Abstract
Viruses exploit different strategies to escape immune surveillance, including the introduction of mutations in cytotoxic T-lymphocyte (CTL) epitopes. The sequence of these epitopes is critical for their binding to major histocompatibility complex (MHC) class I molecules and recognition by specific CTLs, both of which interactions may be lost by mutation. Sequence analysis of the nucleoprotein gene of influenza A viruses (H3N2) isolated in The Netherlands from 1989 to 1999 revealed two independent amino acid mutations at the anchor residue of the HLA-B27-specific CTL epitope SRYWAIRTR (383 to 391). A R384K mutation was found in influenza A viruses isolated during the influenza season 1989–1990 but not in subsequent seasons. In the influenza season 1993–1994, a novel mutation in the same CTL epitope at the same position was introduced. This R384G mutation proved to be conserved in all influenza A viruses isolated from 1993 onwards. Both mutations R384K and R384G abrogated MHC class I presentation and allowed escape from recognition by specific CTLs.
Cytotoxic T lymphocytes (CTLs) of the CD8+ phenotype control viral infections by recognizing antigenic peptides of viral proteins presented by infected cells in association with major histocompatibility complex (MHC) class I molecules. The interaction of specific CTLs with these complexes may lead to the elimination of infected cells. Viruses exploit several strategies to escape from immune surveillance by CTLs (20, 31). One strategy involves the introduction of amino acid mutations within CTL epitopes or in sequences flanking these epitopes. The flanking sequences are important for cytosolic processing of the viral proteins to yield the CTL epitopes, usually 9-mer peptides, while the epitope sequences themselves are critical both for association with MHC class I molecules and for recognition by virus-specific CTLs. Mutations within or in close proximity to CTL epitopes, therefore, may be accompanied by loss of CTL-mediated lysis of target cells (14, 46). In addition, mutations in CTL epitopes may generate peptides that antagonize CTL function (2, 10, 17, 19, 37). Mutations that affect CTL epitopes, resulting in escape from immune surveillance by specific CTLs, have been described for several viruses causing persistent infections, including lymphocytic choriomeningitis virus (27, 34), Epstein-Barr virus (1, 4, 7, 8), human immunodeficiency virus (HIV) (6, 13, 20, 26, 33, 36), hepatitis B virus (3), and hepatitis C virus (45).
Influenza A viruses, causing acute infections, continuously escape from recognition by virus-neutralizing antibodies as a result of accumulation of mutations in their surface glycoproteins hemagglutinin and neuraminidase (antigenic drift) or by introduction of new subtypes of these glycoproteins (antigenic shift). The more conserved internal proteins of influenza viruses, such as the nucleoprotein (NP) and the matrix protein are important targets for CTLs (12, 24). Mutations in these proteins, which occur less frequently than in the surface glycoproteins, potentially could affect CTL-mediated immune surveillance.
Here we show that mutations at the anchor residue of the HLA-B27-restricted CTL epitope SRYWAIRTR (383 to 391), found in the NP of influenza A (H3N2) viruses isolated between 1989 and 1999 in The Netherlands, abrogate MHC class I presentation and recognition by specific CTLs.
MATERIALS AND METHODS
RNA isolation, RT-PCR, and sequencing.
RNA of 59 influenza A (H3N2) viruses of the influenza season 1989–1990, 16 of the season 1991–1992, 16 of the season 1992–1993, 56 of the season 1993–1994, and 15 of the season 1998–1999 (arbitrarily chosen) obtained from the Dutch National Influenza Centre originating from geographically distinct areas in The Netherlands was isolated using a high-pure RNA isolation kit (Boehringer Mannheim) and dissolved in 50 μl of diethylpyrocarbonate-treated H2O. The RNA was used as template to multiply all eight gene segments in a reverse transcriptase PCR (RT-PCR) using a single primer set. The RT-PCR mixture contained 5 μl of RNA, 10 pmol of M13-uni12 primer (CAGGAAACAGCTATGACCAGCAAAAGCAGG), 10 pmol of M13-uni13 primer (TGTAAAACGACGGCCAGTAGTAGAAACAAGG), 0.01 M dithiothreitol, 0.25 mM deoxynucleoside triphosphates (dNTPs), 10 U of RNasin (Promega), 8 U of avian myeloblastosis virus RT (Promega), and 1.25 U of Pfu polymerase (Stratagene) in a total of 25 μl of 1× Pfu polymerase buffer. The mixture was incubated for 60 min at 42°C followed by 4 min at 95°C, 2 min at 37°C, and 3 min at 72°C and 19 cycles of incubation for 1 min at 95°C, 1 min at 50°C, and 3 min at 72°C. The resulting cDNAs were used as template in an NP-specific PCR (nucleotides [nt] 696 to 1243). One microliter of template was added to 25 μl of reaction mixture containing 5 pmol of NP696 primer (TGCTTATGAGAGAATGTGCAA), 5 pmol of NP1243 primer (TCTGTTGGTTGGTGTTTCCTCC), 1.5 mM MgCl2, 20 μM dNTPs, and 2.5 U of Taq polymerase (Promega) in a total of 25 μl of 1× Taq polymerase buffer. The mixture was incubated for 2 min at 95°C followed by 1 min at 50°C and 3 min at 72°C and 29 cycles of incubation for 1 min at 95°C, 30 s at 50°C, and 3 min at 72°C. Amplified DNA was diluted 1:5, and 10 μl was added to 10 μl of sequencing reaction mixture (DYEnamic ET terminator cycle sequencing premix kit; Amersham Pharmacia Biotech, Inc.) containing 10 pmol of NP696 primer. The resulting mixture was incubated for 30 s at 95°C, 15 s at 45°C, and 2 min at 60°C for a total of 30 cycles. Then, 2 μl of 3 M NaAc (pH 4.8) and 80 μl of absolute ethanol were added followed by incubation on ice for 15 min and centrifugation at 2,400 × g for 30 min. Pellets were resuspended in 3 μl of sample buffer, and 0.8 μl was loaded onto a sequence gel followed by automatic sequencing (ABI sequencer). Phylogenetic analysis was performed using DNAML software (Phylip version 3.5).
Isolation and analysis of NP-specific CTL clones.
