Abstract
Long non-coding RNAs (lncRNAs) have gathered significant attention due to their pivotal role in plant growth, development, and biotic and abiotic stress resistance. Despite this, there is still little understanding regarding the functions of lncRNA in these domains in the tea plant (Camellia sinensis), mainly attributable to the insufficiencies in gene manipulation techniques for tea plants. In this study, we designed a novel strategy to identify evolutionarily conserved trans-lncRNA (ECT-lncRNA) pairs in plants. We used highly consistent base sequences in the exon-overlapping region between trans-lncRNAs and their target gene transcripts. Based on this method, we successfully screened 24 ECT-lncRNA pairs from at least two or more plant species. In tea, as observed in model plants such as Arabidopsis, alfalfa, potatoes, and rice, there exists a trans-lncRNA capable of forming an ECT-lncRNA pair with transcripts of the 12-oxophytodienoate reductase (OPR) family, denoted as the OPRL/OPR pair. Considering evolutionary perspectives, the OPRL gene cluster in each species likely originates from a replication event of the OPR gene cluster. Gene manipulation and gene expression analysis revealed that CsOPRL influences disease resistance by regulating CsOPR expression in tea plants. Furthermore, the knockout of StOPRL1 in Solanum tuberosum led to aberrant growth characteristics and strong resistance to fungal infection. This study provides insights into a strategy for the screening and functional verification of ECT-lncRNA pairs.
Introduction
Non-coding RNAs (ncRNAs) play pivotal roles in numerous biological processes across organisms [1–3]. Long non-coding RNAs (lncRNAs) constitute a subclass of ncRNAs characterized by transcripts exceeding 200 nucleotides in length and poor protein-coding potential. They are commonly classified based on their genomic localization and mode of gene expression regulation, either in cis or trans. For example, lncRNAs can be categorized as natural antisense transcripts (NATs), overlapping lncRNAs (OT-lncRNAs), long intergenic non-coding RNAs (lincRNAs), and intronic non-coding RNAs (incRNAs) [4, 5].
Identifying functional non-coding transcripts from databases containing more lncRNAs poses a significant challenge [6]. LncRNAs exhibit diverse roles in physiological processes within plants, engaging in chromosome silencing, genomic imprinting, chromatin modification, transcriptional activation and interference, control of alternative splicing, regulation of protein translation and transport, and regulation of miRNA function [7]. Several functional roles of lncRNAs in plants have been documented. For example, cold-assisted intronic non-coding RNA (COLDAIR) regulates histone methylation within the chromatin region of flowering locus c (FLC), a pivotal flowering gene encoding a protein crucial for flowering inhibition, thereby regulating vernalization in Arabidopsis [8]. Additionally, the lncRNA ELENA1, in conjugation with the mediator of RNA polymerase II transcription subunit 19a-like protein (MED19a), collectively promotes the expression of pathogenesis-related gene1 (PR1) in response to pathogenic infections in plants. Mechanistically, ELENA1 impedes fibrillarin (FIB2) binding to MED19a, facilitating FIB2 release from the PR1 promoter and augmenting PR1 expression [9].
Reports on functional lncRNAs in tea plants are rare, with only a few studies using sequencing data for analysis [10, 11]. The unique characteristics of lncRNAs, including weak sequence conservation, low expression levels, strong tissue and organ specificity, and inducible expression, present challenges in accurately estimating their abundance in animals and plants. Moreover, their weak sequence conservation hinders identifying lncRNA functions in non-model plants, such as tea plants, which lack a robust genetic transformation system. Currently, the Agrobacterium-mediated transient gene expression system and antisense oligodeoxynucleotide (AsODN)-based gene expression inhibition are widely used for functional verification of lncRNA gene pairs in tea plants [12, 13].
Jasmonic acid (JA) and its derivatives are crucial regulators in plant defense mechanisms against insects and necrotic pathogens [14, 15]. The biosynthesis of JA initiates within the chloroplasts, where the substrate α-linolenic acid (C18:3) undergoes catalysis by 13 lipoxygenases (13-LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC), leading to the formation of 12-oxo-phytodienoic acid (12-OPDA), an intermediate compound in JA synthesis. Subsequently, 12-OPDA translocates to the peroxisome and is enzymatically converted into JA by the catalytic action of OPDA reductase (OPR). OPR plays a pivotal role as a critical enzyme in synthesizing JA [16, 17].
Phylogenetic tree analysis facilitates the classification of OPR within the various plant populations into seven distinct subgroups [18]. Furthermore, plant OPRs can be categorized into two subtypes: OPRI and OPRII. Members of the OPRI subfamily preferentially catalyze cis-(−)-OPDA as a substrate, while those of the OPRII subfamily promote the reduction of cis-(+)-OPDA [19, 20]. OPRII subfamily members are primarily responsible for JA synthesis in plants. For example, Arabidopsis OPR3, belonging to the OPRII subfamily, exhibits a gene mutation resulting in merely 1/500 of the JA content of the wild type (WT), leading to male-sterility phenotypes in Arabidopsis [21, 22]. Although OPRI subfamily members are believed to play a secondary role in JA synthesis, they are involved in plant resistance regulation [23]. However, the mechanism by which OPRI subfamily members regulate plant resistance remains unclear. Literature reports suggest that OPR2, an OPRI subfamily in Arabidopsis thaliana, can convert the direct JA precursor, 4,5-didehydro-JA, into JA, thereby enhancing plant resistance to pathogens and insect feeding [22]. In this study, evolutionary conservative trans-lncRNA (ECT-lncRNA) pairs were identified based on conserved sequences in the overlapping regions between lncRNAs and their target genes in tea plants and conserved sequences in the overlapping regions of lncRNAs between tea plants and other plant species. Several OPRL/OPR pairs were identified from tea plants, Arabidopsis, alfalfa, potato, and rice, in which the OPR belonged to the OPRI subfamily. Experimental findings demonstrate that OPRLs influence the growth, development, and disease resistance of tea and potato plants subjected to gene manipulation. Overall, this study elucidates the regulatory mechanism of plant OPRI subfamily members and confirms their involvement in regulating plant resistance, growth, and development.
Results
Prediction of ECT-lncRNA pairs in plants
To predict the ECT-lncRNA pairs in plants, we first compiled transcript data of lncRNA and mRNAs from stress-treated tea materials in our research group, as detected by a sequencing company. Additionally, transcript data of lncRNAs and mRNAs from various plant species were obtained from public databases, such as CANTATAdb and NCBI. A total of 5101 lncRNAs were identified from 48 000 transcripts from the treated tea seedlings. Through lncRNA–mRNA interaction analysis, 3313 cis-lncRNAs and 2904 trans-lncRNAs were identified. Detailed information on the identified cis-lncRNAs and trans-lncRNAs is provided in Supplementary Data Table S1.
Based on the cDNA sequence consistency between lncRNAs and target genes, 24 candidate gene pairs were screened from 2904 tea trans-lncRNAs and mRNA data (Table 1). The standard for 24 candidate gene pairs is that there is a highly consistent overlap region of base sequences between lncRNA and target gene exons, and the consistency of base sequences in the overlap region is >80%. These ECT-lncRNA pairs accounted for only 0.8% of the total trans-lncRNAs in tea plants. In order to verify the authenticity of ECT-lncRNAs in tea plants, edna we cloned these 24 ECT-lncRNAs, of which 20 were cloned by us (Supplementary Data Fig. S1). Furthermore, following the same prediction procedure, using 24 candidate gene pairs in tea plants as bait sequences for screening, based on the consistency of their overlapping region bases, their orthologous genes were found in at least one plant species (Table 2).
Table 1.
ECT-lncRNA pairs screened from tea plant genome data.
