Abstract
Herpes simplex virus type 1 (HSV-1) capsids are initially assembled with an internal protein scaffold. The scaffold proteins, encoded by overlapping in-frame UL26 and UL26.5 transcripts, are essential for formation and efficient maturation of capsids. UL26 encodes an N-terminal protease domain, and its C-terminal oligomerization and capsid protein-binding domains are identical to those of UL26.5. The UL26 protease cleaves itself, releasing minor scaffold proteins VP24 and VP21, and the more abundant UL26.5 protein, releasing the major scaffold protein VP22a. Unlike VP21 and VP22a, which are removed from capsids upon DNA packaging, we demonstrate that VP24 (containing the protease domain) is quantitatively retained. To investigate factors controlling UL26 capsid incorporation and retention, we used a mutant virus that fails to express UL26.5 (ΔICP35 virus). Purified ΔICP35 B capsids showed altered sucrose gradient sedimentation and lacked the dense scaffold core seen in micrographs of wild-type B capsids but contained capsid shell proteins in wild-type amounts. Despite C-terminal sequence identity between UL26 and UL26.5, ΔICP35 capsids lacking UL26.5 products did not contain compensatory high levels of UL26 proteins. Therefore, HSV capsids can be maintained and/or assembled on a minimal scaffold containing only wild-type levels of UL26 proteins. In contrast to UL26.5, increased expression of UL26 did not compensate for the ΔICP35 growth defect. While indirect, these findings are consistent with the view that UL26 products are restricted from occupying abundant UL26.5 binding sites within the capsid and that this restriction is not controlled by the level of UL26 protein expression. Additionally, ΔICP35 capsids contained an altered complement of DNA cleavage and packaging proteins, suggesting a previously unrecognized role for the scaffold in this process.
Herpes simplex virus (HSV) virions are multilayered, and their assembly requires several steps (reviewed in references 21 and 59). The double-stranded DNA viral genome is enclosed within a well-ordered protein capsid. A more amorphous layer of proteins referred to as the tegument surrounds the capsid. Lying outside of the tegument, the outermost layer of the virion consists of a lipid envelope containing viral glycoproteins.
In the nucleus of infected cells, viral capsids are initially assembled with an internal protein core or scaffold. Packaging of replicated viral DNA into these preformed capsids involves the processing of scaffold proteins by the scaffold-associated protease and release of the scaffold proteins from the capsid. In a poorly understood process requiring seven additional viral genes, genome-length DNA is cleaved from larger-than-unit-length concatamers and packaged into capsids. Capsids containing viral DNA subsequently acquire tegument and envelope to become mature virions.
Three types of intracellular capsids (A, B, and C capsids), are routinely isolated from infected cells by sucrose gradient sedimentation (17, 45). The structures of A, B, and C capsid shells are indistinguishable (3, 67), but the internal contents differ. C capsids contain the viral genome and are the precursors to infectious virions (45). B capsids lack viral DNA and instead contain the proteolytically processed forms of the internal scaffold proteins (35, 36, 45, 50). A capsids lack both internal scaffold proteins and viral DNA (17, 45) and may be the products of abortive attempts at DNA packaging (57).
A fourth type of capsid, the procapsid, has recently been identified as a precursor to A, B, and C capsids (37, 38, 40). Procapsids are the earliest form of capsid observed during in vitro capsid assembly reactions (37). Upon prolonged incubation at room temperature, the spherical, unstable procapsid shell undergoes structural transformations that render it indistinguishable from more stable, angular A, B, and C capsids (37). Procapsids are not typically seen during wild-type herpesvirus infection, presumably because they are transient intermediates in the in vivo capsid maturation pathway. In the absence of the scaffold-associated protease procapsids accumulate, suggesting that protease cleavage of the internal capsid scaffold proteins controls the transition from unstable procapsid to mature capsid during herpesvirus infection (40).
Recent studies have shed light on the highly ordered protein composition and overall structure of the mature capsid shell. The capsid is an icosahedron (69) composed of four proteins: VP5 (virion protein 5, encoded by the UL19 gene), VP19c (UL38), VP23 (UL18), and VP26 (UL35) (8, 10, 51). The number of copies of each capsid protein is strictly defined by the capsid's rigid symmetry. Five- and six-membered rings of VP5 form the ring-like penton and hexon subunits of the capsid and are attached to one another by tripartite protein complexes, triplexes, composed of one copy of VP19c and two copies of VP23 (39, 67). Six-membered rings of VP26 reside on the distal tips of the 150 hexons but not on the 12 pentons of A, B, and C capsids (4, 73). VP26 is absent from procapsids isolated from HSV-infected cells (40) and does not colocalize with capsid proteins in the nucleus until after capsid angularization (5).
In contrast to the highly ordered capsid shell, the structure of the internal protein scaffold is less well defined. Although the scaffold displays some radial symmetry that suggests an ordered internal structure, the core almost entirely lacks the rigid icosahedral symmetry of the capsid shell (66, 72). The scaffold core is visible as a dark ring in electron micrographs of capsids. The diameter of the scaffold ring is larger in procapsids, containing unprocessed scaffold proteins, than in B capsids, containing processed scaffold proteins. Procapsids also contain higher levels of scaffold proteins than B capsids (40).
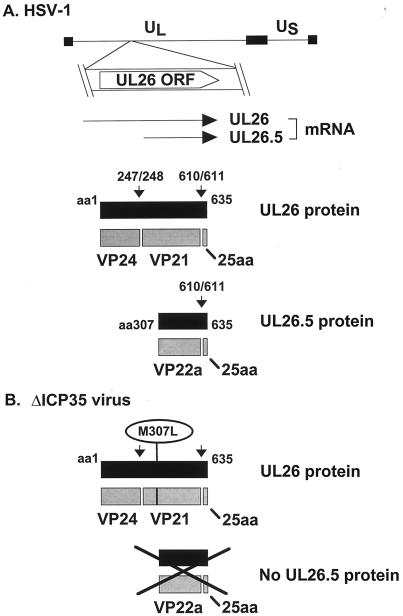
The protein composition of the B capsid scaffold has been extensively studied. The scaffold is composed of the products of the overlapping UL26 and UL26.5 mRNA transcripts (20, 30, 50) (Fig. 1A), and a similar gene arrangement is found in all herpesviruses examined to date (reviewed in reference 16). UL26 and UL26.5 encode proteins with identical C termini containing oligomerization (11, 44) and VP5-binding (22, 41) domains. Unique N-terminal sequences of UL26 encode a serine protease domain (27–29) followed by a 59-amino-acid (aa) linker region that connects the protease domain to the oligomerization domain. The scaffold proteins are initially incorporated into capsids in an unprocessed form (23, 48, 52, 64). During capsid maturation, the UL26-encoded protease processes itself to release the N-terminal protease domain (VP24) (Fig. 1A). The protease also processes both itself and the UL26.5-encoded protein at an additional site near the C terminus to release a 25-aa peptide that serves to tether the scaffold to the capsid shell (22, 41). The products of protease cleavage found within intracellular capsids are VP24 and VP21 (10, 47, 68), resulting from cleavage of the UL26 protein, and VP22a (36, 50), resulting from cleavage of the UL26.5 protein (Fig. 1A).
FIG. 1.
HSV-1 UL26 and UL26.5 gene products. (A) The UL26 open reading frame is indicated below its location in the prototype orientation of the HSV-1 genome (shown with thin line representing unique long [UL] and unique short [US] regions bounded by terminal repeated sequences, represented by thick bars). UL26 and UL26.5 mRNA transcripts and protein products are also depicted. Primary translation products are represented by solid boxes. Gray boxes represent proteolysis products. Vertical arrows indicate the sites of proteolytic processing of the scaffold proteins (14). Numbers indicate aa residues at the N and C termini of the polypeptides or the sites of proteolytic processing. (B) Structure of the ΔICP35 mutant virus and its predicted protein products. Mutation of Met-307 of UL26 to Leu prevents translation of the UL26.5 protein (31). A black X covers the products whose translation is prevented by the M307L mutation.
Despite the fact that the C-terminal 329 aa of UL26 are identical to those of UL26.5, both containing the same C-terminal VP5-binding domain, there is a marked difference in the incorporation of UL26 and UL26.5 proteins into capsids. The UL26 proteins VP24 and VP21 are present at approximately 10-fold-lower levels within B capsids than the UL26.5 protein VP22a (39). It has been suggested that this difference results from the reduced expression of UL26 relative to UL26.5 in infected cells (22, 53). Alternatively, as proposed by Tatman et al. (61), sequences unique to UL26 may control its reduced level of incorporation into capsids, since recombinant capsids formed within insect cells contained apparently wild-type levels of UL26 and UL26.5 proteins.
