhaps through the recruitment of a histone acetyltransferase enhances transcriptional activity. El Kharroubi et al. (13) have also shown that activation of the integrated proviral HIV genome requires alteration of the local chromatin via acetylation of the nucleosome adjacent to the start site. Our findings imply that recruitment of HDAC by YY1 might prevent such changes in the local chromatin, maintaining the nucleosome in a deacetylated state and inhibiting HIV expression.
We propose a dynamic model of HIV LTR regulation that would allow the establishment of virological latency in rare CD4 T cells. Following T-cell activation necessary for viral entry, reverse transcription, and other steps of the viral life cycle which lead to proviral integration, the rare activated cell avoids apoptosis or viral or immune-mediated destruction. Dampening of LTR expression by YY1 and LSF may play an important role at this stage. This cell then follows pathways that typically reestablish the resting, memory state. The HIV-1 LTR remains silent due to the predominant effects of repressor molecules, resulting in an inaccessible chromatin structure about the LTR. This cell may later exit virological latency if it encounters stimuli that increase nuclear levels of NF-κB, again changing LTR chromatin structure. Levels of the viral activator Tat then increase within the cell, driving the equilibrium toward viral expression.
Further study of cellular factors that establish or maintain the rare latent state of HIV infection may yield novel techniques to manipulate HIV expression, allow better understanding of the HIV replication in T lymphocytes, and lead to specific therapies directed at the quiescent reservoir of HIV infection.
ACKNOWLEDGMENTS
We thank Laurel Matey and Randall Merling for excellent technical assistance. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HLCD4-CAT from Barbara K. Felber and George N. Pavlakis, pNL4-3 from Malcolm Martin, and pYU-2 from Beatrice Hahn and George Shaw.
This work was supported by an Ortho-McNeil Young Investigator award from the IDSA and NIH grants AI 41366 and AI 45297 to D.M.M.; Medical Research Council of Canada grant MT-9186 to J.R.D., an MRC Senior Scientist; NIH grant GM53874 to Y.S.; and a DFCI/Sandoz Discovery grant to U.H.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and non-human cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austen M, Lüscher B, Lüscher-Firzlaff J M. Characterization of the transcriptional regulator YY1. J Biol Chem. 1997;272:1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- 3.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 4.Bushmeyer S, Park K, Atchison M L. Characterization of functional domains within the multifunctional transcription factor, YY1. J Biol Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- 5.Chiang C-M, Roeder R G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–535. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 6.Chun T-W, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano R F. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;12:1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 7.Chun T-W, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, Kuo Y H, Brookmeyer R, Zeiger M A, Barditch-Crovo P, Siliciano R F. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 8.Chun T-W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciminsale V, Felber B K, Campbell M, Pavlakis G N. A bioassay for HIV-1 based on Env-CD4 interaction. AIDS Res Hum Retroviruses. 1990;6:1281–1287. doi: 10.1089/aid.1990.6.1281. [DOI] [PubMed] [Google Scholar]
- 10.Cowell I G. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 11.Davie J R. Covalent modifications of histones: expression from chromatin templates. Curr Opin Genet Dev. 1998;8:173–178. doi: 10.1016/s0959-437x(98)80138-x. [DOI] [PubMed] [Google Scholar]
- 12.Dignam J D. Preparation of extracts from higher eukaryotes. Methods Enzymol. 1990;182:194–203. doi: 10.1016/0076-6879(90)82017-v. [DOI] [PubMed] [Google Scholar]
- 13.El-Kharroubi A, Piras G, Zensen R, Martin M A. Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol Cell Biol. 1998;18:2535–2544. doi: 10.1128/mcb.18.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 15.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 16.Flanagan J R. Autologous stimulation of YY1 transcription factor expression: role of an insulin-like growth factor. Cell Growth Differ. 1995;6:185–190. [PubMed] [Google Scholar]
