Abstract
Adverse early-life experiences (ELA) affect a majority of the world's children. Whereas the enduring impact of ELA on cognitive and emotional health is established, there are no tools to predict vulnerability to ELA consequences in an individual child. Epigenetic markers including peripheral-cell DNA-methylation profiles may encode ELA and provide predictive outcome markers, yet the interindividual variance of the human genome and rapid changes in DNA methylation in childhood pose significant challenges. Hoping to mitigate these challenges we examined the relation of several ELA dimensions to DNA methylation changes and outcome using a within-subject longitudinal design and a high methylation-change threshold.
DNA methylation was analyzed in buccal swab/saliva samples collected twice (neonatally and at 12 months) in 110 infants. We identified CpGs differentially methylated across time for each child and determined whether they associated with ELA indicators and executive function at age 5. We assessed sex differences and derived a sex-dependent ‘impact score’ based on sites that most contributed to methylation changes.
Changes in methylation between two samples of an individual child reflected age-related trends and correlated with executive function years later. Among tested ELA dimensions and life factors including income to needs ratios, maternal sensitivity, body mass index and infant sex, unpredictability of parental and household signals was the strongest predictor of executive function. In girls, high early-life unpredictability interacted with methylation changes to presage executive function. Thus, longitudinal, within-subject changes in methylation profiles may provide a signature of ELA and a potential predictive marker of individual outcome.
Keywords: Early-life stress, Methylomics, Executive control, Within-subject design, Stress, DNA methylation, Epigenetics, Adverse childhood experiences, Biomarkers, Precision medicine
Highlights
-
•
Changes in methylation profiles within an individual child can largely eliminate variance.
-
•
Alteration in these methylation profiles can predict outcomes in a manner dependent on the child's sex.
-
•
We suggest a "polyepigenomic score" based on our findings, which has the potential for clinical utility.
1. Introduction
Early-life experiences may exert a profound cumulative impact on lifespan trajectories of mental and physical health. Unsurprisingly, a robust body of work has focused on the contribution of salient early-life experiences, and especially of early-life adversity (ELA) to cognitive and mental health outcomes. In cohorts from diverse countries, socioeconomic levels and cultures, such studies have typically focused on the level of adversity, the cumulative impact of different types of adversity, and the distinct impacts of dimensions of adversity such as deprivation vs threat (Monk et al., 2016a; Callaghan and Tottenham, 2016; McLaughlin et al., 2019; Herzog and Schmahl, 2018; Luby et al., 2013, 2019; Gee et al., 2013; Palacios-Barrios and Hanson, 2019; Lebel et al., 2016; Sandman et al., 2015; Anda et al., 2006; Wade et al., 2019; Farah, 2018; Sheridan and McLaughlin, 2014). This strong literature supports the roles of ELA and its specific dimensions in predisposing individuals to physical, cognitive and mental health disorders (Moffitt et al., 2011; Richmond-Rakerd et al., 2021; Ursache and Noble, 2016). However, the predictive value of ELA to a vulnerability to mental and physical health problems applies well at the population level, yet is little better than chance for an individual child (Baldwin et al., 2021). Therefore, a significant unmet challenge remains in our ability to predict for an individual child whether they will be vulnerable or resilient to physical, cognitive or mental health problems.
Whereas trauma, poverty and abuse early in life significantly increase the risk of experiencing poorer cognition and mental health throughout the lifespan (Brown et al., 1995; Eriksson et al., 2014; Lupien et al., 2009/04; Chen and Baram, 2016; Novick et al., 2018; Raymond et al., 2018; Short and Baram, 2019), the significant amount of variance unaccounted for in child developmental outcomes has led to a search for additional potential sources of adversity that might have been missed. Such additional dimension of adversity, which explains some of the variance in cognitive and emotional outcomes, is unpredictability of early parental care behaviors and home environment. Initially detected in experimental animal models of early-life adversity (Molet et al., 2014, 2016; Rice et al., 2008), unpredictable sequences of parental care behaviors have emerged as an important potential predictor of susceptibility to later cognitive and emotional deficits (Groh et al., 2012). Specifically, in rodent models, resource scarcity elicited fragmented and unpredictable sequences of maternal care during a sensitive developmental period. In turn, these aberrant sensory signals to the developing brain influenced brain circuit maturation by altering selective microglial pruning of neuronal synapses (Bolton et al., 2022), leading to significant impairments of cognitive functions (Brunson et al., 2005; Ivy et al., 2010; Short et al., 2020) and reward behaviors (Molet et al., 2016; Bolton et al., 2018; Levis et al., 2021; Birnie et al., 2023). In human studies, this additional novel ELA was characterized initially by measuring unpredictable sequences of sensory signals that the caregiver transmits to the infant and child (such as speech, touch or visual cue). The concept was then extended to other proximate sources of signals to the developing brain, including also the household and environment. The multiple sources and timescales of unpredictability are measured using the Questionnaire on the Unpredictability of Childhood (QUIC) (Glynn et al., 2019; Lindert et al., 2022). The contribution of unpredictable signals from caregivers and home environment (“unpredictability”) to children's cognitive and emotional functions has now been established across diverse populations (Glynn et al., 2018, 2019, 2021; Davis et al., 2017, 2019a, 2022; Holmberg et al., 2022), and remains robust upon inclusion in the statistical models of established adverse experiences. Unpredictable parental and environmental signals to the infant impact the maturation of connectivity, measured using magnetic resonance imaging (Granger et al., 2021; Jirsaraie et al., 2023), and presage emotional problems also in adulthood (Spadoni et al., 2022).
Sex differences in ELA outcomes have been recognized: women endorsing depression, anxiety or addiction are more likely to report ELA compared with men (Capusan et al., 2021; Gershon et al., 2008; Hyman et al., 2006, 2008; Lansford et al., 2010; Marsh et al., 2018; Najavits et al., 1997; Peltier et al., 2019; Shand et al., 2011; Widom et al., 1995; Levis et al., 2022). However, whether or not such sex differences emerge prior to puberty and whether females are more vulnerable already in childhood has not been resolved, providing an impetus to examine the role of sex in the current studies (Hankin et al., 2016).
Predicting the impact of ELA on later life mental health involves three elements: the nature of the insult(s), an appropriate, universally applicable outcome, and an accessible reliable marker, detectable already early in life, that correlates robustly with the outcome measures. Here we assessed several ELA dimensions including unpredictability, and chose children's ability to regulate behavior and attention, effortful control, as an outcome measure, as it associates with a high level of executive function (Moffitt et al., 2011; Hankin et al., 2016; Caspi et al., 1998; White et al., 1994; Fergusson et al., 2013). In turn, executive function during childhood is one of the most robust predictors of cognitive and emotional outcomes and of success throughout life. Importantly, effortful control is highly influenced by early-life experiences (Birnie et al., 2023; Spadoni et al., 2022). We employed changes in DNA methylation as a potential marker of the impact of ELA, because such changes are known to be highly sensitive to environmental influences (Smith et al., 2020; Katrinli et al., 2022). The levels of methylation of specific DNA nucleotides and their changes throughout development have been a topic of extensive study (Horvath, 2013; Hannum et al., 2013; Horvath and Raj, 2018; Maegawa et al., 2010; McGill et al., 2022; Knight et al., 2016; Suarez et al., 2018; Jovanovic et al., 2017; Wikenius et al., 2019). DNA methylation patterns correlate with chronological age, providing ‘epigenetic’ or ‘DNA-methylation’ clocks (Horvath, 2013; Hannum et al., 2013; Knight et al., 2016). Adversity throughout life (Jovanovic et al., 2017; McEwen et al., 2020; Musci et al., 2023; Quach et al., 2017; Dammering et al., 2021), as well as mental and physical disease states (Danese and McEwen, 2012) seem to accelerate this ‘epigenetic age’ and even to predict the timing of death (Horvath and Raj, 2018; Musci et al., 2023).
However, the use of DNA methylation as a potential predictor of ELA impact on development has been challenging. Whereas at the population level, DNA methylation ‘signatures’ of adversity are apparent at both individual timepoints and across time (Suarez et al., 2018; Jovanovic et al., 2017; Dammering et al., 2021), their role as a predictive marker for an individual child has been limited by the high levels of inter-individual differences of the human genome and its DNA methylation patterns. In addition, methylation risk scores for the outcomes of ELA grapple with highly dynamic changes of DNA methylation early in life (∼4 fold more rapid than in adults) (Alisch et al., 2012) involving both augmented methylation and demethylation, depending on which CpG sites are examined (Wikenius et al., 2019; McEwen et al., 2020; Alisch et al., 2012). Thus, identifying DNA methylation patterns and changes that associate with ELA and predict outcome for a given child has continued to present a challenge, potentially requiring setting robust criteria for methylation changes, which allow detection of adversity effects beyond those of age (Knight et al., 2016; Suarez et al., 2018; McEwen et al., 2020; Dammering et al., 2021; Alisch et al., 2012).
Here we capitalized on our preclinical studies that had employed a within-subject design and a high-threshold criterion for DNA methylation changes, allowing distinguishing the effects of ELA from those of age (Jiang et al., 2019). In the current study we examined DNA methylation changes between two samples obtained from the same child (in the neonatal period and at one year of age) and assessed methylation changes, then correlated these changes with several dimensions of ELA as well as with executive function at age 5 years. We identify a novel set of DNA methylation changes that correlate with aspects of ELA and with outcome in a sex-specific manner and may thus be useful as a future predictive marker.
2. Material and methods
2.1. Participants
Study participants were 126 infants enrolled before birth, part of a longitudinal study evaluating the role of early experiences in cognitive and emotional development. All study procedures were approved by the Institutional Review Board for Protection of Human Subjects at Chapman University and the University of California-Irvine. Each mother provided written, informed consent for herself and her child. Demographic information for the cohort appears in Table 1.
Table 1.
Demographic characteristics of sample.
| F (N = 50) | M (N = 60) | |
|---|---|---|
| Race (n, %) | ||
| African American or Black | 0 (0) | 1 (Callaghan and Tottenham, 2016) |
| Asian | 5 (Luby et al., 2013) | 4 (Palacios-Barrios and Hanson, 2019) |
| Latinx | 22 (Glynn et al., 2018) | 27 (Granger et al., 2021) |
| Multi-Ethnic | 13 (Molet et al., 2014) | 12 (Eriksson et al., 2014) |
| Non-Hispanic White | 10 (Eriksson et al., 2014) | 16 (Molet et al., 2016) |
| Income Needs Ratio | ||
| IQR | 382 | 475.5 |
| Median | 244 | 252 |
| QUIC | ||
| IQR: | 2 | 2 |
| Median | 1 | 0.5 |
| Mean (±sd) | 1.45 (±1.85) | 1.32 (±1.87) |
| Maternal Sensitivity | ||
| IQR: | 1.25 | 1.25 |
| Median | 9.5 | 9.25 |
| Mean (±sd) | 9.35 (±0.85) | 9.28 (±0.99) |
| Maternal Depression | ||
| IQR: | 5.25 | 5.75 |
| Median | 5.17 | 4.5 |
| Mean (±sd) | 5.66 (±3.78) | 5.87 (±4.14) |
| Age of EC Assessment (months) | ||
| IQR | 10.75 | 11 |
| Median | 61.5 | 62 |
| Mean (±sd) | 62.31 (±5.80) | 62.48 (±6.22) |
| Effortful Control Score | ||
| IQR | 0.65 | 0.83 |
| Median | 5.08 | 5.20 |
| Mean (±sd) | 5.05 (±0.54) | 5.17 (±0.51) |
Notes: Ethnicity is per maternal report. IQR signifies interquartile range.
Note: Child race/ethnicity determined by parent report.
Five samples were removed because of low sequencing reads (<9 million), seven samples were excluded due to high variability in the number of sequenced sites, and four samples were removed after participants were diagnosed with significant learning impairments. Analysis was performed on remaining 110 samples (m = 60, f = 50)
2.2. Income to needs ratio
Family income-to-needs ratio (INR) was calculated by dividing total annual household income by the appropriate U.S. Census Bureau poverty threshold based on family size. According to those guidelines, 42.6 percent of the families were living below 200% (a ratio of 2.0) of the federal poverty line. These families living below 200% of the federal poverty line are considered low income and would, for example, qualify for the Supplemental Nutrition Assistance Program (SNAP). The average percent of households with children who were living in poverty (<200%) in the United States in 2022 was 37%, with New Hampshire having the lowest rates (20%) and Mississippi having the highest (51%). California ranks 27th with a statewide rate of 35% and thus, the rate in our sample is similar and representative. However, it is also worth noting that the participants in this study reside in Southern California, which is a relatively expensive area of residence in an expensive state, while the US census income-to-needs ratio standards described above are based on national living standards. Using metrics that adjust for the cost of living in the county of residence (Glasmeier; Foundation TAEC), a median income to needs ratio of 2.44 in this study cohort corresponds to 70% of the families living below the living wage level.