In round-bottomed microtiter plates, 1,000 peripheral blood mononuclear cells (PBMCs) of a selected donor (HLA-A01, A03, B07, B2705, Cw02, or Cw07) were stimulated twice, with an interval of 1 week, with 2.5 × 104 gamma-irradiated (30 min, 3,000 rads) autologous phytohemagglutinin-stimulated PBMCs pulsed with the peptide SRYWAIRTR (an HLA-B27-restricted CTL epitope of the influenza A virus NP [amino acids (aa) 383 to 391]). The cells were cultured in RPMI 1640 medium containing l-glutamine (2 mM), streptomycin (100 μg/ml), penicillin (100 IU/ml), 2-mercaptoethanol (2 × 10−5 M), interleukin-2 (50 U/ml), and 10% pooled human serum at 37°C and 5% CO2. One week after the second stimulation, expanded cells were analyzed for peptide-specific CTL activity. Cells from wells showing CTL activity were cloned by limiting dilution (0.3, 1, and 3 cells per well) and stimulated nonspecifically by adding 3 × 104 APD B-lymphoblastoid cell line (B-LCL) cells, 3 × 104 BSM B-LCL cells, and 6 × 105 allogeneic PBMCs (which were all gamma irradiated); 1 μg of phytohemagglutinin and 50 U of interleukin-2 per ml (44). After incubation for 2 weeks, clones showing CTL activity were stimulated specifically with gamma-irradiated peptide-pulsed autologous PBMCs. After incubating the clones for 12 days, they were stimulated nonspecifically as described above in 75-cm2 flasks. After 2 weeks, cells were harvested, aliquoted, and stored at −135°C until use. These CTLs will be referred to as the NP/B27 CTL clone. An HLA-A3-restricted CTL clone specific for the influenza A virus NP epitope ILRGSVAHK (aa 265 to 273) was kindly provided by W. Biddison, National Institutes of Health (NIH), Bethesda, Md., and will be referred to as the NP/A3 CTL clone. The phenotype of both CTL clones was determined by fluorescence-activated cell sorting (FACS) analysis using monoclonal antibodies specific for CD3, CD4, and CD8, and their specificity and HLA restriction were confirmed in CTL assays. To this end, 106 cells of an Epstein-Barr virus-transformed B-LCL of an HLA-A3- and -B27-positive donor and mismatched (HLA-A3- and -B27-negative) B-LCL cells were incubated with the peptide ILRGSVAHK or SRYWAIRTR (10 μM) for 1 h at 37°C and used as target cells in CTL assays with the respective CTL clones as effectors.
Preparation of target cells.
B-LCL cells (106) of one donor (HLA-A3 and -B27 positive) were incubated with the peptide ILRSGVAHK (HLA-A3), SRYWAIRTR (HLA-B27), or SKYWAIRTR or SGYWAIRTR (HLA-B27 mutant peptide) at a concentration giving the highest specific lysis (10 μM for the ILRGSVAHK peptide and 1 μM for the SRYWAIRTR and mutant peptides). In addition, the same B-LCL cells were infected with recombinant vaccinia viruses (RVV) expressing the NP of influenza virus A/Puerto Rico/8/34;H1N1 (A/PR/8/34) (kindly provided by B. Moss, NIH), A/Netherlands/018/94;H3N2 (A/Neth/18/94) (generated essentially as previously described [38]), or a control vaccinia virus (VSC65), each at a multiplicity of infection of 10. Also, B-LCL cells were infected with the influenza A virus A/PR/8/34; A/Netherlands/651/89;H3N2 (A/Neth/651/89), having an R384K mutation; or A/Neth/18/94, having an R384G mutation in the NP gene. Cells were cultured in RPMI 1640 medium containing l-glutamine (2 mM), streptomycin (100 μg/ml), penicillin (100 IU/ml), and 10% fetal bovine serum at 37°C and 5% CO2. After 16 h of incubation, cells were washed and used as target cells in CTL assays with the NP/A3 and NP/B27 CTL clones as effector cells.
CTL assays.
Target cells (B-LCL cells) were labeled for 1 h with 75 μCi of Na2[51Cr]O4 in RPMI 1640 medium. Cells were washed three times in culture medium (see above) and resuspended in this medium at a concentration of 104 cells/50 μl. Effector cells (CTLs) were suspended in this medium at a concentration of 2.5 × 104, 5 × 104, or 1 × 105 cells/100 μl (effector-to-target [E:T] ratios, 2.5:1, 5:1, and 10:1). Fifty microliters of target cells was incubated either with 100 μl of medium (spontaneous release), with 100 μl of 10% Triton X-100 (maximum release), or with 100 μl of effector cells (experimental release) for 4 h at 37°C. Supernatants were harvested, and radioactivity was measured by gamma counting. The percentage of specific lysis was calculated as 100 × (experimental release − spontaneous release)/(maximum release − spontaneous release). CTL assays were performed in triplicate per target per E:T ratio.
Functional analysis of wild-type and mutant NP.
The NP coding sequences of influenza virus A/Hong Kong/2/68;H3N2 (A/HK/2/68), representing wild-type virus, and A/Neth/18/94 (R384G mutant) were amplified by PCR using pBluescript plasmids containing the NP genes of both viruses as templates with a NotI forward primer, CAGCGGCCGCATGGCGTCCCAAGGC, and an XhoI reverse primer, CACTCGAGTTAATTGTCGTACTCCTCTGC (restriction endonuclease recognition sequences are underlined, and start and stop codons of the NP gene are in boldface). PCRs were performed with 10 ng of plasmid DNA, 10 pmol of each of the primers, 1.5 mM MgCl2, 20 μM dNTPs, and 5 U of Pfu polymerase in a total of 100 μl of 1× Pfu polymerase buffer. This mixture was heated for 3 min at 94°C followed by a total of 20 cycles consisting of 1 min at 94°C, 2 min at 50°C, and 4 min at 72°C. The PCR products were cloned as NotI-XhoI fragments in a modified version of the eukaryotic expression plasmid pcDNA3 (Invitrogen) followed by large-scale production of plasmid DNA and purification by CsCl gradient centrifugation according to standard methods.