| LncRNA | Target genes | |||||
|---|---|---|---|---|---|---|
| Target ID | Sequence identity with lncRNA (%) | Annotation | Sequence identity in overlapping region between lncRNAs and their target genes (%) | Sequence consistency a | ||
| 1 | LTCONS_00008922 | TEA022935.1 | 23 | Magnesium transporter MRS2–4-like | 97 | + |
| 2 | LTCONS_00007788 | TEA012272.1 | 29 | Eukaryotic initiation factor 4A-2 | 99 | + |
| 3 | LTCONS_00044543 | TEA004681.1 | 22 | Zinc finger CCCH domain-containing protein 53 isoform X1 | 97 | + |
| 4 | LTCONS_00026852 | TEA025907.1 | 20 | 12-Oxophytodienoate reductase | 99 | + |
| 5 | LTCONS_00004664 | TEA022107.1 | 10 | Histone H2B.3 | 100 | − |
| 6 | LTCONS_00016955 | TEA012518.1 | 41 | Protein NUCLEAR FUSION DEFECTIVE 6 | 99 | + |
| 7 | LTCONS_00064414 | TEA033352.1 | 25 | Elongation factor 1-alpha-like | 88 | − |
| 8 | LTCONS_00064957 | TEA000254.1 | 23 | Transmembrane 9 superfamily member 4-like | 91 | + |
| 9 | LTCONS_00005358 | TEA000289.1 | 45 | Cytochrome b-c1 complex subunit Rieske-4 | 98 | + |
| 10 | LTCONS_00049549 | TEA007641.1 | 14 | Vesicle-associated membrane protein 721-like | 96 | + |
| 11 | LTCONS_00010505 | TEA009846.1 | 17 | Dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase | 99 | − |
| 12 | LTCONS_00051279 | TEA022308.1 | 35 | Haloacid dehalogenase-like hydrolase domain-containing protein 3 | 96 | + |
| 13 | LTCONS_00021103 | TEA008317.1 | 14 | Alanine—glyoxylate aminotransferase 2 homolog 2 mitochondrial-like | 98 | + |
| 14 | LTCONS_00041093 | TEA000506.1 | 19 | LIMR family protein At5g01460 | 92 | + |
| 15 | LTCONS_00041460 | TEA024759.1 | 20 | Proteasome subunit alpha type-1-B | 100 | − |
| 16 | LTCONS_00049579 | TEA022706.1 | 37 | GDSL esterase/lipase | 100 | + |
| 17 | LTCONS_00063928 | TEA008207.1 | 16 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4-like isoform X1 | 85 | + |
| 18 | LTCONS_00002846 | TEA012270.1 | 43 | Calmodulin-like | 97 | + |
| 19 | LTCONS_00007220 | TEA005414.1 | 31 | 40S ribosomal protein S6 | 91 | + |
| 20 | LTCONS_00037250 | TEA004050.1 | 39 | Mitochondrial outer membrane protein porin of 36 kDa | 100 | + |
| 21 | LTCONS_00050008 | TEA010642.1 | 12 | Basic leucine zipper 34-like | 99 | + |
| 22 | LTCONS_00050164 | TEA015820.1 | 21 | BI1-like protein | 89 | − |
| 23 | LTCONS_00053896 | TEA032488.1 | 29 | 2-Alkenal reductase (NADP(+)-dependent)-like | 98 | + |
| 24 | LTCONS_00053641 | TEA028539.1 | 35 | 14-3-3-like protein | 99 | + |
a+, Consistency; −, complementarity.
Table 2.
ECT-lncRNA pairs screened from other species.
| LncRNA from tea plants | Orthologous lncRNA (species) | Target genes of species | ||||
|---|---|---|---|---|---|---|
| Target ID | Sequence identity with lncRNA (%) | Sequence identity of overlapping region (%) | Sequence consistency a | |||
| 1 | LTCONS_00008922 | CNT2038389 (Malus domestica) | XM_029094701.1 | 31 | 99 | − |
| CNT2092531 (Solanum tuberosum) | XM_006349290.2 | 20 | 100 | + | ||
| 2 | LTCONS_00007788 | CNT20227177 (Theobroma cacao) | XM_007012261.2 | 39 | 100 | − |
| CNT2072931 (Populus trichocarpa) | XM_024604078.1 | 18 | 100 | − | ||
| CNT2080641 (Oryza nivara) | XM_026026114.1 | 45 | 100 | − | ||
| 3 | LTCONS_00044543 | CNT20189851 (Prunus persica) | XM_020556848.1 | 13 | 98 | − |
| 4 | LTCONS_00026852 | CNT20208736 (Medicago truncatula) | XM_003610710.4 | 31 | 98 | − |
| CNT20208735 (Medicago truncatula) | 31 | 98 | − | |||
| CNT2091126 (Solanum tuberosum) | XM_006367626 | 57 | 94 | − | ||
| CNT20180321 (Arabidopsis thaliana) | NM_106318 | 19 | 97 | − | ||
| CNT20229132 (Oryza sativa) | AK108079 | 20 | 97 | − | ||
| 5 | LTCONS_00004664 | CNT2033313 (Glycine max) | NM_001360367.2 | 59 | 100 | − |
| CNT2047077 (Chenopodium quinoa) | XM_021860512.1 | 38 | 96 | − | ||
| CNT20237064 (Vitis vinifera) | XM_019220518.1 | 46 | 95 | − | ||
| 6 | LTCONS_00016955 | CNT2043167 (Malus domestica) | XM_008383619.2 | 37 | 100 | + |
| CNT2043166 (Malus domestica) | XM_008383620.3 | 45 | 100 | + | ||
| CNT2043165 (Malus domestica) | XM_008383621.3 | 19 | 97 | + | ||
| CNT2043164 (Malus domestica) | 36 | 100 | + | |||
| CNT2043163 (Malus domestica) | 42 | 100 | + | |||
| CNT2039564 (Malus domestica) | XM_008383620.3 | 36 | 100 | + | ||
| CNT2038026 (Malus domestica) | XM_008379084.3 | 42 | 100 | + | ||
| CNT2038025 (Malus domestica) | XM_008379085.3 | 45 | 100 | + | ||
| 7 | LTCONS_00064414 | CNT2035352 (Malus domestica) | XM_008382323.3 | 44 | 96 | − |
| CNT20171807 (Brassica oleracea) | XM_013748622.1 | 53 | 100 | − | ||
| CNT2044767 (Malus domestica) | XM_029099752.1 | 32 | 100 | − | ||
| CNT20145755 (Brassica napus) | XM_013860892.3 | 44 | 97 | − | ||
| CNT208893 (Cucumis sativus) | XM_004138916.3 | 69 | 97 | − | ||
| CNT208892 (Cucumis sativus) | 61 | 97 | − | |||
| CNT207653 (Cucumis sativus) | 59 | 94 | − | |||
| CNT205932 (Cucumis sativus) | XM_051090445.1 | 48 | 98 | − | ||
| 8 | LTCONS_00064957 | CNT2074390 (Populus trichocarpa) | XM_006384061.3 | 50 | 100 | − |
| CNT2074389 (Populus trichocarpa) | 44 | 100 | − | |||
| CNT2038576 (Malus domestica) | XM_008376813.3 | 25 | 100 | − | ||
| 9 | LTCONS_00005358 | CNT20111926 (Brassica rapa) | XM_009127664.3 | 62 | 97 | − |
| CNT20143858 (Brassica napus) | XM_003757908.1 | 58 | 98 | − | ||
| 10 | LTCONS_00049549 | CNT20116043 (Brassica rapa) | XM_009134663.3 | 31 | 100 | − |
| CNT20149407 (Brassica napus) | XM_013878762.3 | 21 | 91 | − | ||
| 11 | LTCONS_00010505 | CNT2042500 (Malus domestica) | XM_029089082.1 | 35 | 95 | + |
| CNT2038101 (Malus domestica) | 21 | 98 | − | |||
| 12 | LTCONS_00051279 | CNT2044449 (Malus domestica) | XM_008394181.3 | 23 | 97 | + |
| CNT2044448 (Malus domestica) | 28 | 97 | + | |||
| 13 | LTCONS_00021103 | CNT20237770 (Vitis vinifera) | XM_002267751.4 | 27 | 98 | − |
(Continued)
Table 2.
Continued
| LncRNA from tea plants | Orthologous lncRNA (species) | Target genes of species | ||||
|---|---|---|---|---|---|---|
| Target ID | Sequence identity with lncRNA (%) | Sequence identity of overlapping region (%) | Sequence consistency a | |||
| 14 | LTCONS_00041093 | CNT2037034 (Malus domestica) | XM_008357492.3 | 21 | 100 | + |
| 15 | LTCONS_00041460 | CNT20238040 (Vitis vinifera) | XM_002281912.3 | 31 | 100 | − |
| 16 | LTCONS_00049579 | CNT2043334 (Malus domestica) | ||||
| 17 | LTCONS_00063928 | CNT2028772 (Manihot esculenta) | XM_021786738.1 | 13 | 96 | + |
| 18 | LTCONS_00002846 | CNT20180106 (Arabidopsis lyrata) | XM_021024651.1 | 43 | 100 | − |
| 19 | LTCONS_00007220 | CNT2075529 (Populus trichocarpa) | XM_006382832.3 | 29 | 100 | − |
| 20 | LTCONS_00037250 | CNT2072920 (Populus trichocarpa) | XM_006378092.3 | 61 | 100 | − |
| 21 | LTCONS_00050008 | CNT2030544 (Manihot esculenta) | XM_021763170.2 | 30 | 100 | + |
| 22 | LTCONS_00050164 | CNT20237038 (Vitis vinifera) | XM_002279332.4 | 31 | 100 | − |
| 23 | LTCONS_00053896 | CNT2072341 (Populus trichocarpa) | XM_035033563.1 | 17 | 96 | − |
| 24 | LTCONS_00053641 | CNT20238707 (Vitis vinifera) | XM_002262766.4 | 26 | 100 | − |
+, Consistency; −, complementarity.