An additional distinction between UL26 and UL26.5 proteins occurs upon DNA packaging. Although the bulk of the internal scaffold is absent from DNA-containing capsids, VP24 has been detected in all forms of capsids and in mature virions (3, 17, 58, 60), suggesting that its fate might differ from that of the other scaffold proteins. Indeed, quantitative Western blots presented in this paper demonstrate that unlike the levels of VP21 and VP22a, the level of VP24 does not change upon scaffold release or DNA packaging.
To further investigate whether capsid incorporation of UL26 proteins is strictly controlled, we used a mutant virus (ΔICP35 virus [31]) which fails to express UL26.5 (Fig. 1B). Growth of the ΔICP35 virus is impaired, although capsids are formed during ΔICP35 virus infection (31). The ΔICP35 virus forms small plaques on noncomplementing cells and displays a multiplicity of infection (MOI)-dependent 100- to 1,000-fold reduction in progeny virus (31). Since the ΔICP35 virus does not express the most abundant scaffold protein, VP22a, but can still direct the assembly of capsids, ΔICP35 mutant B capsids contain a scaffold composed entirely of UL26 proteins. Study of these mutant capsids has allowed examination of the factors controlling the incorporation of VP24 and VP21 in the absence of VP22a. Additionally, since the ΔICP35 virus also displays a defect in DNA packaging (31), we examined the content of packaging proteins associated with these mutant capsids.
MATERIALS AND METHODS
Cells and viruses.
Vero cells and the stably transformed Vero cell lines 35J (31) and BMS-MG22 (15) were grown in minimal essential medium (Life Technologies, Inc.) supplemented with 10% heat-inactivated fetal bovine serum (Sigma). The medium for 35J and BMS-MG22 cells was also supplemented with G418 (500 μg/ml). HSV-1 (strain KOS) was grown in Vero cells. The ΔICP35 and Prb viruses were propagated in 35J cells (31, 33), and the m100 virus was propagated in BMS-MG22 cells as previously described (15). For complementation assays, Vero cells were plated 1 day prior to transfection at a density of 500,000 cells per well of a six-well plate. Plasmid DNA was transfected (1 μg per well of cells) using the Lipofectamine Plus kit (Life Technologies), according to the manufacturer's directions. At 20 h after transfection, cells were superinfected with the relevant virus. After 2 h of adsorption at 37°C, infected cells were washed once with pH 3.0 glycine buffer (100 mM glycine, 137 mM NaCl, 5 mM KCl, 0.68 mM CaCl2) to inactivate residual extracellular virus. At 24 h postinfection, infectious virus was released from cells by one freeze-thaw cycle (−80°C and 37°C) and sonication in an ice-water bath (Branson Sonifier 250; 30 bursts, 50% duty, setting 5). Virus production was measured by titration in duplicate on Vero cells and on the appropriate complementing cell line. The complementation index is expressed as [virus obtained after transfection of test plasmid (titer of resulting virus on complementing cells) minus (titer on Vero cells)] divided by [virus obtained after transfection of appropriate vector plasmid (titer of resulting virus on complementing cells) minus (titer on Vero cells)].
Expression of recombinant scaffold proteins.
The M307L mutation was introduced into a plasmid containing UL26 (pRB4090), kindly provided by Bernard Roizman (29), by a modification of the PCR mutagenesis procedure used by Gao et al. (15) using Pfu polymerase (Stratagene) as described below. To make the pCMV-UL26(M307L) plasmid, the UL26 gene was first mutated by PCR using primer 1 (5′-GGGAAGCGGCCGCCATATGGCAGCCGATGCC-3′; adds additional 5′ NotI and NdeI sites containing an initiation codon) and primer 2 (5′-CGGGGTTTAAAGGGGGCAGTACCG-3′, containing the Met-to-Leu mutation at aa 307). The 0.9-kb product was then used as a 5′ primer in a second PCR along with primer 3 (5′-GGAATTCAGCGGGCCCCCATCATCTG-3′; adds an additional 3′ EcoRI site after the UL26 stop codon). The UL26 gene containing the mutation was then PCR amplified using primer 4 (5′-GGGAAGCGGCCGCCATATGG-3′) and primer 3. The amplified 1.9-kb fragment (UL26 gene with the M307L mutation) was inserted into the pIRESPURO vector (Clontech) at the NotI and EcoRI sites. PCR-amplified C-terminal sequences were replaced with a BsrGI-EcoRI fragment subcloned from pRB4090, and the PCR-amplified N terminus was verified by nucleotide sequencing. To make pCMV-preVP21, the N terminus of the protease gene was deleted from pCMV-UL26(M307L) by digestion with NdeI and PinAI and replaced with a fragment from plasmid pBac-preVP21, containing the additional methionine and N terminus of VP21, so that the final clone contains a methionine followed by amino acids 248 to 635 of UL26. The pBac-preVP21 clone was made by subcloning a 1.2-kb ApoI-KpnI fragment from pRB4090 into pVL1392 (Invitrogen) and then insertion of a linker (containing an initiation codon) at the BglII and ApoI sites of the resulting plasmid. The linker was made by annealing two oligonucleotides (5′-GATCTCATATGAGCGAGA-3′ and 5′-AATTTCTCGCTCATATGA-3′) which contain a 5′ BglII site, an NdeI site containing the initiation Met, and an ApoI site.
Capsid isolation.
Cells were infected at a multiplicity of infection of 10 PFU per cell. At 20 to 23 h postinfection, cells were collected by centrifugation at 1,000 × g for 10 min and washed once in Dulbecco's phosphate-buffered saline (D-PBS; 8 g of NaCl, 2.16 g of Na2HPO4 · 7H2O, 0.2 g of KCl, and 0.2 g of KH2PO4 per liter). The cell pellet was resuspended in an equal volume of 2× lysis buffer (65) containing 1 M NaCl, 20 mM Tris (pH 7.6), 2 mM EDTA, 2% Triton X-100, and protease inhibitors (2 mM Pefabloc SC [Boehringer-Mannheim]), 10 μg of Bestatin [Sigma] per ml, 10 μg of antipain [Sigma] per ml, and 1 complete protease inhibitor cocktail tablet [Boehringer-Mannheim]) per 25 ml of lysis buffer. The cell pellet was lysed by freeze-thawing three times, and the lysate was precleared by centrifugation at 8,000 rpm in the Beckman SW28 rotor. Capsids were partially purified by two centrifugation steps. In the first step, capsids were centrifuged through a 35% (wt/vol) sucrose cushion prepared in TNE (500 mM NaCl, 20 mM Tris [pH 7.6], 1 mM EDTA). The capsid pellet was then resuspended in TNE, and capsids were purified on 20 to 50% (wt/vol) sucrose gradients centrifuged at 24,000 rpm in the SW28 rotor for 60 min, essentially as previously described (65). Capsids were visualized as light-scattering bands and either collected by side puncture and aspiration into a syringe or fractionated from the bottom of the tube using a Buchler Auto Densiflow II.
Electron microscopy.
Purified capsids were pelleted by dilution in D-PBS and centrifugation for 1 h at 24,000 rpm in the Beckman SW28 rotor at 4°C. The resulting capsid pellet was processed for electron microscopy as previously described (31).
SDS-PAGE and Western blots.
Samples for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were precipitated by the addition of trichloroacetic acid to 10% (final volume), incubation on ice for 10 min, and centrifugation at 12,000 × g for 20 min. Pellets were resuspended in loading buffer (200 mM Tris [pH 8.8], 100 mM dithiothreitol, 2% SDS, 10% glycerol), boiled for 3 min, and separated by electrophoresis in 4 to 20% polyacrylamide gradient gels (Bio-Rad) for analysis of total proteins by Coomassie staining. For Western blotting, different types of gels were run, as follows: 12% polyacrylamide for analysis of VP5, VP23, VP24, VP21, VP22a, UL15, and UL25; 4 to 20% polyacrylamide for analysis of UL6; and 7.5% polyacrylamide for analysis of UL28. Gels were electrophoretically transferred to nitrocellulose blots, which were washed twice in TBS (20 mM Tris, 500 mM NaCl [pH 7.5]) and blocked for 60 min in blocking buffer (TBS plus 0.2% nonfat dry milk). Primary antibodies were diluted in blocking buffer plus 0.1% Tween-20. Antibodies were added to blots for 2 h at the following dilutions: monoclonal antibody (MAb) 13-183 against VP5 (Advanced Biotechnologies Inc.) at 1:1,000; MAb 1D2 against VP23 (40) at 1:2,000; MAb MCA406 (Serotec Inc.) against VP21/VP22a at 1:10,000; MAb 9-2 against VP24 at 1:1,000 (A. K. Sheaffer, A. J. C. Evans, C. DiIanni, and S. Weinheimer, unpublished data); polyclonal antibody (PAb) CL9 (26) against UL6 at 1:1,000; PAb ID1 (24) against UL25 at 1:1,000; and PAb H85 against UL28 (63) at 1:1,000. Alkaline phosphatase-conjugated goat anti-mouse or anti-rabbit immunoglobulin G secondary antibodies (Bio-Rad) were added to blots for 2 h at a 1:4,000 dilution in blocking buffer plus 0.1% Tween-20. Secondary antibodies were detected using the Immunstar chemiluminescent detection kit (Bio-Rad) and exposure of blots to Kodak XAR-5 film as directed by the manufacturer. Proteins were quantitated by densitometry of the resulting films using the Molecular Dynamics Personal Densitomer SI and ImageQuant software (version 4.0). To ensure that exposures were within the linear range of detection, serial dilutions of each sample were quantitated.