The respective plasmids were used for transfection into 293T cells. Plasmid DNA (1.5 μg) was mixed with equal amounts of the plasmids pHMG-PB1, pHMG-PB2, and pHMG-PA (encoding the polymerase proteins PB1, PB2, and PA, respectively; kindly provided by P. Palese, Mount Sinai School of Medicine, New York, N.Y. [35]) and 0.5 μg of plasmid RF419 (constructed essentially as described previously [29]), from which the green fluorescent protein (GFP) gene flanked with the influenza A virus noncoding region of the NS gene segment is transcribed in a negative orientation. This plasmid mixture was transfected into 293T cells as described previously (32). One day after transfection, cells were subjected to FACS analysis. Cells transfected with plasmid pcDNA3 without cloned NP sequences served as a negative control, while cells transfected with plasmid pEGFP-N1 (encoding enhanced GFP; Clontech) served as a positive control.
Nucleotide sequence accession numbers.
Nucleotide sequences have been submitted to GenBank and can be retrieved by the following accession numbers: AF225709 to AF225764 (influenza season 1993–1994), AF225765 to AF225823 (influenza season 1989–1990), AF225824 to AF225839 (influenza season 1991–1992), AF225840 to AF225855 (influenza season 1992–1993), and AF225856 to AF225869 and AF226872 (influenza season 1998–1999).
RESULTS
NP gene sequences of influenza A (H3N2) viruses.
Sequence analysis of the NP genes of influenza A (H3N2) viruses isolated in The Netherlands from 1989 to 1999 was performed. The region of the NP genes sequenced encompasses aa 240 to 391 (or nt 720 to 1175) and harbors four previously described CTL epitopes: aa 265 to 273, aa 338 to 347, aa 380 to 388, and aa 383 to 391, presented by HLA-A3, -B37, -B8, and -B27 molecules, respectively (9, 15, 25, 42). As shown in Table 1, differences in the nucleotide sequences of viruses isolated in the same season were observed. Overall, 46 different nucleotide sequences were identified in 162 viruses isolated from 1989 to 1999. Nevertheless, within one season viruses were closely related, as is shown in the maximum likelihood tree in Fig. 1. The amino acid sequences found in the influenza A viruses isolated in the respective influenza seasons are shown in Fig. 2, and nucleotide mutations underlying differences in these amino acid sequences are shown in Table 2. Eleven different amino acid sequences were identified, and phylogenetic analysis based on these amino acid sequences revealed essentially the same distances between the recent influenza virus isolates (1993 to 1998) and older isolates (Fig. 1). In the influenza season 1989–1990, 13 out of 59 isolated viruses had an R384K mutation affecting both the HLA-B8 and HLA-B27 epitopes (Fig. 2). In fact, R384 is the anchor residue of the HLA-B27 epitope and critical for association with MHC class I molecules. This R384K mutation was not found in subsequent seasons. However, in the season 1993–1994, a novel mutation at the same position in these CTL epitopes was found. This R384G mutation was present in all 56 viruses tested of this season and maintained in all viruses tested of the 1998–1999 season (Fig. 2). In all mutant viruses, the G at position 384 was coded for by the same codon (Table 2). The abrupt introduction of the R384G mutation in 1993–1994 was accompanied by two other amino acid mutations in the sequenced NP region, S259L and E375G. These mutations are located in close proximity to the HLA-A3 and the HLA-B8 and HLA-B27 epitopes, respectively (Fig. 2). We also sequenced several viruses isolated between 1994 and 1998 which all showed the R384G, S259L, and E375G mutations (data not shown). In contrast to the overlapping HLA-B8 and HLA-B27 epitopes, the HLA-A3 epitope ILRGSVAHK (aa 265 to 273) proved to be conserved. Only three silent mutations were found in this epitope in 3 out of the 162 viruses tested (data not shown). Likewise, no mutations were found that affected the HLA-B37 epitope (338 to 347).
TABLE 1.
Virus variants and number of each variant observed per influenza seasona
| Season (n) | Variantb | No. of variant |
|---|---|---|
| 1989–1990 (59) | 1 | 18 |
| 2 | 14 | |
| 3 | 8 | |
| 4 | 7 | |
| 5 | 3 | |
| 6 | 1 | |
| 7 | 1 | |
| 8 | 1 | |
| 9 | 1 | |
| 10 | 1 | |
| 11 | 1 | |
| 12 | 1 | |
| 13 | 1 | |
| 14 | 1 | |
| 1991–1992 (16) | 15 | 8 |
| 16 | 3 | |
| 17 | 3 | |
| 18 | 1 | |
| 19 | 1 | |
| 1992–1993 (16) | 20 | 7 |
| 21 | 4 | |
| 22 | 1 | |
| 23 | 1 | |
| 24 | 1 | |
| 25 | 1 | |
| 26 | 1 | |
| 1993–1994 (56) | 27 | 35 |
| 28 | 3 | |
| 29 | 2 | |
| 30 | 2 | |
| 31 | 2 | |
| 32 | 1 | |
| 33 | 1 | |
| 34 | 1 | |
| 35 | 1 | |
| 36 | 1 | |
| 37 | 1 | |
| 38 | 1 | |
| 39 | 1 | |
| 40 | 1 | |
| 41 | 1 | |
| 42 | 1 | |
| 43 | 1 | |
| 1998–1999 (15) | 44 | 11 |
| 45 | 3 | |
| 46 | 1 |
The nucleotide sequence of the NP gene (nt 720 to 1175) of each virus of a particular season was compared to the consensus sequence of that season.
Variant numbers correspond with the numbers shown in Fig. 1.
FIG. 1.
Maximum likelihood tree based on nucleotide sequences of the NP gene. Part of the NP genes (nt 720 to 1175) of 162 influenza A (H3N2) viruses isolated from 1989 to 1999 was sequenced and subjected to phylogenetic analysis. Included in the figure are the influenza viruses A/Texas/1/77, A/Memphis/5/80, and A/Beijing/353/89. The numbers shown in the figure correspond to the numbers shown in Table 1. The insert represents a protein distance tree (Protdist, Fitch) based on the NP sequence of the representative influenza virus strains. The letter code used for the respective amino acid sequences corresponds to that of Fig. 2.