NATs are coding or non-coding RNAs with sequence complementarity to other transcripts (sense transcripts), potentially regulating the expression of their sense partner(s) at either the transcriptional or post-transcriptional level. Depending on their genomic origins, NATs can be classified into cis-acting and trans-acting NATs [24, 25]. While cis-NATs have been extensively studied, research on trans-NATs is still rare [26–28]. In the 24 ECT-lncRNA pairs and 49 homologous ECT-lncRNA pairs, there are 5 and 37 trans-NAT pairs in tea plants (Table 1) and other plants (Table 2), respectively. Even among the homologous ECT-lncRNA pairs, some may be trans-NAT pairs, while others are not, e.g. 12-oxophytodienoate reductase trans-lncRNA pairs.
Sequence alignment results revealed very low cDNA sequence consistency between trans-lncRNAs and their target genes (10–45%) in tea plants. However, the sequence consistency of the overlapping region between trans-lncRNAs and their target genes was very high (85–100%) (Table 1). Similarly, the sequence alignment results showed very low sequence consistency between orthologous trans-lncRNAs and their target genes, ranging from 13% to 69%. Nevertheless, in the overlapping region the sequence consistency between trans-lncRNAs and their target genes was very high, ranging from 91 to 100% (Table 2).
Functional enrichment analysis of the 24 ECT-lncRNA pairs revealed that genes involved in plant growth and development included magnesium transporter MRS2-4-like, eukaryotic initiation factor 4A-2, zinc finger CCCH domain-containing protein, and calmodulin-like proteins. Additionally, genes associated with plant resistance response included 12-oxophytodienoate reductase, cytochrome b-c1 complex subunit Rieske-4, vesicle-associated membrane protein, calmodulin-like proteins, and 14-3-3-like proteins (Table 1).
Evolutionary conservation of OPRL/OPR pairs in plants
In the preceding results, our focus lies on the evolutionarily conserved 12-oxophytodienoate reductase–trans-lncRNA (OPRL-trans-lncRNA) pairs present in tea plants, Arabidopsis, potato, alfalfa, and monocotyledonous rice, where the mRNA transcript in these pairs is predicted to encode OPRs, pivotal components of the jasmonate signaling pathway. These 12-oxophytodienoate reductase–trans-lncRNA pairs are hereafter referred to as OPRLs/OPRs.
The gene sequences and chromosomal locations of OPRs and OPRLs, spanning algae, gymnosperms, and angiosperms, were acquired from genomic data provided by NCBI (Supplementary Data Tables S2 and S3). While OPRs were detected across algal, gymnosperm, and angiosperm genomes, OPRLs were exclusively identified in angiosperms, including monocotyledonous rice, dicotyledonous Arabidopsis, alfalfa, horseradish, potato, and tea tree. No responsive OPRLs were observed in algae and gymnosperms.
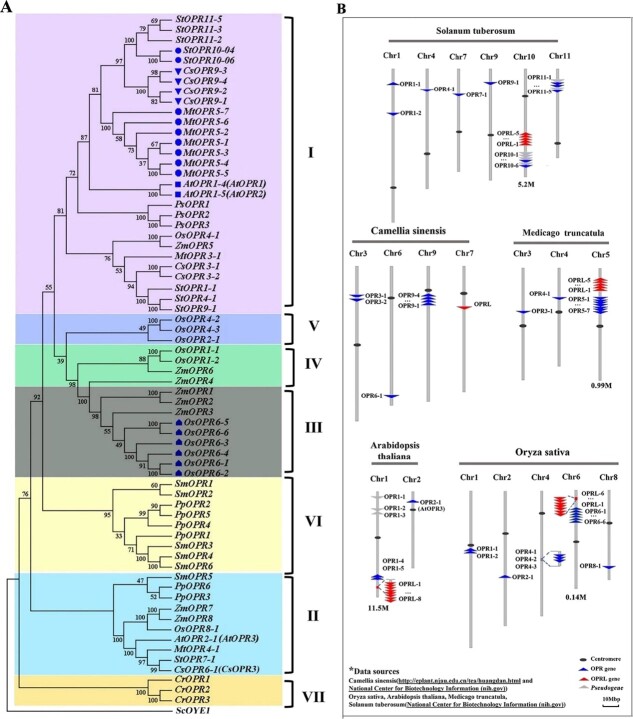
According to the phylogenetic analysis, the OPR family was divided into seven subgroups across algae, gymnosperms, and angiosperms (Fig. 1A). Within tea plants, the seven identified OPR genes were distributed between the first (belonging to the OPRI subfamily) and second subgroups (belonging to the OPRII Subfamily). Chromosome mapping revealed that OPRs and OPRLs in tea plants, rice, Arabidopsis, alfalfa, and potato were dispersed across chromosomes (Fig. 1B). Notably, four OPRs in tea plants (CsOPR9-1, CsOPR9-2, CsOPR9-3, and CsOPR9-4) were found in tandem on chromosome 9, with CsOPR6-1, a member of the OPRII subfamily member (CsOPR6-1), a direct homolog of AtOPR3, located on chromosome 6. CsOPR9-1/2/3/4, members of the OPRI subfamily, constitute the OPR portion of the OPRL/OPR pairs. Similarly, OPR genes in other plants are either clustered or individually distributed on different chromosomes. In contrast, the OPR component of the OPRL/OPR pairs is situated on chromosomes as gene clusters. Thus, the OPR portion of the OPRL/OPR pairs in various plants belongs to the OPRI subfamily and is organized as gene clusters on chromosomes.
Figure 1.
Phylogenetic analysis of OPR genes in 10 representative plants and chromosome mapping of five pairs of 12-oxophytodienoate reductase (OPR) and OPRL. A Neighbor joining and minimalist analysis methods were used to construct a phylogenetic tree using the MEGA5.0 software. The tree consists of seven groups, each highlighted in a different color. Sequence information is provided in Supplementary Data Table S2. B Chromosome localization of OPR/OPRL pairs in monocotyledons (Oryza sativa) and dicotyledons (Solanum tuberosum, Camellia sinensis, Medicago truncatula, and Arabidopsis thaliana). Location information is provided in Supplementary Data Table S3.
Chromosome mapping unveiled that OPRLs were arranged on chromosomes as tandem gene clusters comprising five to eight OPRL genes in all plants except the tea plant. The OPRL clusters in all plants, excluding tea plants, and their target OPR clusters were positioned adjacent to the same chromosome. The distance between OPRs and OPRLs varied from 0.14 to 11.5 Mb across different plants. Moreover, the length of OPRL sequences exhibited inconsistency among plants, with the shortest sequence identified in A. thaliana (227 bp) and the most extended sequence observed in the tea plant (CsOPRL; 1125 bp). Utilizing sequence alignment results (Supplementary Data Fig. S2), each OPRL was paired with its corresponding target OPR to form an OPRL/OPR pair (Supplementary Data Table S4). Notably, based on highly conserved overlapping region sequences, CsOPRL was found to form OPRL/OPR pairs with CsOPR9-1/2/3/4. Furthermore, the alignment results indicated that the base sequences in the overlapping regions of eight AtOPRLs were entirely consistent with those of AtOPR1-4 (AtOPR1) and exhibited high consistency (96%) with AtOPR1-5 (AtOPR2) and relatively low sequence consistency (40%) with AtOPR2-1 (AtOPR3). This suggests that AtOPRLs might regulate the function of AtOPR1 and AtOPR2 rather than AtOPR3. Sequence alignment was conducted between OPRs and OPRLs to determine the location of the overlapping region in OPRL/OPR pairs (Supplementary Data Fig. S3) (Fig. 2A). In A. thaliana, eight AtOPRLs overlapped with AtOPR1 or AtOPR2 at the N-terminus, with the overlapping region situated in the first and second exon regions spanning the AtOPR1 transcript or AtOPR2 transcript. It is worth noting that there are no introns in the DNA sequence of the AtOPRL genes (Supplementary Data Table S3), unlike the DNA sequence of the AtOPR1 or AtOPR2 genes.
Figure 2.