RESULTS
Composition and fate of the internal scaffold.
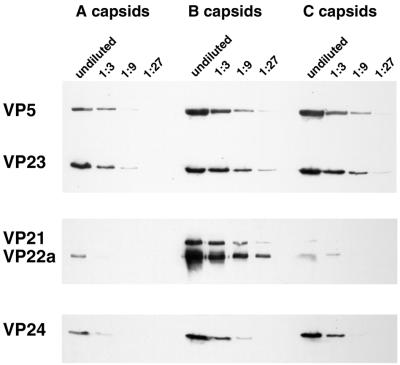
The proteins that form the internal scaffold of HSV-1 B capsids are shown in Fig. 1A. Although several studies showed that VP24 was present in HSV-1 B capsids (10, 35, 39, 47, 68) or A and C capsids (3), only the work of Gibson and Roizman (17) and Stevenson et al. (60) showed that VP24 was present in A, B, and C capsids. However, none of these studies quantitatively compared its levels in the three capsid types. We sought to determine if VP24 was quantitatively retained in capsids after export of the bulk of the scaffold and packaging of viral DNA. A quantitative presence of VP24 in all three capsid types would be consistent with active retention rather than a more passive residual presence due to inefficient export. To compare the levels of VP24 present in wild-type A, B, and C capsids, we purified capsids from HSV-1 (strain KOS)-infected cells by sucrose gradient sedimentation, as described in Materials and Methods. Threefold dilutions of each capsid type were separated by SDS-PAGE, and replicate gels were analyzed by Western blotting (Fig. 2). In order to standardize the amounts of each capsid type, one blot was probed with antisera against capsid shell proteins VP5 and VP23 (Fig. 2, upper panel). Since the A, B, and C capsid shells are identical (3), equal amounts of VP5 and VP23 are present in all three forms. To analyze the fates of the internal scaffold proteins, a second blot was probed with antibody against an epitope common to both VP21 and VP22a (Fig. 2, center panel), and a third blot was probed with antibody against VP24 (Fig. 2, lower panel). In agreement with published data, B capsids contained the highest levels of scaffold proteins VP21 and VP22a, with only trace amounts of these proteins remaining in both A and C capsid preparations. Scans of autoradiographic exposures from similar blots revealed that B capsids contained approximately 5- to 10-fold-higher levels of VP22a than VP21 (unpublished data), in agreement with previous reports (35, 39).
FIG. 2.
VP24 is present at similar levels in all forms of intracellular HSV-1 capsids. Western blots of A, B, and C capsids purified by sucrose gradient sedimentation from HSV-1 (KOS)-infected Vero cells are shown. Threefold dilutions of capsids were run in parallel. Replicate Western blots were probed with MAbs directed against VP5 and VP23 (upper panel), MAb MCA406, which recognizes VP21 and VP22a (center panel), and MAb 9-2, directed against VP24 (lower panel). Chemiluminescent Western blots were exposed to Kodak XAR-5 film, and exposures were prepared for presentation using a Kodak DCS400 digital camera and Adobe PhotoShop 5.0.
In contrast to the bulk of the internal scaffold, which is essentially absent from A and C capsids, we found that VP24 is present in all three types of capsids, in agreement with previous reports (17, 60). Furthermore, the data in Fig. 2 show that the amount of VP24 was virtually identical in all three capsid types. These results suggest that VP24 is specifically and quantitatively retained in capsids when the bulk of the internal scaffold is lost upon DNA packaging. Thus, the behavior of VP24 in capsids resembles that of the capsid shell proteins VP5 and VP23 more closely than the behavior of the scaffold proteins VP21 and VP22a.
Characterization of capsids lacking the major scaffold protein, VP22a.
To investigate how the relative levels of VP24, VP21, and VP22a are controlled within the capsid, we used the ΔICP35 mutant virus (31). This virus contains a mutation which changes the initial methionine of VP22a to leucine (M307L mutation), preventing translation of the UL26.5 transcript (Fig. 1A and B). Although the most abundant scaffold protein, VP22a, is absent from the ΔICP35 virus-infected cells, a significant number of capsids are assembled (31). Virus yields, however, are substantially reduced from wild-type levels (31, 53).
We reasoned that capsids made during infection with the ΔICP35 virus might contain increased, compensatory amounts of UL26 scaffold proteins. If VP24 formed a structural, regulated component of the capsid shell, however, even selective pressure caused by the absence of UL26.5 protein would not result in increased incorporation of UL26 proteins.
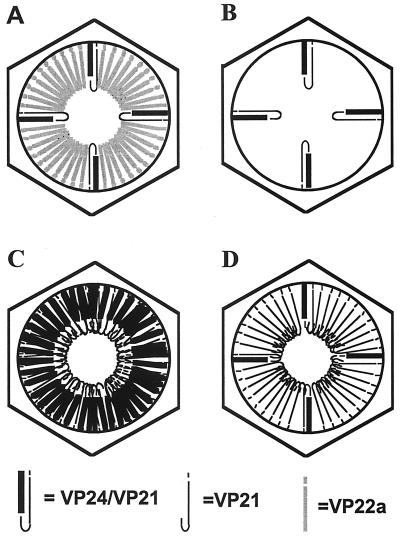
Figure 3A shows a model representing a wild-type B capsid containing 10-fold-higher levels of the UL26.5 product (VP22a) than of the UL26 products (VP24 and VP21); this model includes features depicted in models presented by others (49, 59, 64). Models B through D show hypothetical compositions of the scaffold found in ΔICP35 capsids. Model B predicts that the UL26 proteins, VP21 and VP24, bind only at specific sites within the capsid and that these sites are distinct from those occupied by the UL26.5 protein. This model predicts that ΔICP35 capsids contain a low, wild-type copy number of the VP24 and VP21 proteins. Models C and D predict that the UL26 proteins are incorporated at higher levels within the capsid to compensate for the absence of VP22a. If model C is correct, we expected to find an increased amount of both VP24 and VP21 by virtue of their linkage prior to protease processing. If model D is correct, we expected to find a low, wild-type copy number of VP24 and an increased level of VP21 to compensate for the lack of VP22a. Model D predicts that cleavage outside of the capsid might allow an increased amount of VP21 to enter the capsid after its release from VP24 or that cleavage of extra UL26 inside capsids might be accompanied by retention of high levels of VP21 but not of VP24 in sites normally occupied by VP22a.
FIG. 3.
Models of potential scaffold proteins of the ΔICP35 mutant virus capsids. Diagram A represents the scaffold composition of B capsids from wild-type HSV-1 infection. Diagrams B to D represent three different models for the potential scaffolds of ΔICP35 mutant virus capsids. The capsid shell is represented by a hexagon. The different potential scaffolds are drawn within the capsid shells. Black scaffold lines depict UL26 products, including VP24 (thick line) and VP21, and 25 aa (thin lines). Grey scaffold lines depict UL26.5 products (VP22a and 25 aa). Model B predicts that within ΔICP35 mutant capsids, the UL26 proteins VP21 and VP24 bind only at specific sites within the capsid and that these sites are distinct from those occupied by the UL26.5 protein. Models C and D predict that the UL26 proteins VP21 and VP24 are incorporated at higher levels within the mutant capsids to compensate for the absence of the UL26.5 protein, VP22a. Model C predicts an increased amount of both UL26 proteins, VP24 and VP21, by virtue of their linkage prior to protease processing. Model D predicts an increased amount of VP21 inside ΔICP35 capsids due either to import of pre-VP21 upon cleavage outside of capsids or to export of VP24 after cleavage of excess UL26 protein inside of capsids.