FIG. 2.
Amino acid sequences of the NP (aa 240 to 391) of influenza A (H3N2) viruses isolated from 1989 to 1999. The consensus sequence of each season is shown in the upper rows, whereas variant sequences are shown in the lower rows. CTL epitopes are underlined and shown in boldface. Amino acid differences between seasons are shown in boldface, while differences within a season are shown in boldface italic. All mutations are marked with an arrow. The number in parentheses refers to the number of isolates showing that sequence.
TABLE 2.
Amino acid mutations in the influenza virus NP and the corresponding nucleotide mutations
| Season | Amino acid mutation | Nucleotide mutation | No.a |
|---|---|---|---|
| 1989–1990 | K293R | AAA→AGA | 1 |
| T350S | ACC→TCC | 8 | |
| M374I | ATG→ATA | 1 | |
| E375G | GAA→GGA | 1 | |
| R384K | AGG→AAG | 13 | |
| 1991–1992 | N287S | AAT→AGT | 4 |
| G290D | GGC→GAC | 4 | |
| 1992–1993 | S287N | AGT→AAT | 2 |
| D290G | GAC→GGC | 2 | |
| 1993–1994 | V299L | GTG→TTG | 2 |
| R361T | AGA→ACA | 2 | |
| R384G | AGG→GGGb | 56 | |
| 1998–1999 | R384G | AGG→GGGb | 15 |
| R389T | AGG→ACG | 1 |
Number of viruses containing the mutation; see also Fig. 2.
Since all viruses from the influenza season 1993–1994 onwards contained the R384G mutation, the codon encoding the original R384 in the NP of influenza viruses of the previous season (1992–1993) was taken for comparison.
Recognition of influenza A virus NP by specific CTL clones.
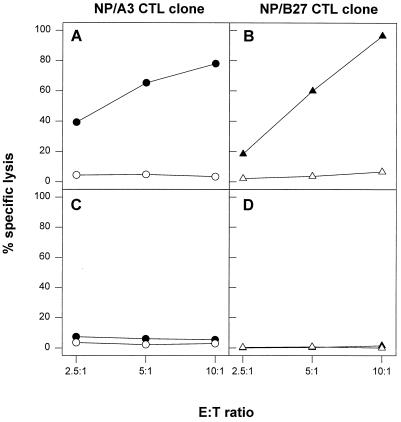
An HLA-B27-restricted CTL clone, designated NP/B27, with specificity for the NP epitope SRYWAIRTR (aa 383 to 391), was generated. This CTL clone lysed matched target cells, pulsed with peptide SRYWAIRTR (Fig. 3B and D). As a control, an HLA-A3-restricted CTL clone, designated NP/A3, with specificity for the conserved NP epitope ILRGSVAHK (aa 265 to 273) was used. As shown in Fig. 3A and C, this CTL clone lysed matched target cells pulsed with the corresponding peptide. The phenotype of both CTL clones as determined by FACS analysis was CD3+ CD4− CD8+ (data not shown).
FIG. 3.
Confirmation of specificity and HLA restriction of CTL clones. HLA-A3- and -B27-positive (A and B) and HLA-A3- and -B27-negative (C and D) B-LCL cells were incubated with the HLA-A3-specific peptide ILRGSVAHK (solid circles) or the HLA-B27-specific peptide SRYWAIRTR (solid triangles) or left untreated (open circles and open triangles, respectively) followed by incubation with the NP/A3 (A and C) or NP/B27 (B and D) CTL clone. CTL assays were performed in triplicate at three E:T ratios. Mean percentages of specific lysis are shown.
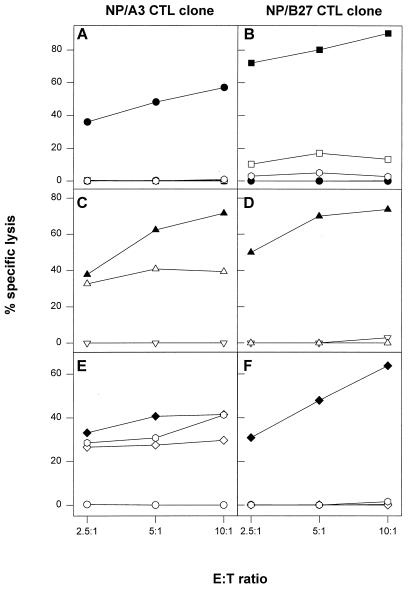
The effect of the R384K and R384G mutations in the HLA-B27-specific NP epitope SRYWAIRTR on CTL-mediated lysis was first studied with synthetic peptides. The NP/B27 CTL clone lysed target cells pulsed with the peptide SRYWAIRTR, whereas control untreated target cells or cells pulsed with the mutant peptide SKYWAIRTR or SGYWAIRTR were not recognized by this CTL clone (Fig. 4B). Next, cells infected with influenza A viruses having the respective mutations in the NP were used as target cells. Target cells infected with influenza virus A/PR/8/34, A/Neth/651/89, or A/Neth/18/94 were all recognized by the NP/A3 CTL clone (Fig. 4E). However, the NP/B27 CTL clone lysed only target cells infected with influenza virus A/PR/8/34, which had the nonmutated epitope, and failed to recognize target cells infected with influenza virus A/Neth/651/89 or A/Neth/18/94, which had the R384K or the R384G mutation, respectively (Fig. 4F). These data were further confirmed using RVV expressing the NP of A/PR/8/34 (nonmutated epitope) or A/Neth/18/94 (R384G mutant epitope). The NP/A3 CTL clone recognized target cells infected with RVV expressing NP of A/PR/8/34 or A/Neth/18/94 equally well and failed to recognize target cells infected with a control vaccinia virus (Fig. 4C). The NP/B27 CTL clone, however, recognized target cells infected with RVV expressing NP of A/PR/8/34 but failed to recognize target cells infected with RVV expressing NP of A/Neth/18/94 (Fig. 4D).
FIG. 4.