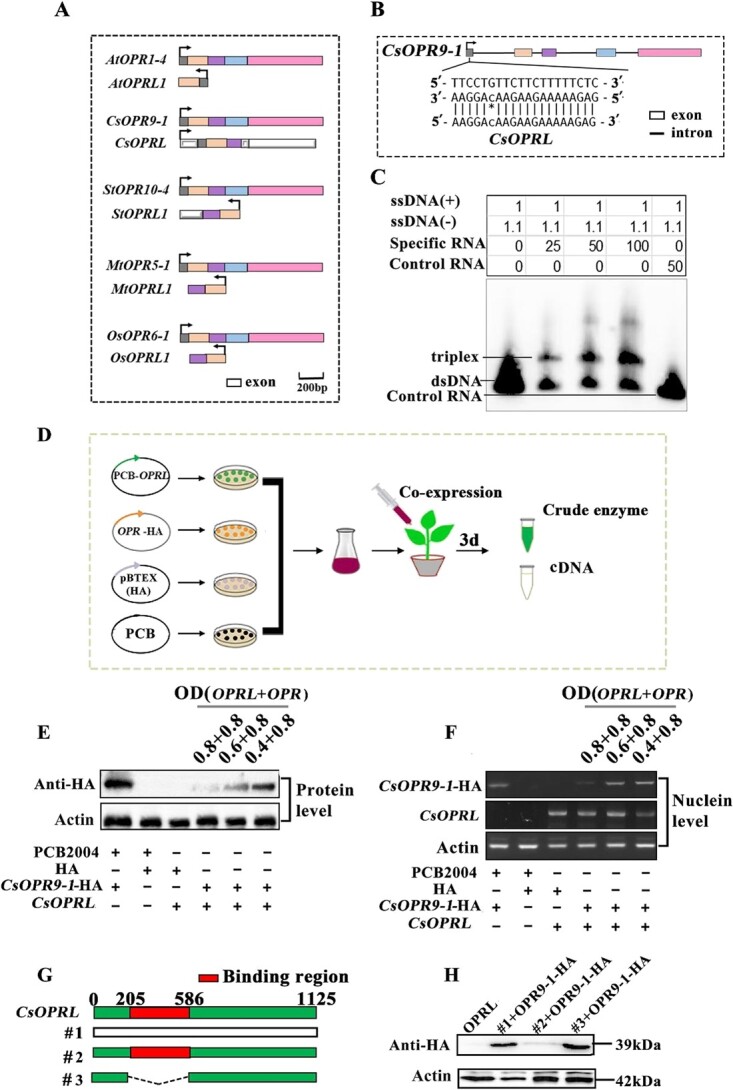
Sequence analysis of OPRL/OPR pairs and identification of mechanisms through which CsOPRL regulates CsOPR9-1. A Structure diagram of OPRL/OPR pairs in five plants. B, C Site of formation of RNA–DNA triplexes in tea plants and gel formation map of EMSA in vitro. D Co-expression of OPRL/OPR pairs. E, F Western blotting and RT–PCR analysis of the interaction between OPRs and OPRLs in tea plants. G–H CsOPRL was truncated and recombined to further validate its key functional region.
In Medicago truncatula, five MtOPRLs and seven MtOPRs were found to overlap at the N-terminus. The overlapping region encompassed the second and third exons of two MtOPRs and exhibited high conservation (95%). Similarly, in Solanum tuberosum, five StOPRLs and two StOPRs overlapped in the second and third exon regions at the N-terminus, with a sequence similarity of 90%. Additionally, the second and third exon regions of six OsOPRLs and six OsOPRs overlapped at the N-terminus in Oryza sativa, with a sequence similarity of 92%. In the tea plant, OPRL overlapped with OPR9-1/2/3/4 at the N-terminus, spanning the first, second, and third exon regions. A schematic diagram illustrating the overlapping regions between OPRs and OPRLs in different plants is presented in Fig. 2A. Given the similarity of these overlapping regions and their comparable chromosomal locations across different plants, it is plausible that the OPRL/OPR pairs are conserved in angiosperms.
To confirm the reliability of the OPRL sequences mentioned above, we cloned the DNA sequence of OPRL from the tea plant. We discovered that the overlapping region between OPRL and OPR9-1/2/3/4 contained the first, second, and third exons and an intron sequence (Supplementary Data Fig. S3).
Based on these findings, we hypothesize that the OPRL clusters in plants may originate from incomplete residues of OPR clusters following replication, potentially representing pseudogenes with impaired functions or newly functionalized genes.
Mechanism underlying the interaction between CsOPRL and CsOPR9-1
We hypothesize that the single-stranded RNA of OPRLs and one of the two DNA strands of OPRs form a complementary chain in the overlapping region, thus creating a triplex that impedes the transcription of the target OPRs. To test this hypothesis, Triplexator [29], a predictor of RNA–RNA and RNA–DNA interactions, was utilized to predict the sites where OPRs and OPRLs may form triple helices in different species. However, the results revealed inconsistency in the sites where OPRs and OPRLs formed triple helices among species (Supplementary Data Fig. S1).
A 20-bp sequence in the overlying region of CsOPRL predicted to form a triplex with CsOPR9-1 was synthesized to validate the prediction accuracy (Fig. 2B). The presence of this triplex was confirmed in samples containing specific RNA and the sense and antisense strands of DNA of CsOPR9-1, with its content increasing proportionally with RNA concentration (Fig. 2C). These findings suggest that CsOPRL can bind to the DNA of CsOPR9-1, thereby suppressing its transcription.
To confirm the regulatory effects of CsOPRL on CsOPR9-1 expression in vivo, we constructed two expression vectors encoding CsOPR9-1-HA and CsOPRL, respectively, and transiently co-expressed them in N. benthamiana using the cauliflower mosaic virus (CaMV)-35S promoter (Fig. 2D). Western blotting and semi-quantitative RT–PCR revealed a gradual increase in the protein and mRNA expression of CsOPR9-1-HA with decreasing CsOPRL concentration (Fig. 2E and F). These results suggest that CsOPRL inhibits the expression of CsOPR9-1 and affects its protein expression.
To identify the site of action of CsOPRL, the overlapping region (205th–586th base) of CsOPRL was truncated (Fig. 2G). Subsequently, empty (1#), complete CsOPRL (2#), and truncated CsOPRL (3#) expression vectors were co-expressed with CsOPR9-1-HA in N. benthamiana using the 35S promoter. Western blotting revealed the absence of CsOPR9-1-HA in the processing area with the overlapping region (CsOPRL [2#]) (Fig. 2H). These results indicate that the inhibitory effects of CsOPRL on CsOPR9-1-HA expression were attenuated after the overlapping region was truncated, suggesting that CsOPRL and CsOPR9-1 interact in the overlapping region.
Confirmation of the regulatory effects of CsOPRL on CsOPR9-1 function using a heterologous expression system
To verify the function of CsOPR9-1, the 35S promoter of the CaMV was used to induce heterologous overexpression of CsOPR9-1 and CsOPRL in the model plant A. thaliana. Subsequently, plants co-expressing CsOPR9-1 and CsOPRL were obtained through hybridization (Supplementary Data Fig. S4). RT–PCR analysis revealed a significant reduction in the transcriptional level CsOPR9-1 in plants co-expressing CsOPR9-1 and CsOPRL.
Morphological observations of plant cultures indicated that CsOPR9-1-overexpressing plants exhibited significantly smaller sizes and shorter root lengths than the WT, indicating inhibited growth (Fig. 3A–C). However, these characteristics did not significantly differ among control plants, plants expressing CsOPRL alone, and plants co-expressing CsOPRL and CsOPR9-1. Subsequently, we assessed the JA and JA isoleucine (ile) contents in Arabidopsis. We observed a significant increase in JA and JA-ile contents in CsOPR9-1-overexpressing plants compared with control plants. In contrast, no significant change was observed in JA and JA-ile in CsOPRL-overexpressing and hybrid plants (Fig. 3D).
Figure 3.
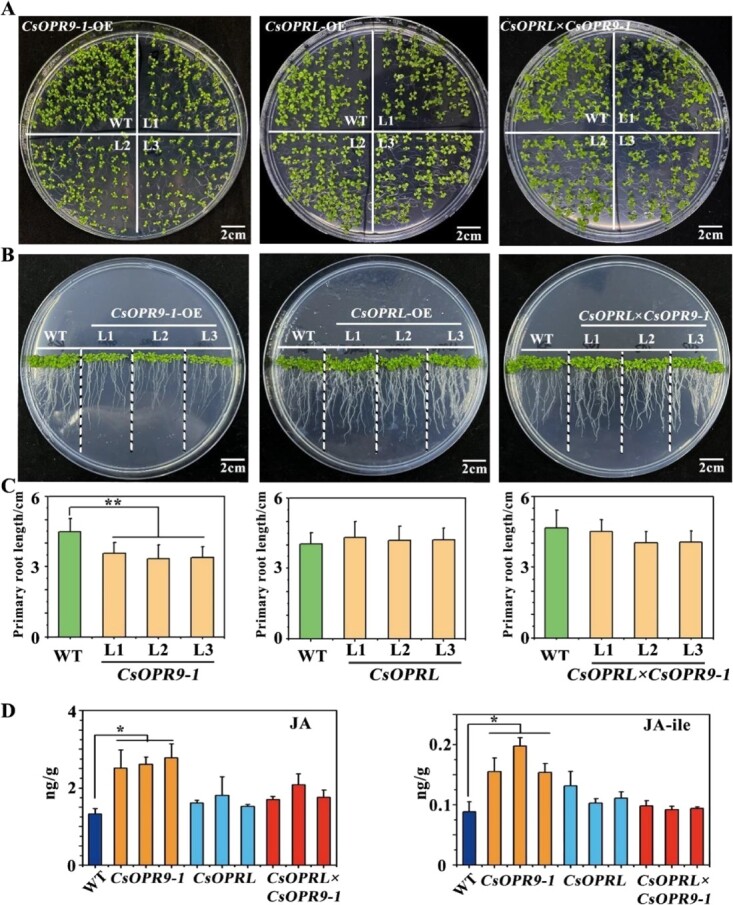
Exogenous overexpression of CsOPRL can restore the inhibitory effects of CsOPR9-1 on plant development in A. thaliana. A, B Growth phenotypes of the primary leaves and roots of 7-day-old Arabidopsis seedlings (scale bar = 2 cm). C, D Physiological indicators and JA and JA-ile contents of CsOPRL, CsOPR9-1, and CsOPRL × CsOPR9-1.