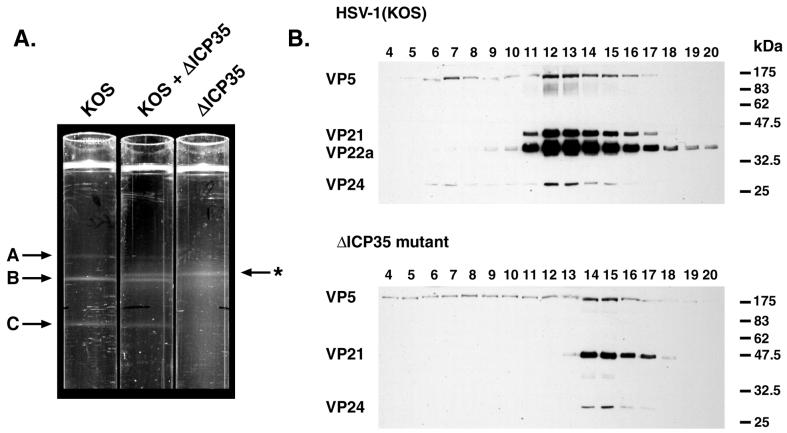
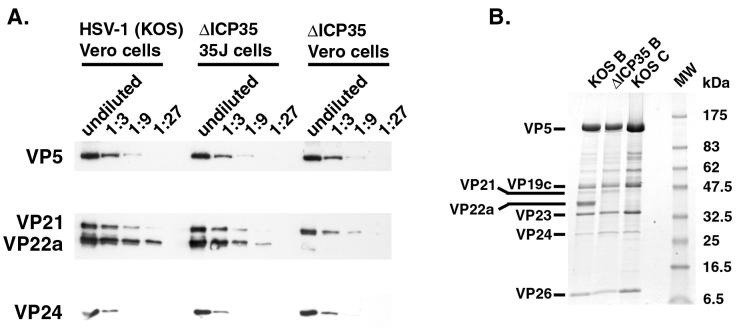
Capsids from ΔICP35 virus-infected cells were compared to those from wild-type HSV-1 (strain KOS)-infected cells. As previously reported, B capsids and a dramatically reduced number of C capsids were produced during infection with the ΔICP35 virus (31). In multiple experiments, we noticed that infection with the mutant virus produced 5- to 10-fold fewer B capsids than a wild-type infection (as judged by light-scattering bands obtained upon sucrose gradient sedimentation) (unpublished observations). Moreover, mutant B capsids appeared to sediment more slowly through sucrose gradients. To confirm this difference in sedimentation, ΔICP35 capsids were sedimented through a sucrose gradient either alone or in combination with wild-type capsids (Fig. 4A). Mutant B capsids formed a light-scattering band distinct from that of wild-type B capsids, suggesting a structural difference between the two capsid types.
FIG. 4.
Altered sucrose gradient sedimentation of mutant capsids. (A) The positions of light-scattering bands corresponding to wild-type A, B, and C capsids and ΔICP35 mutant B capsids (marked with an asterisk) are indicated by arrows adjacent to three 20 to 50% (wt/vol) sucrose gradients centrifuged in parallel. The gradients contain lysates from HSV-1 (KOS)- and ΔICP35-infected cells, as indicated. (B) Fractionated sucrose gradients of HSV-1 and ΔICP35 mutant virus-infected cells were analyzed by Western blotting with antibodies against VP5, VP21/VP22a, and VP24, as indicated. Fractions were collected beginning from the bottom of the gradient (fraction 1) to the top (fraction 20). Fractions 4 through 20 are shown. The positions of molecular size standards electrophoresed in parallel are indicated on the right (in kilodaltons). Photographs were prepared for presentation using a Kodak DCS400 digital camera and Adobe PhotoShop 5.0.
To verify that the light-scattering band contained capsids, sucrose gradient fractions were collected and analyzed by Western blotting (Fig. 4B). The wild-type B capsid peak (fractions 12 and 13 of the HSV-1 KOS gradient) contained the major capsid protein VP5 and the scaffold proteins VP21, VP22a, and VP24. As expected, the ΔICP35 B capsid peak (fractions 14 and 15) contained VP5, VP21, and VP24 but only trace amounts of VP22a. The trace amounts of VP22a are presumed to be revertants in the virus stocks, as previously reported (31). In agreement with the position of the light-scattering band in Fig. 4A, the peak of mutant B capsid proteins was shifted in position within the sucrose gradient with respect to wild-type B capsids.
The scaffold content of mutant capsids was quantitated to determine which model in Fig. 3 most closely represents the structure of ΔICP35 capsids. When similar amounts of mutant and wild-type capsids were compared (Fig. 5A), the levels of both VP24 and VP21 were unchanged from those of wild-type B capsids. The reduced level of VP22a incorporation into ΔICP35 capsids could be fully complemented by growth of the virus in a complementing cell line expressing UL26.5 (35J cells) (31).
FIG. 5.
ΔICP35 mutant capsids contain wild-type levels of VP21 and VP24. (A) Dilutions of HSV-1 KOS B capsids and ΔICP35 mutant B capsids grown on Vero cells or 35J cells (expressing pre-VP22a), as indicated, were analyzed by Western blotting with antibodies against VP5, VP21/22a, and VP24. (B) Aliquots of wild-type B capsids (labeled KOS B) and C capsids (labeled KOS C) and ΔICP35 mutant capsids (ΔICP35 B) were separated on an SDS–4 to 20% PAGE gel and stained with Coomassie brilliant blue. Molecular size standards (lane MW) were electrophoresed in parallel, and the positions of the individual standards are shown. Western blots and the Coomassie-stained gel were photographed and prepared for presentation using a Kodak DCS400 digital camera and Adobe PhotoShop 5.0.
ΔICP35 capsids were also compared to wild-type B capsids by Coomassie-stained SDS-PAGE (Fig. 5B). The capsid shell and scaffold proteins in mutant capsids appeared unchanged except for the absence of VP22a. Scans of Coomassie-stained gels confirmed that the amounts of VP21 and VP24 were not increased in mutant capsids to compensate for the absence of VP22a (unpublished observations). Figure 5B also shows that the ΔICP35 mutant capsids did not contain other, unforeseen changes in capsid protein composition.
These results demonstrate that model B of Fig. 3 correctly predicts the scaffold composition of the ΔICP35 capsids. This result is significant since it demonstrates that herpesvirus capsids can be maintained with a minimal internal scaffold composed of only UL26 products. Additionally, the number of UL26 scaffold proteins within the capsid is strictly maintained in the face of strong selective pressure caused by the absence of the major scaffold protein, VP22a.
Mutant capsids lack typical B capsid scaffold cores.
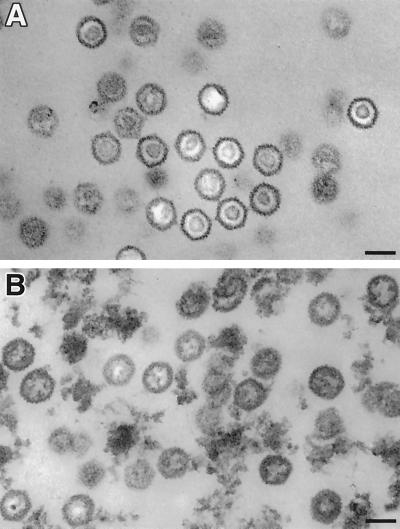
The aberrant migration of ΔICP35 capsids in sucrose gradients suggested that these capsids were structurally different from wild-type B capsids. To analyze this further, we examined purified mutant and wild-type capsids by electron microscopy. Electron micrographs of negatively stained samples showed that ΔICP35 capsids were similar in size and shape to wild-type capsids (W. W. Newcomb and J. C. Brown, unpublished observations). To optimally visualize the internal scaffold, thin-section electron microscopy was also used (Fig. 6). Wild-type B capsids contained the typical dense internal ring characteristic of processed internal scaffold proteins (Fig. 6A). In contrast, mutant capsids contained a much less dense and apparently less organized internal structure (Fig. 6B). These capsids resemble those previously seen in sections of ΔICP35-infected cells (31), confirming that sucrose gradient-stable capsids accurately represented those seen in cells.
FIG. 6.
ΔICP35 mutant capsids contain aberrant internal cores. Electron micrographs of thin sections of sucrose gradient-purified B capsids obtained from (A) HSV-1 KOS-infected cells and (B) ΔICP35 mutant virus-infected cells. Arrows indicate representative capsids. Bars, 100 nm. Negatives were scanned and prepared for presentation using Adobe PhotoShop 5.0.
The results in Fig. 4 through 6 suggested that the structure of ΔICP35 capsids is altered. However, the alteration appeared to be manifested from a change in only the VP22a scaffold protein and not from a measurable change in the capsid shell.
Increased UL26 expression does not compensate for the lack of the major scaffold protein.
The results above suggest that the capsid scaffold includes a defined amount of the UL26 products VP24 and VP21. The controlled incorporation of these proteins into the capsid could result from binding to a site distinct from that of UL26.5. This hypothesis is supported by the specific, quantitative retention of VP24 in all capsid forms. Alternatively, the lower level of UL26 expression than of UL26.5 could account for its reduced representation in capsids. We attempted to directly analyze the scaffold protein content of ΔICP35 capsids made in cells overexpressing UL26. However, efforts to isolate a cell line constitutively overexpressing full-length UL26(M307L) were unsuccessful, presumably because the protein is toxic to the cell.