- 17.Galvin K M, Shi Y. Multiple mechanisms of transcriptional repression by YY1. Mol Cell Biol. 1997;17:3723–3732. doi: 10.1128/mcb.17.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia J A, Wu F K, Mitsuyasu R, Gaynor R B. Interactions of cellular proteins involved in the transcriptional regulation of human immunodeficiency virus. EMBO J. 1987;6:3761–3770. doi: 10.1002/j.1460-2075.1987.tb02711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geisberg J V, Lee W S, Berk A J, Ricciardi R P. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc Natl Acad Sci USA. 1994;91:2488–2492. doi: 10.1073/pnas.91.7.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 21.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 22.Huang H-C, Sundseth R, Hansen U. Transcription factor LSF binds two variant bipartite sites within the SV40 late promoter. Genes Dev. 1990;4:287–298. doi: 10.1101/gad.4.2.287. [DOI] [PubMed] [Google Scholar]
- 23.Inouye C J, Seto E. Relief of YY1-induced transcriptional repression by protein-protein interaction with the nucleolar phosphoprotein B23. J Biol Chem. 1994;270:15187–15193. [PubMed] [Google Scholar]
- 24.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 25.Johnson C A, Turner B M. Histone deacetylases: complex transducers of nuclear signals. Semin Cell Dev Biol. 1999;10:179–188. doi: 10.1006/scdb.1999.0299. [DOI] [PubMed] [Google Scholar]
- 26.Jones K A, Luciw P A, Duchange N. Structural arrangements of transcription control elements within the 5′-untranslated leader regions of the HIV-1 and HIV-2 promoters. Genes Dev. 1988;2:1101–1114. doi: 10.1101/gad.2.9.1101. [DOI] [PubMed] [Google Scholar]
- 27.Kadonaga J T. Purification of sequence-specific binding proteins by DNA affinity chromatography. Methods Enzymol. 1991;208:10–23. doi: 10.1016/0076-6879(91)08004-2. [DOI] [PubMed] [Google Scholar]
- 28.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 29.Kato H, Horikoshi M, Roeder R G. Repression of HIV-1 transcription by a cellular protein. Science. 1991;251:1476–1479. doi: 10.1126/science.2006421. [DOI] [PubMed] [Google Scholar]
- 30.Kurokawa M, Mitani K, Irie K, Matsuyama T, Takahashi T, Chiba S, Yazaki Y, Matsumoto K, Hirai H. The oncoprotein Evi-1 represses TGF-beta signaling by inhibiting Smad3. Nature. 1998;394:92–96. doi: 10.1038/27945. [DOI] [PubMed] [Google Scholar]
- 31.Laherty C D, Yang W-M, Davie J-M, Sun J R, Seto E, Eisenmann R N. Histone deacetylase associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 32.Lee J S, Galvin R H, See K M, Wang J, Shi Y. Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 1995;23:925–931. doi: 10.1093/nar/23.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J-S, Galvin K M, Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc Natl Acad Sci USA. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J-S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 35.Levine M, Manley J L. Transcriptional repression of eukaryotic promoters. Cell. 1989;59:405–408. doi: 10.1016/0092-8674(89)90024-x. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Chen H Y, Davie J R. Properties of chicken erythrocyte histone deacetylase associated with the nuclear matrix. Biochem J. 1996;314:631–637. doi: 10.1042/bj3140631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Kappes J C, Conway J A, Price R W, Shaw G M, Hahn B H. Complete nucleotide sequence, genomic organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol. 1991;59:284–291. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim L C, Swendenmann S L, Scheffrey M. Molecular cloning of the α-globin transcription factor CP2. Mol Cell Biol. 1992;12:828–835. doi: 10.1128/mcb.12.2.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1997;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 40.Malim M H, Fenrick R, Ballard D W, Hauber J, Bohnlein E, Cullen B R. Functional characterization of a complex protein-DNA-binding domain located within the human immunodeficiency virus type 1 long terminal repeat leader region. J Virol. 1989;63:3213–3219. doi: 10.1128/jvi.63.8.3213-3219.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margolis D M, Somasundaran M, Green M R. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol. 1994;68:905–910. doi: 10.1128/jvi.68.2.905-910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy L, Kao H-Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 43.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 44.Parada C A, Yoon J B, Roeder R G. A novel LBP-1-mediated restriction of HIV-1 transcription at the level of elongation in vitro. J Biol Chem. 1995;270:2274–2283. doi: 10.1074/jbc.270.5.2274. [DOI] [PubMed] [Google Scholar]
- 45.Park K, Atchison M L. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3′ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc Natl Acad Sci USA. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pazin M J, Sheridan P L, Cannon K, Cao Z, Keck J G, Kadonaga J T, Jones K A. NF-kappa B-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- 47.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 48.Ratnasabapathy R, Sheldon M, Johal L, Hernandez N. The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev. 1990;4:2061–2074. doi: 10.1101/gad.4.12a.2061. [DOI] [PubMed] [Google Scholar]