2.3. Maternal sensitivity
At 6 and 12-months postpartum, maternal sensitivity was evaluated using a coding scheme developed for the National Institute for Child Health and Development (NICHD) Study for Early Child Care and Youth Development (NICHD Early Care Research Network, 1999). This paradigm is an objective, behaviorally-based laboratory assessment tool for studying maternal behavior that is well-validated and predictive of the quality of mother-child attachment (NICHD Early Care Research Network, 1999; Brooks-Gunn et al., 2002). Mother-child pairs were videotaped in a semi-structured 10-min play session, in which mothers are given a standard set of age-appropriate toys and told to play with their infant as they would at home. Following the NICHD procedure (Egeland and Hiester, 1993), a composite rating of quality of maternal care is created by summing ratings of sensitivity to non-distress, maternal positive regard, and intrusiveness (reverse-coded). Twenty percent of the tapes were selected at random, without coder knowledge, and double-coded to obtain an index of inter-rater reliability, which averaged 88% across the two assessments. The correlation between 6 and 12 months was 0.36 and so they were combined to create a single composite measure, to provide a consistent measure of this dimension of maternal care throughout the first postnatal year. Separate analyses of the correlation of maternal sensitivity at 6 months or 12 months with effortful control are provided in the Supplemental Materials (Figs. S–3).
2.4. Maternal depressive symptoms
At 2, 6, and 12 months postpartum maternal depressive symptoms were measured with the 10-item Edinburgh Postnatal Depression Scale (EPDS) (Cox et al., 1987). Possible scores on this scale range from 0 to 30, with a score of 10 or more indicating probable minor depression and 13 or more likely major depression. In the present study, 38 percent of the mothers scored above the threshold for minor depressive episode and 18 percent scored in the probable major depressive range for at least one of the postpartum assessments. Correlation coefficient between maternal depression 2 months and 6 months: R = 0.66; 2 months and 12 months: R = 0.59; 6 months and 12 months: R = 0.57.
In view of these high correlations, the scores at the three time-points for each mother were averaged to provide a consistent measure of maternal depressive symptoms throughout the first postnatal year.
2.5. Unpredictability
Unpredictability of signals from the caretaker(s) has been identified by us as an important dimension of early life adversity (Glynn et al., 2019; Lindert et al., 2022; Davis et al., 2017, 2019a, 2022), and these findings have been confirmed and extended by others (Holmberg et al., 2022; Xu et al., 2023). Here, we assessed unpredictability of the early environment with the Questionnaire of Unpredictability in Childhood (QUIC (Glynn et al., 2019; Davis et al., 2019a; Davis and Glynn, 2023; Glynn et al., 2023);). The original self-report version of the QUIC is a 38-item questionnaire that assesses exposure to unpredictability in social, emotional and physical domains of a child's environment. It displays excellent psychometric properties (α = 0.89; test-retest reliability = 0.91) and is associated with observational measures of parental and household unpredictability. For the purposes of this study, the parent-report preschool version of the QUIC was employed (Glynn et al., 2023). This version predicts both behavioral observations and parent-report of child effortful control (Davis et al., 2019a; Davis and Glynn, 2023). Items and endorsement rates are included as Supplemental Tables S–1.
2.6. Child effortful control
Effortful control is widely considered an informative measure of executive function in children. Notably, in large prospective studies, effortful control in young children was an excellent predictor of school performance and of success later in life (Moffitt et al., 2011; Hankin et al., 2016; Caspi et al., 1998; White et al., 1994; Fergusson et al., 2013). Here, at roughly five years of age, effortful control measured with the Child Behavior Questionnaire (CBQ) (Rothbart et al., 2001) among the 90 participants who had attended the 5 year follow up visit. This the widely used and developmentally appropriate parent report instrument exhibits strong internal reliability and validity(Worobey and Blajda, 1989; Goldsmith and Campos, 1990; Gartstein and Rothbart, 2003); (Worobey and Blajda, 1989; Goldsmith and Campos, 1990; Gartstein and Rothbart, 2003), good stability over time (Putnam et al., 2006; Davis et al., 2019b), and consistency between parent report and observations in both the home and laboratory (Rothbart et al., 2000; Kochanska et al., 1996). Additionally bolstering its validity, this instrument was developed to reduce the possibility of reporting bias by asking about specific behaviors in defined situations, rather than asking about more global judgments about a child's temperament or behaviors.
2.7. Buccal swab collection
Infant DNA samples were collected via buccal swab from newborns (Mage = 2.6 weeks, SD = 0.92) and again at one year (Mage = 12.4 months, SD = 0.52) using the DNA Genotek Oragene Discover (DNA Genotek Cat# OGR-575) kit.
2.8. Isolation and quantification of DNA for making reduced representation bisulfite sequencing (RRBS) libraries from human buccal swab/saliva
The Buccal swab/saliva samples were incubated at 50 °C for 2 h. Next, 1/25 volume of prepIT-L2P (DNA Genotek Cat# PT-L2P-45) was added, samples were incubated on ice for 10 min and centrifuged at room temperature to collect the supernatant. Genomic DNA was prepared from this supernatant using the Quick gDNA kit (Zymo Research, Cat# D3025) following the manufacturer's protocol. The quantity of double-stranded DNA was analyzed using Qubit.
RRBS libraries were prepared from 200 ng of genomic DNA digested with MspI restriction enzyme and then extracted with ZR-DNA Clean & Concentrator™-5 kit (Zymo Research, Cat# D4014). According to Illumina's specified guidelines, fragments were ligated to pre-annealed adapters containing 5′-methylcytosine instead of cytosine (www.illumina.com). Adaptor-ligated fragments were then bisulfite-treated using the EZ DNA Methylation-Lightning™ Kit (Zymo Research, Cat# D5459). Preparative-scale PCR (16 cycles) was performed with Illumina index primers, and the resulting products were purified with DNA Clean & Concentrator for sequencing. Amplified RRBS libraries were quantified and qualified by Qubit, Bioanalyzer (Agilent), and Kapa library quant (Kapa systems, Cat# 07960140001) and then sequenced with paired-end 100 bp on the Illumina Nova-seq platform. Based on initial experiments, we chose a depth of 25 million for newborns and 50 million for one-year-olds to gain an average of 10 million mapped reads on the human genome for all samples. Samples were sequenced in batches and no batch control was employed.
2.9. RRBS processing and detection of differentially methylated sites (DMSs)
Adaptor and low-quality reads were trimmed and filtered using Trim Galore! 0.4.3 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/, RRID:SCR_011847) with the parameter “--fastqc –stringency 5–rrbs –length 30 –non_directional.” Reads were aligned to the human genome (hg38) by using Bismark 0.16.3 ((Putnam et al., 2006) RRID:SCR_005604) with “---non_directional” mode. CpG sites were called by “bismark_methylation_extractor” function from Bismark. Sites with coverage of more than 10 were accepted for further study. Differential methylation sites (DMSs) were first called using Methylkit (R 4.0.5) ((Akalin et al., 2012) RRID:SCR_005177) to identify sites with a minimum ±5% change (Jiang et al., 2019) between sample A (neonatal) and B (one year of age) using single CpG sites. We chose a 5% methylation change as such a change distinguished the influence of ELA in a preclinical study (Jiang et al., 2019), and because it is larger than is expected from changes related to age alone during the first year of life, when DNA methylation is rapidly changing (Wikenius et al., 2019; McEwen et al., 2020; Alisch et al., 2012). A site that was identified as having a minimum 5% change in methylation in any individual was then analyzed further as follows: The differential methylation test for individual i at site j generated a p-value is p_ij. Then we summarize all of the tests at site j by combining the p-values across the n individuals using the formula: . That combination statistic was considered the test statistic for site j and yields a site-specific p-value (Dai et al., 2014). A site that passed a Benjamini-Hochberg false discovery rate of q = 0.1 (leading to a p value cutoff = 0.00005) was considered a DMS (Dai et al., 2014). This approach generated 14,037 unique sites for further analysis. See supplemental methods figure SM-1.
Distance from Transcription Start Site (TSS) and Gene Ontology Analyses were performed using the Genomic Regions Enrichment of Annotations Tool (GREAT) (Mclean et al., 2010).
2.10. Calculation of DNA methylation level/percentage and delta methylation
The methylation percentage/level was calculated as the ratio of the methylated read counts over the sum of both methylated and unmethylated read counts for a single CpG site or across CpGs for a region. The delta methylation was calculated using the log2 transformation of the ratio of methylation level in the B sample (mB) and the methylation level in the A sample (mA), defined as log2 ((mB + 0.1)/(mA + 0.1)) (Jiang et al., 2019). The addition of 0.1 to the numerator and denominator addresses the possibility of zero methylation in one or both samples. Increased methylation in the B sample relative to the A sample is shown as a positive value, whereas decreased methylation in B is shown as a negative value.
2.11. Principal component analysis
From the above, we identified 14,037 DMS which we included for further analyses. Principal Component Analysis (PCA) using the prcomp (RRID:SCR_014676) function using R version 4.0.2. ((Wickham, 2016) RRID:SCR_001905) was used as a data reduction technique. PCA analyses were carried out for the A samples, the B samples and the changes in methylation (delta methylation values).
2.12. Impact score calculation
Impact scores were calculated using an adapted computational method (Joseph et al., 2018) for calculating polygenic risk scores. From the DMS set, for each individual site, the change in methylation, a binarized QUIC variable (QUICbin) and their interaction were used to predict effortful control at 5 years (EC ∼ “change in methylation by site” * QUICbin). We used a binarized QUIC variable because, although the version used here has 17 items, it was rare for individuals to endorse more than a few. Of the 90 subjects with QUIC and effortful control data, 45% had a raw score of 0, 23% had a raw score of 1, and 32% had a raw score of 2 or more (with values ranging from 2 to 8, reflecting more unpredictable childhoods). The choice to characterize 2 or more as “high” unpredictability provided a sufficient number of individuals to allow for comparison across the two groups.
In addition, because there are few observations larger than two in this sample, there is not a great deal of data on which to infer whether there would be a linear relationship between effortful control, methylomics and QUIC score. Finally, there is reason to suspect that over the range of the QUIC, one might expect a threshold effect in that larger values beyond some point may not indicate an additional degree of unpredictability.
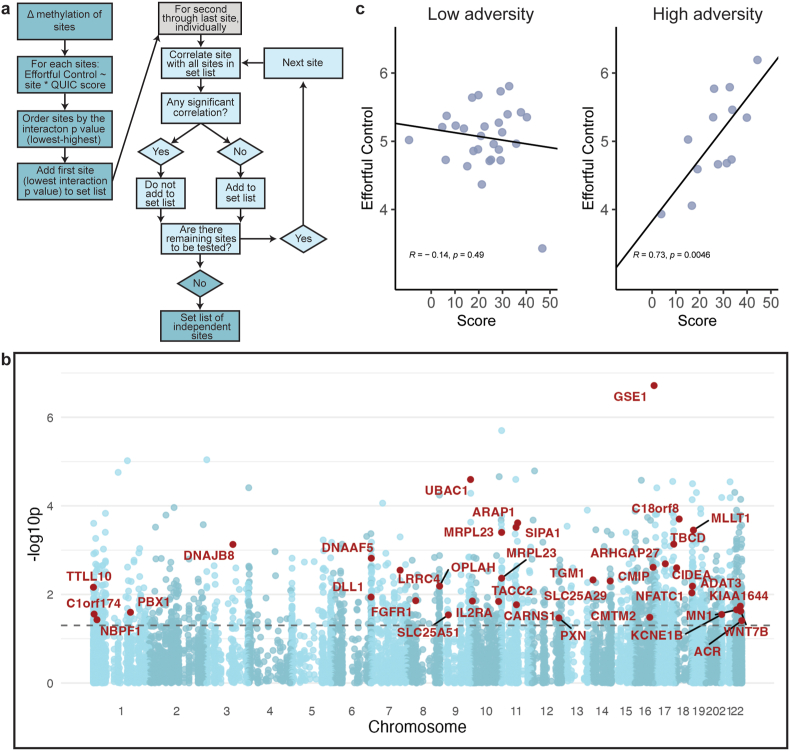
Sites were then ranked by the p value associated with the interaction coefficient and the top predictive site was added to the set list to be included in the model. The second most predictive site through the last site (using p < 0.05 as a criterion) were then considered sequentially, with each being correlated against sites already included in the set list. Any sites that were not significantly correlated with those already in the list were then added to the set list (Fig. 6A). Genes associated with the selected sites were identified using Genomics Regions Enrichment Annotations Tool (GREAT, RRID:SCR_005807) (Mclean et al., 2010), on the GRCh38 assembly to the single nearest gene within 1000 kb. Subject numbers: females = 41, males = 49. 13 ‘low’ females; 28 ‘high’. 16 ‘low’ males; 33 ‘high’.
Fig. 6.