To determine whether the relative expression levels of UL26 and UL26.5 control their incorporation into capsids, we decided to overexpress these proteins during ΔICP35 infection. Since it was unclear if another HSV promoter could be used to exogenously express protein at levels as high as those from the UL26.5 promoter and it was known that transient expression from the human cytomegalovirus major immediate-early promoter afforded high levels of complementation of protease and scaffold mutant viruses (53), we chose a similar approach. We constructed a plasmid, pCMV-UL26(M307L), which expresses increased levels of UL26 (containing the M307L mutation of the UL26.5 initiation codon in the ΔICP35 virus) under the control of the human cytomegalovirus major immediate-early promoter. In addition, we constructed a plasmid to overexpress the C-terminal 388 aa of UL26, known as pre-VP21 [plasmid pCMV-preVP21(M307L)]. Plasmid pCMV-preVP21(M307L) lacks N-terminal VP24 sequences and represents a UL26 product resulting from cleavage at the site between VP24 and VP21 (aa 247-248; Fig. 1) but not the site at the C terminus of the protein (aa 610-611; Fig. 1). Previously published experiments showed that pre-VP21 efficiently complemented the growth of a mutant virus that lacked both VP22a expression and the C-terminal 25-amino-acid peptide (the Prb virus [33]). Based on these experiments, it was hypothesized that complementation could result from high levels of pre-VP21 replacing VP22a as the major scaffold protein.
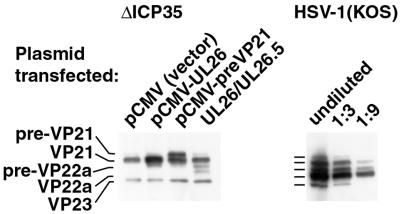
We performed transient complementation experiments with the plasmids to measure the effect of increased UL26 expression. Transfection and subsequent infection with the ΔICP35 virus resulted in increased expression of the desired proteins (Fig. 7). Expression of pCMV-UL26 resulted in largely processed products and the same minor amount of unprocessed UL26 as in the superinfection alone (Fig. 7 and data not shown). In contrast, expression of pCMV-VP21 resulted in a high level of both processed and unprocessed protein. Perhaps these differences arose from a higher efficiency of autocleavage by UL26 than that of trans-cleavage of VP21. To confirm that the overexpressed proteins were functional, we tested their ability to complement two previously described protease mutant viruses. Growth of the m100 virus, which fails to express full-length UL26 (15), was efficiently complemented by the pCMV-UL26(M307L) plasmid (unpublished observations). Likewise, the Prb virus lacking the C-terminal 25 aa of UL26 (33) was efficiently complemented by the pCMV-preVP21 plasmid, confirming its ability to function in capsid maturation (unpublished observations).
FIG. 7.
Transient overexpression of UL26(M307L) and pre-VP21(M307L). Cells were transfected with plasmids expressing the indicated proteins and subsequently infected with the ΔICP35 virus, as described in Materials and Methods. In the left panel, cell lysates were examined for the expressed proteins by Western blotting with antisera against VP21/VP22a scaffold protein and the VP23 shell protein (for standardization of virus infection). The UL26/UL26.5 lane indicates expression of UL26 and UL26.5 under control of their native promoters from plasmid pRB4090 (29). For comparison, the right panel contains threefold dilutions of a wild-type-infected cell lysate. The Western blot was photographed using a Kodak DCS400 digital camera and Adobe PhotoShop 5.0.
Further complementation experiments were performed to determine if overexpression of UL26 could relieve the growth defect of the ΔICP35 virus. Although the growth defect in the ΔICP35 virus is nonlethal (31), reproducible levels of transient complementation can be achieved (Table 1). Transfection of a plasmid containing the UL26.5 gene under its native promoter (pUC-ICP35) (31) resulted in marked complementation ΔICP35 virus growth. Transfection of the pCMV-preVP21(M307L) plasmid resulted in a similar level of complementation. In contrast, transfection of pCMV-UL26(M307L) resulted in only low levels of complementation of the ΔICP35 virus. Thus, pre-VP22a and pre-VP21 were able to efficiently complement the growth of the ΔICP35 mutant virus, while the full-length UL26(M307L) protein was not. To prove that a failure to complement by overexpression of UL26 was not a result of autocleavage and loss of the C-terminal 25-aa peptide from UL26 prior to capsid assembly, we tested the ability of the pCMV-UL26(M307L) plasmid to complement the growth of the Prb virus. In addition to lacking the C-terminal 25 aa of UL26, the Prb virus also contains the M307L mutation. We found that the pCMV-UL26(M307L) plasmid partially complemented growth of the Prb virus to the level of growth expected from the M307L virus, confirming that autocleavage does not remove the C-terminal 25 aa from the UL26 protein (data not shown). These results suggest that linkage of VP21 to VP24 in the UL26 protein limits its ability to functionally replace the most abundant scaffold protein, VP22a.
TABLE 1.
Transient complementation of ΔICP35 virus growth
| Plasmid transfected | Complementation indexa
|
||
|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | |
| pUC18 vectorb | 1.0 | 1.0 | 1.0 |
| pUC-ICP35c,d | 20.0 | 54.5 | 16.8 |
| pIRESPuro vectorc | 1.0 | 1.0 | 1.0 |
| pCMV-UL26(M307L) | 1.9 | 6.1 | 2.0 |
| pCMV-preVP21(M307L) | 14.3 | 33.3 | 10.7 |
Complementation index determined as detailed in Materials and Methods. The multiplicities of superinfection were 0.3, 1.0, and 1.0 for experiments 1, 2, and 3, respectively.
pUC18 served as the control vector for pUC-ICP35c,d (31).
pIRESPuro (Clontech) served as the control vector for pCMV-UL26(M307L) and pCMV-preVP21(M307L) (this report).
Alteration in the scaffold core influences capsid association of a DNA-packaging protein.
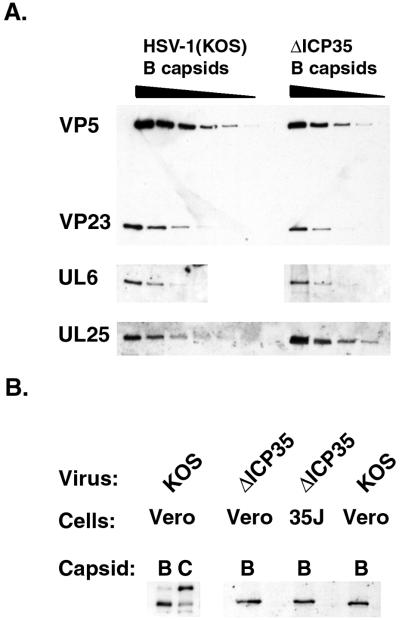
Several of the proteins essential for viral DNA cleavage and packaging are found associated with HSV-1 capsids (1, 34, 42, 43, 55, 62, 71). We theorized that the lack of a dense scaffold core might alter the ability of DNA cleavage and packaging proteins to associate with capsids. To test this hypothesis, we assayed for these proteins by Western blotting (Fig. 8). The UL6, UL15, and UL25 proteins, all of which are known to be associated with one or more forms of intracellular capsids, were detected on mutant B capsids (Fig. 8A and B). The sizes and relative amounts of both UL6 and UL15 were similar to those detected on wild-type capsids. The size species of UL15 protein differ between wild-type B and C capsids (55, 71). The UL15 proteins detected in mutant B capsid preparations with our antisera resemble those previously described on wild-type B capsids but not C capsids (71) (Fig. 8B). The UL28 protein is found predominantly on wild-type B but not C capsids (62, 71). We were able to detect the UL28 protein in ΔICP35 mutant capsids at levels similar to those found in wild-type B capsids (data not shown). In contrast to these similarities with wild-type capsids, the amount of UL25 protein associated with ΔICP35 mutant capsids was increased (Fig. 8A). To assess the level of UL25 increase and its reproducibility, we quantitated Western blots of three separate preparations of ΔICP35 mutant capsids as described in Materials and Methods (Table 2). Capsid samples were standardized to VP5, and the relative amounts of VP23 and UL25 present in each capsid preparation were determined. The levels of UL25 associated with ΔICP35 mutant capsids were approximately 2.8- to 4-fold greater than those associated with wild-type capsids. This increase in UL25 association with mutant capsids was not observed when the mutant virus was grown on the complementing cell line (35J cells) (unpublished observations). To confirm that the increased association of UL25 with mutant capsids was not a result of increased protein expression levels within mutant infected cells, we also analyzed total infected cell lysates (Table 2). The level of UL25 within mutant-infected cells was not increased and therefore could not explain the increased association of UL25 with mutant capsids. In summary, the absence of the major scaffold protein resulted in an increase in the amount of capsid-associated UL25 protein but not of the UL6, UL15, or UL28 proteins.
FIG. 8.
Proteins necessary for the cleavage and packaging of viral DNA are present on ΔICP35 capsids. (A) Twofold dilutions of wild-type B capsids and ΔICP35 B capsids were analyzed by Western blotting with antibodies to VP5 and VP23 (upper panel), the UL6 protein (center panel), and the UL25 protein (lower panel). (B) Wild-type B and C capsids and ΔICP35 mutant capsids were analyzed by Western blotting with an antiserum raised against the UL15 protein. The left panel shows the reactivity of B and C capsids with the UL15 serum. The right panel shows capsids from ΔICP35 virus grown on Vero cells and complementing 35J Vero cells that express UL26.5. Western blots were photographed using a Kodak DCS400 digital camera and Adobe PhotoShop 5.0.