Effect of mutations in the HLA-B27 epitope on CTL-mediated lysis of target cells. (A and B) HLA-A3- and -B27-positive B-LCL cells of one donor were incubated with the HLA-A3-specific peptide ILRGSVAHK (solid circles), the HLA-B27-specific peptide SRYWAIRTR (solid squares), or the HLA-B27 mutant peptide SGYWAIRTR (open squares) or SKYWAIRTR (open hexagons) or left untreated (open circles) and used as targets in CTL assays with the NP/A3 (A) or NP/B27 (B) CTL clone as effector. (C and D) The same B-LCL cells were infected with a control vaccinia virus (open inverted triangles), a vaccinia virus expressing the NP of A/PR/8/34 (solid triangles), or a vaccinia virus expressing NP of A/Neth/18/94 (open triangles) followed by incubation with the NP/A3 (C) or NP/B27 (D) CTL clone. (E and F) Also, B-LCL cells were infected with influenza virus A/PR/8/34 (solid diamonds), A/Neth/18/94 (open diamonds), or A/Neth/651/89 (open hexagons) or left untreated (open circles) followed by incubation with the NP/A3 (E) or NP/B27 (F) CTL clone. CTL assays were performed in triplicate at three E:T ratios. Mean percentages of specific lysis are shown.
Functional analysis of NP sequences.
In order to determine whether the R384G mutation affected the function of the NP, a eukaryotic expression plasmid encoding the NP of A/HK/2/68 (having an R at position 384) or the NP of A/Neth/18/94 (having a G at position 384) was cotransfected with expression plasmids encoding the three polymerase proteins of influenza A virus (PB1, PB2, and PA) and a plasmid expressing GFP RNA in the context of an influenza A virus NS gene segment. Others have shown previously that such negative-sense RNA molecules can serve as templates for production of cRNA and mRNA in the presence of functional NP and polymerase proteins, ultimately resulting in synthesis of the encoded protein (28). GFP synthesis was measured by FACS analysis (Table 3). The percentage of positive cells and mean fluorescence did not differ significantly between cells transfected with the NP gene of A/HK/2/68 and those transfected with the NP gene of A/Neth/18/94, indicating that both NPs were equally functional. Furthermore, influenza viruses A/HK/2/68 and A/Neth/18/94 yielded comparable virus titers in MDCK cells, indicating that both viruses replicated equally well (data not shown).
TABLE 3.
Functional analysis of wild-type and mutant NPa
| NP | % positive cells | Mean fluorescence |
|---|---|---|
| Negative control | 0.3 | 433 |
| A/HK/2/68 | 26.4 | 2,002 |
| A/Neth/18/94 | 23.3 | 1,944 |
| Positive control | 95.0 | 6,383 |
293T cells were transfected with a mixture of plasmids including pHMG-PB1, pHMG-PB2, pHMG-PA, and RF419 and a plasmid encoding NP of A/HK/2/68 or A/Neth/18/94. Cells transfected with plasmid pcDNA3 served as a negative control, while cells transfected with plasmid pEGFP-N1 were used as a positive control. Percentage of cells showing GFP expression and mean fluorescence of 293T cells were measured by FACS analysis. Results of a representative experiment are shown.
DISCUSSION
In the present paper, we show that an R384K or R384G mutation in the HLA-B27-specific epitope SRYWAIRTR (383 to 391) of influenza A virus NP abrogates MHC class I presentation and recognition by specific CTLs. In peptides that associate with HLA-B27, the second residue is often an arginine (R), and this so-called anchor residue is critical for binding to HLA-B27 molecules (18, 22, 23, 40, 43). Mutations at this position are accompanied by loss of binding to HLA-B27 and hence loss of the activity of specific CTLs. This has previously been demonstrated for the CTL epitope KRWIILGLNK (263 to 272) in the HIV type 1 (HIV-1) Gag protein: exchanging R264 for K or G diminished binding to HLA-B27 and lysis of peptide-pulsed target cells, with the R264G mutation having the greatest effect (30). We here show that cells pulsed with mutant peptides, having identical mutations at the anchor residue of the epitope SRYWAIRTR of the influenza A virus NP (see Table 4 for comparison), were not lysed by HLA-B27-restricted CTLs. In addition, we show that cells infected either with influenza A viruses or with vaccinia virus expressing mutant NP were no longer recognized by specific CTLs.
TABLE 4.
Comparison of wild-type and mutant HLA-B27-restricted CTL epitopes of influenza A virus NP with those of the HIV-1 Gag protein
| HLA-B27 epitope | Amino acid sequencea | Reference(s) |
|---|---|---|
| HIV-1 Gag (263–272) | KRWIILGLNK | 5 |
| HIV-1 Gag mutant 1 | KKWIILGLNK | 13, 30b |
| HIV-1 Gag mutant 2 | KGWIILGLNK | 30b |
| Influenza virus NP (383–391) | SRYWAIRTR | 15 |
| Influenza virus NP mutant 1 | SKYWAIRTR | Present paper |
| Influenza virus NP mutant 2 | SGYWAIRTR | Present paper |
Mutations are shown in boldface.
In vitro-generated mutant.
The R384K mutation was found in several isolates of the influenza season 1989–1990, but not in later seasons. This mutation was previously found in two viruses isolated in 1971 and 1972 (41). In contrast, the R384G mutation was found in all influenza A virus isolates from the influenza season 1993–1994 onwards. A search in the influenza virus sequence database (Los Alamos National Laboratory) and in the literature revealed that from the introduction of H3N2 viruses in 1968 until the 1993 epidemics, all virus isolates (except for the R384K mutant viruses mentioned above) had the nonmutated HLA-B27 epitope SRYWAIRTR. Since most viruses have been selected for sequencing based on antigenic properties of their hemagglutinin, we assume that the NP sequences of influenza viruses in this database are random with regard to CTL epitopes. Of note, the R384G mutation has never been found in H1N1 and H2N2 viruses. Since we sequenced influenza viruses that were isolated from patients living in geographically distinct areas in The Netherlands, it is unlikely that all viruses originated from a single source. Moreover, in the region of the NP that was sequenced (representing 152 aa) differences were found between viruses isolated within a single season. Interestingly, the R384G mutation has also been found in influenza A (H3N2) viruses isolated in Japan after 1993, although viruses lacking the R384G mutation cocirculated in this area after 1993 (21).