Impact scores identify individuals vulnerable to poor outcomes following the unpredictability dimension of early-life adversity. A) Flow chart of the computation method used to select highly contributing sites. The clumping and thresholding method identifies sites that appear to have a significant interaction but do not correlate highly with other sites that have already been selected. B) Manhattan plot representing the differentially methylated sites (blue) and the distribution across the chromosomes and the corresponding significance score (-log10p) of each site interacting with QUIC to predict effortful control in females. Sites in red are those selected via the clumping and thresholding algorithm. Dotted line is at p = 0.05. C) The significant interaction of QUIC and risk score calculated from top sites according to our model predicts effortful control at 5 years in females who have experienced more adversity n: low = 28, high = 13. Line represents linear regression. R and p represent Pearson correlation for subgroup. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.13. Statistical analyses
All analysis were performed using R 4.0.2 in RStudio (RStudio Team, 2020). Sample preparation and analysis and quality control was performed ‘blind’. Correlations were calculated using Pearson correlation. A comparison of two group means was performed using Student's t-test. To implement the regression models with interactions, the QUIC scores were converted into binary numbers (QUICbin), with scores greater than one considered ‘high’ and scores of 0 or 1 considered ‘low’ for the reasons described above. Linear regression was performed (EC ∼ QUIC * change in methylation). Figures were made using ggplot2 (RRID:SCR_014601) in R 4.0.2 (Wickham, 2016). Heatmaps were created in R 4.0.2 using ComplexHeatmap ((Gu et al., 2016) RRID:SCR_017270) using row normalization.
3. Results
3.1. Methylation profiles distinguish neonatal samples from those obtained at one year of age
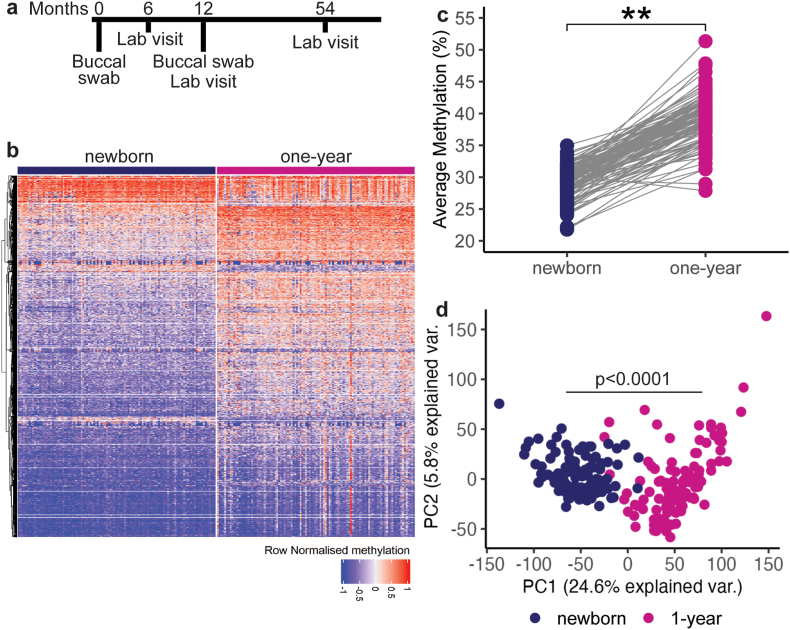
To determine how methylation changes may inform future outcomes, two buccal swabs/saliva samples were collected for each infant: the first during the first month of life and the second ∼ one year (Fig. 1A). In parallel, we collected demographic information on mothers and infants, including infant sex, birthweight, maternal report of infant ancestry, maternal body mass index pre- and postpartum depressive symptoms and income to needs ratio (INR). We also assessed maternal sensitivity and measures of unpredictability in the infant's environment (Table 1). In infants, we tested cognitive and emotional development during the first year of life and at five years (Fig. 1A).
Fig. 1.
Methylation is influenced by age. A) Timeline of sample collection and assessments in 110 infants. B) Heatmap depicting distinct patterns of methylation distinguishing DNA methylation profiles from newborn and 1-year old children. C) Average percentage methylation at selected sites increases with age. D) Using PCA, the first principal component, explaining 25% of the variance, accounts for the age of sample collection. **p < 0.001, bars represent mean, lines represent individual sites.
The analytic workflow of the DNA is depicted in the schematic in SI appendix (Fig. SM1). Briefly, after mapping each of the two samples from 110 infants to the human genome, we accepted samples with 10 million reads and a coverage of ≥10 reads, identifying 1,744,215 methylated sites per newborn- and 1,743,344 per one-year sample. We defined differentially methylated sites as those with methylation changes ≥±5% and significantly different between the two samples. We chose a 5% methylation change with the hope that such a change would distinguish the influence of ELA from that of age alone, as in our preclinical study (Jiang et al., 2019). A site that was endorsed as significantly methylated (see extended methods in the appendix) and that passed a Benjamini-Hochberg false discovery rate of q = 0.1 (leading to a p value cutoff = 0.00005) was considered a DMS (Dai et al., 2014). This approach generated 14,037 unique sites for further analysis.
DNA methylation was age dependent (Fig. 1B and C) (Hannum et al., 2013; Horvath and Raj, 2018; Wikenius et al., 2019; McEwen et al., 2020; Alisch et al., 2012). There was an average 10% increase in methylation levels for the differentially methylated sites between birth and one year (t (Riley et al., 2018) = 31.8, p < 0.001), consistent with described trends for increased methylation in CpG sites over the first postnatal year (Knight et al., 2016; Suarez et al., 2018; Jovanovic et al., 2017; Wikenius et al., 2019; Musci et al., 2023). Principal Component Analysis (PCA) of all samples revealed that the first principal component, accounting for 24.8% of variation in methylation levels, distinguished between the early and late samples (Fig. 1D), in accord with our preclinical study (Jiang et al., 2019) and prior reports (Wikenius et al., 2019). Notably, sex, maternal BMI or infant birthweight did not separate upon PCA analyses (SI appendix Tables S-2, S-3, S-4 and Figs. S–1).
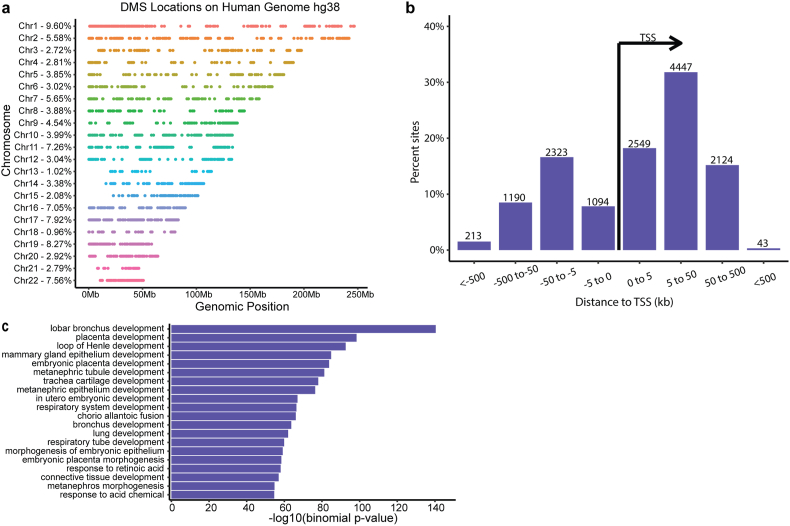
The 14,037 differentially methylated sites resided on all autosomal chromosomes (Fig. 2A). 13,983 localized to within 1000bp of a transcription start site (TSS; Fig. 2B), and associated with 2764 unique genes. Gene ontology (GO) analyses demonstrated a striking abundance of genes involved in development (Fig. 2C). In contrast, 14 sites, corresponding to 8/42 genes identified by (Wikenius et al., 2019) to distinguish 6 week-from 52 week-old Norwegian infants were differentially methylated in our cohort too. This finding is interesting in view of the homogenous ethnicity and SES status of the 214 Norwegian infants (Wikenius et al., 2019) versus the diverse ancestry and SES levels in our 110 subjects.
Fig. 2.
localization and gene ontology of the sites differentially methylated (DMS) between neonatal and one year old samples. (A). Chromosomal distribution of the DMS demonstrates that the reside on all autosomes. Numbers on the left denote the percentage of the overall DMS that localize to each chromosome. (B) Alignment of the DMS with genes and their structures: 13983 of the 14037 DMS localized to within 1000 kb of transcription a start site (TSS), and these 13983 DMS associated with 2764 unique genes. (C) Gene ontology identified developmental processes as the key theme of genes associated with DMS between 10-day old and one year old samples of the same child. n = 110 infants.
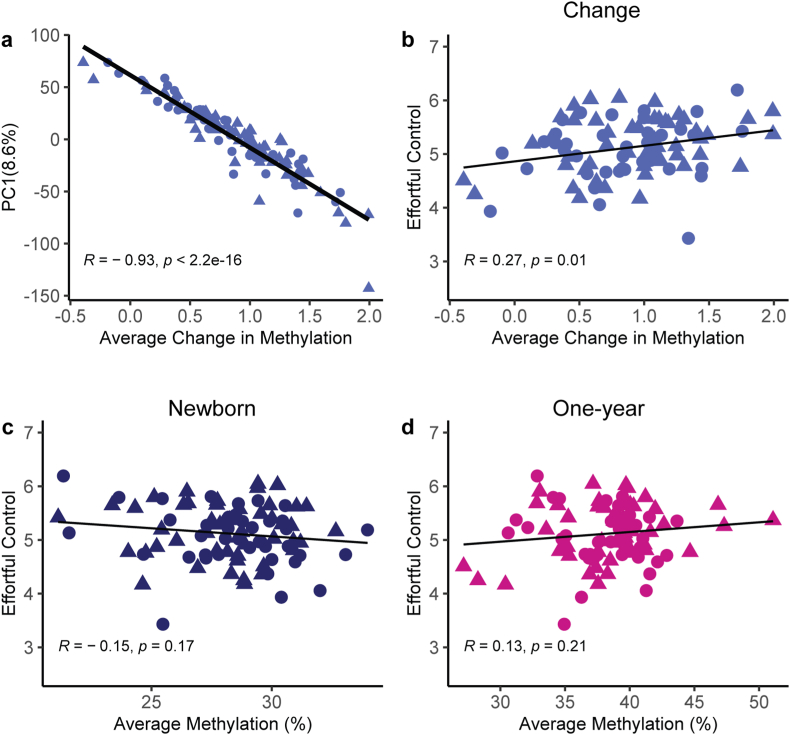
3.2. Methylation changes across time in individual infants, but not methylation at a single time-point, predict effortful control
PCA on the change in methylation (delta methylation) between samples A and B from the same infant showed that the first component (accounting for 8.6% of variance) correlated highly (R = −0.93, p < 2.2 × 10−16) with the average change in methylation (Fig. 3A). The first PCA component also represented the average of the methylation values when PCA was performed on samples A and B separately (R = 0.92, p < 1 × 10−6 and R = −0.81, p < 1 × 10−6 respectively). Therefore, further analyses used the average delta methylation values (for analyses involving both time points) and the average methylation values (for analyses involving a single time point).
Fig. 3.
Methylation-changes of individual infants between the ages of 10 days and one year predict effortful control at 5 years. A) The first component of the principal component analysis (PCA) of methylation changes in the ∼14,000 differentially methylated sites reflect the average change in methylation (n = 110). B) Average methylation changes from newborn to one year of age of an individual child predicts effortful control performance at five years of age (n = 90). C) Average percent methylation in newborns does not predict outcome. D) Similarly, average percent methylation at one year of age does not predict outcome. Note that analogous results were observed when using all 1.74 million methylated sites, as shown in the Supplemental Fig.S-3. Points represent individual samples, circles = females, triangles = males. Line represents linear regression. R represents Pearson correlation coefficient.
We determined the relationship of changes in methylation and effortful control at age five because effortful control is a reliable predictor of cognitive and emotional outcomes (Moffitt et al., 2011; Hankin et al., 2016; Caspi et al., 1998; White et al., 1994; Fergusson et al., 2013; Barry et al., 2022; Taylor et al., 2019; Johnson et al., 2022; Nigg, 2017). The average change in methylation of an individual child over the first year associated with the age 5 effortful control (R = 0.27, p = 0.01), accounting for ∼7.3% of variance in the task (Fig. 3B). In contrast, there was no significant association with effortful control of the average methylation in newborn (R = −0.15, p = 0.17) (Fig. 3C) or in one-year samples (R = 0.13, p = 0.21; Fig. 3D). Similarly, average methylation in all 1.7 million methylated CpGs in either samples A or B did not correlate with effortful control (R = -0.069, p = 0.52 and R = −0.067, p = 0.53 respectively). In contrast, the average delta methylation of all 1.7 million methylated sites between the two samples correlated with the 5-year effortful control, though more weakly than for the differentially methylated sites (R = 0.22, p = 0.036; SI appendix Figs. S–3). Together, these findings suggest that the change in—delta—methylation of an individual child over in the first year of life may provide a better indication of the impact of early-life experiences on methylation, vs. methylation levels at a single timepoint.