Confirmation of CsOPR9-1-induced increase in resistance to pathogenic fungal infection in tea plants
The JA pathway is known to play a pivotal role in the resistance of tea plants to fungal infection [30, 31]. OPRs serve as crucial enzymes catalyzing the synthesis of JA, which branches into two pathways: the OPR3-0dependent main pathway and the OPR2-catalyzed OPR3-independent alternative pathway [22].
Anthracnosis was induced in tea leaves by injecting them with the TYDY-2 strain, and transcriptome sequencing was used to assess the expression of CsOPRs. Four OPRs detected were significantly upregulated: CsOPR9-1, CsOPR9-2, CsOPR3-1, and CsOPR3-2. On day 6 of inoculation, the expression of these genes was 158.45, 30.04, 4.44, and 4.63 times higher compared with the control group. Concurrently, several JA marker genes, such as MYC2, PDF1.2, and JAZ5, were also upregulated to varying extents (Fig. 4A). These findings suggest an upregulation of the JA synthesis pathway in tea plants with anthracnosis, predominantly evidenced by the upregulation of CsOPR9-1, the target gene of CsOPRL. Additionally, qPCR analysis revealed upregulation of CsOPR9s, CsOPR3-1, and CsOPR6-1, with CsOPR9-1 showing notably higher expression than the others. In contrast, CsOPRL expression was downregulated (Fig. 4A). It is worth noting that CsOPR6-1 upregulated gene expression 5-fold after 6 days of inoculation, while CsOPR9-1 was upregulated by as much as 35-fold.
Figure 4.
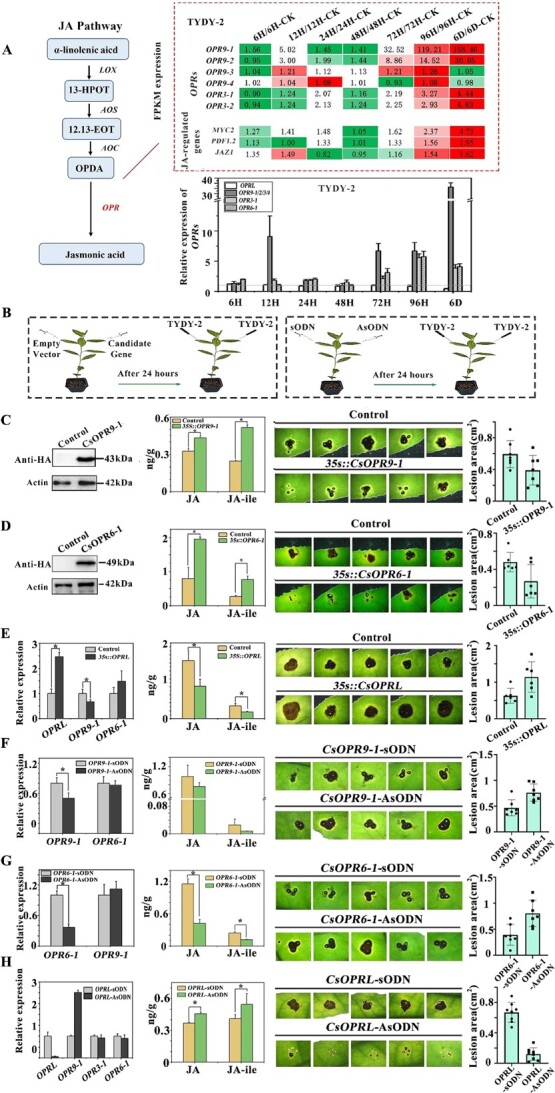
Role of CsOPRs and CsOPRL in resistance of tea plants to anthracnosis. A Schematic diagram of CsOPRs in the jasmonic acid pathway. Changes in CsOPR and JA marker gene expression were examined in leaves with anthracnosis via transcriptomic sequencing and fluorescence quantitative detection. B Effects of transient gene expression mediated by Agrobacterium tumefaciens and gene silencing mediated by AsODNs on the resistance of tea plants to anthracnosis. C–E Effects of Agrobacterium-mediated transient expression of CsOPRs and CsOPRL on the resistance of tea plants to anthracnosis and accumulation patterns of JA and JA-ile in tea plants. The recording time of lesion size was 6 dpi (scale bar = 2 mm). F–H Effects of gene silencing induced by AsODNs on the resistance of tea plants to anthracnosis and accumulation patterns of JA and JA Ile in tea plants. The recording time of lesion size was 6 dpi (scale bar = 2 mm).
The effects of exogenous methyl jasmonate (MeJA) and the JA synthesis inhibitor diethyldithiocarbamic acid (DIECA) on tea plants were examined to validate the role of JA in enhancing tea plant resistance to anthracnose (Supplementary Data Fig. S5A). The results demonstrated that exogenous MeJA significantly reduced the size of infected spots on leaves. In contrast, the JA synthesis inhibitor DIECA increased infected spots, indicating the crucial role of the JA pathway in tea plant resistance to fungal infection. Supplementary Data Fig. S5B displays the effect of MeJA treatment and JA synthesis inhibitor DIECA on the expression of CsOPR and CsOPRL genes in tea plants using fluorescence. The results showed that exogenous MeJA treatment promoted the expression of CsOPR9s, CsOPR3-1, and CsOPR6-1, with the upregulation of CsOPR9-1 expression earlier than the promotion of CsOPR3-1 and CsOPR6-1. Conversely, exogenous MeJA treatment inhibited the expression of CsOPRL, while the effects of JA synthesis inhibitor DIECA were the opposite. This result indicates that the expression of the CsOPRL gene is regulated by JA levels in tea plants.
To elucidate the roles of CsOPR9-1, CsOPR6-1, and CsOPRL genes in tea plant resistance to anthracnose, we used Agrobacterium-mediated transient overexpression technology and antisense oligonucleotide-mediated gene silencing technology (Fig. 4B). Agrobacterium strains containing CsOPR9-1-HA, CsOPR6-1-HA, and CsOPRL genes were injected into tea leaves, followed by inoculation with the TYDY-2 strain to induce disease. Subsequently, we assessed the expression levels of corresponding genes and proteins by immunoblotting with HA antigen (Fig. 4C–E). Our results revealed a significant increase in the expression level of CsOPR9-1 in the experimental group compared with the control, accompanied by a slight increase in JA and JA-ile content and a significant reduction in the area of leaf spots (Fig. 4C). Moreover, the accumulation of JA and JA-ile in plants transiently expressing CsOPR6-1 was significantly higher than that in plants transiently overexpressing CsOPR9-1 (Fig. 4D), potentially due to CsOPR6-1’s involvement in the main pathway of JA synthesis in tea plants. Additionally, in tea plant leaves transiently overexpressing CsOPRL, the expression level of the CsOPR9-1 gene was significantly downregulated. In contrast, CsOPR6-1 expression remained largely unaffected (Fig. 4E). This suggests that CsOPRL modulates CsOPR9-1 expression without affecting CsOPR6-1. Furthermore, in tea plant leaves transiently overexpressing CsOPRL, compared with the control group, the content of JA and JA-ile decreased, and the area of leaf spots on the leaves increased significantly (Fig. 4E).
The gene expression inhibition test results mediated by AsODNs yielded anticipated outcomes. Specifically, when the expression of CsOPR9-1 and CsOPR6-1 genes was interfered with, the experimental group exhibited significantly larger lesion spots compared with the control group, accompanied by a decrease in the contents of JA and JA-ile (Fig. 4F and G). Conversely, interfering with the expression of CsOPRL led to a significant increase in the expression level of CsOPR9-1. In contrast, the expression levels of CsOPR6-1 and CsOPR3-1 genes remained relatively unchanged, further indicating that CsOPR9-1 is the target of CsOPRL in tea plants, whereas CsOPR6-1 and CsOPR3-1 are not (Fig. 4H). In tea seedlings in which the expression of CsOPRL was interfered with, the contents of JA and JA-ile slightly increased, but the area of leaf spots on the leaves decreased significantly compared with the control group (Fig. 4H). Additionally, genes involved in the JA synthesis pathway, including LOX, AOS, and AOC, exhibited upregulation (Supplementary Data Fig. S6).
These findings suggest that the JA pathway plays a role in enhancing the resistance of tea plants to fungal infection. Moreover, CsOPRL contributes to anthracnose resistance by modulating the expression of CsOPR9-1 and influencing the synthesis of JA.
Confirmation of StOPRL function in S. tuberosum
The function of StOPRL was determined in S. tuberosum to determine whether OPRL/OPR pairs exhibit similar functions across different plant species.