The R384G mutation found in the influenza season 1993–1994 was accompanied by two other amino acid mutations in the NP, S259L and E375G. Also, in the Japanese strains containing the R384G mutation (see above) the same accompanying mutations were found, which may indicate a more global spread of these viruses. The S259L mutation is only 6 aa N terminal of the HLA-A3 epitope ILRGSVAHK and, therefore, could have affected processing of this peptide. However, our results show that this mutation did not have an effect on MHC class I presentation of the HLA-A3 epitope. Although a G at position 384 was always accompanied by an L at position 259 and a G at position 375, the latter two amino acids have previously also been found with an R at position 384, indicating that the R384G mutation is not forced by the other two mutations or vice versa and that the mutations observed are not mutually compensatory. In addition, in the influenza season 1989–1990 we obtained a virus isolate having a G at position 375 and an R at position 384 of the NP.
The consequence of a mutation at the anchor residue with respect to virus escape from immune surveillance by CTLs has been demonstrated previously (13). The R264K mutation in the HIV-1 Gag HLA-B27 epitope KRWIILGLNK (263 to 272) was accompanied by progression to AIDS in HIV-1-infected patients who showed strong CTL responses against the nonmutated epitope. Although the role of CTLs in protection from influenza virus infection is still controversial, CTLs are likely to contribute to virus clearance and inhibition of virus spread (39). Therefore, mutations at the anchor residue of the influenza A virus NP epitope SRYWAIRTR may have implications for HLA-B27-positive influenza virus-infected patients. At this point, it is not clear what the consequences of the R384G mutation are with respect to MHC class I binding and/or recognition by CTLs of the HLA-B8 epitope.
The observation that the R384G mutation was conserved in all sequenced influenza A (H3N2) viruses isolated after 1993 in The Netherlands suggests that this mutation is advantageous to the virus. We did not find differences between a wild-type virus and an R384G mutant virus with respect to replication properties in vitro. In addition, in transfection experiments, we have shown that an RNA molecule that resembles an influenza A virus gene segment was equally well transcribed and translated in the presence of wild-type NP and mutant NP. Since the R384G mutation completely abrogates the recognition of the HLA-B27 epitope by specific CTLs, influenza A viruses harboring this mutation may escape from immunity mediated by virus-specific CTLs. HLA-B27-positive individuals constitute approximately 8% of the Caucasian population, which is predominant in The Netherlands. The immune pressure mediated by CTLs in these individuals, which recognize the wild-type HLA-B27 epitope in the NP, may have contributed to the emergence and continued circulation of escape mutant viruses. The mutant virus may have emerged from the quasispecies of influenza viruses in HLA-B27-positive individuals. Since the R384G mutation did not impose functional constraints on the NP, a selective pressure in 8% of the individuals may have been sufficient to drive the selection process. At present, it is unknown whether the HLA-B27 epitope is immunodominant. Conceivably, this would favor the emergence of the R384G mutant virus. Little is known about the in vivo rate of attack of target cells by specific CTLs: an infected cell may be recognized by one CTL but not by another at the same time, allowing the virus to escape from the action of one CTL clone. Once having emerged into the human population, viruses with the R384G mutation are fully replication competent and ultimately have replaced the original virus having the nonmutated epitope.
In contrast to the HLA-B8 and -B27 epitope, the HLA-A3 epitope proved to be conserved; we found only three silent mutations out of 162 sequenced influenza A (H3N2) viruses isolated over 10 years despite a higher prevalence of the HLA-A3 allele in the human population. With the exception of one virus having an I265V mutation (41), influenza virus sequence database searches (including H1N1, H2N2, and H3N2 viruses isolated over more than 60 years) also did not reveal amino acid mutations within this epitope. A possible explanation is that mutations in this region of the NP are not tolerated or are less well tolerated by the virus because of functional constraints. For example, an R267A mutation in the HLA-A3 epitope has been shown elsewhere to affect RNA binding by the NP (11). Recently, a second HLA-B27 epitope in the NP (174 to 184) has been described (16). However, a sequence database search revealed that this HLA-B27 epitope is completely conserved.
CTL escape mutants have been shown to arise in individuals persistently infected with virus, e.g., HIV, as a result of continuous immune pressure mediated by CTLs. Influenza A viruses cause acute infections, affecting a large percentage of individuals each year, and therefore may be considered as persisting in the human population. We have provided epidemiological and immunological evidence for antigenic drift in the influenza A virus NP, possibly as a result of immune pressure mediated by CTLs. Thus, in addition to the introduction of mutations in the surface glycoproteins allowing escape from antibody-mediated immunity, the introduction of mutations in CTL epitopes may be a strategy exploited by influenza A viruses to escape from CTL-mediated immunity. This would be the first example of CTL-mediated antigenic drift in a virus that causes an acute infection.
ACKNOWLEDGMENTS
Part of this work was supported by the Foundation for Respiratory Virus Infections, Notably Influenza (SRVI).
We acknowledge W. Biddison, NIH, Bethesda, Md., for providing us with the NP/A3 CTL clone; B. Moss, NIH, for providing us with RVV expressing the NP of A/PR/8/34; and P. Palese, Mount Sinai School of Medicine, New York, N.Y., for providing us with the HMG-PB1, PB2, and PA expression plasmids. Finally, we thank Ger van der Water for continuous support.