3.3. Unpredictability associates with executive function outcomes at age five years
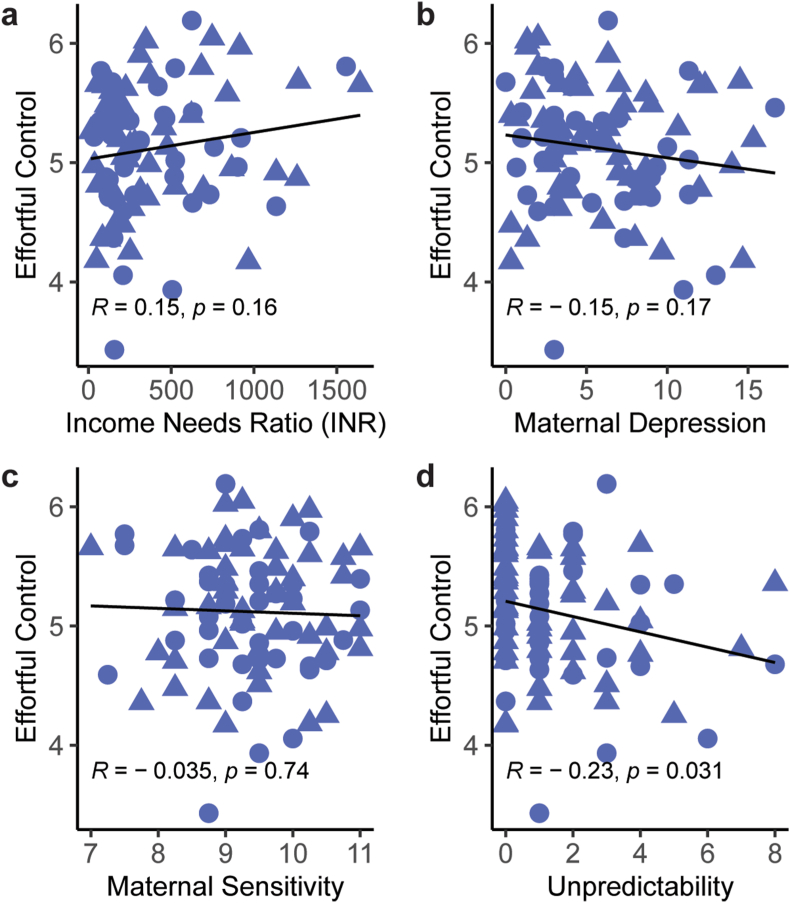
What dimension(s) of ELA predict effortful control? We investigated income-to-needs ratio (INR), maternal depressive symptoms, maternal sensitivity, and unpredictability of caregivers and environment. In our sample, there were weak correlations of INR (R = 0.15, p = 0.16) (Fig. 4A) and maternal depressive symptoms (R = −0.15, p = 0.13) with effortful control (Fig. 4B), and none for maternal sensitivity (R = −0.035, p = 0.15) (Fig. 3C). These results persisted when we analyzed maternal sensitivity separately at 6 and 12 months (R = 0.037. p = 0.73 [6 months]; R = −0.086, p = 0.45 [12 months]; Figs. S–2). The strongest correlation was with unpredictability (R = −0.23, p = 0.031) (Fig. 4D), such that high levels of unpredictability associated with lower effortful control, in accord with prior work (Gee et al., 2013; Holmberg et al., 2022). This suggests that unpredictability is a meaningful dimension of ELA and high levels of unpredictability portend poor effortful control at age five.
Fig. 4.
Unpredictability, assessed using the QUIC, portends functional outcomes at 5 years. A) Income/needs ratio (INR) has a weak association with effortful control in our sample of individuals. B) Correlation of maternal depressive symptoms with effort control suggests a weak negative association in these individuals. C) Measures of maternal sensitivity do not correlate with effortful control at 54 months of age in this sample. D) Unpredictability measured using the Questionnaire of Unpredictability in Childhood (QUIC) is inversely correlated with effortful control score at 54 months. Points represent individual samples, circles = females, triangles = males. Line represents linear regression (n = 90). R represents Pearson correlation coefficient.
3.4. Unpredictability during the first year of life alters the relation of DNA methylation and later outcomes in girls
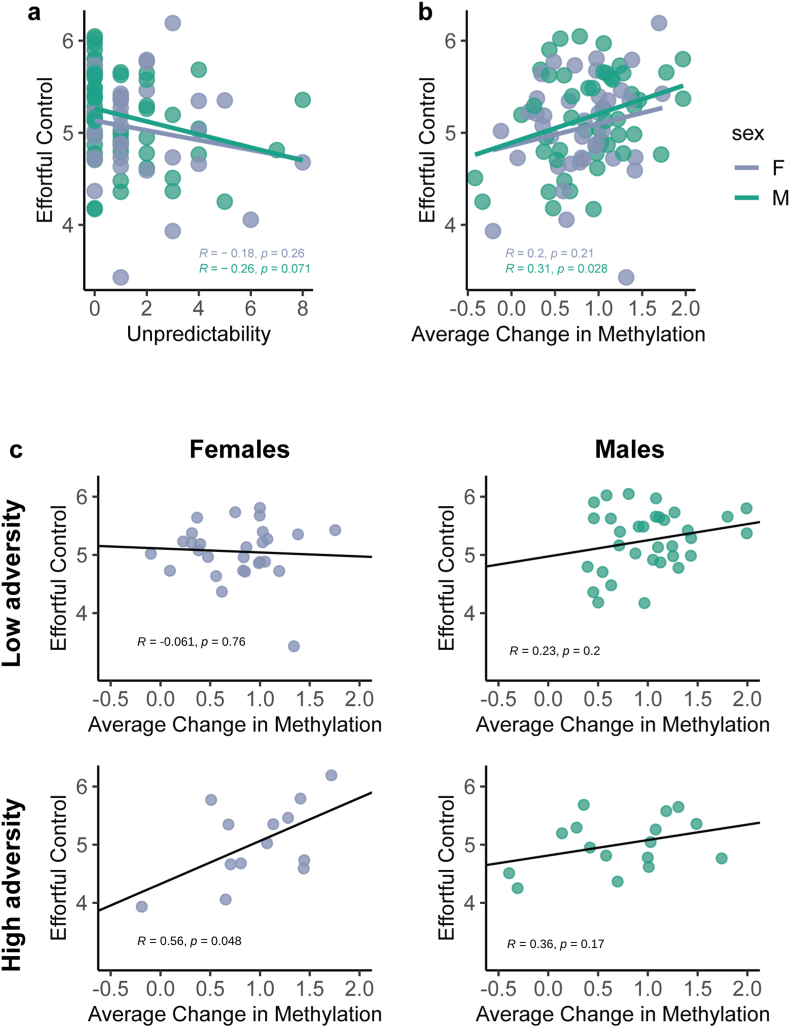
Only a subset of individuals experiencing ELA have negative outcomes and identifying individuals who are most at risk is vital to providing targeted interventions. Therefore, we probed the relationship between unpredictability, an ELA dimension that correlated with outcome, and differential DNA methylation. There was no direct correlation of the change in methylation between samples A and B with unpredictability (R = −0.07, p = 0.51) for the whole cohort. However, given the influence of sex on developmental trajectories (Riley et al., 2018; Brenhouse and Andersen, 2011), DNA methylation (Cisternas et al., 2020; Govender et al., 2022) and outcomes following ELA (Moffitt et al., 2011; McGill et al., 2022; Eisenberg et al., 2001; Chapple and Johnson, 2007), we analyzed the interaction of DNA methylation changes and ELA on effortful control, considering sex (Fig. 5A and B).
Fig. 5.
Unpredictability portends child development and may interact with methylation changes over time. A) Unpredictability assessed using the QUIC predicts effortful control at five years of age to a similar degree in both males and females. B) The change in methylation over the first year of life also predicts effortful control at the same age to the same degree in both sexes. n: females = 41, males = 49. C) There is an interaction between unpredictability and change in methylation in females only: for females who experience high unpredictability, the change in methylation over the first year of life predicts effortful control (please see Table 2 for descriptions of significance). In contrast, there is no such interaction observed in males. n: females low = 13, females high = 28, males low = 16, males high = 33. Points represent individual samples, purple = females, green = males. Line represents linear regression. F = females, M = Male. R and p represent Pearson correlation for subgroup. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We defined subjects as either experiencing high (QUIC >1) or low unpredictability (QUIC ≤1). We used a binary indicator (QUICbin) because of the 90 subjects with QUIC and effortful control data 61 (28 female) had a raw score of 0 or 1, and 29 (13 female) had a raw score ≥2. Binarization provided enough individuals for comparison across the two groups. A linear regression model including unpredictability, average delta methylation and the interaction was used to predict age 5 effortful control for each sex and for sexes combined (Table 2). In females, there was a significant interaction between methylation-change and unpredictability (p = 0.038), a main effect of unpredictability (p = 0.046) and an effect of average delta methylation (p = 0.016; Fig. 5C). These results suggest that high unpredictability during the first postnatal year may alter the degree to which changes in differentially methylated sites in females predict effortful control at 5 years. In males, there were no significant interactions or main effects among the tested parameters. Together, these observations imply that sex might influence how unpredictability interacts with changes in DNA methylation and this interaction may have a predictive value for risk in girls.
Table 2.
Regression models testing associations between methylation changes, unpredictability and effortful control at 5 years of age. n = 90 (41 females). CI = 95% confidence interval.
| Model | Predictors | Estimate | CI | p |
|---|---|---|---|---|
| Combined sexes | ||||
| (intercept) | 4.67 | 4.504,4.840 | <0.0001 | |
| mean change in methylation | 0.42 | 0.256,0.590 | 0.013 | |
| QUIC[low] | 0.34 | 0.113,0.562 | 0.14 | |
| Interaction | −0.25 | −0.474, −0.027 | 0.27 | |
| Girls | ||||
| (intercept) | 4.33 | 3.681,4.973 | <0.0001 | |
| mean change in methylation | 0.74 | 0.147,1.333 | 0.016 | |
| QUIC[low] | 0.78 | 0.015,1.553 | 0.046 | |
| Interaction | −0.81 | −1.570,0.048 | 0.038 | |
| Boys | ||||
| (intercept) | 4.81 | 4.621,5.001 | <0.0001 | |
| mean change in methylation | 0.27 | 0.059,0.473 | 0.21 | |
| QUIC[low] | 0.16 | −0.134,0.459 | 0.59 | |
| Interaction | −0.01 | −0.279,0.301 | 0.97 | |
3.5. Calculating an impact score as a predictive marker of individuals susceptible to early-life adversity
The association of delta DNA methylation and unpredictability with executive function at age five led us to consider an approach for identifying the potential contributions of specific sites (and their respective genes) to the overall relationship. Combined, such strong-effect sites might serve to construct predictive (“polyepigenic”) risk scores for vulnerability to adverse outcomes. We used the data from females because the interactions were present in female samples only. We adopted a polygenic risk score clumping and thresholding method (Joseph et al., 2018; Choi et al., 2020), ran a linear regression using each of the differentially methylated sites and the binarized QUIC score to predict effortful control, and assessed the significance of the interaction (Fig. 6A). The algorithm identified 37 ‘significant’ sites (p < 0.05; Fig. 6B–Table 3). By summing delta methylation between samples A and B at these 37 sites in females, we created an impact score. Analyzing the girls who had experienced greater and lesser degrees of unpredictability separately, the impact score predicted an individual girl's effortful control at age 5 in girls who had experienced greater unpredictability (score × QUICbin interaction; R2 = 0.20, p = 0.0016; Fig. 6C).
Table 3.