StOPRL1 and its target gene, StOPR10-4, were cloned for subsequent functional validation. The findings revealed that StOPRL1 suppressed the expression of StOPR10-4 and decreased its protein content (Fig. 5A and B).
Figure 5.
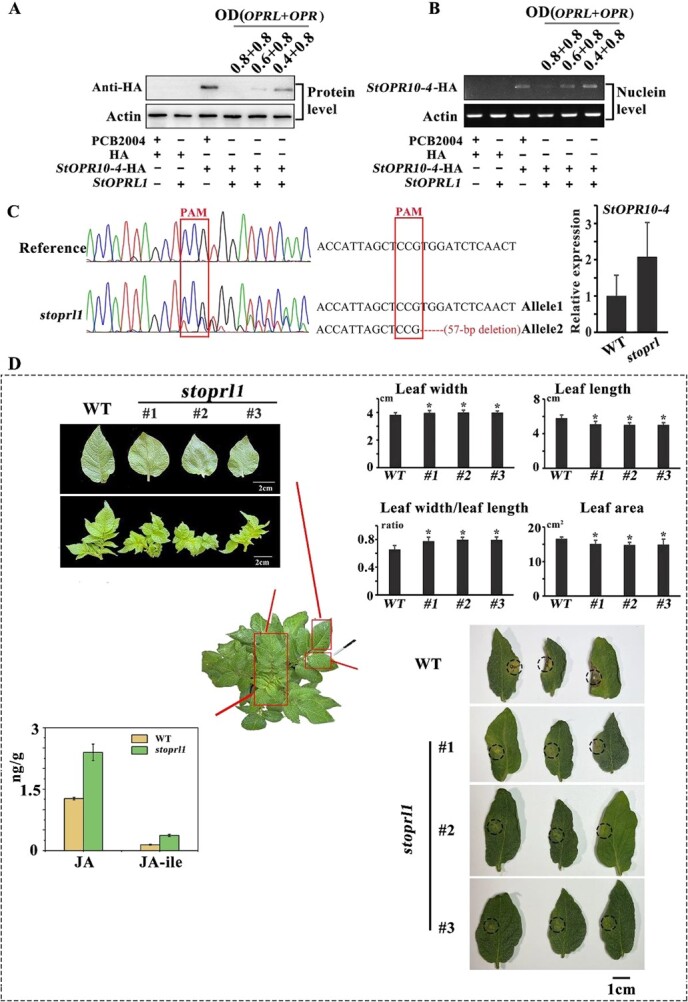
Role of StOPRL1 in the growth and development of potato plants and their resistance response to anthracnosis. A, B Western blotting and RT–PCR analysis of the interaction between OPR and OPRL in potato plants. C Cas9-mediated knockout and StOPR10-4 expression analysis of StOPRL1 in potatoes. D Physiological indicators, JA and JA-ile contents, and disease resistance in the potato knockout line stoprl1.
Furthermore, StOPRL-knockout plants (stoprl1) were produced using Cas9 technology to verify the function of StOPRL1. Quantitative real-time PCR (qRT–PCR) results demonstrated a 1-fold increase in the expression of StOPR10-4 in stoprl1 plants (Fig. 5C). The primary leaves of stoprl1 plants displayed enhanced curling and shrinking, accompanied by near accumulation of JA and JA-ile. Additionally, the terminal lobule exhibited shorter leaf length, increased width, and smaller leaf area. Compared with plants with WT, stoprl1 plants exhibited significantly higher resistance to anthracnosis and a smaller lesion area (Fig. 5D), However, since the stoprl1 mutant strains in this repeat experiment showed almost no disease, only a slight yellowing phenotype, it was not possible to measure the lesion area. These findings suggest that StOPRL, similar to CsOPRL in tea plants, exerts a regulatory role in plant resistance.
Discussion
Identification of OPRL/OPR pairs in angiosperms
From an evolutionary perspective, OPR genes exhibit a relatively ancient origin, being distributed across various plant groups, including green algae, bryophytes, lycopene-rich plants, gymnosperms, monocotyledonous plants, and dicotyledonous plants. However, the expansion of the OPR family in terrestrial plants has occurred extensively through diverse mechanisms, such as cascading repetitive events [18]. Our study revealed through phylogenetic tree analysis that OPR genes experiencing tandem repeat gene amplification events are primarily concentrated within the first and third subgroups, forming gene clusters on chromosomes (Fig. 1A).
Sequence alignment between OPRs and OPRLs and their chromosomal locations suggests that the identified group of OPRL clusters may have originated from residual OPR genes following replication events (Fig. 1B). The conservation of OPRL/OPR pairs across angiosperms, observed in dicotyledonous plants like Arabidopsis, tea, alfalfa, and potatoes, and monocotyledonous plants like rice, is noteworthy. OPRLs exhibit a clustered distribution in the genomes of most studied plants, except for tea plants, which are located on the same chromosome as the OPR gene cluster (Fig. 1B). This suggests a replication event of the OPR gene cluster may be on the same chromosome, possibly resulting in partial sequence loss of the gene. Mosses, lycophytes, and gymnosperms do not display similar OPRL clusters. Overall, these findings imply that replication events leading to the formation of the OPR gene cluster likely occurred before the differentiation of monocotyledons and dicotyledons.
Functional modifications after gene replication, such as pseudogene formation [32, 33], sub-functionalization [34], new functionalization [35], and sub-neural functionalization [36], may have altered the functional constraints between gene clusters within the gene family. The observed sequence consistency between OPRLs and OPRs may have originated from OPR replication events and subsequently undergone new functionalization.
OPRL demonstrates the regulatory role of OPR target gene
From an evolutionary perspective, OPR family genes undergo varying degrees of variation in their introns, exons, and critical amino acid residues in response to long-term selective stress. These changes inevitably impact the function of OPR genes, resulting in subgroup-specific functions [18].
OPRs can be classified into OPR-I and OPR-II based on their substrate specificity in Arabidopsis [22]. Studies have shown that AtOPRs belonging to subfamily I preferentially catalyze 9R,13R-OPDA [37], whereas AtOPR2-1 belonging to subfamily II catalyzes the reduction of 9S,13S-OPDA to form OPC 8:0 [22, 38]. Additionally, OPR members of subfamily II may participate in the JA biosynthesis pathway, while those in subfamily I may be involved in the defense signaling pathway. Subfamily III is exclusive to monocotyledonous plants and likely plays a crucial role in defense signaling and mitogen-activated protein kinase pathways [39].
The OPRs investigated in this study belong to subfamilies I and III. Gene manipulation experiments in tea plants demonstrated that CsOPR9-1 and CsOPRL are involved in regulating the resistance of tea plants to anthracnosis (Fig. 4). Additionally, OPRL knockout resulted in abnormal growth and altered resistance response in potato plants (Fig. 5), suggesting that OPRL regulates plant growth, development, and resistance.
OPRL regulates the expression of OPR genes through the formation of RNA–DNA triplexes
The mechanisms of action of lncRNAs are diverse. They can act as signaling molecules to bind to transcription factors and participate in various regulatory processes or signaling pathways, thereby regulating the spatiotemporal expression of protein-coding genes [40]. Many lncRNAs reside in chromatin and can interact with proteins to promote or inhibit their binding to the target DNA region. Moreover, lncRNAs can serve as molecular scaffolds [8] and interact with various proteins to form ribonucleoprotein complexes. Specific sites within lncRNAs can bind to certain regulatory molecules, affecting several biological processes [41]. Some lncRNAs can act as molecular sponges for miRNAs, blocking their interaction with downstream target genes and indirectly regulating target gene function [42]. On the one hand, lncRNAs can be targeted by miRNAs to produce small interfering RNAs [42, 43]. On the other hand, lncRNAs serve as the source of miRNAs or regulate miRNA accumulation or activity at transcriptional and post-transcriptional levels [44]. In trans-lncRNAs, forming RNA–DNA triplets may represent a common feature of chromatin interaction for recognizing target genes by lncRNAs [45]. Using triad prediction software, we predicted that OPRLs and OPRs formed triple helices in the overlapping region of OPRLs located at the N-terminus (Supplementary Data Fig. S2). The band shift in the electrophoretic mobility shift assay indicated that OPRLs targeted chromatin by forming triple helices (Fig. 2C).
Materials and methods
Plant materials and chemical reagents
The Camellia sinensis var. sinensis cultivar ‘Zhongcha 108’ was used in this study and cultivated in the tea experimental field of Anhui Agricultural University (117.27 E; 31.86 N). Additionally, N. benthamiana, A. thaliana (Col-0), and S. tuberosum L. (cultivar ‘E Shu6’) plants were cultivated in a greenhouse under controlled conditions at 23°C with a 16/8-h photoperiod.
MeJA and DIECA, an inhibitor of JA synthesis, were purchased from Hefei Yili Biotechnology Co., Ltd. MeJA and DIECA were administered for exogenous hormone treatment at concentrations of 300 μM and 10 mM, respectively.