REFERENCES
- 1.Apolloni A, Moss D, Stumm R, Burrows S, Suhrbier A, Misko I, Smidt C, Sculley T. Sequence variation of cytotoxic T cell epitopes in different isolates of Epstein-Barr virus. Eur J Immunol. 1992;22:183–189. doi: 10.1002/eji.1830220127. [DOI] [PubMed] [Google Scholar]
- 2.Bertoletti A, Sette A, Chisari F V, Penna A, Levrero M, De Carli M, Fiaccadori F, Ferrari C. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature. 1994;369:407–410. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- 3.Bertoletti A, Costanzo A, Chisari F V, Levrero M, Artini M, Sette A, Penna A, Giuberti T, Fiaccadori F, Ferrari C. Cytotoxic T lymphocyte response to a wild type hepatitis B virus epitope in patients chronically infected by variant viruses carrying substitutions within the epitope. J Exp Med. 1994;180:933–943. doi: 10.1084/jem.180.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrows J M, Burrows S R, Poulsen L M, Sculley T B, Moss D J, Khanna R. Unusually high frequency of Epstein-Barr virus genetic variants in Papua New Guinea that can escape cytotoxic T-cell recognition: implications for virus evolution. J Virol. 1996;70:2490–2496. doi: 10.1128/jvi.70.4.2490-2496.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buseyne F, McChesney M, Porrot F, Kovarik S, Guy B, Riviere Y. Gag-specific cytotoxic T lymphocytes from human immunodeficiency virus type 1-infected individuals: gag epitopes are clustered in three regions of the p24 gag protein. J Virol. 1993;67:694–702. doi: 10.1128/jvi.67.2.694-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couillin I, Culmann-Penciolelli B, Gomard E, Choppin J, Levy J P, Guillet J G, Saragosti S. Impaired cytotoxic T lymphocyte recognition due to genetic variations in the main immunogenic region of the human immunodeficiency virus 1 NEF protein. J Exp Med. 1994;180:1129–1134. doi: 10.1084/jem.180.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Campos-Lima P O, Gavioli R, Zhang Q J, Wallace L E, Dolcetti R, Rowe M, Rickinson A B, Masucci M G. HLA-A11 epitope loss isolates of Epstein-Barr virus from a highly A11+ population. Science. 1993;260:98–100. doi: 10.1126/science.7682013. [DOI] [PubMed] [Google Scholar]
- 8.de Campos-Lima P O, Levitsky V, Brooks J, Lee S P, Hu F, Rickinson A B, Masucci M G. T-cell responses and virus evolution: loss of HLA A11-restricted CTL epitopes in Epstein-Barr virus isolates from highly A11-positive populations by selective mutation of anchor residues. J Exp Med. 1994;179:1297–1305. doi: 10.1084/jem.179.4.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiBrino M, Tsuchida T, Turner R V, Parker K C, Coligan J E, Biddison W E. HLA-A1 and HLA-A3 T cell epitopes derived from influenza virus proteins predicted from peptide binding motifs. J Immunol. 1993;151:5930–5935. [PubMed] [Google Scholar]
- 10.Dong T, Boyd D, Rosenberg W, Alp N, Takiguchi M, McMichael A, Rowland-Jones S. An HLA-B35-restricted epitope modified at an anchor residue results in an antagonist peptide. Eur J Immunol. 1996;26:335–339. doi: 10.1002/eji.1830260210. [DOI] [PubMed] [Google Scholar]
- 11.Elton D, Medcalf L, Bishop K, Harrison D, Digard P. Identification of amino acid residues of influenza virus nucleoprotein essential for RNA binding. J Virol. 1999;73:7357–7367. doi: 10.1128/jvi.73.9.7357-7367.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotch F, McMichael A, Smith G, Moss B. Identification of viral molecules recognized by influenza-specific human cytotoxic T lymphocytes. J Exp Med. 1987;165:408–416. doi: 10.1084/jem.165.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goulder P J, Phillips R E, Colbert R A, McAdam S, Ogg G, Nowak M A, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael A J, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;2:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 14.Hahn Y S, Hahn C S, Braciale V L, Braciale T J, Rice C M. CD8+ T cell recognition of an endogenously processed epitope is regulated primarily by residues within the epitope. J Exp Med. 1992;176:1335–1341. doi: 10.1084/jem.176.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huet S, Nixon D F, Rothbard J B, Townsend A, Ellis S A, McMichael A J. Structural homologies between two HLA B27-restricted peptides suggests residues important for interaction with HLA B27. Int Immunol. 1990;2:311–316. doi: 10.1093/intimm/2.4.311. [DOI] [PubMed] [Google Scholar]
- 16.Jameson J, Cruz J, Ennis F A. Human cytotoxic T-lymphocyte repertoire to influenza A virus. J Virol. 1998;72:8682–8689. doi: 10.1128/jvi.72.11.8682-8689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jameson S C, Carbone F R, Bevan M J. Clone-specific T cell receptor antagonists of major histocompatibility complex class I-restricted cytotoxic T cells. J Exp Med. 1993;177:1541–1550. doi: 10.1084/jem.177.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jardetzky T S, Lane W S, Robinson R A, Madden D R, Wiley D C. Identification of self-peptides bound to purified HLA-B27. Nature. 1991;353:326–329. doi: 10.1038/353326a0. [DOI] [PubMed] [Google Scholar]
- 19.Klenerman P, Rowland-Jones S, McAdam S, Edwards J, Daenke S, Lalloo D, Koppe B, Rosenberg W, Boyd D, Edwards A, Giangrande P, Phillips R E, McMichael A J. Cytotoxic T-cell activiy antagonized by naturally occurring HIV-1 Gag variants. Nature. 1994;369:403–407. doi: 10.1038/369403a0. [DOI] [PubMed] [Google Scholar]
- 20.Koup R A. Virus escape from CTL recognition. J Exp Med. 1994;180:779–782. doi: 10.1084/jem.180.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindstrom S E, Hiromoto Y, Nerome R, Omoe K, Sugita S, Yamazaki Y, Takahashi T, Nerome K. Phylogenetic analysis of the entire genome of influenza A (H3N2) viruses from Japan: evidence for genetic reassortment of the six internal genes. J Virol. 1998;72:8021–8031. doi: 10.1128/jvi.72.10.8021-8031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madden D R, Gorga J C, Strominger J L, Wiley D C. The structure of HLA-B27 reveals nonamer self-peptides bound in an extended conformation. Nature. 1991;353:321–325. doi: 10.1038/353321a0. [DOI] [PubMed] [Google Scholar]