Top sites contributing to the impact score.
| Gene | Site | p value | R2 |
|---|---|---|---|
| GSE1 | chr16:85650857–85650858 | <0.0001 | 0.49 |
| UBAC1 | chr9:136017659-136017660 | <0.0001 | 0.34 |
| C18orf8 | chr18:23524804–23524805 | 0.00020 | 0.27 |
| ARAP1 | chr11:72754843–72754844 | 0.00024 | 0.27 |
| SIPA1 | chr11:65642488–65642489 | 0.00031 | 0.25 |
| MLLT1 | chr19:6270257–6270258 | 0.00035 | 0.28 |
| MRPL23 | chr11:1951274–1951275 | 0.00040 | 0.24 |
| TBCD | chr17:82795360–82795361 | 0.00073 | 0.22 |
| DNAJB8 | chr3:128421485-128421486 | 0.00074 | 0.25 |
| DNAAF5 | chr7:779363-779364 | 0.0015 | 0.18 |
| ARHGAP27 | chr17:45405991–45405992 | 0.0020 | 0.18 |
| CMIP | chr16:81431947–81431948 | 0.0024 | 0.17 |
| CIDEA | chr18:12278873–12278874 | 0.0025 | 0.16 |
| LRRC4 | chr7:128030495-128030496 | 0.0028 | 0.16 |
| MRPL23 | chr11:1951752–1951753 | 0.0043 | 0.20 |
| TGM1 | chr14:24259744–24259745 | 0.0047 | 0.13 |
| SLC25A29 | chr14:100297661–100297662 | 0.0050 | 0.13 |
| OPLAH | chr8:144052079-144052080 | 0.0049 | 0.12 |
| ADAT3 | chr19:1912103–1912104 | 0.0065 | 0.12 |
| TTLL10 | chr1:1145182-1145183 | 0.0069 | 0.15 |
| NFATC1 | chr18:79485573–79485574 | 0.0092 | 0.12 |
| DLL1 | chr6:170227779-170227780 | 0.012 | 0.11 |
| FGFR1 | chr8:38554659-38554660 | 0.014 | 0.10 |
| IL2RA | chr10:6036875–6036876 | 0.014 | 0.09 |
| TACC2 | chr10:122085904–122085905 | 0.014 | 0.11 |
| CARNS1 | chr11:67411863–67411864 | 0.017 | 0.14 |
| KIAA1644 | chr22:44286585–44286586 | 0.018 | 0.07 |
| MN1 | chr22:27797185–27797186 | 0.022 | 0.14 |
| WNT7B | chr22:45972341–45972342 | 0.039 | 0.10 |
| PBX | chr1:164576643-164576644 | 0.025 | 0.12 |
| C1orf174 | chr1:4161521-4161522 | 0.028 | 0.10 |
| KCNE1B | chr21:8434920–8434921 | 0.028 | 0.10 |
| SLC25A51 | chr9:37938671-37938672 | 0.029 | 0.08 |
| CMTM2 | chr16:66579316–66579317 | 0.033 | 0.05 |
| PXN | chr12:120263326–120263327 | 0.034 | 0.09 |
| NBPF1 | chr1:16725298-16725299 | 0.037 | 0.18 |
| ACR | chr22:50730818–50730819 | 0.039 | 0.09 |
The 37 DMS comprising the impact score belonged to 36 genes. GO- and gene network analyses revealed no significantly enriched terms. In addition, comparing these sites to sites implicated in the Horvath (Horvath and Raj, 2018) and Pediatric epigenetic clocks (McEwen et al., 2020) indicated that these DMS are unique rather than reflecting a biological clock.
4. Discussion
The principal findings presented here are: 1) The use of a longitudinal, within-subject approach identifies changes in methylation over the first postnatal year as a feasible tool which may allow better prediction of later-life executive function versus methylation profiles at a single timepoint. Other important analysis variables include the timing of the adversity and the different analytic approaches employed. 2) Exposures to a higher degree of the unpredictability ELA dimension in early life correlate with poorer effortful control. 3) The interaction of this ELA with changes in methylation is sex-dependent: In girls in our cohort, unpredictability interacts with change in methylation to presage effortful control, suggesting that it may alter the relationship of DNA methylation and subsequent outcomes. 4) A tentative impact score, created using the change in methylation in girls, may provide a predictive marker of the influence of high levels of early-life unpredictability on the future outcome of an individual child. This score should be validated in future studies as a potential indicator of risk.
Identifying individuals with a high risk of developing cognitive and emotional problems after sustaining pre-or early postnatal adversity is important: such a discovery will allow targeting preventative and interventional strategies to those who need them most. Indeed, a number of investigative groups and consortia have aimed to employ DNA methylation profiles of blood or buccal swab cells of infants and children as a correlate of ELA and a predictor of the subsequent outcomes (Suarez et al., 2018; Jovanovic et al., 2017; McEwen et al., 2020; Musci et al., 2023; Dammering et al., 2021; Houtepen et al., 2016; Martins et al., 2021; Maddox et al., 2018).
DNA methylation levels vary with age, and normative patterns and rates of these changes have been established, providing epigenetic, DNA methylation (DNAm) clocks (Horvath, 2013; McEwen et al., 2020). Deviations from this ‘clock’, and especially acceleration of DNAm vs chronological ages, have been considered predictive of aging and disease (Hannum et al., 2013). The rate of DNA methylation change is rapid early in life, and a body of work has focused on creating and harmonizing DNAm clocks that are optimal for infants and children (McGill et al., 2022; Knight et al., 2016; Wikenius et al., 2019; McEwen et al., 2020; Musci et al., 2023; Dammering et al., 2021) and on the influence of ELA on modulating pediatric epigenetic clocks (Suarez et al., 2018; Jovanovic et al., 2017; McEwen et al., 2020; Dammering et al., 2021). Additional clocks have been identified for specific early-life epochs including the use of cord blood to assess gestational age (Knight et al., 2016) and buccal swabs to probe the first year of life (Wikenius et al., 2019). More recently a comparison of seven pediatric clocks highlighted the heterogeneity of sites identified across studies: of 2587 CpGs, 2206 (>80%) were specific to a single clock (Fang et al., 2023).
In agreement with previous work (Pérez et al., 2019; Urdinguio et al., 2016; Wang et al., 2012; Martino et al., 2011), we find that the changes in overall average methylation between one-year old and neonatal samples is largely positive (higher methylation). Methylation changes over time during childhood are bi-directional (Wikenius et al., 2019; McEwen et al., 2020; Alisch et al., 2012). Wilkenius et al., found increased methylation in 36 of 42 sites that change significantly during the first postnatal year, in accord with the current study (Wikenius et al., 2019). They considered this augmented methylation surprising because higher methylation tends to predict reduced gene expression, and speculated that the putative reduction in gene expression during the first year of life might be compensatory to explosive gene expression in utero (Wikenius et al., 2019). It is unclear whether or not the overall increase in methylation observed in the current study is beneficial.
The majority of studies to date have employed profiles of DNA methylation in samples obtained at a single time point. While many groups have focused on postnatal epoch of adversity (e.g., 74,125,126,146–147), important work has aimed to probe the impact of prenatal stress on neuropsychiatric outcomes (e.g., 149,150) and the timing of adversity may differentially influence outcomes. Dunn et al. (2019) and Lussier et al. (2023) aimed to identify sensitive periods using longitudinal approaches and focusing specifically on the timing and dimension of ELA in influencing methylation profiles. They identified ages 3–5 as a putative postnatal sensitive period for methylation effects of ELA, yet they did not assess ELA prenatally or during the first postnatal year. The current longitudinal study suggests that the first year of life may also be an important sensitive period for the effects of certain ELA dimensions (e.g., unpredictability) on DNA methylation and neurodevelopmental outcomes.
We used a longitudinal, or ‘within subject’ approach (Jiang et al., 2019; van der Wal et al., 2020; Bjornsson et al., 2008; Rutten et al., 2018; Snijders et al., 2020) and sampled infants at a relatively short interval--one year--which mitigated the potential dilution of an epigenomic ‘signature’ of ELA by subsequent life events This approach also minimized large variances in methylation among individuals. We examined for the well-established effects of age on methylation profiles (Suarez et al., 2018; Jovanovic et al., 2017; Jiang et al., 2019; Dunn et al., 2019; van der Wal et al., 2020; Czamara et al., 2021; Johansson et al., 2013). We found that the change in methylation profile of an individual child between the first month of life and one year of age was superior at predicting neurodevelopment at five years compared with a methylation profile derived from a single timepoint.
We quantified several ELA types during the interval year between the two samples and their relative contribution to changes in DNA methylation and effortful control at age five. In addition to poverty (income-to-needs ratio), and maternal depressive symptoms and sensitivity, we tested the unpredictable signals from the caretaker and household. This ELA dimension of adversity has emerged in our work (Glynn et al., 2018, 2019, 2021; Davis et al., 2017, 2019a, 2022; Granger et al., 2021; Spadoni et al., 2022; Noroña-Zhou et al., 2020; Glynn and Baram, 2019) and independently, in work by others (Holmberg et al., 2022; Xu et al., 2023; Szepsenwol et al., 2022; McGinnis et al., 2022; Doom et al., 2016) as a contributor to cognitive outcomes including effortful control (Davis et al., 2017, 2019a, 2022; Xu et al., 2023; Howland et al., 2021) and emotional (Davis et al., 2019a; Glynn et al., 2018; Glynn and Baram, 2019; Doom et al., 2016; Gillespie and Rao, 2022) outcomes in children, adolescents and adults. The neurobiological basis for the detrimental effects of unpredictable environmental signals on brain development are not fully understood. In both humans and experimental models, sensory input from the environment (e.g., light patterns, patterns of tones) are required for appropriate maturation of the respective brain circuits. In experimental models, unpredictable patterns of sensory inputs may impact brain circuit maturation by disrupting the selective microglial pruning of synapses (Bolton et al., 2022; Birnie and Baram, 1979).
Here, both lesser changes in methylation during the first year of life and high levels of maternal unpredictability predicted poorer effortful control at age five. In addition, sex-dependent interactions of methylation changes and unpredictability were observed in girls. The discovery of sex effects of ELA on methylation profiles and outcome already prior to puberty is intriguing. In adults, a more rapid epigenetic ageing has been reported in women (Horvath et al., 2016; Simpkin et al., 2016), and a more rapid epigenetic ageing in adolescent girls (but not boys) with a history of ELA (Tang et al., 2020). Greater epigenetic ageing in girls than in boys experiencing ELA has been reported (Dammering et al., 2021). In contrast, looking at the effect of prenatal exposure to maternal depression on DNA-methylation age of newborns, Suarez et al., identified an effect in boys but not girls (Suarez et al., 2018), and a similar male vulnerability was observed by McGill et al., for maternal anxiety (McGill et al., 2022), and by work from our group (Sandman et al., 2013). Studies of selective vulnerabilities of males to prenatal stress are buttressed by work in experimental models demonstrating similar male vulnerability (Bale, 2016). For postnatal stress, both Dammering et al. (2021) and our own studies (Davis and Glynn, 2023) suggest greater vulnerability in girls. Together, the combined body of work suggests that sex effects can be detected prior to puberty, and that different types of adversity and its timing, i.e., the developmental age in which adversity takes place, may influence which sex is more affected.
We used reduced representation bisulfite sequencing (RRBS), whereas others have employed bisulfite conversion and genomic DNA methylation profiling using the Illumina HumanMethylation450 BeadChip which assesses DNA methylation levels at >480,000 CpG sites (Rutten et al., 2018; Snijders et al., 2020; Czamara et al., 2021; Johansson et al., 2013). Still others used targeted sequencing of specific sites (Bjornsson et al., 2008). All these methods have assets and limitations: RRBS only samples ∼5% of the genome, but includes ∼95% of gene-related CpG sites. In our hands, it uncovered methylation at ∼1.74 million CpGs, well in the range of the Illumina chip approach. In contrast, the use of methylation panels allows sequencing assessment of predefined sites, including known, stress-dependent genes (Non et al., 2016; Monk et al., 2016b) yet does not allow discovery of novel sites as markers. Hence, we believe RRBS provides a compromise between targeted sequencing and a whole genome approach.
The discovery of a robust association of unpredictability, changes in methylation and effortful control in girls led us to probe whether specific differentially methylated sites could be used to create an individually predictive impact score. Whereas risk scores typically require cohorts with a minimum of 100 subjects (Choi et al., 2020), these size recommendations are based on the use of each subject as a unitary entity within a population. Here, the ‘delta methylation’ approach compares each individual to themselves, reducing the effect of population variance on diluting effect sizes.
Previous studies examining biomarkers of childhood adversity have yielded mixed results, emphasizing the need for considering and aligning predictive algorithms (Non et al., 2016; Rubens et al., 2023; Monk et al., 2016b; Conradt et al., 2018). A comprehensive systematic review of the association of DNA methylation with ELA describes the top 11 genes featured in the literature (Rubens et al., 2023). However, none of these genes were identified in our predictive impact score, as the included studies often fail to consider adversity subtypes or timing. Studies by Dunn and Lussier employ a similar predictive model to that developed in our work. Their model evaluates the impact of the recency, accumulation, and timing of multiple adversities on the variance in DNA methylation at a given time point. In contrast, our study design investigates how unpredictability, as a form of adversity, influences changes in methylation over a year to predict outcomes. Comparing the key methylation sites reported by Dunn et al. and the updated analysis by Lussier et al. with those identified in our study reveals no overlap in genes with altered methylation. This underscores the necessity for targeted biomarkers capable of accounting for the specific impact of different dimensions of ELA and individual variance.
There are several limitations to our study, the primary being the cohort size. Epigenetic and genetic studies often include tens-hundred thousand subjects, providing power that our cohort (Govender et al., 2022) does not permit. In addition, parsing the group by sex further reduces sample size, with a risk of overfitting. We acknowledge this issue and note that studies of similar size can provide important and innovative information (Jovanovic et al., 2017). In addition, we aimed to address the cohort size in part with the use of a longitudinal within-subject design, enabling assessment of DNA changes within an individual over time rather than a cross section comparison of different groups, which is more sensitive to random effects and overfitting in small samples. Capitalizing on this design, we attempt to generate a polyepigenetic impact score, and note that this score has yet to be validated because the size of the current cohort did not permit splitting into training and testing subsets (Choi et al., 2020). Thus, validation of the current impact score requires larger naïve datasets. Nevertheless, we suggest that the technologies and approaches presented here provide valuable insights into the potential of using the interaction of ELA dimensions and methylation changes across defined epochs as potential indicators of the impact (‘epigenetic scar’) of adversity on an individual child, with significant predictive promise.
