Abstract
Loss-of-function variants in CCM1/KRIT1, CCM2/MGC4607, and CCM3/PDCD10 genes are identified in the vast majority of familial cases with multiple cerebral cavernous malformations. However, genomic DNA sequencing combined with large rearrangement screening fails to detect a pathogenic variant in 5% of the patients. We report a family with two affected members harboring multiple CCM lesions, one with severe hemorrhages and one asymptomatic. No causative variant was detected using DNA sequencing of the three CCM genes, CNV detection analysis, and RNA sequencing. However, a loss of heterozygosity in CCM2 was observed on cDNA sequences in one of the two affected members, which strongly suggested that this locus might be involved. Whole genome sequencing (WGS) identified a balanced structural variant on chromosome 7 with a breakpoint interrupting the CCM2 gene, preventing normal mRNA synthesis. These data underline the importance of WGS in undiagnosed patients with typical multiple CCM.
Subject terms: Genetic testing, Whole genome amplification
Introduction
Cerebral cavernous malformation (CCM - OMIM# 116860) are vascular malformations mostly located within the CNS that occur as a sporadic or familial, autosomal dominant, condition [1]. Familial cases present most often with multiple lesions and are due to loss of function variants in one of the 3 CCM genes: CCM1/KRIT1 (Krev Interaction Trapped 1) [* 604214], CCM2/MGC4607 (encoding a protein named malcavernin) [* 607929] or CCM3/PDCD10 (programmed cell death 10) [* 609118] [2]. Pathogenic variants are identified in one of the three CCM genes in over 95% of familial CCM cases [3, 4]. A small part of familial cases remains however without identified mutation. Here we report a family in whom no mutation was identified by standard technics. Whole-genome sequencing then identified a large genomic structural variant, involving the CCM2 gene.
Description of patients
Index Patient II-1: A 24-year-old woman was admitted because of a recent, subacute episode of gait unsteadiness associated with unusual headaches. Clinical examination disclosed central right facial palsy and cerebellar ataxia with left upper limb ataxia and dysarthria. Brain CT-scan and MRI disclosed a recent hemorrhage of the right mesencephalon associated with multiple other CCM lesions. She subsequently presented several episodes of recurrent hemorrhages of the same CCM lesion resulting in severe left hemiplegia associated with a marked anarthria. She died at the age of 49 from complications of being bedridden.
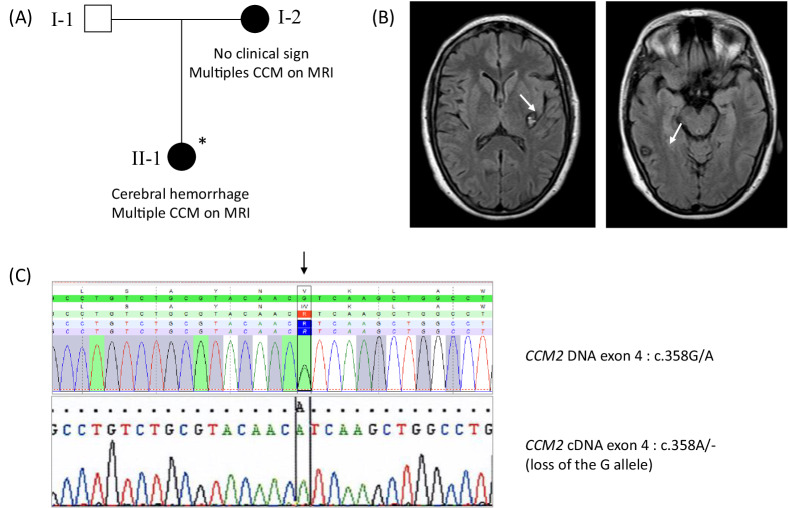
Her mother (I-2) was asymptomatic but cerebral MRI showed multiple CCM lesions (Fig. 1).
Fig. 1. Description of the patients and sequencing.
A Pedigree: Black symbols indicate individuals with CCM on cerebral MRI. B Cerebral MRI of patient I-2: axial FLAIR images showing right temporal cavernoma and another close to the left lenticular nucleus, the arrows indicate CCM lesions. C DNA and cDNA sequences for patient I-2: the SNP located in exon 4 (NM_031443.4(CCM2):c.358G > A - rs11552377) is absent in the cDNA sequences of a PCR product obtained with primers located in exon 2 (forward) and exon 5 (reverse) of CCM2.
Methods and results
Patient II-1 underwent sequencing of the three CCM genes and quantitative multiplex PCR of short fluorescent fragments (QMPSF) was performed to search for large rearrangements as described previously [5]. Sequencing of the cDNA was also performed and did not detect any anomaly. The same analyses were done in a second step for her mother. No mutation was detected but a loss of heterozygosity was observed on cDNA (rs11552377 located in exon 4 of CCM2 was present on genomic DNA sequences and not detected on cDNA sequences), suggesting the implication of CCM2 in the pathology of this family (Fig. 1).
Genome sequencing was conducted on AURAGEN platform for Patient I-2. Whole genome sequencing was performed following the recommendations of the France Genomic Medicine Plan (PFGM 2025). Whole blood extracted genomic DNA was sequenced according to standard procedures for a PCR-Free genome on a NovaSeq6000 instrument (Illumina). Sequencing data were aligned to the GRCh38p13 full assembly using bwa 0.7+. Variants were called by several algorithms including GATK4+, Bcftools1.10+, Manta1.6+, CNVnator0.4+, and annotated using the variant effect predictor. Detected variants were prioritized using in-house procedures. (Further details are available on request on www.auragen.fr.).