- 49.Romerio F, Gabriel M N, Margolis D M. Repression of human immunodeficiency virus type 1 through the novel cooperation of human factors YY1 and LSF. J Virol. 1997;71:9375–9382. doi: 10.1128/jvi.71.12.9375-9382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seto E, Lewis B, Shenk T. Interaction between transcription factors Sp1 and YY1. Nature. 1993;365:462–464. doi: 10.1038/365462a0. [DOI] [PubMed] [Google Scholar]
- 51.Sheridan P L, Mayall T P, Verdin E, Jones K A. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 1997;11:3327–3340. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Y, Lee J S, Galvin K M. Everything you have ever wanted to know about Yin Yang 1. 1997. Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 53.Shi Y, Seto E, Chang L-S, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 54.Shirra M K, Hansen U. LSF and NTF-1 share a conserved DNA recognition motif yet require different oligomerization states to form a stable protein-DNA complex. J Biol Chem. 1998;273:19260–19268. doi: 10.1074/jbc.273.30.19260. [DOI] [PubMed] [Google Scholar]
- 55.Shirra M K, Zhu Q, Huang H-C, Pallas D, Hansen U. One exon of the human LSF gene includes conserved regions involved in novel DNA-binding and dimerization motifs. Mol Cell Biol. 1994;14:5076–5087. doi: 10.1128/mcb.14.8.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shrivastava A, Calame K. An analysis of genes regulated by the multifunctional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shrivastava A, Saleque S, Kalpana G V, Artandi S, Goff S P, Calame K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science. 1993;262:1889–1892. doi: 10.1126/science.8266081. [DOI] [PubMed] [Google Scholar]
- 58.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 59.Sun J M, Chen H Y, Moniwa M, Samuel S, Davie J R. Purification and characterization of chicken erythrocyte histone deacetylase 1. Biochemistry. 1999;38:5939–5947. doi: 10.1021/bi982633k. [DOI] [PubMed] [Google Scholar]
- 60.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 61.Usheva A, Shenk T. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994;76:1115–2182. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 62.Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 63.Verdin E. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J Virol. 1991;65:6790–6799. doi: 10.1128/jvi.65.12.6790-6799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verdin E, Paras P, Jr, Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volker J L, Rameh L E, Zhu Q, DeCaprio J, Hansen U. Mitogenic stimulation of resting T cells causes rapid phosphorylation of the transcription factor LSF and increased DNA-binding activity. Genes Dev. 1997;11:1435–1446. doi: 10.1101/gad.11.11.1435. [DOI] [PubMed] [Google Scholar]
- 66.Webster L B, Ricciardi R P. trans-dominant mutants of E1A provide genetic evidence that the zinc finger of the trans-activating domain binds a transcription factor. Mol Cell Biol. 1991;11:4287–4296. doi: 10.1128/mcb.11.9.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams R S, Johnston S A, Riedy M, DeVit M J, McElligot S G, Sanford J C. Introduction of foreign genes into tissues of living mice by DNA-coated microprojectiles. Proc Natl Acad Sci USA. 1991;88:2726–2730. doi: 10.1073/pnas.88.7.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolffe A P. New insights into chromatin function in transcriptional control. FASEB J. 1992;6:3354–3361. doi: 10.1096/fasebj.6.15.1464369. [DOI] [PubMed] [Google Scholar]
- 69.Wong J K, Hezareh M, Gunthard H F, Havlit D V, Ignacio C I, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 70.Wu F K, Garcia J A, Harrich D, Gaynor R B. Purification of the human immunodeficiency virus type 1 enhancer and TAR binding proteins EBP-1 and UBP-1. EMBO J. 1988;7:2117–2130. doi: 10.1002/j.1460-2075.1988.tb03051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang W M, Inouye C, Seto E. Cyclophilin A and FKBP12 interact with YY1 and alter its transcriptional activity. J Biol Chem. 1995;270:15187–15193. doi: 10.1074/jbc.270.25.15187. [DOI] [PubMed] [Google Scholar]
- 72.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 74.Yoon J-B, Li G, Roeder R G. Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol Cell Biol. 1994;14:1776–1785. doi: 10.1128/mcb.14.3.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Iratni R, Bromage-Erdjument H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 76.Zhong F, Swendeman S L, Popik W, Pitha P M, Sheffery M. Evidence that levels of the dimeric cellular transcription factor CP2 play little role in the activation of the HIV-1 long terminal repeat in vivo or following superinfection with herpes simplex virus type 1. J Biol Chem. 1994;269:21269–21276. [PubMed] [Google Scholar]
- 77.Zhou Q, Gendrich R W, Engel D A. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J Virol. 1995;69:4323–4330. doi: 10.1128/jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]