5. Data sharing Statement
Data sharing will comply with National Institute of Health guidelines. Epigenomic datasets will be submitted to the appropriate databanks and will be made available on request.
CRediT authorship contribution statement
Annabel K. Short: Writing – original draft, Methodology, Investigation, Conceptualization. Ryan Weber: Writing – review & editing, Methodology, Formal analysis, Data curation. Noriko Kamei: Investigation, Data curation. Christina Wilcox Thai: Investigation, Formal analysis, Data curation. Hina Arora: Writing – review & editing, Formal analysis, Data curation. Ali Mortazavi: Writing – review & editing, Supervision, Methodology, Formal analysis, Data curation. Hal S. Stern: Writing – review & editing, Validation, Supervision, Formal analysis, Data curation. Laura Glynn: Writing – review & editing, Writing – original draft, Supervision, Methodology, Investigation, Formal analysis, Data curation. Tallie Z. Baram: Writing – review & editing, Writing – original draft, Supervision, Project administration, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Supported by NIH NIMH P50 MH096889, a Precision Medicine Initiative grant from the State of California (OPR20141), Bren Foundation, and a generous gift from Syntropy Technologies LLC. The authors thank the families who participated in this project and our dedicated staff.
Handling editor: Prof R Lawrence Reagan
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2024.100652.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alisch R.S., Barwick B.G., Chopra P., Myrick L.K., Satten G.A., Conneely K.N., et al. Age-associated DNA methylation in pediatric populations. Genome Res. 2012;22(4):623–632. doi: 10.1101/gr.125187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anda R.F., Felitti V.J., Bremner J.D., Walker J.D., Whitfield Ch, Perry B.D., et al. The enduring effects of abuse and related adverse experiences in childhood. Eur Arch Psychiatry Clin Neurosci. 2006;256(3):174–186. doi: 10.1007/s00406-005-0624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin J.R., Caspi A., Meehan A.J., Ambler A., Arseneault L., Fisher H.L., et al. Population vs individual prediction of poor health from results of adverse childhood experiences Screening. JAMA Pediatr. 2021;175(4):385. doi: 10.1001/jamapediatrics.2020.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale T.L. The placenta and neurodevelopment: sex differences in prenatal vulnerability. Dialogues Clin. Neurosci. 2016;18(4):459–464. doi: 10.31887/DCNS.2016.18.4/tbale. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry K.R., Hanson J.L., Calma‐Birling D., Lansford J.E., Bates J.E., Dodge K.A. Developmental connections between socioeconomic status, self‐regulation, and adult externalizing problems. Dev. Sci. 2022;25(6) doi: 10.1111/desc.13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnie M.T., Baram T.Z. Principles of emotional brain circuit maturation. Science. 1979;376(6597):1055–1056. doi: 10.1126/science.abn4016. 2022 Jun 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnie M.T., Short A.K., de Carvalho G.B., Taniguchi L., Gunn B.G., Pham A.L., et al. Stress-induced plasticity of a CRH/GABA projection disrupts reward behaviors in mice. Nat. Commun. 2023;14(1):1088. doi: 10.1038/s41467-023-36780-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson H.T., Sigurdsson M.I., Fallin M.D., Irizarry R.A., Aspelund T., Cui H., et al. Intra-individual change over time in DNA methylation with familial Clustering. JAMA. 2008;299(24):2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton J.L., Molet J., Regev L., Chen Y., Rismanchi N., Haddad E., et al. Anhedonia following early-life adversity involves aberrant interaction of reward and anxiety circuits and is Reversed by partial Silencing of amygdala Corticotropin-Releasing Hormone gene. Biol Psychiatry. 2018;83(2):137–147. doi: 10.1016/j.biopsych.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton J.L., Short A.K., Othy S., Kooiker C.L., Shao M., Gunn B.G., et al. Early stress-induced impaired microglial pruning of excitatory synapses on immature CRH-expressing neurons provokes aberrant adult stress responses. Cell Rep. 2022;38(13) doi: 10.1016/j.celrep.2022.110600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenhouse H.C., Andersen S.L. Developmental trajectories during adolescence in males and females: a cross-species understanding of underlying brain changes. Neurosci. Biobehav. Rev. 2011;35(8):1687–1703. doi: 10.1016/j.neubiorev.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks-Gunn J., Han W.J., Waldfogel J. Maternal Employment and child cognitive outcomes in the first three Years of life: the NICHD study of early child care. Child Dev. 2002;73(4):1052–1072. doi: 10.1111/1467-8624.00457. [DOI] [PubMed] [Google Scholar]
- Brown A.S., Susser E.S., Lin S.P., Neugebauer R., Gorman J.M. Increased risk of affective disorders in males after second Trimester prenatal exposure to the Dutch Hunger Winter of 1944–45. Br. J. Psychiatry. 1995;166(5):601–606. doi: 10.1192/bjp.166.5.601. [DOI] [PubMed] [Google Scholar]
- Brunson K.L., Kramár E., Lin B., Chen Y., Colgin L.L., Yanagihara T.K., et al. Mechanisms of late-onset cognitive decline after early-life stress. J. Neurosci. 2005;25(41):9328–9338. doi: 10.1523/JNEUROSCI.2281-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan B.L., Tottenham N. The Neuro-environmental Loop of plasticity: a cross-species analysis of parental effects on emotion Circuitry development following typical and adverse caregiving. Neuropsychopharmacology. 2016;41(1):163–176. doi: 10.1038/npp.2015.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capusan A.J., Gustafsson P.A., Kuja-Halkola R., Igelström K., Mayo L.M., Heilig M. Re-examining the link between childhood maltreatment and substance use disorder: a prospective, genetically informative study. Mol Psychiatry. 2021;26(7):3201–3209. doi: 10.1038/s41380-021-01071-8. [DOI] [PubMed] [Google Scholar]
- Caspi A., Moffitt T.E., Entner Wright B.R., Suva P.A. Early failure in the labor market: childhood and adolescent predictors of unemployment in the transition to adulthood. Am Sociol Rev. 1998;63(3):424–451. [Google Scholar]
- Chapple C.L., Johnson K.A. Gender differences in impulsivity. Youth Violence Juv. Justice. 2007;5(3):221–234. [Google Scholar]
- Chen Y., Baram T.Z. Toward understanding how early-life stress reprograms cognitive and emotional brain networks. Neuropsychopharmacology. 2016;41(1):197–206. doi: 10.1038/npp.2015.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.W., Mak T.S.H., O'Reilly P.F. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 2020;15(9):2759–2772. doi: 10.1038/s41596-020-0353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisternas C.D., Cortes L.R., Bruggeman E.C., Yao B., Forger N.G. Developmental changes and sex differences in DNA methylation and demethylation in hypothalamic regions of the mouse brain. Epigenetics. 2020;15(1–2):72–84. doi: 10.1080/15592294.2019.1649528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt E., Adkins D.E., Crowell S.E., Monk C., Kobor M.S. An epigenetic pathway approach to investigating associations between prenatal exposure to maternal mood disorder and newborn neurobehavior. Dev. Psychopathol. 2018;30(3):881–890. doi: 10.1017/S0954579418000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.L., Holden J.M., Sagovsky R. Detection of postnatal depression. Br. J. Psychiatry. 1987;150(6):782–786. doi: 10.1192/bjp.150.6.782. [DOI] [PubMed] [Google Scholar]
- Czamara D., Tissink E., Tuhkanen J., Martins J., Awaloff Y., Drake A.J., et al. Combined effects of genotype and childhood adversity shape variability of DNA methylation across age. Transl. Psychiatry. 2021;11(1):88. doi: 10.1038/s41398-020-01147-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H., Leeder J.S., Cui Y. A modified generalized Fisher method for combining probabilities from dependent tests. Front. Genet. 2014;5(FEB):32. doi: 10.3389/fgene.2014.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammering F., Martins J., Dittrich K., Czamara D., Rex-Haffner M., Overfeld J., et al. The pediatric buccal epigenetic clock identifies significant ageing acceleration in children with internalizing disorder and maltreatment exposure. Neurobiol Stress. 2021;15 doi: 10.1016/j.ynstr.2021.100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese A., McEwen B.S. Adverse childhood experiences, allostasis, allostatic load, and age-related disease. Physiol. Behav. 2012;106(1):29–39. doi: 10.1016/j.physbeh.2011.08.019. [DOI] [PubMed] [Google Scholar]
- Davis E.P., Glynn L. The power of Predictability: patterns of signals in early life shape neurodevelopment and mental health trajectories. JCPP (J. Child Psychol. Psychiatry) 2023 doi: 10.1111/jcpp.13958. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E.P., Stout S.A., Molet J., Vegetabile B., Glynn L.M., Sandman C.A., et al. Exposure to unpredictable maternal sensory signals influences cognitive development across species. Proc. Natl. Acad. Sci. USA. 2017;114(39):10390–10395. doi: 10.1073/pnas.1703444114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E.P., Korja R., Karlsson L., Glynn L.M., Sandman C.A., Vegetabile B., et al. Across continents and demographics, unpredictable maternal signals are associated with children's cognitive function. EBioMedicine. 2019;46:256–263. doi: 10.1016/j.ebiom.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E.P., Korja R., Karlsson L., Glynn L.M., Sandman C.A., Vegetabile B., et al. Across continents and demographics, unpredictable maternal signals are associated with children's cognitive function. EBioMedicine. 2019;46:256–263. doi: 10.1016/j.ebiom.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E.P., McCormack K., Arora H., Sharpe D., Short A.K., Bachevalier J., et al. Early life exposure to unpredictable parental sensory signals shapes cognitive development across three species. Front. Behav. Neurosci. 2022;20:16. doi: 10.3389/fnbeh.2022.960262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doom J.R., Vanzomeren-Dohm A.A., Simpson J.A. Early unpredictability predicts increased adolescent externalizing behaviors and substance use: a life history perspective. Dev. Psychopathol. 2016;28(4pt2):1505–1516. doi: 10.1017/S0954579415001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn E.C., Soare T.W., Zhu Y., Simpkin A.J., Suderman M.J., Klengel T., et al. Sensitive periods for the effect of childhood adversity on DNA methylation: results from a prospective, longitudinal study. Biol Psychiatry. 2019;85(10):838–849. doi: 10.1016/j.biopsych.2018.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeland B., Hiester M. Institute of Child Development, University of Minnesota; 1993. Teaching Task Rating Scales. [Google Scholar]
- Eisenberg N., Cumberland A., Spinrad T.L., Fabes R.A., Shepard S.A., Reiser M., et al. The relations of regulation and emotionality to children's externalizing and internalizing problem behavior. Child Dev. 2001;72(4):1112–1134. doi: 10.1111/1467-8624.00337. [DOI] [PubMed] [Google Scholar]
- Eriksson M., Räikkönen K., Eriksson J.G. Early life stress and later health outcomes-findings from the Helsinki Birth Cohort Study. Am. J. Hum. Biol. 2014;26(2):111–116. doi: 10.1002/ajhb.22502. [DOI] [PubMed] [Google Scholar]
- Fang F., Zhou L., Perng W., Marsit C.J., Knight A.K., Cardenas A., et al. Evaluation of pediatric epigenetic clocks across multiple tissues. Clin Epigenetics. 2023;15(1):142. doi: 10.1186/s13148-023-01552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah M.J. Socioeconomic status and the brain: prospects for neuroscience-informed policy. Nat. Rev. Neurosci. 2018;19(7):428–438. doi: 10.1038/s41583-018-0023-2. [DOI] [PubMed] [Google Scholar]
- Fergusson D.M., Boden J.M., Horwood L.J. Childhood self-control and adult outcomes: results from a 30-year longitudinal study. J. Am. Acad. Child Adolesc. Psychiatry. 2013;52(7):709–717.e1. doi: 10.1016/j.jaac.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Foundation TAEC. Kids Count Data Center. 2022 [cited 2024 Jan 4]. CHILDREN BELOW 200% POVERTY IN UNITED STATES. .