TABLE 2.
UL25 protein levels in ΔICP35 mutant capsids and infected cellsa
| Protein | Relative protein level (% of control)
|
|||||
|---|---|---|---|---|---|---|
| Expt 1
|
Expt 2
|
Expt 3
|
||||
| Capsids | Cells | Capsids | Cells | Capsids | Cells | |
| VP5 | 100 | NDb | 100 | 100 | 100 | 100 |
| VP23 | 125 | ND | 77 | 85 | 73 | 66 |
| UL25 | 279 | ND | 403 | 134 | 287 | 76 |
Protein levels were determined as described in Materials and Methods and are presented as the percentage of each protein in ΔICP35 capsids (capsids) or total infected cell lysates (cells) relative to that found in HSV-1 KOS B capsids or HSV-1 KOS total infected-cell lysates, examined in parallel. Comparisons between samples and experiments were performed by setting levels for HSV-1 KOS to 100%. Comparisons within samples were performed by setting levels of VP5 to 100%.
ND, not done.
DISCUSSION
The results presented in this paper point toward the existence of a mechanism to control the differential incorporation and retention of the UL26 and UL26.5 scaffold proteins in HSV-1 capsids. The copy number of UL26 products VP21 and VP24 within ΔICP35 B capsids was strictly maintained, even in the face of selective pressure exerted by the lack of UL26.5 expression. ΔICP35 virus growth is significantly more impaired at low MOIs, leading to the hypothesis that increased levels of UL26 at high MOIs partially complement the UL26.5 defect (31). However, mutant capsids isolated from high-MOI infections contained only wild-type levels of UL26 proteins. Moreover, transient-complementation results (Table 1) indicated that increased expression of UL26 did not relieve the ΔICP35 growth defect, consistent with the model that the incorporation of UL26 proteins into capsids is independent of protein expression levels. Differences between the capsid association of the UL26 and UL26.5 proteins are also evident upon examination of DNA-containing capsids. Although the UL26.5 protein VP22a and the C-terminal product of UL26, VP21, are absent from capsids containing DNA, the N-terminal protease domain of UL26, VP24, is present in quantitatively similar amounts in capsids containing DNA (C capsids), containing scaffold (B capsids), or lacking both DNA and scaffold (A capsids).
Our results are consistent with a model in which distinct, specific binding sites exist within the capsid for the UL26 and UL26.5 proteins. The C-terminal 25-aa tail common to both UL26 and UL26.5 proteins has been shown to bind VP5 (22, 41). This interaction is thought to anchor the scaffold proteins to the capsid shell and is required to produce sealed capsids (23, 33, 64). However, our transient-complementation experiments revealed that despite possessing the entire UL26.5 protein within its C terminus, full-length UL26 did not efficiently complement the growth of the ΔICP35 virus. In contrast to full-length UL26, overexpressed pre-VP21 did functionally complement ΔICP35 growth. Based upon these results, we hypothesize that sequences within VP24 either prevent binding of the UL26 protein to UL26.5-specific sites within the capsid or direct binding to a distinct, UL26-specific binding site.
Because our complementation experiments did not involve direct examination of the scaffold content of capsids, we can only conclusively state that overexpression of UL26 does not compensate for the growth defect of the ΔICP35 virus. However, these results are consistent with the idea that protein expression levels are not involved in regulation of UL26 incorporation into capsids. An alternative explanation for the failure of UL26 overexpression to complement ΔICP35 growth is that increased UL26 inhibits capsid assembly or packaging. It has been observed upon assembly of HSV-1 capsids in recombinant-baculovirus-infected cells that the absence of UL26.5 results in large numbers of incomplete shells and few intact capsids (61, 65). We also cannot exclude the formal possibility that UL26 overexpression could result in capsids that are unable to package DNA or otherwise mature.
We find that capsids lacking VP22a are structurally unique. In electron micrographs, the external shell of ΔICP35 capsids appeared normal, but the dense internal core characteristic of wild-type HSV-1 capsids was absent and sucrose gradient sedimentation of ΔICP35 capsids was altered. Capsids similar in appearance to those of ΔICP35 are also seen upon infection of insect cells with recombinant baculoviruses expressing HSV-1 capsid shell and UL26 proteins (61, 65). Therefore, only a minimal scaffold composed of wild-type amounts of UL26 proteins was sufficient for the formation and maintenance of HSV-1 capsid shells.
The simplest interpretation of the above data is that mutant capsids were actually assembled with the same minimal internal scaffold. However, the idea that capsids can be assembled with a limited number of scaffold molecules is not entirely consistent with the model for capsid assembly proposed by Newcomb et al. (38, 40). This model predicts that capsid subunits, each composed of a VP5 molecule in complex with two scaffolding molecules, are added one at a time to the growing capsid shell. Our results with the ΔICP35 virus imply that a majority of capsid subunits can polymerize without interacting with a scaffolding protein. Precedent exists for the polymerization of capsid shell proteins in the absence of scaffold proteins. Desai et al. reported that in the complete absence of UL26 and UL26.5 proteins, the capsid shell proteins VP5, VP19c, VP23, and VP26 could organize into higher-order structures or “sheets” containing defined hexons (13). Similar structures were also found upon expression of HSV-1 capsid shell proteins in the recombinant baculovirus system (61, 65). Additionally, aberrant HSV-1 capsid particles lacking both UL26 and UL26.5 scaffold proteins and VP23 have been constructed in the baculovirus system and found to be altered in size and symmetry (54). Perhaps a scaffolding protein need only be present at certain crucial points within the growing capsid shell to allow proper size, symmetry, and closure of the capsid. The fact that ΔICP35 capsids are not altered in size or symmetry demonstrates that UL26 can competently perform these scaffolding functions alone. It is possible that the model proposed by Newcomb et al. is operative under optimal conditions during wild-type virus infection but that assembly of ΔICP35 capsids operates by a less efficient, alternative pathway under conditions of limited scaffold availability. Alternatively, the finding that procapsids contain somewhat higher levels of scaffold proteins than B capsids (40) raises the possibility that the ΔICP35 capsids are initially assembled with higher levels of UL26 proteins. Further structural analyses of ΔICP35 capsids may provide insight into the types of scaffolding protein and capsid shell interactions that are minimally essential for proper capsid assembly.
Separate capsid binding sites for UL26 and UL26.5 proteins could also explain the retention of VP24 in wild-type C capsids after DNA packaging. It has been proposed that retention of VP24 within C capsids might result from either the lack of an export signal or the presence of a specific capsid retention signal (49). We favor the latter explanation, as we have found that if cleavage between VP24 and VP21 is prevented (A247S virus [32]), uncleaved UL26 protein containing both the VP24 and VP21 proteins is retained in C capsids (A. K. Sheaffer and D. J. Tenney, unpublished observations). These results support the existence of a unique signal that dictates retention of VP24 and is dominant over the signal in VP21 that directs its export upon DNA packaging. In addition, a recent publication by Desai and Person (12) demonstrates that a scaffold mutation which decreases the level of VP24 retained within capsids can be overcome by second-site suppressor mutations within the N terminus of VP5. This observation is consistent with the presence of a distinct capsid-binding site for VP24, as suggested by the authors (12).
While our findings allude to a separate binding site for VP24, its precise location within the capsid remains elusive. There are technical difficulties in obtaining mature, angular, sucrose-stable capsids with and without VP24 for definitive structural comparisons, since VP24 protease activity is required for their generation (37, 38, 40, 48, 66). Recently, however, reconstructions of procapsids generated by infection with HSV-1 protease mutant viruses have allowed comparison of capsids containing an inactive, temperature-sensitive form of the UL26 protein with those lacking the UL26 protein (40). These studies revealed additional density near the center of procapsids containing UL26, suggesting that in the procapsid the unique N terminus of UL26 may be directed towards the center of the capsid (40). This orientation of UL26 does not suggest a simple explanation for its retention within C capsids, as the N terminus would not be in close proximity to a binding site on the interior of the capsid shell. It is possible, however, that binding of VP24 to the capsid shell occurs only after cleavage of the UL26 protein. In fact, this hypothesis is consistent with the observation that removal of the C-terminal 25 aa from UL26 is required for the quantitative retention of VP24 within angular capsids (46), perhaps because this cleavage is required to generate the binding site for VP24. If this hypothesis is correct, the capsid incorporation of UL26 protein and the retention of VP24 may be controlled by two distinct and as yet poorly understood mechanisms.