Infection with pathogenic fungi
A pathogenic isolate (TYDY-2) belonging to Colletotrichum camelliae was used to infect the leaves of tea and potato plants. The TYDY-2 strain was initially cultured on potato glucose agar (PDA) medium at 28°C for 5 days, following which the spores were collected via centrifugation at 6000 xg/min for 10 min. Subsequently, the collected spores were resuspended in sterile water, and their concentration was adjusted to 106 spores/ml under microscopic monitoring for subsequent inoculation.
A total of 50 μl of the conidial suspension of TYDY-2 was then inoculated into plant leaves punctured at the upper epidermis using a sterile syringe. Control plants were inoculated with an equivalent volume of sterile distilled water. The inoculated leaves were covered with a film to maintain high humidity and facilitate fungal growth. Following a 24-h incubation period, the film was removed, and any symptoms on the infected leaves were recorded within 6 days post-inoculation. Subsequent to symptom observation, the infected leaves were collected and rapidly frozen. These frozen samples were then used to detect mRNA and lncRNA expression levels through RNA-sequencing (RNA-seq) and qRT–PCR.
Detection of mRNAs and lncRNAs in tea plants using RNA-seq
Transcriptomic sequencing of mRNAs and lncRNAs in TYDY-2-infected leaves was conducted by the Beijing Genomics Institute (BGI). Total RNA was isolated from each sample, with ribosomal RNA removed prior to constructing a chain-specific library. All reconstructed transcripts were matched and aligned with reference genomic data (http://139.196.163.62/). The lncRNAs were identified using CPC software, txCdsPredict software, CNCI software, and the pfam database.
Acquisition of data from lncRNAs of different plant species
The cDNA sequences of lncRNAs from several plant species were downloaded from CANTATAdb, including Malus domestica, S. tuberosum, Theobroma cacao, Populus trichocarpa, Oryza nivara, Corchorus capitularis, Prunus persica, M. truncatula, Glycine max, Vitis vinifera, and A. thaliana. Target gene sequences of lncRNAs were screened and downloaded from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov). Links to various database sites are listed in Supplementary Data Table S4.
Data on the chromosomal location of OPRs and OPRLs across different species, including angiosperms and lower plants, was extracted from the abovementioned plant genome websites (NCBI, https://www.ncbi.nlm.nih.gov; CANTATAdb, http://cantata.amu.edu.pl/index.html).
Gene cloning, qRT–PCR, multiple sequence alignment, and phylogenetic analysis
Total RNA was extracted from tea and potato plant leaves using Fruit-mate (Takara, Dalian, China) and the RNAiso Plus kit (Takara, Dalian, China), followed by reverse transcription to cDNA using PrimeScript-RT-Master Mix (Takara). PCR amplification was conducted using PrimeSTAR Max DNA polymerase (Takara). Real-time PCR was performed using Hieff qPCR SYBR Green Mix (Yeasen). The primer sequences used for PCR are listed in Supplementary Data Table S5.
The PCR products were ligated into the pEASY Blunt Simple Vector (TransGen Biotech, Beijing, China) and transformed into chemically competent DH5α cells (Weidi Biotechnology, Shanghai, China) for subsequent sequence analysis.
Multiple sequence alignment was conducted using DNAMAN software. A phylogenetic tree was constructed (using adjacency statistics) using MEGA 6.0 with a bootstrap test of 1000 replicates. The evolutionary distance was calculated using the Poisson model.
Plasmid construction and heterologous expression of CsOPR9-1 and CsOPRL in A. thaliana
CsOPR9-1 and CsOPRL were expressed in A. thaliana, and hybrid plants were generated through crossing. Primer sequences with the attB linker were ligated to the complete open reading frames of CsOPR9-1 and CsOPRL via in vitro PCR amplification. The resulting PCR products were purified using the Gateway BP cloning enzyme mixture, cloned into the entry vector pDONR207 (laboratory of Xiangchengbin, USTC), and then transferred into the expression vector pCB2004 (laboratory of Xiangchengbin, USTC) using the Gateway LR cloning enzyme system. The expression vectors containing the target genes (pCB2004-CsOPR9-1 and pCB2004-CsOPRL) were subsequently introduced into the expression strain GV3101 (laboratory of Xiangchengbin, USTC). The primer sequences are listed in Supplementary Data Table S5.
The recombinant plasmids pCB2004-CsOPR9-1 and pCB2004-CsOPRL were chemically transformed into GV3101. Plants overexpressing CsOPR9-1 and CsOPRL were generated via Agrobacterium-mediated transformation of WT A. thaliana (Col-0). Following homozygous culture to the T3 generation, plants overexpressing CsOPR9-1 and CsOPRL were hybridized to produce hybrid plants.
Electrophoretic mobility shift assay
The electrophoretic mobility shift assay was conducted to detect the triplex formation of CsOPR9-1 with CsOPRL, following a previously reported method with some modifications [46]. Single nucleotide strands of the predicted triple helix formation site in CsOPR9-1, the RNA strand with the specific sequence of the predicted binding domain in CsOPRL, and control RNA were procured from General Biosystems Company. Initial double-stranded hybridization involved single-stranded cDNA in hybridization buffers (10 mM Tris–HCl + 50 mM NaCl +10 mM MgCl2; pH 7.4) at 95°C for 5 min and then cooled to room temperature. Subsequently, 200 nM double-stranded DNA was incubated with single strands of RNA at varying concentrations in a hybridization buffer at 60°C for 1 h and then cooled to room temperature to form a triple helix. The 4-μl reaction mixture was analyzed on 15% polyacrylamide gels and stained with GelRed for 35 min. The triplex sequences are listed in Supplementary Data Table S5.
Use of Agrobacterium-mediated transient transformation in N. benthamiana to verify the relationship between OPRs and OPRLs
As previously described, tobacco leaves underwent Agrobacterium-mediated co-transformation to confirm the regulatory relationship between lncRNAs and mRNAs [47]. Overnight-grown transformed Agrobacterium strains (pCB2004-CsOPRL, pCB2004-StOPRL1, CsOPR9-1-HA, and StOPR10-4-HA) were injected into the leaves of N. benthamiana when the OD at 600 nm reached 0.8. The cultures were centrifuged, and the resulting pellet was resuspended twice in a solution containing 10 mM MgCl2, 10 mM MES, and 100 μM acetosyringone (pH 5.6). The suspension cultures were then injected into the leaves of well-watered 6-week-old plants. Leaf samples were harvested 2 days post-injection, and gene expression was analyzed via western blotting and qRT–PCR, as described in the sections above. Each experiment was repeated at least three times.
Antisense oligonucleotide-mediated gene suppression and infection of tea plants with pathogenic fungi
Candidate AsODNs targeting CsOPRL and CsOPR9-1/2/3/4 were selected using SOLIGO software and synthesized by General Biosystems Company. A total of 1 ml of 100-μM AsODN solution was injected into tea seedlings, while tea seedlings injected with sense oligonucleotides (sODNs) served as control. Further experiments were conducted after 48 h of incubation, or the samples were frozen in liquid nitrogen for quantitative gene expression analysis. All primer sequences are listed in Supplementary Data Table S5.
Subsequently, tea plants were infected with pathogenic fungi following a 24-h incubation period, and symptoms were observed after an appropriate number of days.
Agrobacterium-mediated transient gene expression and infection of tea plants with pathogenic fungi
Agrobacterium-mediated transient gene expression was induced in tea plants following the protocol described in a previous study [12]. Cultured Agrobacterium strains (pCB2004 CsOPRL, CsOPR9-1-HA) were grown overnight until the OD at 600 nm reached 0.8. The bacterial cells were injected into the leaves of tea plants and resuspended twice in the suspended solution, as described previously. After a 2-h incubation at room temperature, the tea leaves were injected with the bacterial cells and grown in a greenhouse for 72 h. Subsequently, quantitative analysis was performed to evaluate gene expression.
Following a 24-h incubation period, tea plants were infected with pathogenic fungi, and symptoms were observed after an appropriate number of days.
Knockout of StOPRL1 via CRISPR/Cas9 technology
A specific genomic RNA (gRNA) targeting StOPRL1 was selected using the online tool CRISPR-P (http://cbi.hzau.edu.cn/crispr/). The target sgRNA expression box was constructed using the intermediate vector pYLgRNA-AtU3d and assembled into the pYLCRISPR/Cas9 vector. The recombinant plasmid, confirmed by correct sequencing results, was transformed into Escherichia coli. Subsequently, the recombinant plasmid was transformed into Agrobacterium for further experimentation.
The CRISPR/Cas9-StOPRL1 expressing plasmid was transformed into potato via Agrobacterium-mediated transformation [48]. Transgenic potato plants were selected based on hygromycin resistance. DNA from positive plants was extracted for specific fragment amplification and subsequently sent for sequencing. The sequencing results were analyzed using SnapGene software. All primer sequences are listed in Table S5.
Acknowledgements
We thank the National Natural Science Foundation of China (32372756, U21A20232).