Genome sequencing identified a large rearrangement on the short arm of chromosome 7 at the CCM2 locus as suspected. Split reads sequences allowed to characterize the rearrangement that involved three breakpoints; the first one was located in the intron 2 of CCM2. The fragment of 267 kb bounded by the breakpoints 1 and 2 is inverted and inserted at the third breakpoint located 1.5 Mb further along chromosome 7 at 7q12.3. A sequence of 12 nucleotides is added at the first break point: seq[GRCh38] NC_000007.14:g.45043702_46521017delins[AGAAGGAAATTT;45310743_46521014;45043709_45310738inv] (Fig. 2). This structural variant leads to the interruption of intron 2 of CCM2 and prevents the synthesis of normal mRNA. This variant explains the presence of CCM lesions in the patients. No other gene is interrupted by this rearrangement.
Fig. 2. Characterization of the structural variant.
A: IGV visualization of short read sequencing showing the split reads at a breakpoint. B Location and schematic representation of the structural variant on chromosome 7; CCM2 gene is in black. C Agarose gel electrophoresis of PCR products obtained with primers located on both sides of the third breakpoint for both patients II-1 and I-2 (lanes 1 and 2), the father I-1 (lane 3), and a normal control (lane 4). D Sanger sequence was performed for patient II-1 at the third breakpoint (3rd BP PCR).
A specific PCR was designed with primers on either side of the third breakpoint and resulted in the amplification of a PCR product in both patients. Amplification was absent in the healthy father of patient II (I-1) and a normal control. (Fig. 2).
Discussion
Here, we report a novel structural variant affecting CCM2 ; this variant was identified by WGS in a patient for whom DNA sequencing of the three CCM genes, cDNA sequencing, and CNV detection were negative. Although cDNA sequence did not detect any variant or splicing anomaly, the loss of heterozygosity of an exonic SNP in the cDNA was a major indication of the involvement of the CCM2 gene that was confirmed by WGS. Very few cases of structural variants have been reported so far in CCM genes: one inversion of CCM2 exon 1 [6] and one interchromosomal insertion in CCM2 exon 6 [7]. The performance of WGS and the use of tools like Manta software are indicated for the detection of structural variants in negative screening patients, particularly in familial forms [8].
Inversions are a type of structural variants that are difficult to analyze owing to their balanced nature and the location of breakpoints, often within complex repeated regions. Introns 1 and 2 of CCM2 are large and contain numerous SINE and LINE repeats, which could favor rearrangements. Such an anomaly could explain the missing molecular diagnosis of CCM.
Acknowledgements
We thank the family for participating in this study. This research was made possible through access to the data generated by the France Genomic Medicine Plan 2025.
Author contributions
FR designed the work that led to the submission, acquired data, and played an important role in interpreting the results. AC, PL, and XA acquired data and revised the manuscript. TG acquired the data. NC played an important role in interpreting the results and revised the manuscript. ETL revised the manuscript.
Funding
No financial assistance was received in support of the study.
Data availability
The datasets generated during the current study are available from the corresponding author.
Competing interests
The authors declare no competing interests.
Ethical approval
Study participants or legal representatives gave written informed consent for clinical testing, research use and publication. The analyses were performed in accordance with French regulations and the principles of the Declaration of Helsinki.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Labauge P, Denier C, Bergametti F, Tournier-Lasserve E. Genetics of cavernous angiomas. Lancet Neurol. 2007;6:237–44. doi: 10.1016/S1474-4422(07)70053-4. [DOI] [PubMed] [Google Scholar]
- 2.Riant F, Bergametti F, Ayrignac X, Boulday G, Tournier-Lasserve E. Recent insights into cerebral cavernous malformations: the molecular genetics of CCM. FEBS J. 2010;277:1070–5. doi: 10.1111/j.1742-4658.2009.07535.x. [DOI] [PubMed] [Google Scholar]
- 3.Denier C, Labauge P, Bergametti F, Marchelli F, Riant F, Arnoult M, et al. Genotype–phenotype correlations in cerebral cavernous malformations patients. Ann Neurol. 2006;60:550–6. doi: 10.1002/ana.20947. [DOI] [PubMed] [Google Scholar]
- 4.Spiegler S, Najm J, Liu J, Gkalympoudis S, Schröder W, Borck G, et al. High mutation detection rates in cerebral cavernous malformation upon stringent inclusion criteria: one-third of probands are minors. Mol Genet Genom Med. 2014;2:176–85. doi: 10.1002/mgg3.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riant F, Cecillon M, Saugier-Veber P, Tournier-Lasserve E. CCM molecular screening in a diagnosis context: novel unclassified variants leading to abnormal splicing and importance of large deletions. Neurogenetics. 2013;14:133–41. doi: 10.1007/s10048-013-0362-0. [DOI] [PubMed] [Google Scholar]
- 6.Spiegler S, Rath M, Hoffjan S, Dammann P, Sure U, Pagenstecher A, et al. First large genomic inversion in familial cerebral cavernous malformation identified by whole genome sequencing. Neurogenetics. 2018;19:55–9. doi: 10.1007/s10048-017-0531-7. [DOI] [PubMed] [Google Scholar]
- 7.Pilz RA, Schwefel K, Weise A, Liehr T, Demmer P, Spuler A, et al. First interchromosomal insertion in a patient with cerebral and spinal cavernous malformations. Sci Rep. 2020;10:6306. doi: 10.1038/s41598-020-63337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang ZD, Du J, Lam H, Abyzov A, Urban AE, Snyder M, et al. Identification of genomic indels and structural variations using split reads. BMC Genomics. 2011;12:375. doi: 10.1186/1471-2164-12-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available from the corresponding author.