- Gartstein M.A., Rothbart M.K. Studying infant temperament via the Revised infant behavior questionnaire. Infant Behav. Dev. 2003;26(1):64–86. [Google Scholar]
- Gee D.G., Gabard-Durnam L.J., Flannery J., Goff B., Humphreys K.L., Telzer E.H., et al. Early developmental emergence of human amygdala–prefrontal connectivity after maternal deprivation. Proc. Natl. Acad. Sci. USA. 2013;110(39):15638–15643. doi: 10.1073/pnas.1307893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon A., Minor K., Hayward C. Gender, victimization, and psychiatric outcomes. Psychol. Med. 2008;38(10):1377–1391. doi: 10.1017/S0033291708003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie M.L., Rao U. Relationships between depression and executive functioning in adolescents: the Moderating role of unpredictable home environment. J. Child Fam. Stud. 2022;31(9):2518–2534. doi: 10.1007/s10826-022-02296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasmeier AK. Massachusetts Institute of Technology. 2023 [cited 2023 Oct 5]. Living Wage Calculator. .
- Glynn L.M., Baram T.Z. The influence of unpredictable, fragmented parental signals on the developing brain. Front. Neuroendocrinol. 2019 doi: 10.1016/j.yfrne.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn L.M., Howland M.A., Sandman C.A., Davis E.P., Phelan M., Baram T.Z., et al. Prenatal maternal mood patterns predict child temperament and adolescent mental health. J. Affect. Disord. 2018;228(June 2017):83–90. doi: 10.1016/j.jad.2017.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn L.M., Stern H.S., Howland M.A., Risbrough V.B., Baker D.G., Nievergelt C.M., et al. Measuring novel antecedents of mental illness: the Questionnaire of Unpredictability in Childhood. Neuropsychopharmacology. 2019;44(5):876–882. doi: 10.1038/s41386-018-0280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn L.M., Davis E.P., Luby J.L., Baram T.Z., Sandman C.A. A predictable home environment may protect child mental health during the COVID-19 pandemic. Neurobiol Stress. 2021;14 doi: 10.1016/j.ynstr.2020.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn L.M., Stern H.S., Baram T.Z., Davis E.P. Chapman University Digital Commons; 2023. Questionnaire of Predictability in Childhood Questionnaire Manual. [Google Scholar]
- Goldsmith H.H., Campos J.J. The structure of temperamental fear and pleasure in infants: a psychometric perspective. Child Dev. 1990;61(6):1944–1964. [PubMed] [Google Scholar]
- Govender P., Ghai M., Okpeku M. Sex-specific DNA methylation: impact on human health and development. Mol. Genet. Genom. 2022;297(6):1451–1466. doi: 10.1007/s00438-022-01935-w. [DOI] [PubMed] [Google Scholar]
- Granger S.J., Glynn L.M., Sandman C.A., Small S.L., Obenaus A., Keator D.B., et al. Aberrant maturation of the Uncinate Fasciculus follows exposure to unpredictable patterns of maternal signals. J. Neurosci. 2021;41(6):1242–1250. doi: 10.1523/JNEUROSCI.0374-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh A.M., Roisman G.I., van Ijzendoorn M.H., Bakermans-Kranenburg M.J., Fearon R.P. The significance of insecure and disorganized attachment for children's internalizing symptoms: a meta-analytic study. Child Dev. 2012;83(2):591–610. doi: 10.1111/j.1467-8624.2011.01711.x. [DOI] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Hankin B.L., Snyder H.R., Gulley L.D. Developmental Psychopathology. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2016. Cognitive risks in developmental psychopathology; pp. 1–74. [Google Scholar]
- Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., Sadda S., et al. Genome-wide methylation profiles reveal Quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog J.I., Schmahl C. Adverse childhood experiences and the consequences on neurobiological, psychosocial, and Somatic Conditions across the lifespan. Front Psychiatry. 2018;9:420. doi: 10.3389/fpsyt.2018.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg E., Kataja E.L., Davis E.P., Pajulo M., Nolvi S., Lahtela H., et al. Unpredictable maternal sensory signals in caregiving behavior are associated with child effortful control. PLoS One. 2022;17(12) doi: 10.1371/journal.pone.0279384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S., Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- Horvath S., Gurven M., Levine M.E., Trumble B.C., Kaplan H., Allayee H., et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17(1):171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtepen L.C., Vinkers C.H., Carrillo-Roa T., Hiemstra M., van Lier P.A., Meeus W., et al. Genome-wide DNA methylation levels and altered cortisol stress reactivity following childhood trauma in humans. Nat. Commun. 2016;7(1) doi: 10.1038/ncomms10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howland M.A., Sandman C.A., Davis E.P., Stern H.S., Phelan M., Baram T.Z., et al. Prenatal maternal mood entropy is associated with child neurodevelopment. Emotion. 2021;21(3):489–498. doi: 10.1037/emo0000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman S.M., Garcia M., Sinha R. Gender specific associations between types of childhood maltreatment and the onset, Escalation and severity of substance Use in cocaine dependent adults. Am. J. Drug Alcohol Abuse. 2006;32(4):655–664. doi: 10.1080/10623320600919193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman S.M., Paliwal P., Chaplin T.M., Mazure C.M., Rounsaville B.J., Sinha R. Severity of childhood trauma is predictive of cocaine relapse outcomes in women but not men. Drug Alcohol Depend. 2008;92(1–3):208–216. doi: 10.1016/j.drugalcdep.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy A.S., Rex C.S., Chen Y., Dubé C., Maras P.M., Grigoriadis D.E., et al. Hippocampal dysfunction and cognitive impairments provoked by chronic early-life stress involve excessive activation of CRH receptors. J. Neurosci. 2010;30(39):13005–13015. doi: 10.1523/JNEUROSCI.1784-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Kamei N., Bolton J.L., Ma X., Stern H.S., Baram T.Z., et al. Intra-individual methylomics detects the impact of early-life adversity. Life Sci. Alliance. 2019;2(2) doi: 10.26508/lsa.201800204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirsaraie R.J., Palma A.M., Small S.L., Sandman C.A., Davis E.P., Baram T.Z., et al. Prenatal exposure to maternal mood entropy is associated with a Weakened and Inflexible salience network in adolescence. Biol Psychiatry Cogn Neurosci Neuroimaging. 2023 doi: 10.1016/j.bpsc.2023.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson Å., Enroth S., Gyllensten U. Continuous aging of the human DNA methylome throughout the human lifespan. Suter CM. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0067378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.B., Voegtline K.M., Ialongo N., Hill K.G., Musci R.J. Self-control in first grade predicts success in the transition to adulthood. Dev. Psychopathol. 2022;24:1–13. doi: 10.1017/S0954579421001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph P.V., Wang Y., Fourie N.H., Henderson W.A. A computational framework for predicting obesity risk based on optimizing and integrating genetic risk score and gene expression profiles. PLoS One. 2018;13(5) doi: 10.1371/journal.pone.0197843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic T., Vance L.A., Cross D., Knight A.K., Kilaru V., Michopoulos V., et al. Exposure to Violence accelerates epigenetic aging in children. Sci. Rep. 2017;7(1):8962. doi: 10.1038/s41598-017-09235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katrinli S., Maihofer A.X., Wani A.H., Pfeiffer J.R., Ketema E., Ratanatharathorn A., et al. Epigenome-wide meta-analysis of PTSD symptom severity in three military cohorts implicates DNA methylation changes in genes involved in immune system and oxidative stress. Mol Psychiatry. 2022;27(3):1720–1728. doi: 10.1038/s41380-021-01398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight A.K., Craig J.M., Theda C., Bækvad-Hansen M., Bybjerg-Grauholm J., Hansen C.S., et al. An epigenetic clock for gestational age at birth based on blood methylation data. Genome Biol. 2016;17(1):206. doi: 10.1186/s13059-016-1068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanska G., Murray K., Jacques T.Y., Koenig A.L., Vandegeest K.A. Inhibitory control in young children and its role in emerging Internalization. Child Dev. 1996;67(2):490–507. [PubMed] [Google Scholar]
- Lansford J.E., Dodge K.A., Pettit G.S., Bates J.E. Does physical abuse in early childhood predict substance Use in adolescence and early adulthood? Child. Maltreat. 2010;15(2):190–194. doi: 10.1177/1077559509352359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C., Walton M., Letourneau N., Giesbrecht G.F., Kaplan B.J., Dewey D. Prepartum and postpartum maternal depressive symptoms are related to children's brain structure in preschool. Biol Psychiatry. 2016;80(11):859–868. doi: 10.1016/j.biopsych.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Levis S.C., Bentzley B.S., Molet J., Bolton J.L., Perrone C.R., Baram T.Z., et al. On the early life origins of vulnerability to opioid addiction. Mol Psychiatry. 2021;26(8):4409–4416. doi: 10.1038/s41380-019-0628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis S.C., Baram T.Z., Mahler S.V. Neurodevelopmental origins of substance use disorders: Evidence from animal models of early‐life adversity and addiction. Schmidt M., editor. Eur. J. Neurosci. 2022;55(9–10):2170–2195. doi: 10.1111/ejn.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindert N.G., Maxwell M.Y., Liu S.R., Stern H.S., Baram T.Z., Poggi Davis E., et al. Exposure to unpredictability and mental health: validation of the brief version of the Questionnaire of Unpredictability in Childhood (QUIC-5) in English and Spanish. Front. Psychol. 2022:13. doi: 10.3389/fpsyg.2022.971350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby J., Belden A., Botteron K., Marrus N., Harms M.P., Babb C., et al. The effects of poverty on childhood brain development. JAMA Pediatr. 2013;167(12):1135. doi: 10.1001/jamapediatrics.2013.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby J.L., Tillman R., Barch D.M. Association of timing of adverse childhood experiences and caregiver support with Regionally specific brain development in adolescents. JAMA Netw. Open. 2019;2(9) doi: 10.1001/jamanetworkopen.2019.11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien S.J., McEwen B.S., Gunnar M.R., Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat. Rev. Neurosci. 2009;10(6):434–445. doi: 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- Lussier A.A., Zhu Y., Smith B.J., Simpkin A.J., Smith A.D.A.C., Suderman M.J., et al. Sensitive periods for the effect of childhood adversity on DNA methylation: updated results from a prospective, longitudinal study. Biological Psychiatry Global Open Science. 2023;3(3):567–571. doi: 10.1016/j.bpsgos.2022.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox S.A., Kilaru V., Shin J., Jovanovic T., Almli L.M., Dias B.G., et al. Estrogen-dependent association of HDAC4 with fear in female mice and women with PTSD. Mol Psychiatry. 2018;23(3):658–665. doi: 10.1038/mp.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S., Hinkal G., Kim H.S., Shen L., Zhang L., Zhang J., et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20(3):332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J.C., Park K., Lin Y.A., Bersamira C. Gender differences in trends for heroin use and nonmedical prescription opioid use, 2007–2014. J. Subst. Abuse Treat. 2018;87:79–85. doi: 10.1016/j.jsat.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino D.J., Tulic M.K., Gordon L., Hodder M., Richman T.R., Metcalfe J., et al. Evidence for age-related and individual-specific changes in DNA methylation profile of mononuclear cells during early immune development in humans. Epigenetics. 2011;6(9):1085–1094. doi: 10.4161/epi.6.9.16401. [DOI] [PubMed] [Google Scholar]
- Martins J., Czamara D., Sauer S., Rex-Haffner M., Dittrich K., Dörr P., et al. Childhood adversity correlates with stable changes in DNA methylation trajectories in children and converges with epigenetic signatures of prenatal stress. Neurobiol Stress. 2021;15 doi: 10.1016/j.ynstr.2021.100336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen L.M., O'Donnell K.J., McGill M.G., Edgar R.D., Jones M.J., MacIsaac J.L., et al. The PedBE clock accurately estimates DNA methylation age in pediatric buccal cells. Proc. Natl. Acad. Sci. USA. 2020;117(38):23329–23335. doi: 10.1073/pnas.1820843116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M.G., Pokhvisneva I., Clappison A.S., McEwen L.M., Beijers R., Tollenaar M., et al. Reply to: Crossing the “birth Border” for epigenetic effects. Biol Psychiatry. 2022;92(4):e25–e26. doi: 10.1016/j.biopsych.2021.12.008. [DOI] [PubMed] [Google Scholar]