- 23.Madden D R, Gorga J C, Strominger J L, Wiley D C. The three-dimensional structure of HLA-B27 at 2,1 Å resolution suggests a general mechanism for tight peptide binding to MHC. Cell. 1992;70:1035–1048. doi: 10.1016/0092-8674(92)90252-8. [DOI] [PubMed] [Google Scholar]
- 24.McMichael A J, Mitchie C A, Gotch F M, Smith G L, Moss B. Recognition of influenza A virus nucleoprotein by human cytotoxic T lymphocytes. J Gen Virol. 1986;67:719–726. doi: 10.1099/0022-1317-67-4-719. [DOI] [PubMed] [Google Scholar]
- 25.McMichael A J, Gotch F M, Rothbard J. HLA B37 determines an influenza virus nucleoprotein epitope recognized by cytotoxic T lymphocytes. J Exp Med. 1986;164:1397–1406. doi: 10.1084/jem.164.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMichael A J, Phillips R E. Escape of human immunodeficiency virus from immune control. Annu Rev Immunol. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. [DOI] [PubMed] [Google Scholar]
- 27.Moskophidis D, Zinkernagel R M. Immunobiology of cytotoxic T-cell escape mutants of lymphocytic choriomeningitis virus. J Virol. 1995;69:2187–2193. doi: 10.1128/jvi.69.4.2187-2193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann G, Watanabe T, Hiroshi I, Watanabe S, Goto H, Gao P, Hughes M, Perez D R, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. J Virol. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann G, Watanabe T, Kawaoka Y. Plasmid-driven formation of influenza virus-like particles. J Virol. 2000;74:547–551. doi: 10.1128/jvi.74.1.547-551.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nietfield W, Bauer M, Fevrier M, Maier R, Holzwarth B, Frank R, Maier B, Riviere Y, Meyerhans A. Sequence constraints and recognition by CTL of an HLA-B27-restricted HIV-1 gag epitope. J Immunol. 1995;154:2188–2197. [PubMed] [Google Scholar]
- 31.Oldstone M B A. How viruses escape from cytotoxic T lymphocytes: molecular parameters and players. Virology. 1997;234:179–185. doi: 10.1006/viro.1997.8674. [DOI] [PubMed] [Google Scholar]
- 32.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retrovirus by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips R E, Rowland-Jones E S, Nixon D F, Gotch F M, Edwards J P, Ogunlesi A O, Elvin J G, Rothbard J A, Bangham C R, Rizza C R, McMichael A J. Human immunodeficiency virus genetic variation that can escape cytotoxic T-cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 34.Pircher H, Moskophidis D, Rohrer U, Bürki K, Hengartner H, Zinkernagel R M. Viral escape by selection of cytotoxic T cell-resistant virus variants in vivo. Nature. 1990;346:629–633. doi: 10.1038/346629a0. [DOI] [PubMed] [Google Scholar]
- 35.Pleschka S, Jaskunas R S, Engelhardt O G, Zürcher T, Palese P, Garcia-Sastres A. A plasmid-based reverse genetics system for influenza A virus. J Virol. 1996;70:4188–4192. doi: 10.1128/jvi.70.6.4188-4192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price D A, Goulder P J, Klenerman P, Sewell A K, Easterbrook P J, Troop M, Bangham C R, Phillips R E. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid S W, McAdam S, Smith K J, Klenerman P, O'Callaghan C A, Harlos K, Jakobsen B K, McMichael A J, Bell J I, Stuart D L, Jones E Y. Antagonist HIV-1 Gag peptides induce structural changes in HLA B8. J Exp Med. 1996;184:2279–2286. doi: 10.1084/jem.184.6.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rimmelzwaan G F, Siebelink K H, Huisman R C, Moss B, Francis M J, Osterhaus A D M E. Removal of the cleavage site of recombinant feline immunodeficiency virus envelope protein facilitates incorporation of the surface glycoprotein in immune-stimulating complexes. J Gen Virol. 1994;75:2097–2102. doi: 10.1099/0022-1317-75-8-2097. [DOI] [PubMed] [Google Scholar]
- 39.Rimmelzwaan G F, Osterhaus A D M E. Cytotoxic T lymphocyte memory: role in cross-protective immunity against influenza? Vaccine. 1995;13:703–705. doi: 10.1016/0264-410x(94)00030-q. [DOI] [PubMed] [Google Scholar]
- 40.Rötzschke O, Falk K, Stevanovic S, Gnau V, Jung G, Rammensee H. Dominant aromatic/aliphatic C-terminal anchor in HLA-B2702 and B2705 peptide motifs. Immunogenetics. 1994;39:74–77. doi: 10.1007/BF00171803. [DOI] [PubMed] [Google Scholar]
- 41.Shu L L, Bean W J, Webster R G. Analysis of the evolution and variation of the human influenza A virus nucleoprotein gene from 1933 to 1990. J Virol. 1993;67:2723–2729. doi: 10.1128/jvi.67.5.2723-2729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutton J, Rowland-Jones S, Rosenberg W, Nixon D, Gotch F, Gao X M, Murray N, Spoonas A, Driscoll P, Smith M, Willis A, McMichael A. A sequence pattern for peptides presented to cytotoxic T lymphocytes by HLA B8 revealed by analysis of epitopes and eluted peptides. Eur J Immunol. 1993;23:447–453. doi: 10.1002/eji.1830230222. [DOI] [PubMed] [Google Scholar]
- 43.Tanigaki N, Fruci D, Vigneti E, Starace G, Rovero P, Londei M, Butler R H, Tosi R. The peptide binding specificity of HLA-B27 subtypes. Immunogenetics. 1994;40:192–198. doi: 10.1007/BF00167079. [DOI] [PubMed] [Google Scholar]
- 44.van de Griend R J, van Krimpen B A, Bol S J L, Thompson A, Bolhuis R L H. Rapid expansion of human cytotoxic T cell clones: growth promotion by a heat-labile serum component and various types of feeder cells. J Immunol Methods. 1984;66:285–298. doi: 10.1016/0022-1759(84)90340-5. [DOI] [PubMed] [Google Scholar]
- 45.Weiner A, Erickson A L, Kansopon J, Crawford K, Muchmore E, Hughes A L, Houghton M, Walker C M. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc Natl Acad Sci USA. 1995;92:2755–2759. doi: 10.1073/pnas.92.7.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yellen-Shaw A J, Wherry E J, Dubois G C, Eisenlohr L C. Point mutation flanking a CTL epitope ablates in vitro and in vivo recognition of a full-length viral protein. J Immunol. 1997;158:3227–3234. [PubMed] [Google Scholar]