The retention of VP24 within mature, DNA-containing capsids suggests that VP24 may be required to carry out an additional function following the cleavage of the internal scaffold proteins. This is consistent with the observation that two separable functions of the UL26 protein, one requiring enzyme activity and the other requiring release of VP24 from VP21, are essential for virus growth (52). It is possible that the association of VP24 with the capsid shell is required for additional processes such as packaging and retention of viral DNA within capsids, recruitment of tegument proteins to the capsid, or egress of the capsid from the nucleus. It has also been suggested that VP24 might play a role in subsequent entry of virions into the host cell (49). Although analysis of a UL26 temperature-sensitive mutant virus revealed no defect in virus entry (48, 49), the analysis of additional mutations within UL26 may be warranted.
One intriguing observation about ΔICP35 virus infection is that although viral DNA is synthesized at wild-type levels, concatameric DNA is not efficiently cleaved to unit length and packaged into capsids (31). In support of this, we found that upon sucrose gradient sedimentation, the intensity of the light-scattering band containing C capsids was reduced relative to that of the B capsid band (Fig. 4A). While further studies are required to fully understand the precise nature of this defect, the data presented in this paper suggest some potential explanations. The results may point to a role for the scaffold in the formation of capsids competent for packaging viral DNA or to a more direct role of the scaffold protein in DNA cleavage and packaging.
One possible scenario for the impairment in DNA packaging is that ΔICP35 capsids are merely less stable than wild-type capsids. This instability, resulting from the presence of a minimal amount of internal scaffold proteins, might render mutant capsids less able to withstand the rigors of packaging viral DNA. Indeed, we found that ΔICP35 mutant capsids did not tolerate repeated banding on sucrose gradients as well as did wild-type B capsids (unpublished observations).
A second possibility is that the close temporal relationship between viral DNA packaging and release of scaffold from the capsid could point to a more dynamic role for the scaffold proteins in DNA packaging. Studies using a virus with a reversible temperature-sensitive mutation in the protease demonstrated that the kinetics of scaffold processing, DNA cleavage, and DNA packaging are indistinguishable (6, 7). Mechanisms for the linkage of these processes have been proposed for bacteriophage P22, which in many respects serves as a model for herpesvirus capsid assembly and DNA packaging (18). Analysis of bacteriophage P22 scaffolding protein mutants resulted in the proposal that the scaffold may sense the entry of DNA into capsids and transmit signals for capsid structural changes (angularization and stabilization) and scaffold egress from the capsid (19). Oligomerization of herpesvirus scaffold proteins (11, 44) might facilitate the transmission of such a signal. The absence of close proximity of adjacent scaffold molecules in the minimal UL26 scaffold of ΔICP35 capsids could lead to the poor transmission of these signals and result in inefficient DNA packaging.
An alternative reason for inefficient DNA packaging during ΔICP35 virus infection is that the defect is at the level of interaction of mutant capsids with other HSV-1 proteins. Seven HSV-1 genes have been found to be essential for the cleavage and stable packaging of viral DNA into capsids: UL6, UL15, UL17, UL25, UL28, UL32, and UL33 (2, 9, 25, 26, 34, 43, 56, 63, 70). Although the functions of these proteins are not yet clear, at least four of the proteins (UL6, UL15, UL25, and UL28) are found associated with capsids (1, 34, 42, 55, 62, 71). A precedent exists for the involvement of bacteriophage scaffolding proteins in the recruitment of DNA cleavage and packaging proteins to the capsid. Mutations within the phage P22 scaffold protein can result in the assembly of capsids that fail to incorporate the portal protein and other DNA-packaging proteins (19). Since the portal protein forms a unique capsid vertex through which viral DNA is packaged, these scaffolding mutant bacteriophages are unable to package viral DNA.
We found that all four known capsid-associated HSV-1 DNA-packaging proteins were associated with capsids lacking the major scaffold protein. In fact, the level of UL25 protein was increased relative to that in wild-type B capsids. This finding is of interest, since the UL25 protein is not essential for DNA cleavage but is hypothesized to maintain stable packaging of cleaved viral DNA within capsids (34). Based upon our observation that ΔICP35 mutant capsids contain decreased amounts of scaffold proteins but increased levels of UL25 protein, we can speculate that perhaps during wild-type infection the scaffold proteins prevent premature association of UL25 protein with capsids. A role for UL25 in sealing packaged DNA within capsids (as suggested earlier [34]) is consistent with our finding that approximately four- to eightfold-higher levels of UL25 protein are found associated with DNA-containing C capsids than with B capsids (unpublished observations). It will be of interest to determine where UL25 binds to the capsid and how it might carry out its role in DNA packaging. In the case of ΔICP35 mutant capsids, perhaps premature association of UL25 might block efficient scaffold release and/or entry of DNA into mutant capsids.
ACKNOWLEDGMENTS
We gratefully acknowledge Rich Colonno for supporting these studies. We thank Bernard Roizman for supplying the pRB4090 plasmid and Nels Pederson for antiserum against the UL28 protein. We thank our colleagues Carmela Lamberti, Dong Yu, Barbara Robertson, and Patrick McCann for reagents and helpful discussions. We especially appreciate insightful conversations about this work with Valerie Preston and Frazer Rixon.
W.W.N. and J.C.B. were supported by NIH grant AI41644 and NSF award MCB9904879, and S.K.W. was supported by National Institutes of Health grant AI 37549.
REFERENCES
- 1.Ali M A, Forghani B, Cantin E M. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology. 1996;216:278–283. doi: 10.1006/viro.1996.0061. [DOI] [PubMed] [Google Scholar]
- 2.Baines J D, Cunningham C, Nalwanga D, Davison A. The U(L)15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the U(L)15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booy F P, Newcomb W W, Trus B L, Brown J C, Baker T S, Steven A C. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway J F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chi J H I, Wilson D W. ATP-dependent localization of the herpes simplex virus capsid protein VP26 to sites of procapsid maturation. J Virol. 2000;74:1468–1476. doi: 10.1128/jvi.74.3.1468-1476.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church G A, Dasgupta A, Wilson D W. Herpes simplex virus DNA packaging without measurable DNA synthesis. J Virol. 1998;72:2745–2751. doi: 10.1128/jvi.72.4.2745-2751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Church G A, Wilson D W. Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol. 1997;71:3603–3612. doi: 10.1128/jvi.71.5.3603-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa R H, Cohen G, Eisenberg R, Long D, Wagner E. Direct demonstration that the abundant 6-kilobase herpes simplex virus type 1 mRNA mapping between 0.23 and 0.27 map units encodes the major capsid protein VP5. J Virol. 1984;49:287–292. doi: 10.1128/jvi.49.1.287-292.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham C, Davison A J. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology. 1993;197:116–124. doi: 10.1006/viro.1993.1572. [DOI] [PubMed] [Google Scholar]
- 10.Davison M D, Rixon F J, Davison A J. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J Gen Virol. 1992;73:2709–2713. doi: 10.1099/0022-1317-73-10-2709. [DOI] [PubMed] [Google Scholar]
- 11.Desai P, Person S. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology. 1996;220:516–521. doi: 10.1006/viro.1996.0341. [DOI] [PubMed] [Google Scholar]
- 12.Desai P, Person S. Second site mutations in the N-terminus of the major capsid protein (VP5) overcome a block at the maturation cleavage site of the capsid scaffold proteins of herpes simplex virus type 1. Virology. 1999;261:357–366. doi: 10.1006/viro.1999.9877. [DOI] [PubMed] [Google Scholar]
- 13.Desai P, Watkins S C, Person S. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J Virol. 1994;68:5365–5374. doi: 10.1128/jvi.68.9.5365-5374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiIanni C L, Drier D A, Deckman I C, McCann P J, Liu F, Roizman B, Colonno R J, Cordingley M G. Identification of the herpes simplex virus-1 protease cleavage sites by direct sequence analysis of autoproteolytic cleavage products. J Biol Chem. 1993;268:2048–2051. [PubMed] [Google Scholar]
- 15.Gao M, Matusick-Kumar L, Hurlburt W, DiTusa S F, Newcomb W W, Brown J C, McCann P J, Deckman I, Colonno R J. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J Virol. 1994;68:3702–3712. doi: 10.1128/jvi.68.6.3702-3712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson W, Hall M R. Assemblin, an essential herpesvirus proteinase. Drug Design Discovery. 1997;15:39–47. [PubMed] [Google Scholar]
- 17.Gibson W, Roizman B. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greene B, King J. In vitro unfolding/refolding of wild type phage P22 scaffolding protein reveals capsid-binding domain. J Biol Chem. 1999;274:16135–16140. doi: 10.1074/jbc.274.23.16135. [DOI] [PubMed] [Google Scholar]
- 19.Greene B, King J. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology. 1996;225:82–96. doi: 10.1006/viro.1996.0577. [DOI] [PubMed] [Google Scholar]