Authors contributions
T.J. performed the experiments. T.J. and T.M.J. analyzed the data. T.T.L. and C.L. provided experimental materials and tested methods. Y.B.H. performed data analysis. T.J. and T.M.J. contributed to the acquisition of reagents and materials and the selection of analytical tools. L.P.G. and T.X. guided the drawing of experimental figures. T.J. drafted the manuscript. Ting Jiang designed the experiments. All authors have read and approved the final manuscript.
Data availability
The raw sequencing data from this study have been deposited in the Genome Sequence Archive in the BIG Data Center (https://bigd.big.ac.cn/), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under the accession number CRA014289.
Conflict of interest
The authors declare no competing interests.
Supplementary data
Supplementary data are available at Horticulture Research online.
Supplementary Material
Contributor Information
Ting Jiang, School of Life Science, Anhui Agricultural University, Hefei 230036 Anhui, China.
Tianming Jiao, State Key Laboratory of Tea Plant Biology and Utilization/Key Laboratory of Tea Biology and Tea Processing of Ministry of Agriculture/Anhui Provincial Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei 230036 Anhui, China.
Yingbang Hu, School of Life Science, Anhui Agricultural University, Hefei 230036 Anhui, China.
Tongtong Li, State Key Laboratory of Tea Plant Biology and Utilization/Key Laboratory of Tea Biology and Tea Processing of Ministry of Agriculture/Anhui Provincial Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei 230036 Anhui, China.
Cheng Liu, State Key Laboratory of Tea Plant Biology and Utilization/Key Laboratory of Tea Biology and Tea Processing of Ministry of Agriculture/Anhui Provincial Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei 230036 Anhui, China.
Yajun Liu, School of Life Science, Anhui Agricultural University, Hefei 230036 Anhui, China.
Xiaolan Jiang, School of Life Science, Anhui Agricultural University, Hefei 230036 Anhui, China.
Tao Xia, State Key Laboratory of Tea Plant Biology and Utilization/Key Laboratory of Tea Biology and Tea Processing of Ministry of Agriculture/Anhui Provincial Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei 230036 Anhui, China.
Li-Ping Gao, School of Life Science, Anhui Agricultural University, Hefei 230036 Anhui, China.
References
- 1. Chen CY. Plant noncoding RNAs: hidden players in development and stress responses. Annu Rev Cell Dev Biol. 2019;35:407–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lin Y, Schmidt BF, Bruchez MP. et al. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018;46:3742–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220:e202009045. 10.1083/jcb.202009045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7 [DOI] [PubMed] [Google Scholar]
- 5. Iyer MK, Niknafs YS, Malik R. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Terai G, Iwakiri J, Kameda T. et al. Comprehensive prediction of lncRNA-RNA interactions in human transcriptome. BMC Genomics. 2016;17:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang X, Wang W, Zhu W. et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20:5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–9 [DOI] [PubMed] [Google Scholar]
- 9. Seo JS, Sun HX, Park BS. et al. ELF18-INDUCED LONG NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell. 2017;29:1024–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wan S, Zhang Y, Duan M. et al. Integrated analysis of long non-coding RNAs (lncRNAs) and mRNAs reveals the regulatory role of lncRNAs associated with salt resistance in Camellia sinensis. Front Plant Sci. 2020;11:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu C, Zhang S, Fu H. et al. Transcriptome and phytochemical analyses provide new insights into long non-coding RNAs modulating characteristic secondary metabolites of oolong tea (Camellia sinensis) in solar-withering. Front Plant Sci. 2019;10:1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y, Jiang C, Yin S. et al. New insights into the function of plant tannase with promiscuous acyltransferase activity. Plant J. 2023;113:576–94 [DOI] [PubMed] [Google Scholar]
- 13. Zhao M, Zhang N, Gao T. et al. Sesquiterpene glucosylation mediated by glucosyltransferase UGT91Q2 is involved in the modulation of cold stress tolerance in tea plants. New Phytol. 2020;226:362–72 [DOI] [PubMed] [Google Scholar]
- 14. Nguyen TH, Goossens A, Lacchini E. Jasmonate: a hormone of primary importance for plant metabolism. Curr Opin Plant Biol. 2022;67:102197 [DOI] [PubMed] [Google Scholar]
- 15. Gupta A, Bhardwaj M, Tran L-SP. Jasmonic acid at the crossroads of plant immunity and Pseudomonas syringae virulence. Int J Mol Sci. 2020;21:7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Borrego EJ, Kolomiets MV. Synthesis and functions of jasmonates in maize. Plants (Basel). 2016;5:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wasternack C, Strnad M. Jasmonates: news on occurrence, biosynthesis, metabolism and action of an ancient group of signaling compounds. Int J Mol Sci. 2018;19:2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li W, Liu B, Yu L. et al. Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. BMC Evol Biol. 2009;9:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schaller F, Weiler EW. Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana. Structural and functional relationship to yeast old yellow enzyme. J Biol Chem. 1997;272:28066–72 [DOI] [PubMed] [Google Scholar]
- 20. Schaller F, Biesgen C, Müssig C. et al. 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta. 2000;210:979–84 [DOI] [PubMed] [Google Scholar]
- 21. Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA. 2000;97:10625–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chini A, Monte I, Zamarreño AM. et al. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat Chem Biol. 2018;14:171–8 [DOI] [PubMed] [Google Scholar]
- 23. Huang PC, Tate M, Berg-Falloure KM. et al. A non-JA producing oxophytodienoate reductase functions in salicylic acid-mediated antagonism with jasmonic acid during pathogen attack. Mol Plant Pathol. 2023;24:725–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu J-Z, Zhang J-L, Zhang W-G. Antisense RNA: the new favorite in genetic research. J Zhejiang Univ B. 2018;19:739–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santini L, Yoshida L, de Oliveira KD. et al. Antisense transcription in plants: a systematic review and an update on cis-NATs of sugarcane. Int J Mol Sci. 2022;23:11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reis RS, Poirier Y. Making sense of the natural antisense transcript puzzle. Trends Plant Sci. 2021;26:1104–15 [DOI] [PubMed] [Google Scholar]
- 27. Wang H, Chua NH, Wang XJ. Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biol. 2006;7:R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deforges J, Reis RS, Jacquet P. et al. Prediction of regulatory long intergenic non-coding RNAs acting in trans through base-pairing interactions. BMC Genomics. 2019;20:601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buske FA, Bauer DC, Mattick JS. et al. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22:1372–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen L, Zhang L, Xiang S. et al. AtWRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J Exp Bot. 2020;72:1473–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Macioszek VK, Jęcz T, Ciereszko I. et al. Jasmonic acid as a mediator in plant response to necrotrophic fungi. Cells. 2023;12:1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wagner A. The fate of duplicated genes: loss or new function? BioEssays. 1998;20:785–8 [DOI] [PubMed] [Google Scholar]
- 33. Lynch M, Conery J. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5 [DOI] [PubMed] [Google Scholar]
- 34. Masuda HP, Nakabashi M, Morgante PG. et al. Evidence for sub-functionalization of tandemly duplicated XPB nucleotide excision repair genes in Arabidopsis thaliana. Gene. 2020;754:144818 [DOI] [PubMed] [Google Scholar]
- 35. Lynch M, O'Hely M, Walsh B. et al. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Breithaupt C, Kurzbauer R, Schaller F. et al. Structural basis of substrate specificity of plant 12-oxophytodienoate reductases. J Mol Biol. 2009;392:1266–77 [DOI] [PubMed] [Google Scholar]
- 38. Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA. 2000;97:10625–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Y, Qin L, Zhao J. et al. SlMAPK3 enhances tolerance to tomato yellow leaf curl virus (TYLCV) by regulating salicylic acid and jasmonic acid signaling in tomato (Solanum lycopersicum). PLoS One. 2017;12:e0172466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ye X, Wang S, Zhao X. et al. Role of lncRNAs in cis- and trans-regulatory responses to salt in Populus trichocarpa. Plant J. 2022;110:978–93 [DOI] [PubMed] [Google Scholar]
- 42. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–83 [DOI] [PubMed] [Google Scholar]
- 43. Guo G, Liu X, Sun F. et al. Wheat miR9678 affects seed germination by generating phased siRNAs and modulating abscisic acid/gibberellin signaling. Plant Cell. 2018;30:796–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Y, Li X, Yang J. et al. Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat Commun. 2020;11:5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mondal T, Subhash S, Vaid R. et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kalwa M, Hänzelmann S, Otto S. et al. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016;44:10631–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yao S, Liu Y, Zhuang J. et al. Insights into acylation mechanisms: co-expression of serine carboxypeptidase-like acyltransferases and their non-catalytic companion paralogs. Plant J. 2022;111:117–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barrangou R, Birmingham A, Wiemann S. et al. Advances in CRISPR-Cas9 genome engineering: lessons learned from RNA interference. Nucleic Acids Res. 2015;43:3407–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data from this study have been deposited in the Genome Sequence Archive in the BIG Data Center (https://bigd.big.ac.cn/), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under the accession number CRA014289.