- McGinnis E.W., Sheridan M., Copeland W.E. Impact of dimensions of early adversity on adult health and functioning: a 2-decade, longitudinal study. Dev. Psychopathol. 2022;34(2):527–538. doi: 10.1017/S095457942100167X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin K.A., Weissman D., Bitrán D. Childhood adversity and neural development: a systematic review. Annu Rev Dev Psychol. 2019;1(1):277–312. doi: 10.1146/annurev-devpsych-121318-084950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt T.E., Arseneault L., Belsky D., Dickson N., Hancox R.J., Harrington H., et al. A gradient of childhood self-control predicts health, wealth, and public safety. Proc. Natl. Acad. Sci. USA. 2011;108(7):2693–2698. doi: 10.1073/pnas.1010076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molet J., Maras P.M., Avishai-Eliner S., Baram T.Z. Naturalistic rodent models of chronic early-life stress. Dev. Psychobiol. 2014;56(8):1675–1688. doi: 10.1002/dev.21230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molet J., Heins K., Zhuo X., Mei Y.T., Regev L., Baram T.Z., et al. Fragmentation and high entropy of neonatal experience predict adolescent emotional outcome. Transl. Psychiatry. 2016;6(1):e702. doi: 10.1038/tp.2015.200. e702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk C., Feng T., Lee S., Krupska I., Champagne F.A., Tycko B. Distress during Pregnancy: epigenetic regulation of placenta Glucocorticoid-related genes and fetal neurobehavior. Am. J. Psychiatr. 2016;173(7):705–713. doi: 10.1176/appi.ajp.2015.15091171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk C., Feng T., Lee S., Krupska I., Champagne F.A., Tycko B. Distress during Pregnancy: epigenetic regulation of placenta Glucocorticoid-related genes and fetal neurobehavior. Am J Psychiatry. 2016;173(7):705–713. doi: 10.1176/appi.ajp.2015.15091171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musci R.J., Raghunathan R.S., Johnson S.B., Klein L., Ladd-Acosta C., Ansah R., et al. Using epigenetic clocks to characterize biological aging in studies of children and childhood exposures: a systematic review. Prev. Sci. 2023 doi: 10.1007/s11121-023-01576-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najavits L.M., Weiss R.D., Shaw S.R. The link between substance abuse and Posttraumatic stress disorder in women: a Research review. Am. J. Addict. 1997;6(4):273–283. [PubMed] [Google Scholar]
- NICHD Early Care Research Network Chronicity of maternal depressive symptoms, maternal sensitivity, and child functioning at 36 months. NICHD Early Child Care Research Network. Dev. Psychol. 1999;35(5):1297–1310. doi: 10.1037//0012-1649.35.5.1297. [DOI] [PubMed] [Google Scholar]
- Nigg J.T. Annual Research Review: on the relations among self-regulation, self-control, executive functioning, effortful control, cognitive control, impulsivity, risk-taking, and inhibition for developmental psychopathology. JCPP (J. Child Psychol. Psychiatry) 2017;58(4):361–383. doi: 10.1111/jcpp.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Non A.L., Hollister B.M., Humphreys K.L., Childebayeva A., Esteves K., Zeanah C.H., Fox N.A., Nelson C.A., Drury S.S. DNA methylation at stress-related genes is associated with exposure to early life institutionalization. Am. J. Phys. Anthropol. 2016;161(1):84–93. doi: 10.1002/ajpa.23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noroña-Zhou A.N., Morgan A., Glynn L.M., Sandman C.A., Baram T.Z., Stern H.S., et al. Unpredictable maternal behavior is associated with a blunted infant cortisol response. Dev. Psychobiol. 2020;62(6):882–888. doi: 10.1002/dev.21964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick A.M., Levandowski M.L., Laumann L.E., Philip N.S., Price L.H., Tyrka A.R. The effects of early life stress on reward processing. J. Psychiatr. Res. 2018;101(October 2017):80–103. doi: 10.1016/j.jpsychires.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios-Barrios E.E., Hanson J.L. Poverty and self-regulation: Connecting psychosocial processes, neurobiology, and the risk for psychopathology. Compr Psychiatry. 2019;90:52–64. doi: 10.1016/j.comppsych.2018.12.012. [DOI] [PubMed] [Google Scholar]
- Peltier M.R., Verplaetse T.L., Mineur Y.S., Petrakis I.L., Cosgrove K.P., Picciotto M.R., et al. Sex differences in stress-related alcohol use. Neurobiol Stress. 2019;10 doi: 10.1016/j.ynstr.2019.100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez R.F., Santamarina P., Tejedor J.R., Urdinguio R.G., Álvarez-Pitti J., Redon P., et al. Longitudinal genome-wide DNA methylation analysis uncovers persistent early-life DNA methylation changes. J. Transl. Med. 2019;17(1):15. doi: 10.1186/s12967-018-1751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam S.P., Gartstein M.A., Rothbart M.K. Measurement of fine-grained aspects of toddler temperament: the early childhood behavior questionnaire. Infant Behav. Dev. 2006;29(3):386–401. doi: 10.1016/j.infbeh.2006.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach A., Levine M.E., Tanaka T., Lu A.T., Chen B.H., Ferrucci L., et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging. 2017;9(2):419–446. doi: 10.18632/aging.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C., Marin M.F., Majeur D., Lupien S. Early child adversity and psychopathology in adulthood: HPA axis and cognitive dysregulations as potential mechanisms. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2018;85:152–160. doi: 10.1016/j.pnpbp.2017.07.015. [DOI] [PubMed] [Google Scholar]
- Rice C.J., Sandman C.A., Lenjavi M.R., Baram T.Z. A novel mouse model for acute and long-lasting consequences of early life stress. Endocrinology. 2008;149(10):4892–4900. doi: 10.1210/en.2008-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond-Rakerd L.S., Caspi A., Ambler A., d'Arbeloff T., de Bruine M., Elliott M., et al. Childhood self-control forecasts the pace of midlife aging and preparedness for old age. Proc. Natl. Acad. Sci. USA. 2021;118(3) doi: 10.1073/pnas.2010211118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J.D., Chen E.E., Winsell J., Davis E.P., Glynn L.M., Baram T.Z., et al. Network specialization during adolescence: Hippocampal effective connectivity in boys and girls. Neuroimage. 2018;175:402–412. doi: 10.1016/j.neuroimage.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart M.K., Ahadi S.A., Evans D.E. Temperament and personality: origins and outcomes. J. Pers. Soc. Psychol. 2000;78(1):122–135. doi: 10.1037//0022-3514.78.1.122. [DOI] [PubMed] [Google Scholar]
- Rothbart M.K., Ahadi S.A., Hershey K.L., Fisher P. Investigations of temperament at three to seven years: the Children's Behavior Questionnaire. Child Dev. 2001;72(5):1394–1408. doi: 10.1111/1467-8624.00355. [DOI] [PubMed] [Google Scholar]
- RStudio Team . 2020. RStudio: Integrated Development Environment for R. Boston, MA. [Google Scholar]
- Rubens M., Bruenig D., Adams J.A.M., Suresh S.M., Sathyanarayanan A., Haslam D., et al. Childhood maltreatment and DNA methylation: a systematic review. Neurosci. Biobehav. Rev. 2023;147 doi: 10.1016/j.neubiorev.2023.105079. [DOI] [PubMed] [Google Scholar]
- Rutten B.P.F., Vermetten E., Vinkers C.H., Ursini G., Daskalakis N.P., Pishva E., et al. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol Psychiatry. 2018;23(5):1145–1156. doi: 10.1038/mp.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandman C.A., Glynn L.M., Davis E.P. Is there a viability-vulnerability tradeoff? Sex differences in fetal programming. J. Psychosom. Res. 2013;75(4):327–335. doi: 10.1016/j.jpsychores.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandman C.A., Buss C., Head K., Davis E.P. Fetal exposure to maternal depressive symptoms is associated with Cortical Thickness in late childhood. Biol Psychiatry. 2015;77(4):324–334. doi: 10.1016/j.biopsych.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shand F.L., Degenhardt L., Slade T., Nelson E.C. Sex differences amongst dependent heroin users: Histories, clinical characteristics and predictors of other substance dependence. Addict. Behav. 2011;36(1–2):27–36. doi: 10.1016/j.addbeh.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan M.A., McLaughlin K.A. Dimensions of early experience and neural development: deprivation and threat. Trends Cogn Sci. 2014;18(11):580–585. doi: 10.1016/j.tics.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short A.K., Baram T.Z. Early-life adversity and neurological disease: age-old questions and novel answers. Nat. Rev. Neurol. 2019;15(11):657–669. doi: 10.1038/s41582-019-0246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short A.K., Maras P.M., Pham A.L., Ivy A.S., Baram T.Z. Blocking CRH receptors in adults mitigates age-related memory impairments provoked by early-life adversity. Neuropsychopharmacology. 2020;45(3):515–523. doi: 10.1038/s41386-019-0562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpkin A.J., Hemani G., Suderman M., Gaunt T.R., Lyttleton O., Mcardle W.L., et al. Prenatal and early life influences on epigenetic age in children: a study of mother–offspring pairs from two cohort studies. Hum. Mol. Genet. 2016;25(1):191–201. doi: 10.1093/hmg/ddv456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.K., Ratanatharathorn A., Maihofer A.X., Naviaux R.K., Aiello A.E., Amstadter A.B., et al. Epigenome-wide meta-analysis of PTSD across 10 military and civilian cohorts identifies methylation changes in AHRR. Nat. Commun. 2020;11(1):5965. doi: 10.1038/s41467-020-19615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders C., Maihofer A.X., Ratanatharathorn A., Baker D.G., Boks M.P., Geuze E., et al. Longitudinal epigenome-wide association studies of three male military cohorts reveal multiple CpG sites associated with post-traumatic stress disorder. Clin Epigenetics. 2020;12(1):1–13. doi: 10.1186/s13148-019-0798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadoni A.D., Vinograd M., Cuccurazzu B., Torres K., Glynn L.M., Davis E.P., et al. Contribution of early-life unpredictability to neuropsychiatric symptom patterns in adulthood. Depress. Anxiety. 2022;39(10–11):706–717. doi: 10.1002/da.23277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez A., Lahti J., Czamara D., Lahti-Pulkkinen M., Knight A.K., Girchenko P., et al. The epigenetic clock at birth: associations with maternal Antenatal depression and child psychiatric problems. J. Am. Acad. Child Adolesc. Psychiatry. 2018;57(5):321–328.e2. doi: 10.1016/j.jaac.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szepsenwol O., Simpson J.A., Griskevicius V., Zamir O., Young E.S., Shoshani A., et al. The effects of childhood unpredictability and harshness on emotional control and relationship quality: a life history perspective. Dev. Psychopathol. 2022;34(2):607–620. doi: 10.1017/S0954579421001371. [DOI] [PubMed] [Google Scholar]
- Tang R., Howe L.D., Suderman M., Relton C.L., Crawford A.A., Houtepen L.C. Adverse childhood experiences, DNA methylation age acceleration, and cortisol in UK children: a prospective population-based cohort study. Clin Epigenetics. 2020;12(1):55. doi: 10.1186/s13148-020-00844-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor Z.E., Evich C.D., Marceau K., Nair N., Jones B.L. Associations between effortful control, cortisol Awakening response, and depressive problems in Latino Preadolescents. J. Early Adolesc. 2019;39(7):1050–1077. doi: 10.1177/0272431618798509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio R.G., Torró M.I., Bayón G.F., Álvarez-Pitti J., Fernández A.F., Redon P., et al. Longitudinal study of DNA methylation during the first 5 years of life. J. Transl. Med. 2016;14(1):160. doi: 10.1186/s12967-016-0913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursache A., Noble K.G. Socioeconomic status, white matter, and executive function in children. Brain Behav. 2016;6(10) doi: 10.1002/brb3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wal S.J., Maihofer A.X., Vinkers C.H., Smith A.K., Nievergelt C.M., Cobb D.O., et al. Associations between the development of PTSD symptoms and longitudinal changes in the DNA methylome of deployed military servicemen: a comparison with polygenic risk scores. Compr Psychoneuroendocrinol. 2020;4 doi: 10.1016/j.cpnec.2020.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M., Fox N.A., Zeanah C.H., Nelson C.A. Long-term effects of institutional rearing, foster care, and brain activity on memory and executive functioning. Proc. Natl. Acad. Sci. USA. 2019;116(5):1808–1813. doi: 10.1073/pnas.1809145116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Liu X., Zhou Y., Xie H., Hong X., Tsai H.J., et al. Individual variation and longitudinal pattern of genome-wide DNA methylation from birth to the first two years of life. Epigenetics. 2012;7(6):594–605. doi: 10.4161/epi.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.L., Moffitt T.E., Caspi A., Bartusch D.J., Needles D.J., Stouthamer-Loeber M. Measuring impulsivity and examining its relationship to delinquency. J. Abnorm. Psychol. 1994;103(2):192–205. doi: 10.1037//0021-843x.103.2.192. [DOI] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Widom C.S., Ireland T., Glynn P.J. Alcohol abuse in abused and neglected children followed-up: are they at increased risk? J. Stud. Alcohol. 1995;56(2):207–217. doi: 10.15288/jsa.1995.56.207. [DOI] [PubMed] [Google Scholar]
- Wikenius E., Moe V., Smith L., Heiervang E.R., Berglund A. DNA methylation changes in infants between 6 and 52 weeks. Sci. Rep. 2019;9(1):1–12. doi: 10.1038/s41598-019-54355-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey J., Blajda V.M. Temperament ratings at 2 weeks, 2 months, and 1 year: differential stablity of activity and emotionality. Dev. Psychol. 1989;25(2):257–263. [Google Scholar]
- Xu Y., Harms M.B., Green C.S., Wilson R.C., Pollak S.D. Childhood unpredictability and the development of exploration. Proc Natl Acad Sci U S A. 2023;120(49) doi: 10.1073/pnas.2303869120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.