- 20.Holland L E, Sandri-Goldin R M, Goldin A L, Glorioso J C, Levine M. Transcriptional and genetic analyses of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J Virol. 1984;49:947–959. doi: 10.1128/jvi.49.3.947-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Homa F L, Brown J C. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 22.Hong Z, Beaudet-Miller M, Durkin J, Zhang R, Kwong A D. Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein. J Virol. 1996;70:533–540. doi: 10.1128/jvi.70.1.533-540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennard J, Rixon F J, McDougall I M, Tatman J D, Preston V G. The 25 amino acid residues at the carboxy terminus of the herpes simplex virus type 1 UL26.5 protein are required for the formation of the capsid shell around the scaffold. J Gen Virol. 1995;76:1611–1621. doi: 10.1099/0022-1317-76-7-1611. [DOI] [PubMed] [Google Scholar]
- 24.Koslowski K M, Shaver P R, Wang X Y, Tenney D J, Pederson N E. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J Virol. 1997;71:9118–9123. doi: 10.1128/jvi.71.12.9118-9123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamberti C, Weller S K. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72:2463–2473. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamberti C, Weller S K. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 27.Liu F Y, Roizman B. Characterization of the protease and other products of amino-terminus-proximal cleavage of the herpes simplex virus 1 UL26 protein. J Virol. 1993;67:1300–1309. doi: 10.1128/jvi.67.3.1300-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu F Y, Roizman B. Differentiation of multiple domains in the herpes simplex virus 1 protease encoded by the UL26 gene. Proc Natl Acad Sci USA. 1992;89:2076–2080. doi: 10.1073/pnas.89.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F Y, Roizman B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F Y, Roizman B. The promoter, transcriptional unit, and coding sequence of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J Virol. 1991;65:206–212. doi: 10.1128/jvi.65.1.206-212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matusick-Kumar L, Hurlburt W, Weinheimer S P, Newcomb W W, Brown J C, Gao M. Phenotype of the herpes simplex virus type 1 protease substrate ICP35 mutant virus. J Virol. 1994;68:5384–5394. doi: 10.1128/jvi.68.9.5384-5394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matusick-Kumar L, McCann III P J, Robertson B J, Newcomb W W, Brown J C, Gao M. Release of the catalytic domain No from the herpes simplex virus type 1 protease is required for viral growth. J Virol. 1995;69:7113–7121. doi: 10.1128/jvi.69.11.7113-7121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matusick-Kumar L, Newcomb W W, Brown J C, McCann P J, Hurlburt W, Weinheimer S P, Gao M. The C-terminal 25 amino acids of the protease and its substrate ICP35 of herpes simplex virus type 1 are involved in the formation of sealed capsids. J Virol. 1995;69:4347–4356. doi: 10.1128/jvi.69.7.4347-4356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newcomb W W, Brown J C. Use of Ar+ plasma etching to localize structural proteins in the capsid of herpes simplex virus type 1. J Virol. 1989;63:4697–4702. doi: 10.1128/jvi.63.11.4697-4702.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 38.Newcomb W W, Homa F L, Thomsen D R, Trus B L, Cheng N, Steven A, Booy F, Brown J C. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J Virol. 1999;73:4239–4250. doi: 10.1128/jvi.73.5.4239-4250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 40.Newcomb W W, Trus B L, Cheng N, Steven A C, Sheaffer A K, Tenney D J, Weller S K, Brown J C. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J Virol. 2000;74:1663–1673. doi: 10.1128/jvi.74.4.1663-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oien N L, Thomsen D R, Wathen M W, Newcomb W W, Brown J C, Homa F L. Assembly of herpes simplex virus capsids using the human cytomegalovirus scaffold protein: critical role of the C terminus. J Virol. 1997;71:1281–1291. doi: 10.1128/jvi.71.2.1281-1291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel A H, MacLean J B. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology. 1995;206:465–478. doi: 10.1016/s0042-6822(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 43.Patel A H, Rixon F J, Cunningham C, Davison A J. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology. 1996;217:111–123. doi: 10.1006/viro.1996.0098. [DOI] [PubMed] [Google Scholar]
- 44.Pelletier A, Dô F, Brisebois J J, Lagacé L, Cordingley M G. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J Virol. 1997;71:5197–5208. doi: 10.1128/jvi.71.7.5197-5208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perdue J L, Cohen J C, Randall C C, O'Callaghan D J. Biochemical studies of the maturation of herpesvirus nucleocapsid species. Virology. 1976;74:194–208. [PubMed] [Google Scholar]
- 46.Person S, Desai P. Capsids are formed in a mutant virus blocked at the maturation site of the UL26 and UL26.5 open reading frames of herpes simplex virus type 1 but are not formed in a null mutant of UL38 (VP19C) Virology. 1998;242:193–203. doi: 10.1006/viro.1997.9005. [DOI] [PubMed] [Google Scholar]
- 47.Person S, Laquerre S, Desai P, Hempel J. Herpes simplex virus type 1 capsid protein, VP21, originates within the UL26 open reading frame. J Gen Virol. 1993;74:2269–2273. doi: 10.1099/0022-1317-74-10-2269. [DOI] [PubMed] [Google Scholar]
- 48.Preston V G, Coates J A, Rixon F J. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J Virol. 1983;45:1056–1064. doi: 10.1128/jvi.45.3.1056-1064.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 50.Rixon F J, Cross A M, Addison C, Preston V G. The products of herpes simplex virus type 1 gene UL26 which are involved in DNA packaging are strongly associated with empty but not with full capsids. J Gen Virol. 1988;69:2879–2891. doi: 10.1099/0022-1317-69-11-2879. [DOI] [PubMed] [Google Scholar]
- 51.Rixon F J, Davison M D, Davison A J. Identification of the genes encoding two capsid proteins of herpes simplex virus type 1 by direct amino acid sequencing. J Gen Virol. 1990;71:1211–1214. doi: 10.1099/0022-1317-71-5-1211. [DOI] [PubMed] [Google Scholar]
- 52.Robertson B J, McCann P J, Matusick-Kumar L, Newcomb W W, Brown J C, Colonno R J, Gao M. Separate functional domains of the herpes simplex virus type 1 protease: evidence for cleavage inside capsids. J Virol. 1996;70:4317–4328. doi: 10.1128/jvi.70.7.4317-4328.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robertson B J, McCann P J, Matusick-Kumar L, Preston V G, Gao M. Na, an autoproteolytic product of the herpes simplex virus type 1 protease, can functionally substitute for the assembly protein ICP35. J Virol. 1997;71:1683–1687. doi: 10.1128/jvi.71.2.1683-1687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saad A, Zhou Z H, Jakana J, Chiu W, Rixon F J. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J Virol. 1999;73:6821–6830. doi: 10.1128/jvi.73.8.6821-6830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salmon B, Baines J D. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of U(L)15-encoded proteins with B capsids requires at least the U(L)6, U(L)17, and U(L)28 genes. J Virol. 1998;72:3045–3050. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salmon B, Cunningham C, Davison A J, Harris W J, Baines J D. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sherman G, Bachenheimer S. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 58.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steven A, Spear P. Herpesvirus capsid assembly and envelopment. In: Chiu W, Burnett R M, Garcea R L, editors. Structural biology of viruses. Oxford, England: Oxford University Press; 1997. pp. 312–351. [Google Scholar]
- 60.Stevenson A J, Morrison E E, Chaudhari R, Yang C C, Meredith D M. Processing and intracellular localization of the herpes simplex virus type 1 proteinase. J Gen Virol. 1997;78:671–675. doi: 10.1099/0022-1317-78-3-671. [DOI] [PubMed] [Google Scholar]
- 61.Tatman J D, Preston V G, Nicholson P, Elliott R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 62.Taus N S, Baines J D. Herpes simplex virus 1 DNA cleavage/packaging: the UL28 gene encodes a minor component of B capsids. Virology. 1998;252:443–449. doi: 10.1006/viro.1998.9475. [DOI] [PubMed] [Google Scholar]
- 63.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomsen D R, Newcomb W W, Brown J C, Homa F L. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J Virol. 1995;69:3690–3703. doi: 10.1128/jvi.69.6.3690-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 67.Trus B L, Newcomb W W, Booy F P, Brown J C, Steven A C. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc Natl Acad Sci USA. 1992;89:11508–11512. doi: 10.1073/pnas.89.23.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weinheimer S P, McCann P J, O'Boyle D R, Stevens J T, Boyd B A, Drier D A, Yamanaka G A, DiIanni C L, Deckman I C, Cordingley M G. Autoproteolysis of herpes simplex virus type 1 protease releases an active catalytic domain found in intermediate capsid particles. J Virol. 1993;67:5813–5822. doi: 10.1128/jvi.67.10.5813-5822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wildy P, Russell W C, Horne R W. The morphology of herpes virus. Virology. 1960;12:204–222. doi: 10.1016/0042-6822(60)90195-1. [DOI] [PubMed] [Google Scholar]
- 70.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu D, Weller S K. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J Virol. 1998;72:7428–7439. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Z H, Macnab S J, Jakana J, Scott L R, Chiu W, Rixon F J. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc Natl Acad Sci USA. 1998;95:2778–2783. doi: 10.1073/pnas.95.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Z H, Prasad B V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]